Abstract
Lower motor neuron diseases (LMNDs) include a large spectrum of clinically and genetically heterogeneous disorders. Studying a large inbred African family, we recently described a novel autosomal recessive LMND variant characterized by childhood onset, generalized muscle involvement, and severe outcome, and we mapped the disease gene to a 3.9-cM interval on chromosome 1p36. We identified a homozygous missense mutation (c.1940 T→C [p.647 Phe→Ser]) of the Pleckstrin homology domain–containing, family G member 5 gene, PLEKHG5. In transiently transfected HEK293 and MCF10A cell lines, we found that wild-type PLEKHG5 activated the nuclear factor κB (NFκB) signaling pathway and that both the stability and the intracellular location of mutant PLEKHG5 protein were altered, severely impairing the NFκB transduction pathway. Moreover, aggregates were observed in transiently transfected NSC34 murine motor neurons overexpressing the mutant PLEKHG5 protein. Both loss of PLEKHG5 function and aggregate formation may contribute to neurotoxicity in this novel form of LMND.
Lower motor neuron diseases (LMNDs) are clinically characterized by progressive paralysis with amyotrophy and loss of deep tendon reflexes and fasciculations, because of motor neuron degeneration in the anterior horn of the spinal cord and the brainstem. Diagnosis is confirmed by electrophysiological or histological evidence of muscle denervation, with normal or subnormal motor nerve conduction velocities and normal sensory potentials. The classic form of autosomal recessive proximal spinal muscular atrophy (MIM 253300) is linked to the SMN1 gene,1 but numerous LMND variants have been described, differing by localization of motor weakness, mode of inheritance, and age at onset.2,3 To date, six other causative genes were reported in pure LMNDs: five with autosomal dominant inheritance (HSPB8/HSP22 and HSPB1/HSP27 in distal hereditary motor neuronopathy type II [d-HMNII {MIM 158590 and 608634}], GARS and BSCL2 in d-HMNV [MIM 600794], and DCTN1 in d-HMNVII [MIM 607641]) and one with autosomal recessive inheritance (IGHMBP2 in d-HMNVI, also known as ”spinal muscular atrophy with respiratory distress” [SMARD {MIM 604320}]).4–9 All encode proteins that are directly or indirectly involved in two important intracellular pathways: axonal transport and RNA processing.10,11
Elsewhere, we described a novel clinical variant of autosomal recessive LMND with childhood onset in a large inbred family originating from Mali.12 Affected individuals presented with generalized muscle weakness and atrophy with denervation and normal sensation. Bulbar symptoms and pyramidal signs were absent. In four of the five affected children, the outcome was severe, with loss of walking and the need for permanent respiratory assistance before adulthood. Genetic analyses with the use of a homozygosity mapping strategy assigned this LMND locus to a 3.9-cM (or 1.5-Mb) interval on chromosome 1p36, between loci D1S508 and D1S2633 (Zmax=3.79 at θ=0.00 at locus D1S253). Here, we report the identification of the Pleckstrin homology domain–containing, family G member 5 gene, PLEKHG5 (GenBank accession number NM_020631), located within the candidate region, which is mutated in this pedigree.
Subjects and Methods
Clinical Study
Elsewhere, we reported the linkage data on the African family.12 Informed consent was obtained from all family members, and the study was approved by the ethics committee of the Catholic University of Louvain.
Genotyping and Mutation Analysis
We isolated genomic DNA (gDNA) from blood samples, using standard protocols. We performed genotyping for microsatellites as described elsewhere.12 We used the Primer3 program to design PCR primers that flanked all exons of candidate genes, on the basis of the chromosome 1 draft human sequence. Details on PLEKHG5 primers are given in table 1. We performed direct sequencing, using the dideoxy chain-termination method (ABI Big Dye 3.1) on a 3100 automated sequencer (ABI Prism [Applied Biosystems]) in accordance with standard procedures and the manufacturer’s recommendations. The mutation was verified bidirectionally on gDNA and cDNA. We extracted whole mRNA from fibroblast cell lines of patients (RNeasy [Qiagen]), and we obtained cDNA products by RT-PCR, using oligo(dT) and random hexamers (Transcriptor First Strand cDNA Synthesis Kit [Roche]).
Table 1. .
PLEKHG5 Primers Used for gDNA and cDNA Sequencing Analyses, Plasmid Constructions, and Real-Time RT-PCR
| Primer Sequence(5′→3′) |
||
| Analysis and Exon (GenBank Accession Number) | Forward | Reverse |
| gDNA: | ||
| Exon 1 (NM_198681) | TCTGTGGTGTTGCTTTCCTG | GCCTGCAAGTGGCTCTTAAA |
| Exon 1 (NM_020631) | TCAGAGTTCCCTTGCAGCTT | GGGACCAGTCACTTCCAGAG |
| Exon 1 (NM_001042663) | TGGAAACTGACCTCGGAGAC | CCCGGAGGAGGTTAGGAG |
| Exon 1 (NM_001042664) | GCGCGGCTACCGTAATTC | TTCTGTCCATCGGTTTAGGG |
| Exon 1 (NM_001042665) | GCTCCACAGTCTCCAAGGTG | GGACTCCACACCCCTACCTC |
| Exon 2 (NM_198681) | TGAAGGGAGGACTGAGGAGA | TCTGTGGATAGCTGGTGCTG |
| Exon 3 (NM_198681) | ATCCAGCAGAGGGGAAACTT | TCCTTATGACGCCCTAGCAC |
| Exon 4 (NM_198681) | TAACAGGCTGTGGTCCCTCT | TCTGCCCATCAGCCTTACTT |
| Exon 5 (NM_198681) | AGACCAGGTACCGTTCGATG | GATCTCCCAGACCTCTGCAC |
| Exon 6 (NM_198681) | AATGAGGGCGGTGAGGAG | TGGCTCCTGCATACATGATT |
| Exon 7 (NM_198681) | CCGCCTGGTTCTAACACAC | AGCATCCAGCAGAGACACCT |
| Exon 8 (NM_198681) | CCTCCTCCACCAGACCAG | CCATTTTCCAGAAGGGACAA |
| Exon 9 (NM_198681) | AGAGATGCAGAGACCCTCCTC | CCAGCTGCTCCATCTTGTCT |
| Exon 10 (NM_198681) | CAGTGGCAGCACCAACACT | GGTAACAGTGGCCTCTTTGG |
| Exon 11 (NM_198681) | TTGCATGCAGGTTGCTTATC | GCAGCCCTTGTCTGACTTTC |
| Exon 12 (NM_198681) | TTCTTATCCGTGGCTGCTGT | CCCTCCTTCCTGGTACCTCA |
| Exon 13 (NM_198681) | GGGTGAGGTACCAGGAAGGA | TCGTAATTCCTCACCCTTGG |
| Exon 14 (NM_198681) | GCCCAAGTGCAGTAAGGAAG | GTCCAGGGTCCCGTCCTC |
| Exon 15 (NM_198681) | GAGGACGGGACCCTGGAC | AGCTTCAGGTCCAGGGTCAT |
| Exon 16 (NM_198681) | AAGTCGGTGCTGAGGAAGAC | GCCAGAGACTGACTCCCATC |
| Exon 17 (NM_198681) | TGCCTAGCTGAGATGGGAGT | GGGACTGCAGAGCTGAGAAC |
| Exon 18 (NM_198681) | GTTCATGGTGGGAGGAGTGT | GGGTACATGGGACAGAATGG |
| Exon 19 (NM_198681) | GTTCATGGTGGGAGGAGTGT | GGGTACATGGGACAGAATGG |
| Exon 20 (NM_198681) | GCTGGGTGGACACCATTTAC | CAGCTACCACGAATGGATCA |
| Exon 21.1 (NM_198681) | CAGGAGGGCAGAGGGTATAA | TAATACCTGGGGCTGGAACA |
| Exon 21.2 (NM_198681) | CCCCACCTGCTCAAGTCTAA | GATGCTCCCAGGCATGAGT |
| Exon 22 (NM_198681) | GAAGAGAGGGTGACCAGAGC | GCAAAGGACTCTTCCCAGTG |
| cDNA: | ||
| Exons 1–3 (NM_198681) | TCTGTGGTGTTGCTTTCCTG | ATAATGCATGGTGCTGTGGA |
| Exons 2–7 (NM_198681) | CCGCTGAAAAGAAGGGACT | CTGTAGGCCTCGAAGGTGAG |
| Exons 6–9 (NM_198681) | GTCCCAGCCATGAAGAAGAA | CAGTGTTGGTGCTGCCACT |
| Exons 8–11 (NM_198681) | CTCCAAGTCCCTGAGTTTGC | ACCCGCAGTTTCCTGATGTA |
| Exons 10–15 (NM_198681) | CTGGGAGGAGGAGTACGATG | GTCTTCCTCAGCACCGACTT |
| Exons 14–18 (NM_198681) | CGCTCTTCAAGCCCTACATC | CAACAGCAGATCCGTGAAGA |
| Exons 17–21 (NM_198681) | TCTGCACCTGGACTTGACAGa | CCAGGCTCTACCACAACCATa |
| Exons 20–21 (NM_198681) | AGCAGGAGGAGGAAGAGGAG | TAATACCTGGGGCTGGAACA |
| Exons 21–22 (NM_198681) | CCCCACCTGCTCAAGTCTAA | CTCCTCCACTCCATCCAGTC |
| Exon 22 (NM_198681) | TGCTTCAGCTACTGCCTCCT | GGGAACTGGGCAGATTCAG |
| RT-PCR: | ||
| Exons 18–20 (NM_198681) | GCTACGGGACCCTGGGTC | TTGCAGCTGGTTCTGGGCA |
Primers used for plasmids constructions.
Plasmid Constructions
PLEKHG5 full-ORF expression clones, containing the complete coding cDNA for isoforms BC042606 or BC015231, were sequence verified (Deutsches Ressourcenzentrum für Genomforschung [RZPD]). To introduce the c.1940 T→C amino acid substitution in the PLEKHG5 plasmids, we amplified cDNA of the patients with primers framing the mutation (table 1). We then restricted the mutated cDNA amplification product and the full-ORF expression clones with two single-cut endonucleases, BstEII and PflMI (New England Biolabs). After ligation, we screened for the presence of the mutated insert, and we verified the absence of additional mutation by sequencing analysis. pCMVlacZ plasmid was constructed by insertion of the Escherichia coli lacZ coding region into the multiple cloning site of pCMX-PL1.13
Cell Culture and Transfection Conditions
HEK293 cells (human embryonic kidney 293 cells) and MCF10A cells (human mammary epithelial cells) were maintained at 37°C in a humidified 5% CO2 atmosphere, in Dulbecco's modified Eagle medium (DMEM) F12 (Invitrogen) supplemented with 5% (for MCF10A) or 10% (for HEK293) horse serum (Cambrex), 100 IU/ml penicillin, and 100 μg/ml streptomycin (Sigma). Also added to the medium for MCF10A cells was 0.1 μg/ml cholera toxin (Calbiochem), 10 μg/ml insulin (Invitrogen), 0.5 μg/ml hydrocortisone (Sigma), and 20 ng/ml human epidermal growth factor (Invitrogen). Before transfection, exponentially proliferating cells were trypsinized, and 1.6×105 MCF10A cells or 2.6×105 HEK293 cells were plated in each well of a 6-well plate. Twenty-four hours after plating, cells were transfected using 3 μl of Fugene 6 (Roche) in accordance with the manufacturer’s instructions. For luciferase-reporter gene assays and real-time RT-PCR analyses, 0.5 μg of the expression vectors was transfected together with 1 μg of the reporter plasmid (nuclear factor κB [NFκB] cis-reporting system [Stratagene]) and with 20 ng of constitutive reporter plasmid (pCMVlacZ) for luciferase activity normalization. Expression vectors were replaced by 50 ng of pFC-MEKK (Stratagene) and 0.5 μg of carrier DNA (pCat), to provide a positive control for NFκB activation. For western-blot analyses and immunofluorescence studies, cells were transfected with 1.5 μg of expression vector DNA.
NSC34 cells (a mouse embryonic spinal cord–neuroblastoma cell line with a motor neuron phenotype, kindly provided by Dr. Neil Cashman, University of Toronto14) were cultured at 37°C under 5% CO2 and 95% air in DMEM supplemented with 10% fetal calf serum. NSC34 cells in a 24-well plate were transfected with either the wild-type or the mutant PLEKHG5 plasmids with use of Lipofectamine (Invitrogen). The transfection efficacy was controlled using cotransfection with a green fluorescent protein (GFP)–expressing plasmid.
Enzymatic Assays
Cells were harvested 48 h after transfection. Lysis and enzymatic activity dosages were performed with the β-Gal Reporter Gene Assay (chemiluminescent) kit (Roche) and the Luciferase Reporter Gene assay (high-sensitivity) kit (Roche).
Gene-Expression Analysis by Quantitative RT-PCR
Total RNA was isolated 48 h after transfection and was purified using the Trizol procedure (Invitrogen) in accordance with the manufacturer’s instructions. cDNA was transcribed from 3 μg of total RNA by use of random hexamers and M-MLV Reverse Transcriptase (Invitrogen), in the presence of RNase inhibitor (Promega). Quantitative real-time RT-PCR amplification was performed on cDNA by use of the qPCR Master Mix Plus for SYBR Green I (Eurogentec) in the presence of PLEKHG5-specific primers (Eurogentec) (table 1). The measurement of the β2 microglobulin gene provided an amplification control that allowed PLEKHG5 expression to be normalized. Each reaction was performed in triplicate, by use of an MX3000P Real-Time PCR System (Stratagene).
Synthesis of Polyclonal Antibody to PLEKHG5
Polyclonal antibodies to PLEKHG5 were obtained by immunization of two rabbits with two synthesized specific peptides (NH2-CYLRVKAPAKPGDEG-CONH2 and NH2-CKVDIYLDQSNTPLSL-CONH2) and were purified on a sepharose column (Covalab). High reactivity of immunopurified antibodies was confirmed by ELISA.
Western-Blot Analysis
Proteins were harvested 48 h after transfection. Cells were washed three times with ice-cold PBS and then were lysed in 0.32 M sucrose or in lysis buffer (10 mM Hepes [pH 7.8], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, and 1 mM dithiothreitol) and 10% Nonidet P40 (Sigma), with the addition of the Protease Inhibitors cocktail (Roche). Cell extracts were boiled for 5 min in Laemmli buffer and were submitted to a 7.5% SDS-PAGE. Proteins were electroblotted onto nitrocellulose membranes. After electroblotting, membranes were treated with 5% dry milk in TBS buffer (50 mM Tris [pH 8.1] and 150 mM NaCl) containing 0.05% Tween-20, for 1 h at room temperature, and then were hybridized overnight at 4°C with anti-PLEKHG5 antibodies (1:1,000) diluted in blocking solution. The antigen-antibody complexes were revealed by secondary incubation with horseradish peroxidase–coupled goat anti-rabbit immunoglobulin G antibodies (1:10,000 [Sigma]). Immunoreactive proteins were visualized using enhanced chemiluminescence reagents (Perkin Elmer). Hybridization with β-actin antibody (1:1,000 [Sigma]) was performed as a control.
Immunocytochemistry and Microscopy
Transfected HEK293 and MCF10A cells were fixed in 4% formaldehyde 48 h after transfection. We permeabilized cells with 1% Triton X-100 in PBS for 15 min and blocked aspecific fixation sites in 5% nonfat milk for 2 h at room temperature. Cells were incubated with anti-PLEKHG5 polyclonal antibodies (1:100) for 1 h and then with fluorescein isothiocyanate–conjugated goat anti-rabbit antibody (1:1,000 [Sigma]) for an additional 1 h. Cells were examined using a Zeiss Axioplan 2 imaging microscope equipped with ISIS 3 software (MetaSystem).
Transfected NSC34 murine cells were treated for PLEKGH5 immunofluorescence with the polyclonal anti-PLEKGH5 antibody (1:100) coupled with diamidino-4′,6-phenylindole-2 dichlorhydrate staining of the nuclei. Cells were incubated with the primary anti-PLEKHG5 antibody (1:100) overnight, followed by a rhodamine (A-594)–conjugated anti-rabbit secondary antibody (1:800 [Invitrogen]) for an additional 1 h. PLEKGH5- and GFP-coexpressing cells were observed using confocal analysis (Leica).
Statistical Analysis
P values were calculated by Welch’s t test, by use of R software (version 2.3.0.).
Results
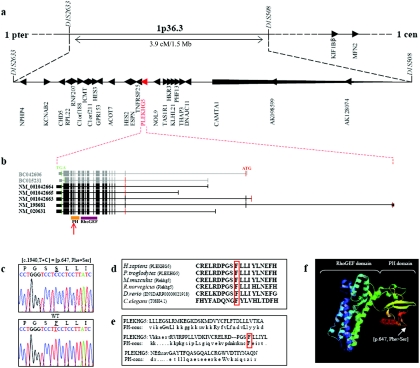
We sequenced the 25 candidate and predicted genes located in the 3.9-cM mapped interval on chromosome 1p36 (NPHP4, KCNAB2, CHD5, RPL22, RNF207, C1orf188, ICMT, C1orf211, HES3, GPR153, ACOT7, HES2, ESPN, TNFRSF25, PLEKHG5, NOL9, TAS1R1, HKR3,KLHL21, PHF13, THAP3, DNAJC11, CAMTA1, AK098599, and AK128074), as well as two additional candidate genes known to cause Charcot-Marie-Tooth disease (CMT) and located very close to this chromosomal region (MFN2 and KIF1Bβ) (fig. 1a). In the five affected members of the Malian family, we found a homozygous mutation (c.1940 T→C) resulting in an amino acid substitution (p.647 Phe→Ser) in the pleckstrin homology (PH) domain of the PLEKHG5 protein (fig. 1c). The mutation was not detected in 300 healthy controls (600 chromosomes), of whom 250 originated from Mali (500 chromosomes). The mutant phenylalanine is highly conserved across species and is a canonical amino acid residue of the PH consensus sequence (fig. 1d and 1e). We then screened PLEKHG5 in a sample of four unlinked families and 16 isolated patients with a close phenotype, but we identified no additional mutations.
Figure 1. .
Physical map of the locus associated with generalized LMND, structure of PLEKHG5, and location of the mutation identified in the African family. a, Location and transcriptional direction of the 27 sequenced genes (UCSC Genome Browser). b, Structure of PLEKHG5 (UCSC Genome Browser). The five isoforms annotated by NCBI genome browser are in black, and the two Mammalian Gene Collection Full ORF mRNAs used for plasmid constructions are in gray. The coding exons are shown as vertical bars, illustrating their approximate size and position. The position of the initiation codon (ATG) at nucleotide +1 is indicated in red, and the position of the stop codon (TGA) is indicated in green. The 5′ and 3′ UTRs are shown as shorter vertical bars. Locations of the PH and RhoGEF domains are also shown (Pfam). The identified homozygous mutation is located in the PH domain (red arrow). c, Electrophoregram of affected individuals compared with a control, showing the homozygous c.1940 T→C (p.647 Phe→Ser) mutation (GenBank accession number NM_020631.2). WT = wild type. d, Phenylalaline at position 647 (boxed in red) highly conserved across species: Homo sapiens, Pan troglodytes, Mus musculus, Rattus norvegicus, Danio rerio, and Caenorhabditis elegans (UCSC Genome Browser). e, Comparison of the PH domain of PLEKHG5 with the PH domain consensus sequence (PH-cons). In this sequence, the eight consensus residues are indicated in capital letters, including the mutant phenylalanine (boxed in red) (Pfam). f, Predictive three-dimensional structure of PLEKHG5 (ModBase). The RhoGEF domain has an α-helical structure, and the PH domain consists of seven β-sheets, followed by a C-terminal amphipathic helix. The mutation occurs in the area of the β5/β6 loop of the PH domain, which may be part of the phospholipids binding site.15
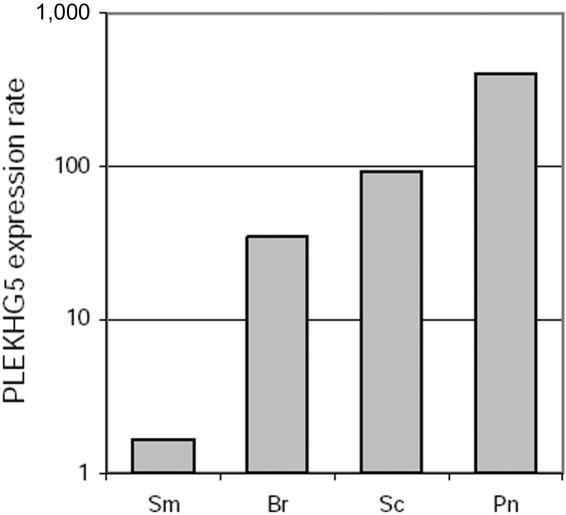
PLEKHG5 spans 53,917 bp on human chromosome 1 and codes for a member of the Dbl protein family that shares a PH domain and a guanine nucleotide exchange factor for Rho protein (RhoGEF) domain.16 Interestingly, it has been suggested elsewhere that the PLEKHG5 protein has an NFκB-activating function.17 Five mRNA isoforms annotated by National Center for Biotechnology Information (NCBI) genome browser and two Mammalian Gene Collection Full ORF mRNAs have been reported to code for proteins that mainly differ by their N-terminal end (GenBank accession numbers NM_020631, NM_198681, NM_001042663, NM_001042664, NM_001042665, BC042606, and BC015231) (fig. 1b). According to microarray databases (UniGene, Expression Profile Viewer), PLEKHG5 is ubiquitously expressed, but predominantly in the peripheral nervous system and brain. Using western blotting and immunofluorescence, we failed to detect endogenous PLEKHG5 protein in various human cells (fibroblastic cell lines, lymphoblastoid cell lines, cultured amniocytes, and cultured primary hepatocytes) (data not shown). However, using real-time quantitative RT-PCR, we confirmed the ubiquitous expression of PLEKHG5 in the human nervous system, with a predominance in peripheral nerve and spinal cord (fig. 2).
Figure 2. .
Expression of PLEKHG5 in the human nervous system. PLEKHG5 expression was evaluated in several human nervous tissues by real-time quantitative RT-PCR. All reactions were performed in triplicate, and PLEKHG5 expression was normalized according to β2 microglobulin expression. Results were expressed as amplification rates in comparison with PLEKHG5 expression in untransfected HEK293 cell line. PLEKHG5 transcripts were detected in all studied tissues, supporting its ubiquitous expression pattern in the human nervous system, with a predominance in peripheral nerve and spinal cord. Sm = skeleton muscle; Br = brain; Sc = spinal cord; Pn = peripheral nerve.
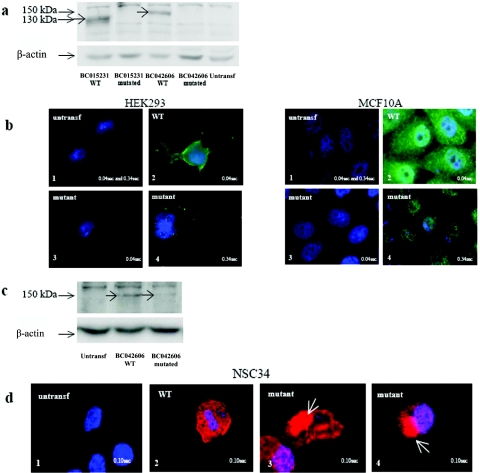
To further investigate the impact of the c.1940 T→C mutation, two isoforms of PLEKHG5 (BC042606 and BC015231) (fig. 1b) were transfected in HEK293 and MCF10A cell lines. The wild-type PLEKHG5 protein was not visualized by western blotting or immunofluorescence before transfection. By contrast, after transfection of expression vectors encoding wild-type BC015231 and BC042606 isoforms, we detected protein fragments of 130 and 150 kDa in cell lysates, using polyclonal rabbit anti-PLEKHG5 antibodies (fig. 3a). In addition, immunofluorescence analysis revealed that wild-type PLEKHG5 proteins were diffusely localized in cytoplasm (fig. 3b). On the contrary, in cells transfected with the corresponding c.1940 T→C mutant constructs, mutant PLEKHG5 proteins were consistently undetectable by western blotting, suggesting an instability of the mutant variants (fig. 3a). By immunofluorescence, mutant PLEKHG5 proteins were not detected with a classic exposure time (0.04 s). However, a light, diffuse cytoplasmic signal could be detected after a longer exposure time (0.34 s) (fig. 3b).
Figure 3. .
Analysis of wild-type (WT) and mutant PLEKHG5 protein expression in untransfected (untransf) and transiently transfected HEK293, MCF10A, and NSC34 cells. a, Analysis by western blotting of PLEKHG5 protein expression in HEK293 cells. PLEKHG5 protein was not detectable in untransfected cells. The 130-kDa and 150-kDa bands were detected in cells overexpressing the BC015231 and BC042606 wild-type isoforms. In contrast, no similar bands were observed in cells transfected with the mutant variants. Similar results were found after distinct protein extraction methods (with 0.32 M sucrose or lysis buffer A; see the “Subjects and Methods” section) and in both HEK293 and MCF10A cells. b, Immunofluorescence assays of PLEKHG5 protein expression in HEK293 and MCF10A cells. b1, There was an absence of signal in untransfected cells, even with a long exposure time (0.34 s). b2, With an exposure time of 0.04 s, cells transfected with wild-type PLEKHG5 showed a diffuse cytoplasmic distribution of PLEKHG5 protein. b3, In cells transfected with the mutant constructs, no protein was visualized with an identical exposure time (0.04 s). b4, Increased exposure time (0.34 s) revealed a weak signal in the nucleus and the cytoplasm. c, Analysis by western blotting of PLEKHG5 protein expression in NSC34 cells. Wild-type PLEKHG5 protein was not detectable in untransfected cells. The 150-kDa band was detected in cells overexpressing the BC042606 wild-type isoform. In contrast, the band intensity was clearly decreased in cells transfected with the BC042606 mutant variant. d, Immunofluorescence assays of PLEKHG5 protein expression in NSC34 cells. The transfection efficacy was controlled using cotransfection with a GFP-expressing plasmid. d1, There was an absence of signal in GFP-negative nontransfected cells. d2, Cells transfected with the wild-type PLEKHG5 plasmid showed a diffuse cytoplasmic distribution of the PLEKHG5 protein. d3 and d4, Presence of large immunoreactive aggregates (white arrows) in the soma of NSC34 motor neurons transfected with the mutant PLEKHG5 plasmids.
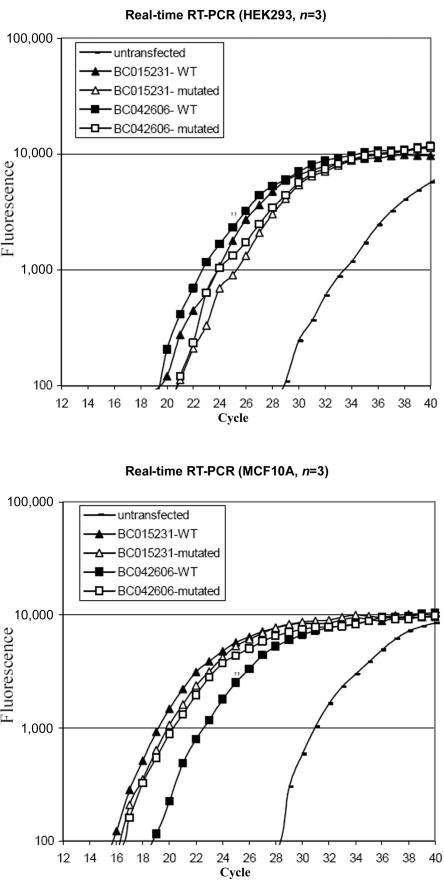
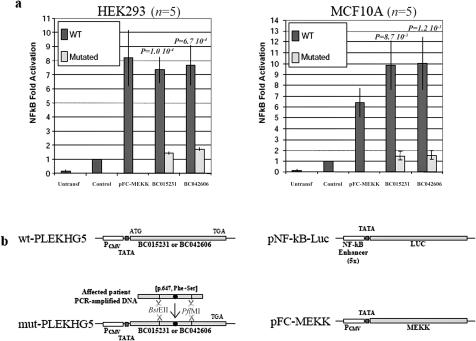
The two isoforms of PLEKHG5 were then tested for their ability to activate the NFκB pathway. HEK293 and MCF10A cell lines were transfected with a luciferase-reporter gene responsive to NFκB, together with expression vectors encoding either wild-type or c.1940 T→C mutated PLEKHG5 cDNAs. Although the PLEKHG5 transcript was barely detected in untransfected control cells by use of real-time quantitative RT-PCR, its abundance increased up to 1,000-fold after transfection in cells transfected with either wild-type or mutant PLEKHG5 variants (fig. 4). Consistently, the luciferase activity was more than sixfold higher in the wild-type PLEKHG5–transfected cells than in the control cells. By contrast, induction of the luciferase activity was markedly reduced in cells transfected with the mutant isoforms (fig. 5a), therefore demonstrating that the amino acid substitution severely impaired PLEKHG5 ability to activate the NFκB pathway. This loss of activity clearly reflects, at least in part, the instability of the mutant protein.
Figure 4. .
Quantification, by real-time RT-PCR, of PLEKHG5 expression levels in untransfected and transfected HEK293 and MCF10A cells. At 48 h after transfection, the cells were harvested and were analyzed by real-time RT-PCR for PLEKHG5 mRNA. The fluorescence was measured in every PCR cycle and was proportional to the accumulation of PCR product. Endogenous expression of PLEKHG5 was detected in untransfected control cells but was markedly increased in transfected cells (up to 1,000-fold, estimated by ΔΔCt method). Overexpression of wild-type (WT) and mutated PLEKHG5 in transfected cells was similar. Triplicate analyses were performed for each sample.
Figure 5. .
Comparison between wild-type (WT) and mutated PLEKHG5 activity on the NFκB signaling pathway. a, Reporter construct consisting of the luciferase gene placed under control of a promoter containing the consensus NFκB binding sites (Stratagene), transfected in HEK293 or MCF10A cells alone (Control), in combination with expression vectors for wild-type and mutated PLEKHG5 proteins (BC015231 and BC042606), or in combination with expression vector for a positive control of NFκB activation (pFC-MEKK). Values are expressed as fold activation over transfection of the reporter plasmid alone. Bars indicate the SD of five independent experiments. The differences between wild-type and mutated PLEKHG5 are statistically significant according to Welch’s t test. b, Schematic representation of the plasmid constructions (see the “Subjects and Methods” section). Untransf. = untransfected. PCMV = CMV promoter; TATA = TATA box.
Finally, we transiently transfected murine motor neuronal NSC34 cells with the two wild-type BC015231 and BC042606 isoforms and the corresponding mutant constructs. Western-blotting experiments confirmed the instability of the mutant PLEKHG5 proteins, which accumulated to much lower level than that of the wild-type proteins (fig. 3c). Detection of the mutant PLEKHG5 proteins by immunofluorescence revealed formation of aggregates in the motor neuron somas close to the nucleus in the majority (60%–70%) of the transfected cells (fig. 3d). This was observed neither in NSC34 cells transfected with the wild-type counterparts nor in nontransfected cells.
In conclusion, transfection experiments using PLEKHG5 variants in distinct cellular models supported instability and loss of NFκB-activating function of the mutant PLEKHG5 proteins and revealed their involvement in aggregate formation in transfected murine motor neuron cells.
Discussion
The mechanism by which the mutation in the PH domain of PLEKHG5 leads to an autosomal recessive, generalized LMND is unclear. PH domains are protein modules of ∼100 aa, found in a wide range of eukaryotic proteins, many of which are involved in cell signaling and cytoskeletal regulation. They play a membrane-anchoring function because of their ability to bind to phosphoinositides.18 In Dbl-family proteins, including PLEKHG5, PH domains also independently contribute to the allosteric regulation of the RhoGEF domain. This latter domain activates GTPases by stimulating the exchange of GDP to GTP, thereby initiating various signaling mechanisms that regulate neuronal shape and plasticity, dendrite growth, synapse formation, and neuronal survival.19–22 Recent experiments show that mutations in the PH domain, impairing phosphoinositide binding, do not systematically affect protein subcellular localization. However, in all cases, these mutations significantly reduce the guanine nucleotide exchange activity of the Dbl proteins.23
Two proteins sharing a PH domain or a PH/RhoGEF domain have already been shown to account for human neurodegenerative diseases: Dynamin 2 (encoded by DNM2) and alsin (encoded by ALS2). Mutations of the PH domain of Dynamin 2 have been reported in CMT, dominant intermediate B (DI-CMTB [MIM 606482]), whereas mutations outside the PH domain are responsible for a dominant myopathic phenotype (centronuclear myopathy [MIM 160150]), suggesting that the PH domain could be specifically involved in motor neuron maintenance.24,25 Mutations in ALS2 have been described in three overlapping autosomal recessive diseases: juvenile amyotrophic lateral sclerosis (ALS2 [MIM 205100]), infantile-onset ascending spastic paraplegia (IAHSP [MIM 607225]), and juvenile primary lateral sclerosis (PLS [MIM 606353]).26–32 The alsin protein is composed of three guanine nucleotide exchange factor domains, which result in Ran, Rho/Rac1, and Rab5 guanine nucleotide exchange activities, involved in motor neuron maintenance, axonal transport, and neurite outgrowth.10,11 The crucial role of the PH/RhoGEF domain for the alsin-mediated neuroprotection against the toxic effect of the mutant form of SOD1 has recently been demonstrated.33,34 Similar to PLEKHG5, endogenous alsin is a low-abundance protein enriched in neural tissues.35 The reported mutations of the ALS2 gene are rarely located in the PH domain. However, all but one of the causative mutations generate alsin protein truncation, often leading to the loss of the PH domain. Except for the recently reported missense G540E mutation, a loss-of-function mechanism was attributed to the causative homozygous mutations in the ALS2 gene. Indeed, when expressed in cultured HEK293 cells, most of the disease-associated mutant forms are unstable and are rapidly degraded by the proteasome, as observed with mutant PLEKHG5.35
The Rho family of GTPases are known to activate the NFκB signaling pathway.36,37 Here, we confirm the NFκB-activating function of PLEKHG5. The NFκB signaling pathway has not been found to be directly involved in human motor neuronopathies. However, its neuronal antiapoptotic role has been documented in various cellular models, and several studies showed that inhibition of the NFκB pathway promoted apoptosis in neurons.38–43 Moreover, down-regulation of NFκB activity has recently been observed in the expanded polyglutamine protein-expressing neuro-2a cells, suggesting that this pathway could be involved in the pathogenesis of polyglutamine-degenerative disorders, including Kennedy disease (MIM 313200), a spinobulbar muscular atrophy.44 Therefore, considering its wide CNS expression pattern and its NFκB-stimulating activity, PLEKHG5 might play a role in neuronal maintenance. Indeed, a mutation in the PH domain leading to the loss of NFκB activation could fail to protect neurons against apoptosis and could compromise the neuronal survival.
Finally, we showed that mutant PLEKHG5 variants formed aggregates in transfected NSC34 murine motor neurons. Interestingly, aggregates were never observed in wild-type–transfected NSC34 cells, and formation of mutant PLEKHG5 aggregates appeared to be specific to the neuronal cell line, because they were not observed in transfected HEK293 and MCF10A cells. This last finding argues against the hypothesis of an artifact of overexpression.45 Animal models or human postmortem histological studies would be useful for evaluating whether this observation in vitro is correlated with intracellular inclusions in vivo. Although the neurotoxic or neuroprotective effect of aggregates is still debated, abnormal aggregation is a common feature in several neurodegenerative diseases, including motor neuron diseases.45–49 In particular, protein aggregation has been recognized as a characteristic change in degenerating motor neurons of amyotrophic lateral sclerosis (MIM 105400) with mutant SOD1.50 Moreover, transfection experiments revealed abnormal aggregation of various mutated proteins involved in autosomal dominant distal hereditary motor neuropathies, such as the small heat-shock proteins 1 and 8 (HSPB1 and HSPB8), the seipin protein (Bscl2), and the dynactin protein (DCTN1).8,9,51–53 In these examples, there is increasing evidence that the misfolded protein and aggregate structures lead to a dominant pathological phenotype by a toxic gain-of-function mechanism or by the combination of both loss of function and toxic gain of function.9,51,53–56 Conversely, specific aggregation of mutant proteins involved in autosomal recessive LMNDs has not yet been reported.
Whether PLEKHG5 is directly or indirectly involved in RNA processing or axonal transport in motor neuron, as postulated for the other LMND-causative genes, remains to be further explored. As already described in neurodegenerative diseases with aggregate accumulation, mutant PLEKHG5 could generate toxic interaction with various intracellular partners, including specific components of the axonal cytoskeleton, leading to the disruption of the anterograde transport pathway in motor neurons.7,8,11,33,51,56–60
In conclusion, we have demonstrated that a form of LMND with childhood onset is caused by a homozygous mutation (c.1940 T→C [p.647 Phe→Ser]) in the PLEKHG5 gene. This mutation caused protein instability, impaired the ability of PLEKHG5 to activate the NFκB pathway in transfected HEK293 and MCF10A cell lines, and eventually led to aggregate formation in a transfected murine NSC34 motor neuron cell line. The observation of a mutation of the PLEKHG5 gene in a motor neuron degenerative disorder suggests that the RhoGEF-mediated NFκB signaling pathway plays an important function in motor neuron maintenance and could be involved in other human neuronopathies as well.
Acknowledgments
This work was supported by the Fonds National Belge de la Recherche Scientifique and the Association Belge contre les Maladies Neuro-Musculaires. We thank Dr. Neil Cashman (University of Toronto), for providing us with the NSC34 cell line; Alain Guillet (Institute of Statistics, Catholic University of Louvain), for statistical analyses; Céline Cailliau (Pediatric Research Unit, Catholic University of Louvain), for technical assistance; and Dr. Jean-Luc Gala (Center for Human Genetics, Catholic University of Louvain), for providing healthy control individuals originating from Mali.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Expression Profile Viewer, http://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.284232
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for mRNAs [accession numbers NM_020631, NM_198681, NM_001042663, NM_001042664, NM_001042665, BC042606, and BC015231)
- ModBase, http://modbase.compbio.ucsf.edu/
- NCBI, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for spinal muscular atrophy, d-HMNII, d-HMNV, d-HMNVII, SMARD, DI-CMTB, centronuclear myopathy, ALS2, IAHSP, PLS, Kennedy disease, and amyotrophic lateral sclerosis)
- Pfam, http://www.sanger.ac.uk/software/Pfam/ (for protein families and domain)
- Primer3, http://frodo.wi.mit.edu/
- RZPD, http://www.rzpd.de/
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–165 10.1016/0092-8674(95)90460-3 [DOI] [PubMed] [Google Scholar]
- 2.Zerres K, Rudnik-Schoneborn S (2003) 93rd ENMC international workshop: non-5q-spinal muscular atrophies (SMA)—clinical picture. Neuromuscul Disord 13:179–183 10.1016/S0960-8966(02)00211-0 [DOI] [PubMed] [Google Scholar]
- 3.Irobi J, De Jonghe P, Timmerman V (2004) Molecular genetics of distal hereditary motor neuropathies. Hum Mol Genet Spec 2 13:R195–R202 10.1093/hmg/ddh226 [DOI] [PubMed] [Google Scholar]
- 4.Grohmann K, Schuelke M, Diers A, Hoffmann K, Lucke B, Adams C, Bertini E, Leonhardt-Horti H, Muntoni F, Ouvrier R, et al (2001) Mutations in the gene encoding immunoglobulin μ-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat Genet 29:75–77 10.1038/ng703 [DOI] [PubMed] [Google Scholar]
- 5.Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, et al (2003) Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet 72:1293–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puls I, Jonnakuty C, LaMonte B, Holzbaur E, Tokito M, Mann E, Floeter M, Bidus K, Drayna D, Oh S, et al (2003) Mutant dynactin in lower motor neuron disease. Nat Genet 33:455–456 10.1038/ng1123 [DOI] [PubMed] [Google Scholar]
- 7.Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, Leung CL, Schagina O, Verpoorten N, Van Impe K, Fedotov V, et al (2004) Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet 36:602–606 10.1038/ng1354 [DOI] [PubMed] [Google Scholar]
- 8.Irobi J, Van Impe K, Seeman P, Jordanova A, Dierick I, Verpoorten N, Michalik A, De Vriendt E, Jacobs A, Van Gerwen V, et al (2004) Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat Genet 36:597–601 10.1038/ng1328 [DOI] [PubMed] [Google Scholar]
- 9.Windpassinger C, Auer-Grumbach M, Irobi J, Patel H, Petek E, Horl G, Malli R, Reed JA, Dierick I, Verpoorten N, et al (2004) Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat Genet 36:271–276 10.1038/ng1313 [DOI] [PubMed] [Google Scholar]
- 10.James PA, Talbot K (2006) The molecular genetics of non-ALS motor neuron diseases. Biochim Biophys Acta 1762:986–1000 [DOI] [PubMed] [Google Scholar]
- 11.Van Den Bosch L, Timmerman V (2006) Genetics of motor neuron disease. Curr Neurol Neurosci Rep 6:423–431 10.1007/s11910-996-0024-9 [DOI] [PubMed] [Google Scholar]
- 12.Maystadt I, Zarhrate M, Leclair-Richard D, Estournet B, Barois A, Renault F, Routon M, Durand M, Lefebvre S, Munnich A, et al (2006) A gene for autosomal recessive lower motor neuron disease with childhood onset maps to 1p36. Neurology 67:120–124 10.1212/01.wnl.0000223834.55225.2d [DOI] [PubMed] [Google Scholar]
- 13.Matis C, Chomez P, Picard J, Rezsohazy R (2001) Differential and opposed transcriptional effects of protein fusions containing the VP16 activation domain. FEBS Lett 499:92–96 10.1016/S0014-5793(01)02532-7 [DOI] [PubMed] [Google Scholar]
- 14.Cashman NR, Durham HD, Blusztajn JK, Oda K, Tabira T, Shaw IT, Dahrouge S, Antel JP (1992) Neuroblastoma × spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn 194:209–221 [DOI] [PubMed] [Google Scholar]
- 15.Blomberg N, Baraldi E, Nilges M, Saraste M (1999) The PH superfold: a structural scaffold for multiples functions. Trends Biochem Sci 24:441–445 10.1016/S0968-0004(99)01472-3 [DOI] [PubMed] [Google Scholar]
- 16.Whitehead IP, Campbell S, Rossman KL, Der CJ (1997) Dbl family proteins. Biochem Biophys Acta 1332:F1–F23 [DOI] [PubMed] [Google Scholar]
- 17.Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, Nagano Y, Doi T, Shimotohno K, Harada T, Nishida E, et al (2003) Large-scale identification and characterization of human genes that activate NF-κB and MAPK signaling pathways. Oncogene 22:3307–3318 10.1038/sj.onc.1206406 [DOI] [PubMed] [Google Scholar]
- 18.Harlan JE, Hajduk PJ, Yoon HS, Fesik SW (1994) Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature 371:168–170 10.1038/371168a0 [DOI] [PubMed] [Google Scholar]
- 19.van Leeuwen FN, Kain HET, van der Kammen RA, Michiels F, Kranenburg OW, Collard JG (1997) The guanine nucleotide exchange factor Tiam1 affects neuronal morphology: opposing roles for the small GTPases Rac and Rho. J Cell Biol 139:797–907 10.1083/jcb.139.3.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estrach S, Schmidt S, Diriong S, Penna A, Blangy A, Fort P, Debant A (2002) The human Rho-GEF trio and its target GTPase RhoG are involved in the NGF pathway, leading to neurite outgrowth. Curr Biol 12:307–312 10.1016/S0960-9822(02)00658-9 [DOI] [PubMed] [Google Scholar]
- 21.May V, Schiller MR, Eipper BA, Mains RE (2002) Kalirin Dbl-homology guanine nucleotide exchange factor 1 domain initiates new axon outgrowths via RhoG-mediated mechanisms. J Neurosci 22:6980–6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nahm M, Lee M, Baek SH, Yoon JH, Kim HH, Lee ZH, Lee S (2006) Drosophila RhoGEF4 encodes a novel RhoA-specific guanine exchange factor that is highly expressed in the embryonic central nervous system. Gene 384:139–144 10.1016/j.gene.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 23.Baumeister MA, Rossman KL, Sondek J, Lemmon MA (2006) The Dbs PH domain contributes independently to membrane targeting and regulation of guanine nucleotide-exchange activity. Biochem J 400:563–572 10.1042/BJ20061020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bitoun M, Maugenre S, Jeannet PY, Lacene E, Ferrer X, Laforet P, Martin JJ, Laporte J, Lochmuller H, Beggs AH, et al (2005) Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet 37:1207–1209 10.1038/ng1657 [DOI] [PubMed] [Google Scholar]
- 25.Züchner S, Noureddine M, Kennerson M, Verhoeven K, Claeys K, De Jonghe P, Merory J, Oliveira SA, Speer MC, Stenger JE, et al (2005) Mutations in the pleckstrin homology domain of dynamin 2 cause dominant intermediate Charcot-Marie-Tooth disease. Nat Genet 37:289–294 10.1038/ng1514 [DOI] [PubMed] [Google Scholar]
- 26.Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, et al (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 10.1038/ng1001-166 [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, et al (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 10.1038/ng1001-160 [DOI] [PubMed] [Google Scholar]
- 28.Eymard-Pierre E, Lesca G, Dollet S, Santorelli FM, di Capua M, Bertini E, Boespflug-Tanguy O (2002) Infantile-onset ascending hereditary spastic paralysis is associated with mutations in the alsin gene. Am J Hum Genet 71:518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devon RS, Helm JR, Rouleaun GA, Leitner Y, Lerman-Sagie T, Lev D, Hayden MR (2003) The first nonsense mutation in alsin results in a homogeneous phenotype of infantile-onset ascending spastic paralysis with bulbar involvement in two siblings. Clin Genet 64:210–215 10.1034/j.1399-0004.2003.00138.x [DOI] [PubMed] [Google Scholar]
- 30.Gros-Louis F, Meijer IA, Hand CK, Dube MP, MacGregor DL, Seni MH, Devon RS, Hayden MR, Andermann F, Andermann E, et al (2003) An ALS2 gene mutation causes hereditary spastic paraplegia in a Pakistani kindred. Ann Neurol 53:144–145 10.1002/ana.10422 [DOI] [PubMed] [Google Scholar]
- 31.Lesca G, Eymard-Pierre E, Santorelli FM, Cusmai R, Di Capua M, Valente EM, Attia-Sobol J, Plauchu H, Leuzzi V, Ponzone A, et al (2003) Infantile ascending hereditary spastic paralysis (IAHSP): clinical features in 11 families. Neurology 60:674–682 [DOI] [PubMed] [Google Scholar]
- 32.Panzeri C, De Palma C, Martinuzzi A, Daga A, De Polo G, Bresolin N, Miller CC, Tudor EL, Clementi E, Bassi MT (2006) The first ALS2 missense mutation associated with JPLS reveals new aspects of alsin biological function. Brain 129:1710–1719 10.1093/brain/awl104 [DOI] [PubMed] [Google Scholar]
- 33.Kanekura K, Hashimoto Y, Niikura T, Aiso S, Matsuoka M, Nishimoto I (2004) Alsin, the product of ALS2 gene, suppresses SOD1 mutant neurotoxicity through RhoGEF domain by interacting with SOD1 mutants. J Biol Chem 279:19247–19256 10.1074/jbc.M313236200 [DOI] [PubMed] [Google Scholar]
- 34.Kanekura K, Hashimoto Y, Kita Y, Sasabe J, Aiso S, Nishimoto I, Matsuoka M (2005) A Rac1/phosphatidylinositol 3-kinase/Akt3 anti-apoptotic pathway, triggered by alsinLF, the product of the ALS2 gene, antagonizes Cu/Zn-superoxide dismutase (SOD1) mutant-induced motoneuronal cell death. J Biol Chem 280:4532–4543 10.1074/jbc.M410508200 [DOI] [PubMed] [Google Scholar]
- 35.Yamanaka K, Vande Velde C, Eymard-Pierre E, Bertini E, Boespflug-Tanguy O, Cleveland DW (2003) Unstable mutants in the peripheral endosomal membrane component ALS2 cause early-onset motor neuron disease. Proc Natl Acad Sci USA 100:16041–16046 10.1073/pnas.2635267100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perona R, Montaner L, Saniger L, Sanchez-Perez I, Bravo R, Lacal JC (1997) Activation of the nuclear factor-κB by Rho, CDC42, and Rac-1 proteins. Genes Dev 11:463–475 10.1101/gad.11.4.463 [DOI] [PubMed] [Google Scholar]
- 37.Montaner S, Perona R, Saniger L, Lacal JC (1998) Multiple signaling pathways lead to the activation of the nuclear factor κB by the Rho family of GTPases. J Biol Chem 273:12779–12785 10.1074/jbc.273.21.12779 [DOI] [PubMed] [Google Scholar]
- 38.Yabe T, Wilson D, Schwartz JP (2001) NFκB activation is required for the neuroprotective effects of pigment epithelium-derived factor (PEDF) on cerebellar granule neurons. J Biol Chem 276:43313–43319 10.1074/jbc.M107831200 [DOI] [PubMed] [Google Scholar]
- 39.Zhu Y, Culmsee C, Klumpp S, Krieglstein J (2004) Neuroprotection by transforming growth factor-β1 involves activation of nuclear factor-κB through phosphatidylinositol-3-OH kinase/Akt and mitogen-activated protein kinase-extracellular-signal regulated kinase 1,2 signaling pathways. Neuroscience 123:897–906 10.1016/j.neuroscience.2003.10.037 [DOI] [PubMed] [Google Scholar]
- 40.Barger S, Moerman A, Mao X (2005) Molecular mechanisms of cytokine-induced neuroprotection: NFκB and neuroplasticity. Curr Pharm Des 11:985–998 10.2174/1381612053381594 [DOI] [PubMed] [Google Scholar]
- 41.Dhandapani K, Wade F, Wakade C, Mahesh V, Brann D (2005) Neuroprotection by stem cell factor in rat cortical neurons involves AKT and NFκB. J Neurochem 95:9–19 10.1111/j.1471-4159.2005.03319.x [DOI] [PubMed] [Google Scholar]
- 42.Kaltschmidt B, Uherek M, Wellmann H, Volk B, Kaltschmidt C (1999) Inhibition of NF-κB potentiates amyloid β-mediated neuronal apoptosis. Proc Natl Acad Sci USA 96:9409–9414 10.1073/pnas.96.16.9409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamatani M, Mitsuda N, Matsuzaki H, Okado H, Miyake S, Vitek MP, Yamaguchi A, Tohyama M (2000) A pathway of neuronal apoptosis induced by hypoxia/reoxygenation: roles of nuclear factor-κB and Bcl-2. J Neurochem 75:683–693 10.1046/j.1471-4159.2000.0750683.x [DOI] [PubMed] [Google Scholar]
- 44.Goswami A, Dikshit P, Mishra A, Nukina N, Jana NR (2006) Expression of expanded polyglutamine proteins suppresses the activation of transcription factor NFκB. J Biol Chem 281:37017–37024 10.1074/jbc.M608095200 [DOI] [PubMed] [Google Scholar]
- 45.Johnston JA, Ward CL, Kopito RR (1998) Aggresomes: a cellular response to misfolded proteins. J Cell Biol 143:1883–1898 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood JD, Beaujeux TP, Shaw PJ (2003) Protein aggregation in motor neuron disorders. Neuropathol Appl Neurobiol 29:529–545 10.1046/j.0305-1846.2003.00518.x [DOI] [PubMed] [Google Scholar]
- 47.Ross CA, Poirier MA (2004) Protein aggregation and neurodegenerative disease. Nat Med Suppl 10:S10–S17 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- 48.Dhib-Jalbut S, Arnold DL, Cleveland DW, Fisher M, Friedlander RM, Mouradian MM, Przedborski S, Trapp BD, Wyss-Coray T, Yong VW (2006) Neurodegeneration and neuroprotection in multiple sclerosis and other neurodegenerative diseases. J Neuroimmunol 176:198–215 10.1016/j.jneuroim.2006.03.027 [DOI] [PubMed] [Google Scholar]
- 49.Gispert-Sanchez S, Auburger G (2006) The role of protein aggregates in neuronal pathology: guilty, innocent, or just trying to help? J Neural Transm Suppl 70:111–117 [DOI] [PubMed] [Google Scholar]
- 50.Bruijn LI, Miller TM, Cleveland DW (2004) Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci 27:723–749 10.1146/annurev.neuro.27.070203.144244 [DOI] [PubMed] [Google Scholar]
- 51.Ackerley S, James PA, Kalli A, French S, Davies KE, Talbot K (2006) A mutation in the small heat-shock protein HSPB1 leading to distal hereditary motor neuronopathy disrupts neurofilament assembly and the axonal transport of specific cellular cargoes. Hum Mol Genet 15:347–354 10.1093/hmg/ddi452 [DOI] [PubMed] [Google Scholar]
- 52.Irobi J, Dierick I, de Corte V, Gettemans J, Robberecht W, Van Den Bosch L, Timmermans JP, De Jonghe P, Timmerman V (2006) 11th International Congress of the World Muscle Society: in vitro expression of small heat shock protein HSPB8 and HSPB1 mutations causing axonal neuropathy. Neuromuscul Disord 16:645–646 10.1016/j.nmd.2006.05.023 [DOI] [Google Scholar]
- 53.Levy JR, Sumner CJ, Caviston JP, Tokito MK, Ranganathan S, Ligon LA, Wallace KE, LaMonte BH, Harmison GG, Puls I, et al (2006) A motor neuron disease–associated mutation in p150Glued perturbs dynactin function and induces protein aggregation. J Cell Biol 172:733–745 10.1083/jcb.200511068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX (1994) Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264:1772–1775 10.1126/science.8209258 [DOI] [PubMed] [Google Scholar]
- 55.Matsumoto G, Stojanovic A, Holmberg CI, Kim S, Morimoto RI (2005) Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J Cell Biol 171:75–85 10.1083/jcb.200504050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsumoto G, Kim S, Morimoto RI (2006) Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J Biol Chem 281:4477–4485 10.1074/jbc.M509201200 [DOI] [PubMed] [Google Scholar]
- 57.Lin H, Zhai J, Canete-Soler R, Schlaepfer WW (2004) 3′ untranslated region in a light neurofilament (NF-L) mRNA triggers aggregation of NF-L and mutant superoxide dismutase 1 proteins in neuronal cells. J Neurosci 24:2716–2726 10.1523/JNEUROSCI.5689-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strey CW, Spellman D, Stieber A, Gonatas JO, Wang X, Lambris JD, Gonatas NK (2004) Dysregulation of stathmin, a microtubule-destabilizing protein, and up-regulation of Hsp25, Hsp27, and the antioxidant peroxiredoxin 6 in a mouse model of familial amyotrophic lateral sclerosis. Am J Pathol 165:1701–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin H, Zhai J, Schlaepfer WW (2005) RNA-binding protein is involved in aggregation of light neurofilament protein and is implicated in the pathogenesis of motor neuron degeneration. Hum Mol Genet 14:3643–3659 10.1093/hmg/ddi392 [DOI] [PubMed] [Google Scholar]
- 60.Lin H, Schlaepfer WW (2006) Role of neurofilament aggregation in motor neuron disease. Ann Neurol 60:399–406 10.1002/ana.20965 [DOI] [PubMed] [Google Scholar]