Abstract
Charcot-Marie-Tooth (CMT) disorders are a clinically and genetically heterogeneous group of hereditary motor and sensory neuropathies characterized by muscle weakness and wasting, foot and hand deformities, and electrophysiological changes. The CMT4H subtype is an autosomal recessive demyelinating form of CMT that was recently mapped to a 15.8-Mb region at chromosome 12p11.21-q13.11, in two consanguineous families of Mediterranean origin, by homozygosity mapping. We report here the identification of mutations in FGD4, encoding FGD4 or FRABIN (FGD1-related F-actin binding protein), in both families. FRABIN is a GDP/GTP nucleotide exchange factor (GEF), specific to Cdc42, a member of the Rho family of small guanosine triphosphate (GTP)–binding proteins (Rho GTPases). Rho GTPases play a key role in regulating signal-transduction pathways in eukaryotes. In particular, they have a pivotal role in mediating actin cytoskeleton changes during cell migration, morphogenesis, polarization, and division. Consistent with these reported functions, expression of truncated FRABIN mutants in rat primary motoneurons and rat Schwann cells induced significantly fewer microspikes than expression of wild-type FRABIN. To our knowledge, this is the first report of mutations in a Rho GEF protein being involved in CMT.
Hereditary motor and sensory neuropathies (HMSNs), commonly referred to as “Charcot-Marie-Tooth (CMT) disease,” are among the most common inherited neurological diseases, with an overall prevalence of about 1–4/10,000.1 Clinically, HMSNs are characterized by progressive muscular and sensory defects starting at the distal extremities, with chronic weakness, pes cavus, and loss of deep tendon reflexes.2 Two main subgroups have been defined on the basis of electrophysiological and histopathological characteristics: the demyelinating form (CMT1) and the axonal form (CMT2). CMT1 can be distinguished from CMT2 by measuring motor nerve–conduction velocities (NCVs): patients affected with CMT1 show reduced NCVs (⩽38 m/s), whereas patients affected with CMT2 show NCVs ⩾38 m/s; the normal value is ⩾48 m/s. Recently, a new group of CMT diseases with electrophysiological and histopathological characteristics overlapping CMT1 and CMT2, referred to as “intermediate CMT,” has been described.3
Genetically, CMT disease is characterized by a great heterogeneity4 (Neuromuscular Disease Center and Inherited Peripheral Neuropathies Mutation Database). All modes of inheritance have been reported. Autosomal recessive demyelinating forms of CMT disease (CMT4) are less frequent, usually of earlier onset, and more severe than the autosomal dominant forms (CMT1), with a fast progression to severe disability leading to higher frequency of wheelchair dependency in the life course.5
Among the 50 loci (30 genes) identified to date, 9 correspond to CMT4, among which 6 genes have already been identified: GDAP1 (MIM 606598)6,7 at chromosome 8q13-q21.1 (CMT4A [MIM 214400])8; MTMR2 (MIM 603557)9 at chromosome 11q22 (CMT4B1 [MIM 601382])10; SBF2/MTMR13 (MIM 607697)11,12 at chromosome 11p15 (CMT4B2 [MIM 604563 and MIM 607739])13; SH3CT2/KIAA1985 (MIM 608206)14 at chromosome 5q23-q33 (CMT4C [MIM 601596])15; NDRG1 (MIM 605262)16 at chromosome 8q24.3 (HMSN-Lom [MIM 601455])17; EGR2 (MIM 129010),18 P0 (MIM 159440),19,20 and PMP22 (MIM 601097)21 (CMT4E [MIM 605523] or Dejerine-Sottas syndrome [MIM 145900]); PRX (MIM 605725)22,23 at chromosome 19q13.3 (CMT4F [MIM 605725]24 and Dejerine-Sottas syndrome); HMSN-Russe (MIM 601455) at chromosome 10q22-q23,25 for which no corresponding mutated gene has yet been identified; and, finally, CMT4H (MIM 609311), which we recently assigned to chromosome 12p11.21-q13.11.26
The roles and function of proteins involved in autosomal recessive CMT (AR-CMT) disease are diverse, including proteins involved in polyposphoinositide signaling27 (myotubularins MTMR2 and MTMR13), mitochondrial fission proteins (GDAP1),28 structural myelin proteins like periaxin (L-PRX), proteins involved in growth arrest and cell differentiation (NDRG1), and proteins with no known function (SH3CT2/KIAA1985). Moreover, several proteins related to GTPase signaling have been identified in CMT disease: mitofusin 2 (MFN2),29 dynamin 2 (DNM2),30 RAB7,31 ARHGEF10,32 and SEPT9.33 Rho GTPases are molecular switches that control a wide variety of signal-transduction pathways in eukaryotic cells, including regulation of the actin and microtubule cytoskeleton and cell polarization, migration, and proliferation.34,35 Along with GTPase-activating proteins (GAPs) and guanine nucleotide dissociation inhibitors (GDIs), Rho GDP/GTP nucleotide exchange factors (Rho GEFs) are essential regulators of Rho GTPases. About 1% of the human genome encodes proteins that either regulate or are regulated by direct interactions with members of the Rho family small GTPases, and more than 85 Rho GEFs, regulating 22 GTPases, are known to date.34 In this article, we report the identification of mutations in FGD4 encoding FGD4/FRABIN (FGD1-related F-actin binding protein [GenBank accession number NP_640334]) in two families from Algeria and Lebanon with members affected with CMT4H. FRABIN is a GEF specific to Cdc42, a member of the Rho family of small GTP-binding proteins (Rho GTPases).36,37
Consistent with these reported functions, expression of truncated FRABIN mutants in rat primary motoneurons and rat Schwann cells induced significantly fewer microspikes than expression of wild-type FRABIN. In all, FRABIN is the sixth protein related to GTPase signaling but the first Rho GEF to be identified in CMT disease.
Material and Methods
Patients
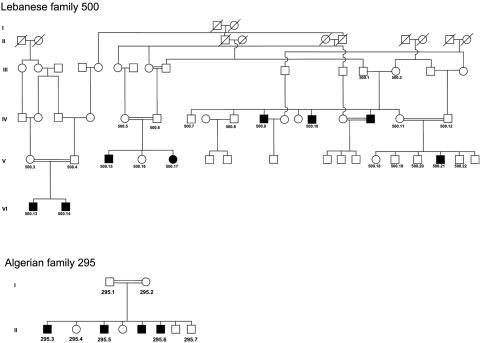
We previously published the localization of CMT4H from one Lebanese family and one Algerian family with CMT4H.26 Patients are affected with early-onset demyelinating neuropathy. For detailed clinical, electrophysiological, and histological descriptions and data, see the work of De Sandre-Giovannoli.26 After informed consent was obtained from all individuals and parents of children included in this study, EDTA blood samples were collected, and genomic DNA was extracted from lymphocytes with the use of standard methods. In all, 29 DNA samples were collected for the study, including 10 from affected individuals (fig. 1). Biopsies of skin and sural nerve samples were performed in Lebanese patient 500.21 under local anesthesia after informed consent was obtained from his parents. All protocols performed in this study complied with the ethics guidelines of the institutions involved.
Figure 1. .
Pedigrees of the Lebanese (500) and Algerian (295) families. Blackened symbols indicate affected individuals.
Mutation Analysis
Exploration of the entire coding sequence of candidate genes (KIF21A [GenBank accession number AY368076], RAPGEF3 [GenBank accession number U78168], and FGD4 [GenBank accession numbers AK057294, BC045552, and AY367054]) was performed in Lebanese patient 500.21 and Algerian patient 295.3 (fig. 1). Intronic primers were designed using Primer3 software. DNA sequences were obtained from the UCSC Genome Browser (May 2004 freeze), by comparison of genomic DNA with cDNA sequences. For FGD4, six additional alternatively spliced exons described in other sequences from the UCSC database were screened for mutations: exons 6a and 16b are present in GenBank sequence BC045552, and exons 12a, 15a, 18, and 19 are present in GenBank sequence AY367054. Primer sequences and annealing temperatures used in PCRs for FGD4 are described in table 1. Those used for KIF21A and RAPGEF3 are available on request. Genomic DNA was amplified for one patient of each family under standard PCR conditions. All PCR-amplified fragments were analyzed by denaturing high-performance liquid chromatography (dHPLC)/WAVE (Transgenomic) and were fluorescently sequenced in both directions for those presenting abnormal elution profiles, with the use of sequencing facilities (MWG Biotech). Conditions for dHPLC analysis are available on request. Chromatograms were compared with reference sequences with the use of Sequencher v4.2 (Gene Codes). The identified c.893T→G and the C.893T→C genomic variants were tested in 216 Lebanese and 108 Algerian control chromosomes by restriction endonuclease digestion with the use of MlnII and MaeII (New England Biolabs), respectively.
Table 1. .
Primer Sequences and Annealing Temperatures for FGD4 Analysis of Coding Sequences at the Genomic and Transcriptional Level[Note]
| Primer Sequence (5′–3′) |
||||
| Analysis and Amplicon | Forward | Reverse | Amplification Product Size (bp) |
Optimal Annealing Temperature (°C) |
| Genomic DNA: | ||||
| Exon 1 | gccttaggagggctggttac | gataatgcccccacacaaac | 296 | 56 |
| Exon 2 | cctgccctttctttctgaac | gtggccatcatttttctggt | 258 | 56 |
| Exon 3 | cacccaggacaccagaattt | cctgccttctcccattgtaa | 292 | 54 |
| Exon 4a | ccaatgttttcatgcttcctt | tgagctaagagtggcagcttt | 282 | 56 |
| Exon 4b | ctcagggtgcacagacttgt | cagggttttccctgctcat | 395 | 55 |
| Exon 5 | gatggctgaaagaaccgagt | aaaacaggcccttccttcat | 329 | 55 |
| Exon 6aa | ccactgctgctgacctttta | aaaacaaaaacccaaaccttca | 257 | 55 |
| Exon 6 | gagcccactatgtgcctagc | cagcatgactgccttaaaca | 272 | 56 |
| Exon 7 | gccactgcactacagtctgg | ggagtagaatgaattttggctagg | 370 | 60 |
| Exon 8 | tcatgcaggtgatggacagt | gaaaccataaggctgcttgg | 337 | 56 |
| Exon 9 | gcacaggaaggacaaagcat | ttcataaacattctttttggctca | 234 | 56 |
| Exon 10 | tgcctgtatgtgatcttgctg | ctcccaaagtgctgggatta | 277 | 60 |
| Exon 11 | cctgatcagtttcccctatttc | gttgcctgattttggaaagc | 275 | 56 |
| Exon 12 | agttggaacaacaggaaagca | cagacagaggctgcagtgag | 231 | 56 |
| Exon 12ab | gcctcttgagtagctgggact | tgaaaattaagtgaacctgttctga | 291 | 55 |
| Exon 13 | tgagaaactttaatgtgtgctttga | gggaaaaggttggagaacaa | 228 | 56 |
| Exon 14 | tccgaaagtgctgggattat | cgtaattggcaccaaaataaa | 383 | … |
| Exon 15 | caaaaatctgccttctatgctt | ttgcagtgagctgagatcgt | 363 | 59 |
| Exon 15ab | gaggctgaggcaggagaat | cccagaacagagcctgaactt | 596 | … |
| Exon 16c | tttggatagtccagggaaaca | attttgagtccgctgtccac | 325 | 56 |
| Exon 17 | ctgtttgggacagtggtagga | tcctgagacctccacaccat | 398 | 56 |
| Exon 18b | ggacacacttaagttttggctca | gactgctcaagtatgttggaagg | 393 | 55 |
| Exon 19b | tgatttgcaattctgtctttcc | gaccatgattgacttcagattttt | 493 | 55 |
| RT-PCR: | ||||
| Exon 5–exon 8 | gcaaactgttggaagaagca | agggaatcaggaggcaattt | 412 | 55 |
| Fragment 1 | tcgctttagttccaccttgc | tccttcatctcatggtgctg | 826 | 59 |
| Fragment 2 | ctacaggactccaggcatagg | tgaagtgttccgagcagcta | 850 | 60 |
| Fragment 3 | gattccctggactggaatga | gaagaaagctgcaca | 847 | 59 |
| Fragment 4 | aaaagagccccaagatggat | tgcatttatgggcctatatttt | 768 | 55 |
| RTQ-PCR: | ||||
| FGD4 (exon 2) | tcagatctcatcagtcgctttg | acagcagactctttcttcaaatca | 74 | … |
| TBP (reference gene 1) | gctggcccatagtgatcttt | cttcacacgccaagaaacagt | 60 | … |
| GUSB (reference gene 2) | cgccctgcctatctgtattc | tccccacagggagtgtgtag | 95 | … |
| Mouse RT-PCRd: | ||||
| Exon 4–exon 8 | atgggattggatacgttgga | aggaatgcgctgaataggc | 498 | 60 |
Note.— Specific primers for RTQ-PCR are also mentioned. All human primers for genomic screening of mutations were selected on the basis of comparison of genomic DNA sequence with GenBank cDNA sequence AK057294. Exon 4 was amplified in two amplicons (4a and 4b).
Alternatively spliced exons found in GenBank sequence BC045552.
Alternatively spliced exons found in GenBank sequence AY367054.
For the longer exon 16 in sequence BC045552, we used reverse primer 5′-gaaaggggacagatgatcca-3′ and an annealing temperature of 55°C.
Primers were designed using GenBank sequence NM_139234.
Transcriptional Analysis
Total RNA was extracted from freshly isolated lymphocytes with the use of either TRIZOL (Invitrogen Life Technologies), on the basis of protocol derived from the work of Chomozynsky and Sacchi,38 or silica-gel RNeasy colums (QIAGEN). cDNAs were obtained from total RNA with use of Superscript I Reverse Transcriptase (Invitrogen Life Technologies) and random primers (Invitrogen Life Technologies), in accordance with the recommendations of the supplier. cDNAs were then PCR amplified as described above. Primers and annealing temperatures are described in table 1. RT-PCR experiments, which gave rise to several fragments, were cloned into PGEM Vector (PROMEGA), in accordance with the supplier’s instructions, and were transformed by electroporation into electrocompetent Escherichia coli cells. Positive clones were selected and amplified by PCR, and fragments were subsequently sequenced as described above.
Real-Time RT-PCR
SYBR Green real-time PCRs (RTQ-PCRs) were realized according to standard protocols in a 20-μl final volume, with the use of primers at a final 500 mM concentration on a Lightcycler 480 (Roche). Of the RNAse H–treated (Invitrogen Life Technologies) cDNAs obtained from reverse transcription of DNAse I–treated total RNA extracted from cultured fibroblasts by use of the High Capacity cDNA Archive Kit (Applied Biosystems), 50 ng were used in each reaction. The expression level of FGD4 was normalized to that of two different genes: TBP (TATA-box binding protein) and GUS-B (β-glucuronidase). Two calibrator samples (normal fibroblast cell lines) were used, and experiments were repeated three times. The FGD4 expression ratio for a sample was calculated as the ratio between the average FGD4 signal in the patient’s fibroblast and the signal from calibrators. Primer sequences can be found in table 1.
Expression Studies
One microgram of each used commercial total RNAs extracted from brain, heart, skeletal muscle, uterus and cerebellum (Ambion-Applied Biosystems), placenta and testis (Clontech) were used in RT-PCRs as described above. Primers located in exons 5 (forward) and 8 (reverse) of FGD4 were used for PCR amplification, which resulted in a 412-bp fragment (see table 1). β-actin was used as a control for normalization with the use of primers 5′-CAATAGTGATGACCTGGCCGT-3′ (forward) and 5′-AGAGGGAAATCGTGCGTGAC-3′ (reverse).
FGD4 expression in different mouse parts of the brain and CNS tissues was studied by use of the ORIGENE Mouse Brain Rapid-Scan Panel. cDNAs obtained from polyA+ RNAs immobilized on a 96-well plate were amplified using either the aforementioned FGD4 primers (table 1) or β-actin control primers, with the use of PCR conditions recommended by the supplier.
Expression Constructs
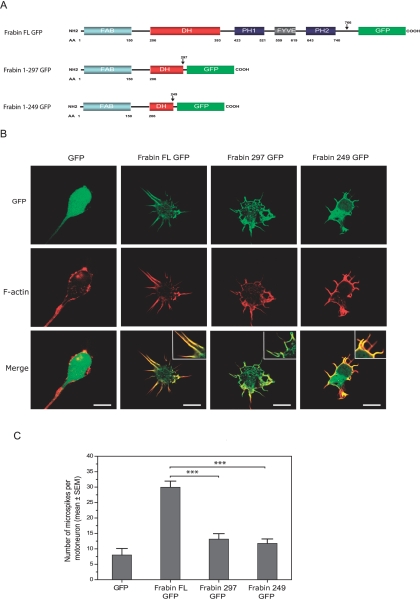
The cDNA for rat wild-type Frabin-GFP was isolated from the pMXII Frabin-GFP plasmid kindly provided by Prof. Takai and was subcloned into an XhoI-restricted pCAGGS expression vector.39 Truncated forms of Frabin were obtained by PCR (Triple master), with the use of pCAGGS Frabin-GFP as template and forward primer Frabin Fw NheI (5′-AAGGAAACTA GCTAGC ACCATGGAGGAGTCTAATCC-3′) together with reverse primer Frabin Rev 297 Not1 (5′-TTCCTTTTTT GCGGCCGC CTTAAGGAATGGTGCCAAC-3′) or Frabin Rev 249 Not1 (5′-TTCCTTTTTT GCGGCCGC CATCTCTGCAGGAAATGAGCCTC-3′) (underlined parts of sequences correspond to endonuclease restriction sites). PCR products were subcloned into pCAGGS-GFP 5′ of the GFP reading frame. The obtained constructs were checked by sequencing.
Cell Culture
Spinal motoneurons were prepared from E14.5 Sprague Dawley rat embryos as described elsewhere.40 Cell pellets were resuspended in electroporation buffer (125 mM NaCl, 5 mM KCl, 1.5 mM MgCl2, 10 mM glucose, and 20 mM HEPES [pH 7.4]) at a density of 50,000 motoneurons per 50 μl and were transferred to 4-mm gap cuvettes (Eppendorf). The cellular suspensions were then incubated for 15 min at room temperature with 5 μg pCAGGS GFP plasmid or the same molar amount of pCAGGS Frabin expression plasmid and were electroporated using a BTX-ECM830 electroporator (Genetronics). Electroporation conditions were 3 square pulses of 5 ms at 200 volts with intervals of 1 s. Motoneurons were cultured on polyornithin/laminin–coated coverslips in supplemented neurobasal medium (Invitrogen Life Technologies) containing 1 ng/ml GDNF, 1 ng/ml BDNF, and 10 ng/ml CNTF.
Primary human fibroblasts and RT4 schwannoma cells were cultured in Dulbecco's modified Eagle medium (DMEM) (Invitrogen Life Technologies) supplemented with 10% fetal calf serum (Invitrogen Life Technologies) and l-glutamine in a 37°C incubator stabilized at 5% CO2. RT4 cell lines are described to be arguably equivalent to primary Schwann cells in vitro.41
Immunostaining and Microscopy
Transduced primary motoneurons and RT4 Schwann cells were fixed at 1 division (DIV) with 4% (vol/vol) formaldehyde, were washed three times with PBS, and were blocked and permeabilized with PBS containing 2% (vol/vol) goat serum, 2% (vol/vol) donkey serum, 0.1% (wt/vol) BSA, 50 mM lysine, and 0.01% (vol/vol) Triton X-100. F-actin was stained for 30 min with Alexa Fluor 546-conjugated phalloidin (diluted 1:80 in blocking buffer [Molecular Probes]). Images of GFP–fluorescent cells were acquired with a Zeiss LSM 510 confocal laser scanning microscope that used a 63× water immersion objective and 3× zoom for motoneurons or 1× zoom for RT4 cells.
In motoneuron cultures, the number of microspikes was determined per individual transduced cell (n=25 neurons per condition). In RT4 Schwann cell cultures, microspike formation was expressed as the percentage of transduced cells bearing more than 10 f-actin–positive protrusions at their peripheries.37 Per condition, >100 transduced RT4 cells were analyzed. Statistical analyses were performed using SigmaStat software (Systat).
At 90% confluency, fibroblast cultures were treated with trypsin-EDTA (0.05% trypsin and 0.53 mM EDTA.4Na [Invitrogen Life Technologies]). After they were counted, 105 cells were plated on 2-well Lab Tek Chamber slides (Nalge Nunc International) and were cultured in DMEM, high-glucose medium supplemented with 10% calf serum, 2 mM l-glutamine, and a mix of penicillin-streptomycine-fungizone. After 24 h of growth in a 37°C incubator stabilized at 5% CO2, cells were washed twice in Dubelcco’s PBS and were fixed in 4% paraformaldehyde.
Cells were then washed for 10 min in PBS and were permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. After they were blocked for 1 h at room temperature in PBS containing 1% BSA, cells were incubated with primary antibodies diluted in the incubation solution for 1 h at room temperature. Primary antibodies used in this study were mouse monoclonal to actin (Chemicon), mouse monoclonal to γ-tubulin (clone GTU-38 [SIGMA]), rabbit polyclonal to pericentrin (abcam), rabbit polyclonal to α-tubulin (abcam), mouse monoclonal to α-tubulin (clone B-5-1-2 [abcam]), and mouse monoclonal to acetylated tubulin (clone 6-11B-1 [SIGMA]), diluted, respectively, at 1:100, 1:1,000, 1:2,000, 1:100, 1:1,000, and 1:2,000. After several washes in PBS, slides were incubated with secondary antibodies diluted in the incubation solution for 1 h at room temperature. Secondary antibodies were from either Jackson (Jackson ImmunoResearch: FITC-conjugated Rabbit antibody against Mouse [1:100], TRITC-conjugated Donkey antibody against Rabbit [1:100], and FITC-conjugated Goat antibody against Mouse) or Molecular Probes (Invitrogen: Alexa Fluor 488 Goat antibody against Mouse or Goat antibody against Rabbit and Alexa Fluor 555 Goat antibody against Mouse or Goat antibody against Rabbit). Cells were then washed three times for 10 min in PBS, were incubated with DAPI (Sigma-Aldrich) at 100 ng/ml for 15 min, and finally were washed three times in PBS. Slides were mounted in Vectashield mounting medium (Vector), were coverslipped, and were sealed. Digitized microphotographs were recorded using a Leica DMR microscope (Leica Microsystems) equipped with a CoolSNAP camera (Princeton).
Results
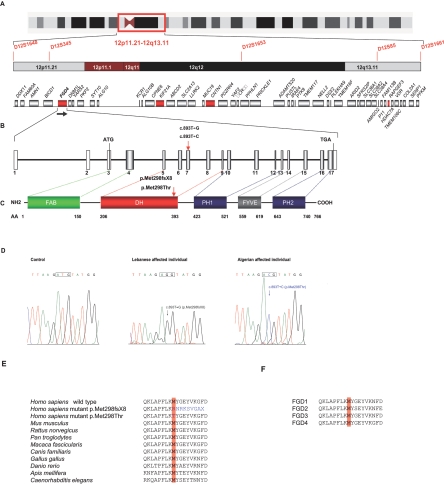
We studied two large consanguineous families affected with CMT4H and originating from Algeria and Lebanon (fig. 1). By use of homozygosity mapping in these families, the locus for CMT4H was previously assigned26 to a 15.8-Mb (11.5-cM) region at chromosome 12p11.21-q13.11, between markers D12S1648 and D12S1661, with a maximum LOD score value of 6.97 (θ=0.001) at marker D12S345. Screening of the coding sequence of several genes (PRPH, CNTN1, RAPGEF3, KIF21A, and FGD4) lying in the candidate interval (fig. 2A), by direct sequencing, allowed us to identify FGD4, encoding FGD4/FRABIN, as a strong candidate (fig. 2B and 2C). Indeed, two homozygous missense variations were found in the two initially linked families: a c.893T→G transversion and a c.893T→C transition in the Lebanese and Algerian families, respectively (fig. 2D). Both mutations segregated perfectly with the phenotype in each pedigree and affected the same nucleotide in FGD4 exon 7. These sequence variations were not found in 216 Lebanese or 108 Algerian control chromosomes and were thus considered to be disease-causing mutations. Multiple alignment of FRABIN with Frabin orthologs from vertebrates and invertebrates indicated that both mutations target a highly conserved amino acid residue (fig. 2E) through evolution. A specific alignment of FRABIN with the four known members of the FGD Rho GEF family in human also demonstrates a perfect conservation of the methionine at position 298 (fig. 2F).
Figure 2. .
CMT4H locus, structure of FGD4 and FRABIN, mutations, and conservation between species. A, Chromosomal region at 12p11.21-q13.11 containing the CMT4H candidate locus, as identified elsewhere,26 covering a 15.8-Mb region located between STR markers D12S1648 and D12S1661. STR markers homozygous by descent are shown in red. The 48 known genes lying in this region are schematically represented, with candidate genes tested in our study indicated in red (FGD4, CNTN1, KIF21A, and RAPGEF3). B, Structure of FGD4. The gene is located at 12p11.21, near the centromere, and is transcribed from telomere to centromere, as indicated by the black arrow. The gene, covering a genomic region of 138,580 bp, as defined by the GenBank reference sequence NM_139241, is composed of 17 exons. The transcript is 2,931 bp long, with a coding sequence (exons 3–17) of 2,301 bp (for mRNA [GenBank accession number AK057294]). The coding region is indicated in gray and UTRs (5′ and 3′) in white. The coding sequence includes exons 3–17. Additional exons found in alternative splice variants are described in the databases but are not represented here. C, Five functional domains presented by FRABIN, encoded by FGD4: FAB, DH presenting a Rho GEF activity, PH1 and PH2, which bind to phosphatidylinositol(3,4,5)P3, and one FYVE domain that interacts with phosphatidylinositol3P. D, Chromatograms showing the two mutations identified in FGD4: the c.893T→G transversion in the Lebanese patients and the c.893T→C transition in the Algerian patient. E and F, The methionine at residue 298, mutated in both patients, which is highly conserved in different species (vertebrates and nonvertebrates), as well as in other members of the human FGD family.
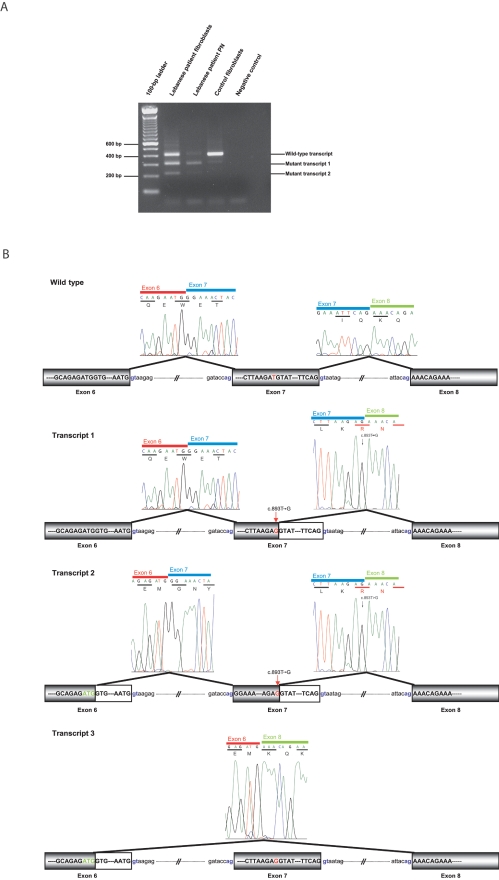
With use of total RNA extracted from lymphoblastoid cell lines or fibroblasts, FGD4 full-length cDNAs were amplified by RT-PCR in patients from both families. In the Algerian patient (295.6) (fig. 1), no abnormal transcript could be identified, which likely ruled out a possible splicing defect associated with this specific mutation and further indicated that the c.893C→T transition is likely to be a missense mutation leading to the replacement of a methionine by a threonine at codon 298 (p.Met298Thr) (fig. 2D). Conversely, in the Lebanese patient (500.21) (fig. 1), transcripts at a size lower than expected were retrieved, which suggested an associated splicing defect (fig. 3A). Indeed, further sequencing revealed a transcript (transcript 1) lacking the last 100 bp of exon 7, resulting from the use of an upstream donor splice site created by the c.893T→G mutation at codon 298 in exon 7, instead of the usual natural consensus 5′ donor site (fig. 3B). The c.893T→G mutation, initially predicted to be a missense p.Met298Arg mutation, is consequently also a splicing mutation predicted to likely result in a truncated protein of 305 aa instead of 766 aa (p.298MetfsX8) (fig. 3B). Persistence of the mutated full-length transcript was observed in the patient’s fibroblasts, whereas it was completely absent in the patient’s peripheral nerve (fig. 3A). Additionally, two other transcripts were also identified in this patient (fig. 3A and 3B): transcript 2, which lacked the last 99 bp of exon 6 in addition to the last 100 bp of exon 7, because of the use of a cryptic donor splice site in exon 6, and transcript 3, identified after cloning of RT-PCR fragments obtained from the patient’s lymphoblast cDNA, in which the last 99 bp of exon 6 and all of exon 7 are deleted (fig. 3B). For transcript 2, two homologous ESTs from different species lacking the end of exon 6 were found in the databases (GenBank accession number AA305646 for Homo sapiens and GenBank accession number CN643653 for Macaca mulatta), suggesting that it might be a naturally produced alternative transcript and that we describe here a mutant form of this transcript. This transcript would encode a 253-aa truncated protein. Transcript 3 was not reported in the databases and might either be produced by the mutation or be another tissue-specific alternative transcript. The predicted protein would harbor an 81-aa inframe deletion in the Dbl homology (DH) domain.
Figure 3. .
The c.893T→G splicing mutation. A, Results of RT-PCR experiments performed on the Lebanese patient’s fibroblasts and peripheral nerve (PN) and in the control fibroblasts, with use of a forward primer located in exon 5 and a reverse primer located in exon 8 (see table 1). In control fibroblasts, a 412-bp band corresponding to the wild-type transcript was obtained. In the Lebanese patient’s fibroblasts, we observed two additional bands corresponding to transcripts 1 and 2 described in panel B. In fibroblasts, a band the size of the wild-type transcript was observed, corresponding to the correctly spliced transcript with a homozygous p.Met298Arg missense mutation. B, Schematic representation of splicing defects caused by the c.893T→G Lebanese mutation observed in panel A. In the first mutant (transcript 1), the GT donor splice site created by the mutation was used instead of the usual consensus 3′ donor site. The transcript deleted from the last 100 bp of exon 7 is predicted to yield a 305-aa truncated protein, and the mutation is described as “p.Met298fsX8.” In transcript 2 (224 bp), in addition to the last 100 bp of exon 7, the last 99 bp of exon 6 are deleted because of the use of a cryptic donor splice site in exon 6. Transcript 3, identified after the cloning of RT-PCR fragments obtained from the patient’s lymphoblast cDNA, lacks the last 99 bp of exon 6 and all of exon 7.
Finally, in the course of full-length cDNA amplification, two other splice variants were identified, neither of which corresponded to the C-terminal splice variants of Frabin described in mouse (GenBank accession number NP_631978).42 In the first one, described in the databases, the entire initiation codon containing exon 3 is deleted. Removal of exon 3 leads to absence of the known AUG initiation codon. ORF predictions (National Center for Biotechnology Information ORF Finder) predict the use of a downstream AUG at codon 94 of the normal protein FRABIN that would lead to a 673-residue protein identical to FRABIN, but deprived of the first FAB domain. The second alternative transcript, produced by a mechanism similar to the one described in exon 6 for transcripts 2 and 3, leads to the deletion of the last 54 bp of exon 16. The predicted protein would present an 18-aa inframe deletion within the PH2 domain.
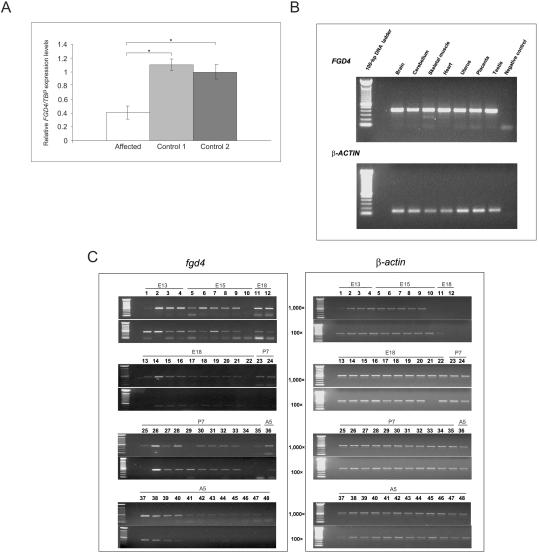
Transcriptional studies were conducted by RTQ-PCR with the use of RNAs extracted from fibroblasts of the Lebanese patient and controls (fig. 4A). This showed a 60% reduction in FGD4 mRNA levels, indicating that the truncated mRNA might be degraded through nonsense-mediated mRNA decay (NMD) and that the corresponding protein might not be produced. Unfortunately, a polyclonal antibody against FRABIN generated in rabbit but failed to detect FRABIN in control tissues (data not shown).
Figure 4. .
Expression analysis. A, RTQ-PCR experiments with use of cDNAs extracted from the patient’s fibroblasts and two controls. TBP was used as the reference gene for normalization. An approximate average twofold underexpression of FGD4 was observed in Lebanese patients with CMT4H as compared with calibrators (P<.05, Wilcoxon-Mann-Whitney test). Similar results were obtained using normalization with GUSB (data not shown). B, Semiquantitative expression of FGD4 in different human adult tissues. β-actin was used for normalization. C, Expression of FGD4 in different mouse nerve tissues. Two cDNA concentrations were used (100× and 1,000×). β-actin was used for normalization. Lanes (L) correspond to the following: L1, embryo day 13 (E13) telencephalon/diencephalons. L2, E13 mesencephalon (midbrain). L3, E13 rhombencephalon (hindbrain). L4, E13 spinal cord. L5, E15 telencephalon. L6, E15 diencephalon. L7, E15 midbrain. L8, E15 pons. L9, E15 medulla. L10, E15 spinal cord. L11, E18 frontal cortex. L12, E18 posterior cortex. L13, E18 entorhinal cortex. L14, E18 olfactory bulb. L15, E18 hippocampus. L16, E18 striatum. L17, E18 thalamus. L18, E18 hypothalamus. L19, E18 midbrain. L20, E18 pons. L21, E18 medulla. L22, E18 spinal cord. L23, postnatal day 7 (P7) frontal cortex. L24, P7 posterior cortex. L25, P7 entorhinal cortex. L26, P7 olfactory bulb. L27, P7 hippocampus. L28, P7 striatum. L29, P7 thalamus. L30, P7 hypothalamus. L31, P7 cerebellum. L32, P7 midbrain. L33, P7 pons. L34, P7 medulla. L35, P7 spinal cord. L36, adult 5-wk (A5) frontal cortex. L37, A5 posterior cortex. L38, A5 entorhinal cortex. L39, A5 olfactory bulb. L40, A5 hippocampus. L41, A5 striatum. L42, A5 thalamus. L43, A5 hypothalamus. L44, A5 cerebellum. L45, A5 midbrain. L46, A5 pons. L47, A5 medulla. L48, A5 spinal cord. FGD4 was expressed in all parts of the brain and in spinal cord, but with stronger expression at embryonic and prenatal stages than at postnatal and adult stages.
FGD4 is known to be ubiquitously expressed, at least in all tested tissues.37 In this study, we have also detected its expression in different human tissues including peripheral nerve (figs. 3A and 4B). Additionally, we checked the Fgd4 expression pattern in mouse nerve tissues at different stages of development and demonstrated that it was expressed in all parts of the brain and in the spinal cord at embryonic, postnatal, and adult stages (fig. 4C). Interestingly, levels of expression were lower in postnatal and adult tissues than in embryonic tissues.
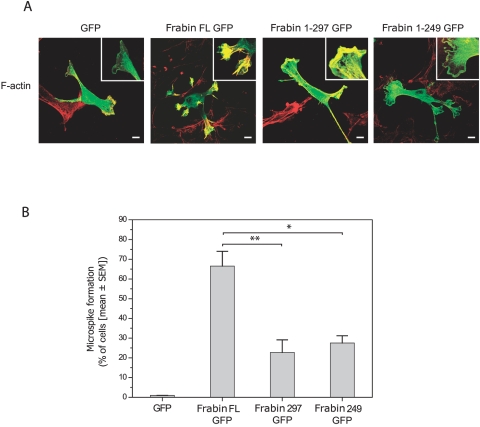
To further explore the functional consequences of the truncating mutation identified, we analyzed, by confocal microscopy, embryonic rat spinal motoneurons and rat RT4 schwannoma cells after transfection with expression vectors encoding GFP-tagged forms of either wild-type rat Frabin (GenBank accession number NP_640356) or truncated forms of Frabin corresponding to p.M298fsX8 (fig. 5A). In electroporated rat motoneurons, wild-type full-length Frabin (i) colocalized with phalloidin-stained f-actin in neurite tips and growth cones (not shown), in line with previous data obtained in other cell types,36,37,43,44 and (ii) induced the formation of filopodia-like microspikes (fig. 5B). Expression of full-length Frabin resulted in a 3.75-fold increase in the mean number of microspikes per motoneuron as compared with GFP-expressing motoneurons (fig. 5B and 5C). When the truncated forms of Frabin were expressed in motoneurons, they still colocalized with f-actin but induced significantly fewer microspikes than did full-length Frabin (fig. 5B and 5C). Moreover, the remaining microspikes acquired an abnormally curled shape (fig. 5B). Similar results were observed in rat RT4 schwannoma cells, in which the expression of wild-type full-length Frabin induced formation of microspikes, whereas the expression of truncated Frabin significantly inhibited this effect (fig. 6).
Figure 5. .
A, Domain organization of rat Frabin constructs. Frabin FL GFP = full-length Frabin C-terminally tagged to GFP. Frabin 1–297 GFP = GFP-tagged Frabin lacking the last 469 aa, mimicking the predicted protein resulting from mutant transcript 1 (see fig. 3); Frabin 1–249 GFP = GFP-tagged Frabin lacking the last 513 aa, mimicking the protein corresponding to mutant transcript 2 (see fig. 3). B, Confocal images of rat motoneurons transduced with GFP or GFP-tagged Frabin expression constructs and labeled at 1 DIV for f-actin using Alexa Fluor 546-conjugated phalloidin. While GFP shows uniform cytoplasmic expression, FL Frabin GFP colocalizes with f-actin in microspikes and induces their formation (see merged images and insets). Note reduced number and abnormal shape of microspikes induced by expression of truncated Frabin constructs. Scale bar = 10 μm. C, Diagram showing reduced microspike formation in motoneurons expressing truncated forms of Frabin as compared with motoneurons expressing full-length Frabin (n=25 cells per condition, P<.001 as assessed by Mann-Whitney rank sum test).
Figure 6. .
A, Confocal images showing cultured rat RT4 schwann cells transfected with expression constructs for GFP, full-length (FL) Frabin-GFP or truncated Frabin-GFP (see fig. 5). Cells are co-labeled for f-actin (in red); details of membrane structures are magnified (see insets). Scale bars = 10 μm. B, Diagram showing reduced microspike formation in RT4 cell cultures transfected with truncated forms of Frabin (Frabin 297 GFP and Frabin 249 GFP) as compared with cultures transfected with Frabin FL GFP (* indicates P=.03 and ** indicates P=.011, as assessed by Student t test).
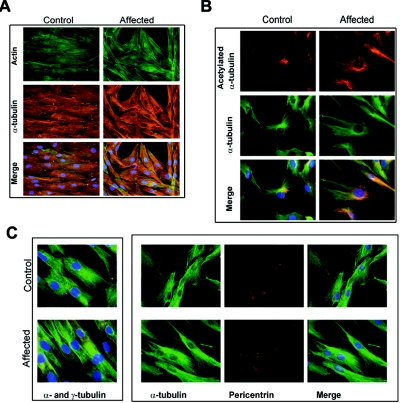
FRABIN is a GEF specific for Cdc42, a Rho GTPase involved in the reorganization of not only actin microfilaments but also the microtubule cytoskeleton.34,45 We consequently tested whether the microfilament (actin-labeling) or microtubule (α-tubulin and acetylated α-tubulin) cytoskeleton or centrosome (γ-tubulin) was disorganized in patient fibroblasts as compared with control fibroblasts. We did not observe any significant abnormalities of the microfilaments or in the distribution and organization of the microtubules (fig. 7).
Figure 7. .
Labeling of microfilament and microtubule cytoskeleton and centrosomes in the control and Lebanese patient (500.21) fibroblasts (see fig. 1). A, Labeling of microfilament (antibody to F-actin) and microtubules (antibody to α-tubulin). No significant differences were observed between the control and patient fibroblasts. B, Labeling of total microtubules (antibody to α-tubulin) and stable microtubules (antibody to acetylated α-tubulin). Stabilized microtubules were denser at centrosomes, but no structural difference was noticed in patients compared with controls. C, Labeling of centrosome with use of antibody to γ-tubulin or pericentrin and antibody to α-tubulin. Centrosome position and structure seemed normal in both patient and control fibroblasts.
Discussion
CMT diseases are a clinically and genetically heterogeneous group of HMSNs, with 50 loci and 30 genes identified to date. These genes encode proteins of various functions: myelin structural proteins; transcription factors involved in myelin gene regulation; proteins involved in protein-sorting synthesis or degradation, transport processes, cytoskeleton and mitochondrial dynamics.46 In this study, we report two mutations in FGD4—the gene encoding FRABIN, an F-actin Rho GEF specific to Cdc4236,37—in patients affected with CMT4H, an autosomal recessive form of demyelinating CMT disease. This 766-aa protein is composed of five functional domains: an N-terminal F-actin binding (FAB) domain, one Dbl homology (DH) domain, two pleckstrin homology (PH) domains, and one cysteine-rich FYVE domain36 (fig. 2C). DH domains were first identified in the Dbl protein and are present in many proteins, where they play a key role in the catalysis of GDP-to-GTP exchange,47 whereas PH and FYVE domains are mainly involved in interactions with different forms of phosphoinositides.48,49
Both mutations in FGD4 described here modify the same nucleotide and affect an evolutionarily highly conserved amino acid residue at position 298 of the FRABIN protein, indicating functional relevance. As this residue lies within the DH domain, responsible for Cdc42 activation, we might postulate that the mutations described here affect or prevent Cdc42 activation, although through different mechanisms. In the Algerian family, the p.Met298Thr mutation might change the conformation of the domain and prevent binding to Cdc42. In the Lebanese family, the p.Met298Arg mutation is in fact a splicing mutation predicted to result in a truncated protein (p.Met298fsX8) lacking not only the terminal part of the DH domain and the first PH domain, which, together with the FAB domain, have been shown to be necessary for Cdc42 activation,43 but also the second PH and FYVE domains, which, together with the DH domain and the first PH domain, are necessary for c-Jun N-terminal kinase (JNK) activation37 and bind to polyphosphoinositides.36 JNK is a member of the stress-activated protein kinases family, a type of mitogen-activated protein kinases described in brain as “microtubule-associated protein kinases” for their phosphorylation of microtubule-associated proteins and other neuronal cytoskeletal proteins. It is one of the key factors involved in morphogenesis in various multicellular organisms and regulates planar cell polarity, cell migration, and apoptosis.50 Suppression of JNK activation due to the p.Met298fsX8 mutation might consequently perturb Schwann cell morphology, polarity, or proliferation or might lead to a lack of activation of transcription factors required for myelination, thus causing aberrant myelination in peripheral nerves. Additionally, in mouse nerve tissues, we found that Fgd4 was expressed in all parts of the brain and in the spinal cord at embryonic, postnatal, or adult stages. Interestingly, levels of expression are lower in postnatal and adult tissues than in embryonic tissues.
This result, together with the facts that FRABIN activates JNK36 and that the JNK pathway is inhibited at the onset of myelination,51 suggests that FGD4 might have a role in early stages of myelination or that its expression leads to inhibition of myelination in prenatal stages. The lack of activation of the JNK pathway may lead to early activation of myelination, which would lead to early, aberrant myelination. This hypothesis is consistent with the observation that the patients present with an early-onset form of CMT with slow progression. Moreover, it has been recently demonstrated that, whereas JNK is present throughout the cell, activated phosphorylated JNK is enriched in the axon throughout development.52 Therefore, with consideration of the close relationship between axons and Schwann cells, the Lebanese mutation in FGD4 might affect myelination through the axon.
In this study, we show that the truncated protein resulting from the Lebanese splicing mutation, if produced, lacks its two PH and FYVE domains, interacting respectively with phosphatidylinositol(3,4,5)P3 (PtdIns(3,4,5)P3) and phosphatidylinositol3P (PtdIns3P). These phospholipids are rare components of membranes and have a role in recruiting proteins to membranes for signal transduction and membrane trafficking as well as for cytoskeleton regulation and apoptosis.49 All these functions might be affected by the mutations found in FGD4. Particularly, it has been described that the signals that affect myelination arising from axons in response to the binding of nerve growth factor to axonal TrkA receptors might involve PI3K and polyphosphoinositides.53 Moreover, MTMR2 and MTMR13, two members of a large family of tyrosine/dual phosphatases, are mutated in CMT4B19 and CMT4B2,12 respectively. These proteins dephosphorylate PtdIns3P and PtdIns3,5P2, which are notably involved in vacuolar transport and membrane trafficking, and recent observations on myotubularin homologues in the nematode Caenorhabditis elegans indicate a role in endocytosis.54 FGD4 might consequently be involved in a signaling pathway involving the myotubularins mutated in other forms of CMT.
As previously reported for CTDP1, in which a mutation lying 389 bp downstream of the donor splice site is responsible for the syndromic neuropathy CCFDN,55 we observed the persistence of the mutated full-length transcript in the fibroblasts and lymphoblasts of the Lebanese patients, even though the mutation is homozygous in patients (fig. 3A). This result suggests that the splice mutation might induce a partial use of the created splicing site and thus explain the clinical intrafamilial heterogeneity observed in the Lebanese family.26 However, the significant reduction in FGD4 expression level evidenced by RTQ-PCR in the Lebanese patient’s cells indicates that the truncated mRNA might be degraded through NMD and that the protein might not be produced. Moreover, FGD4 expression levels in the patient’s peripheral nerve showed a complete absence of the full-length transcript (fig. 3A), consistent with a loss-of-function mutation severely affecting the function of FRABIN, at least in peripheral nerve tissues.
More than 85 GEFs for Rho GTPases have been identified to date,34 and several genes encoding Rho GEFs have been involved in human genetic disorders mainly affecting the CNS or peripheral nervous system. As paradigms, mutations in FGD1 (GenBank accession number NP_004454) and ARHGEF6 (PIX) cause Aarskog syndrome56 and MRX46,57 respectively, two X-linked mental retardation syndromes. As well, a mutation in ARHGEF10 has been associated with slowed NCVs, an endophenotype of demyelinating CMT,32 and ALSIN2 (ALS2) is mutated in juvenile amyotrophic lateral sclerosis58,59 and infantile ascending hereditary spastic paralysis.60
Along with Rho GAPs and Rho GDIs, Rho GEFs are involved in cytoskeleton dynamics through Rho GTPases, the latter having a key role in remodeling actin microfilaments, an essential step for many cell mechanisms, including shape, adhesion, and migration.34,35
Using overexpression of normal and truncated forms of Frabin in embryonic rat spinal motoneurons and rat RT4 schwannoma cells, we show here that cells transfected with expression vectors encoding wild-type rat Frabin colocalize with F-actin in neurite tips and growth cones, in line with previous data obtained in other cell types,36,37,43,44 and induce the formation of filopodia-like microspikes, whereas cells expressing the truncated forms of Frabin still colocalize with F-actin but induce significantly fewer microspikes than does full-length Frabin. Similar results were observed in rat RT4 schwannoma cells, in which the expression of wild-type full-length Frabin induced formation of microspikes, whereas the expression of truncated Frabin significantly inhibited this effect. These data, together with the known requirement of the FAB, DH, and PH domains for microspike formation,37,42,43,61,62 strongly suggest that the p.Met298fsX8 mutation causes disease through a loss-of-function mechanism.
As previously said, FRABIN is a GEF specific to Cdc42, a Rho GTPase involved in the reorganization of actin microfilaments but also the microtubule cytoskeleton.34,45 However, we could not evidence any significant abnormalities of the microfilaments or in the distribution and organization of the microtubules and centrosomes. This may be explained by the fact that the full-length mutated transcript persisted in the patient’s fibroblasts, whereas only the truncated transcript persisted in the patient's peripheral nerves. Moreover, several studies have demonstrated that the roles of FRABIN vary from one cell type to another,36,37,43,44 specifically with respect to which Rho GTPases (Cdc42, Rac, and/or Rho) are indirectly activated and their interrelationships in specific cell types. It is thus likely that FGD4 mutations have different effects in different cell types, and, in particular, that Schwann cells and neurons must be more severely affected. Finally, the affinity of FRABIN for Cdc42 was shown to be weak in comparison with other GEFs such as Dbl.37 Consequently, we cannot exclude the possibility that FGD4 mutations have a weak impact on Cdc42 activation and cytoskeleton organization and that FRABIN instead activates other GTPases and possibly GTPases already known to be involved in CMT diseases, such as MFN2,29 DNM2,30 or RAB7.31
Together, our results demonstrate that mutations in FGD4 cause CMT4H, a rare AR-CMT disease. FRABIN is the sixth protein (after MFN2,29 DNM2,30 RAB7,31 ARHGEF10,32 and SEPT933) related to GTPase signaling but the first Rho GEF to be identified in CMT disease. It is noteworthy that several mechanisms and pathways leading to the pathology remain to be elucidated. However, taking into account the structure and function of FRABIN, as well as the predicted effects of the identified mutations, several hypotheses can be postulated. First, disorganization of the actin/microtubule cytoskeleton might perturb cell division, polarity, and migration and the ensheathment processes by Schwann cells, as well as microtubule-driven mRNA transport in Schwann cells or axons.53 Second, loss of activation of the JNK pathways could lead to an aberrant myelination process in Schwann cells.51 Third, various signaling pathways mediated by polyphosphoinositides—in particular, Schwann cell-axon communications—might be damaged. Overall, FRABIN could play a key role in the proliferation, polarization, and survival of Schwann cells and myelination processes. Like myotubularins, a family of proteins with members found to be mutated in CMT subtypes,9,12 FRABIN seems to be involved in polyphosphoinositide signaling. The identification of FRABIN as a new defective protein in CMT disease is thus a major stimulus for efforts aimed at a better understanding of the pathophysiology of these diseases, as well as the relationships between the Schwann cell and the axon. The identification of the underlying pathogenic mechanisms is under way and, we hope, will provide useful insights into the functional relationships between the different proteins involved in CMT disease.
Acknowledgments
We express our sympathy to the families and gratitude for their full cooperation throughout the study. The authors thank Sandrine Peireira and Daniele Depetris for excellent technical assistance, Marie-Christine Lebart for assistance in the production of a rabbit polyclonal antibody to FRABIN, Prof. Gérard Lefranc for constant support and logistics, Prof. Pierre Cau for helpful comments and criticisms during the study, and Dr. Mike Mitchell for English revisions of this manuscript. We are grateful to Prof. Oliver Hanneman, who kindly provided RT4 schwannoma cell lines, and to Prof. Y. Takai for sharing rat Frabin constructs. We are extremely grateful to Prof. Ruti Parvari and Dina Aranovic for their help in cytoskeleton explorations. This work was financially supported by the Network for Rare Diseases (GIS maladies rares), the Association Française contre les Myopathies (AFM), the Agence Universitaire de la Francophonie, and INSERM. T.H. is supported by a grant from AFM.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih/Genbank/ (for KIF21A [accession number AY368076]; RAPGEF3 [accession number U78168]; FGD4 [accession number NM_139241]; FGD4 mRNA [accession numbers AK057294, BC045552, and AY367054] and ESTs for H. sapiens [accession number AA305646] and Macaca mulatta [accession number CN643653]; Fgd4 [accession number NM_139234]; FGD4 homologs FGD1 [accession number NP_004454], FGD2 [accession number NP_775829], and FGD3 [NP_149077]; and FRABIN proteins for H. sapiens [accession number NP_640334], Rattus norvegicus [accession number NP_640356], Mus musculus [accession number NP_631978], Macaca fascicularis [accession number BAE90450], Caenorhabditis elegans [accession number NP_001023039], Canis familiaris [accession number XP_543741], Pan troglodytes [accession number XP_520721], Gallus gallus [accession number XP_416365], Apis mellifera [accession number XP_394280], and Danio rerio [accession number CAK11116])
- Inherited Peripheral Neuropathies Mutation Database, http://www.molgen.ua.ac.be/CMTMutations/default.cfm
- Neuromuscular Disease Center, http://www.neuro.wustl.edu/neuromuscular/time/hmsn.html (for comprehensive and updated data about CMT diseases)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for GDAP1, CMT4A, MTMR2, CMT4B1, SBF2/MTMR13, CMT4B2, SH3CT2/KIAA1985, CMT4C, NDRG1, HMSN-Lom, EGR2, P0, PMP22, CMT4E, Dejerine-Sottas syndrome, PRX, CMT4F, HMSN-Russe, and CMT4H)
- ORF Finder, http://www.ncbi.nlm.nih.gov/gorf/gorf.html
- Primer3, http://frodo.wi.mit.edu/ (for primer designs)
- UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway
References
- 1.Skre H (1974) Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin Genet 6:98–118 [DOI] [PubMed] [Google Scholar]
- 2.Dyck PJ, Chance P, Lebo R, Carney AJ (1993) Hereditary motor and sensory neuropathies. In: Dyck PJ, Thomas PK, Griffin JW, Low PA, Podulso JF (eds) Peripheral neuropathy. WB Saunders, Philadelphia, pp 1094–1136 [Google Scholar]
- 3.Villanova M, Timmerman V, De Jonghe P, Malandrini A, Rizzuto N, Van Broeckhoven C, Guazzi G, Rossi A (1998) Charcot-Marie-Tooth disease: an intermediate form. Neuromuscul Disord 8:392–393 10.1016/S0960-8966(98)00044-3 [DOI] [PubMed] [Google Scholar]
- 4.Shy ME (2004) Charcot-Marie-Tooth disease: an update. Curr Opin Neurol 17:579–585 10.1097/00019052-200410000-00008 [DOI] [PubMed] [Google Scholar]
- 5.Berciano J, Combarros O (2003) Hereditary neuropathies. Curr Opin Neurol 16:613–622 10.1097/00019052-200310000-00008 [DOI] [PubMed] [Google Scholar]
- 6.Cuesta A, Pedrola L, Sevilla T, Garcia-Planells J, Chumillas MJ, Mayordomo F, Leguern E, Marin I, Vilchez JJ, Palau F (2002) The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat Genet 30:22–24 10.1038/ng798 [DOI] [PubMed] [Google Scholar]
- 7.Baxter RV, Ben Othmane K, Rochelle JM, Stajich J, Hulette C, Dew-Knight S, Hentati F, Ben Hamida M, Bel S, Stenger JE, et al (2002) Ganglioside-induced differentiation-associated protein-1 is mutated in Charcot-Marie-Tooth disease type 4A/8q21. Nat Genet 30:21–22 10.1038/ng796 [DOI] [PubMed] [Google Scholar]
- 8.Ben Othmane K, Hentati F, Lennon F, Ben Hamida C, Blel S, Roses AD, Pericak-Vance MA, Ben Hamida M, Vance JM (1993) Linkage of a locus (CMT4A) for autosomal recessive Charcot-Marie-Tooth disease to chromosome 8q. Hum Mol Genet 2:1625–1628 10.1093/hmg/2.10.1625 [DOI] [PubMed] [Google Scholar]
- 9.Bolino A, Muglia M, Conforti FL, LeGuern E, Salih MA, Georgiou DM, Christodoulou K, Hausmanowa-Petrusewicz I, Mandich P, Schenone A, et al (2000) Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet 25:17–19 10.1038/75542 [DOI] [PubMed] [Google Scholar]
- 10.Bolino A, Brancolini V, Bono F, Bruni A, Gambardella A, Romeo G, Quattrone A, Devoto M (1996) Localization of a gene responsible for autosomal recessive demyelinating neuropathy with focally folded myelin sheaths to chromosome 11q23 by homozygosity mapping and haplotype sharing. Hum Mol Genet 5:1051–1054 10.1093/hmg/5.7.1051 [DOI] [PubMed] [Google Scholar]
- 11.Azzedine H, Bolino A, Taieb T, Birouk N, Di Duca M, Bouhouche A, Benamou S, Mrabet A, Hammadouche T, Chkili T, et al (2003) Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. Am J Hum Genet 72:1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senderek J, Bergmann C, Weber S, Ketelsen UP, Schorle H, Rudnik-Schoneborn S, Buttner R, Buchheim E, Zerres K (2003) Mutation of the SBF2 gene, encoding a novel member of the myotubularin family, in Charcot-Marie-Tooth neuropathy type 4B2/11p15. Hum Mol Genet 12:349–356 10.1093/hmg/ddg030 [DOI] [PubMed] [Google Scholar]
- 13.Othmane KB, Johnson E, Menold M, Graham FL, Hamida MB, Hasegawa O, Rogala AD, Ohnishi A, Pericak-Vance M, Hentati F, et al (1999) Identification of a new locus for autosomal recessive Charcot-Marie-Tooth disease with focally folded myelin on chromosome 11p15. Genomics 62:344–349 10.1006/geno.1999.6028 [DOI] [PubMed] [Google Scholar]
- 14.Senderek J, Bergmann C, Stendel C, Kirfel J, Verpoorten N, De Jonghe P, Timmerman V, Chrast R, Verheijen MH, Lemke G, et al (2003) Mutations in a gene encoding a novel SH3/TPR domain protein cause autosomal recessive Charcot-Marie-Tooth type 4C neuropathy. Am J Hum Genet 73:1106–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeGuern E, Guilbot A, Kessali M, Ravise N, Tassin J, Maisonobe T, Grid D, Brice A (1996) Homozygosity mapping of an autosomal recessive form of demyelinating Charcot-Marie-Tooth disease to chromosome 5q23-q33. Hum Mol Genet 5:1685–1688 10.1093/hmg/5.10.1685 [DOI] [PubMed] [Google Scholar]
- 16.Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P, et al (2000) N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum Genet 67:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalaydjieva L, Hallmayer J, Chandler D, Savov A, Nikolova A, Angelicheva D, King RH, Ishpekova B, Honeyman K, Calafell F, et al (1996) Gene mapping in gypsies identifies a novel demyelinating neuropathy on chromosome 8q24. Nat Genet 14:214–217 10.1038/ng1096-214 [DOI] [PubMed] [Google Scholar]
- 18.Timmerman V, De Jonghe P, Ceuterick C, De Vriendt E, Lofgren A, Nelis E, Warner LE, Lupski JR, Martin JJ, Van Broeckhoven C (1999) Novel missense mutation in the early growth response 2 gene associated with Dejerine-Sottas syndrome phenotype. Neurology 52:1827–1832 [DOI] [PubMed] [Google Scholar]
- 19.Warner LE, Hilz MJ, Appel SH, Killian JM, Kolodry EH, Karpati G, Carpenter S, Watters GV, Wheeler C, Witt D, et al (1996) Clinical phenotypes of different MPZ (P0) mutations may include Charcot-Marie-Tooth type 1B, Dejerine-Sottas, and congenital hypomyelination. Neuron 17:451–460 10.1016/S0896-6273(00)80177-4 [DOI] [PubMed] [Google Scholar]
- 20.Ikegami T, Nicholson G, Ikeda H, Ishida A, Johnston H, Wise G, Ouvrier R, Hayasaka K (1996) A novel homozygous mutation of the myelin P0 gene producing Dejerine-Sottas disease (hereditary motor and sensory neuropathy type III). Biochem Biophys Res Commun 222:107–110 10.1006/bbrc.1996.0705 [DOI] [PubMed] [Google Scholar]
- 21.Roa BB, Dyck PJ, Marks HG, Chance PF, Lupski JR (1993) Dejerine-Sottas syndrome associated with point mutation in the peripheral myelin protein 22 (PMP22) gene. Nat Genet 5:269–273 10.1038/ng1193-269 [DOI] [PubMed] [Google Scholar]
- 22.Guilbot A, Williams A, Ravise N, Verny C, Brice A, Sherman DL, Brophy PJ, LeGuern E, Delague V, Bareil C, et al (2001) A mutation in periaxin is responsible for CMT4F, an autosomal recessive form of Charcot-Marie-Tooth disease. Hum Mol Genet 10:415–421 10.1093/hmg/10.4.415 [DOI] [PubMed] [Google Scholar]
- 23.Boerkoel C, Takashima H, Stankiewicz P, Garcia C, Leber S, Rhee-Morris L, Lupski J (2001) Periaxin mutations cause recessive Dejerine-Sottas neuropathy. Am J Hum Genet 68:325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delague V, Bareil C, Tuffery S, Bouvagnet P, Chouery E, Koussa S, Maisonobe T, Loiselet J, Megarbane A, Claustres M (2000) Mapping of a new locus for autosomal recessive demyelinating Charcot-Marie-Tooth disease to 19q13.1-13.3 in a large consanguineous Lebanese family: exclusion of MAG as a candidate gene. Am J Hum Genet 67:236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers T, Chandler D, Angelicheva D, Thomas PK, Youl B, Tournev I, Gergelcheva V, Kalaydjieva L (2000) A novel locus for autosomal recessive peripheral neuropathy in the EGR2 region on 10q23. Am J Hum Genet 67:664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Sandre-Giovannoli A, Delague V, Hamadouche T, Chaouch M, Krahn M, Boccaccio I, Maisonobe T, Chouery E, Jabbour R, Atweh S, et al (2005) Homozygosity mapping of autosomal recessive demyelinating Charcot-Marie-Tooth neuropathy (CMT4H) to a novel locus on chromosome 12p11.21-q13.11. J Med Genet 42:260–265 10.1136/jmg.2004.024364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laporte J, Blondeau F, Buj-Bello A, Mandel J (2001) The myotubularin family: from genetic disease to phosphoinositide metabolism. Trends Genet 17:221–228 10.1016/S0168-9525(01)02245-4 [DOI] [PubMed] [Google Scholar]
- 28.Niemann A, Ruegg M, La Padula V, Schenone A, Suter U (2005) Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J Cell Biol 170:1067–1078 10.1083/jcb.200507087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, et al (2004) Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet 36:449–451 10.1038/ng1341 [DOI] [PubMed] [Google Scholar]
- 30.Zuchner S, Noureddine M, Kennerson M, Verhoeven K, Claeys K, De Jonghe P, Merory J, Oliveira SA, Speer MC, Stenger JE, et al (2005) Mutations in the pleckstrin homology domain of dynamin 2 cause dominant intermediate Charcot-Marie-Tooth disease. Nat Genet 37:289–294 10.1038/ng1514 [DOI] [PubMed] [Google Scholar]
- 31.Verhoeven K, de Jong P, Coen K, Verpoorten N, Auer-Grumbach M, Kwon J, FitzPatrick D, Schmedding E, De Vriendt E, Jacobs A, et al (2003) Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet 72:722–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verhoeven K, De Jonghe P, Van de Putte T, Nelis E, Zwijsen A, Verpoorten N, De Vriendt E, Jacobs A, Van Gerwen V, Francis A, et al (2003) Slowed conduction and thin myelination of peripheral nerves associated with mutant Rho guanine-nucleotide exchange factor 10. Am J Hum Genet 73:926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhlenbaumer G, Hannibal MC, Nelis E, Schirmacher A, Verpoorten N, Meuleman J, Watts GD, De Vriendt E, Young P, Stogbauer F, et al (2005) Mutations in SEPT9 cause hereditary neuralgic amyotrophy. Nat Genet 37:1044–1046 10.1038/ng1649 [DOI] [PubMed] [Google Scholar]
- 34.Jaffe AB, Hall A (2005) Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 21:247–269 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- 35.Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420:629–635 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- 36.Obaishi H, Nakanishi H, Mandai K, Satoh K, Satoh A, Takahashi K, Miyahara M, Nishioka H, Takaishi K, Takai Y (1998) Frabin, a novel FGD1-related actin filament-binding protein capable of changing cell shape and activating c-Jun N-terminal kinase. J Biol Chem 273:18697–18700 10.1074/jbc.273.30.18697 [DOI] [PubMed] [Google Scholar]
- 37.Umikawa M, Obaishi H, Nakanishi H, Satoh-Horikawa K, Takahashi K, Hotta I, Matsuura Y, Takai Y (1999) Association of Frabin with the actin cytoskeleton is essential for microspike formation through activation of Cdc42 small G protein. J Biol Chem 274:25197–25200 10.1074/jbc.274.36.25197 [DOI] [PubMed] [Google Scholar]
- 38.Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 10.1016/0003-2697(87)90021-2 [DOI] [PubMed] [Google Scholar]
- 39.Jacquier A, Buhler E, Schafer MK, Bohl D, Blanchard S, Beclin C, Haase G (2006) Alsin/Rac1 signaling controls survival and growth of spinal motoneurons. Ann Neurol 60:105–117 10.1002/ana.20886 [DOI] [PubMed] [Google Scholar]
- 40.Henderson CE, Bloch-Gallego E, Camu W (1995) Purification and culture of embryonic spinal motoneurons. In: Cohen J, Wilkin G (eds) Nerve cell culture: a practical approach. Oxford University Press, London, pp 69–81 [Google Scholar]
- 41.Hai M, Muja N, DeVries GH, Quarles RH, Patel PI (2002) Comparative analysis of Schwann cell lines as model systems for myelin gene transcription studies. J Neurosci Res 69:497–508 10.1002/jnr.10327 [DOI] [PubMed] [Google Scholar]
- 42.Ikeda W, Nakanishi H, Takekuni K, Itoh S, Takai Y (2001) Identification of splicing variants of Frabin with partly different functions and tissue distribution. Biochem Biophys Res Commun 286:1066–1072 10.1006/bbrc.2001.5481 [DOI] [PubMed] [Google Scholar]
- 43.Ono Y, Nakanishi H, Nishimura M, Kakizaki M, Takahashi K, Miyahara M, Satoh-Horikawa K, Mandai K, Takai Y (2000) Two actions of frabin: direct activation of Cdc42 and indirect activation of Rac. Oncogene 19:3050–3058 10.1038/sj.onc.1203631 [DOI] [PubMed] [Google Scholar]
- 44.Yasuda T, Ohtsuka T, Inoue E, Yokoyama S, Sakisaka T, Kodama A, Takaishi K, Takai Y (2000) Importance of spatial activation of Cdc42 and rac small G proteins by frabin for microspike formation in MDCK cells. Genes Cells 5:583–591 10.1046/j.1365-2443.2000.00349.x [DOI] [PubMed] [Google Scholar]
- 45.Cau J, Hall A (2005) Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J Cell Sci 118:2579–2587 10.1242/jcs.02385 [DOI] [PubMed] [Google Scholar]
- 46.Niemann A, Berger P, Suter U (2006) Pathomechanisms of mutant proteins in Charcot-Marie-Tooth disease. Neuromolecular Med 8:217–241 10.1385/NMM:8:1:217 [DOI] [PubMed] [Google Scholar]
- 47.Aghazadeh B, Zhu K, Kubiseski TJ, Liu GA, Pawson T, Zheng Y, Rosen MK (1998) Structure and mutagenesis of the Dbl homology domain. Nat Struct Biol 5:1098–1107 10.1038/4209 [DOI] [PubMed] [Google Scholar]
- 48.Haslam RJ, Koide HB, Hemmings BA (1993) Pleckstrin domain homology. Nature 363:309–310 10.1038/363309b0 [DOI] [PubMed] [Google Scholar]
- 49.Gillooly DJ, Simonsen A, Stenmark H (2001) Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochem J 355:249–258 10.1042/0264-6021:3550249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kortenjann M, Shaw PE (1995) The growing family of MAP kinases: regulation and specificity. Crit Rev Oncog 6:99–115 [PubMed] [Google Scholar]
- 51.Jessen KR, Mirsky R (2005) The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci 6:671–682 10.1038/nrn1746 [DOI] [PubMed] [Google Scholar]
- 52.Oliva AA Jr, Atkins CM, Copenagle L, Banker GA (2006) Activated c-Jun N-terminal kinase is required for axon formation. J Neurosci 26:9462–9470 10.1523/JNEUROSCI.2625-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherman DL, Brophy PJ (2005) Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci 6:683–690 10.1038/nrn1743 [DOI] [PubMed] [Google Scholar]
- 54.Laporte J, Bedez F, Bolino A, Mandel JL (2003) Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum Mol Genet 12:R285–R292 10.1093/hmg/ddg273 [DOI] [PubMed] [Google Scholar]
- 55.Varon R, Gooding R, Steglich C, Marns L, Tang H, Angelicheva D, Yong KK, Ambrugger P, Reinhold A, Morar B, et al (2003) Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nat Genet 35:185–189 10.1038/ng1243 [DOI] [PubMed] [Google Scholar]
- 56.Pasteris NG, Cadle A, Logie LJ, Porteous ME, Schwartz CE, Stevenson RE, Glover TW, Wilroy RS, Gorski JL (1994) Isolation and characterization of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell 79:669–678 10.1016/0092-8674(94)90552-5 [DOI] [PubMed] [Google Scholar]
- 57.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, et al (2000) Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet 26:247–250 10.1038/80002 [DOI] [PubMed] [Google Scholar]
- 58.Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, et al (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 10.1038/ng1001-160 [DOI] [PubMed] [Google Scholar]
- 59.Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, et al (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 10.1038/ng1001-166 [DOI] [PubMed] [Google Scholar]
- 60.Eymard-Pierre E, Yamanaka K, Haeussler M, Kress W, Gauthier-Barichard F, Combes P, Cleveland DW, Boespflug-Tanguy O (2006) Novel missense mutation in ALS2 gene results in infantile ascending hereditary spastic paralysis. Ann Neurol 59:976–980 10.1002/ana.20879 [DOI] [PubMed] [Google Scholar]
- 61.Ikeda W, Nakanishi H, Tanaka Y, Tachibana K, Takai Y (2001) Cooperation of Cdc42 small G protein-activating and actin filament-binding activities of frabin in microspike formation. Oncogene 20:3457–3463 10.1038/sj.onc.1204463 [DOI] [PubMed] [Google Scholar]
- 62.Kim Y, Ikeda W, Nakanishi H, Tanaka Y, Takekuni K, Itoh S, Monden M, Takai Y (2002) Association of frabin with specific actin and membrane structures. Genes Cells 7:413–420 10.1046/j.1365-2443.2002.00524.x [DOI] [PubMed] [Google Scholar]