Abstract
Ataxia telangiectasia and Rad3-related (ATR) protein, a kinase that regulates a DNA damage–response pathway, is mutated in ATR-Seckel syndrome (ATR-SS), a disorder characterized by severe microcephaly and growth delay. Impaired ATR signaling is also observed in cell lines from additional disorders characterized by microcephaly and growth delay, including non–ATR-SS, Nijmegen breakage syndrome, and MCPH1 (microcephaly, primary autosomal recessive, 1)–dependent primary microcephaly. Here, we examined ATR-pathway function in cell lines from three haploinsufficient contiguous gene-deletion disorders—a subset of blepharophimosis-ptosis-epicanthus inversus syndrome, Miller-Dieker lissencephaly syndrome, and Williams-Beuren syndrome—in which the deleted region encompasses ATR, RPA1, and RFC2, respectively. These three genes function in ATR signaling. Cell lines from these disorders displayed an impaired ATR-dependent DNA damage response. Thus, we describe ATR signaling as a pathway unusually sensitive to haploinsufficiency and identify three further human disorders displaying a defective ATR-dependent DNA damage response. The striking correlation of ATR-pathway dysfunction with the presence of microcephaly and growth delay strongly suggests a causal relationship.
Ataxia telangiectasia and Rad3-related (ATR) protein is a central component of a DNA damage–response signaling pathway.1,2 ATR (MIM *601215) is mutated in two families displaying Seckel syndrome (SS) (MIM #210600), a disorder characterized by severe microcephaly, proportionate dwarfism, and dysmorphic facial features.3–5 SS is clinically and genetically heterogeneous.6 Significantly, cell lines derived from additional patients with SS, although not harboring mutations in ATR, display ATR-signaling defects.6 Thus, SS can be attributed to defects in ATR signaling, with a subset of patients, designated “ATR-SS,” having mutations in ATR itself. Additionally, three other disorders characterized by microcephaly and growth delay—Nijmegen breakage syndrome (MIM #251260), Fanconi anemia (MIM #227650), and MCPH1 (microcephaly, primary autosomal recessive, 1)–defective primary microcephaly (MIM #251200)—display impaired ATR-signaling responses.7,8 Together, these findings suggest that impaired ATR signaling can impact development, conferring microcephaly and growth delay.
ATR is a phosphoinositol 3-kinase–like kinase (PIKK) that is activated by single-stranded (ss) regions of DNA generated after replication-fork stalling or during the repair of bulky lesions.9 ATR interacts with ATRIP (ATR-interacting protein) and is recruited to ssDNA regions, in part by ATRIP’s ability to bind to replication protein A (RPA), a complex of three subunits, RPA1–3.9–11 The Rad17/Rfc2–5 complex, together with a complex involving Rad9, Rad1, and Hus1, also functions to enhance ATR signaling, by impacting either ATR recruitment or activation.12 Thus, these additional proteins are required for the ATR-signaling response and therefore represent potential candidate genetic defects for SS.6 Hence, defects in these genes may confer “Seckel-like” clinical features.
High-resolution genetic mapping studies have led to the characterization of contiguous-gene-deletion disorders caused by heterozygous microdeletions that result in haploinsufficiency for a single or, more usually, several genes.13,14 It is likely that the clinical manifestations of these disorders arise from the combined impact of haploinsufficiency for multiple genes.15 It is also possible that there are critical genes or pathways sensitive to haploinsufficiency, either alone or when combined with haploinsufficiency for other genes. While investigating disorders exhibiting microcephaly and growth delay, we were struck by the observation that the microdeletion in three such disorders involved haploinsufficiency for genes involved in ATR-pathway function.
Blepharophimosis-ptosis-epicanthus inversus syndrome (BPES [MIM #110100]), which is characterized by small eye sockets (blepharophimosis), drooping eyelids (ptosis), and upward-folding inner eyelids (epicanthus inversus), is an autosomal dominant disorder caused by mutation of the putative forkhead transcription factor FOXL2 (MIM *605597).16,17 Seventeen cases of BPES with heterozygous interstitial deletions of various sizes on chromosome 3q leading to loss of FOXL2 have been documented (reviewed by de Rue et al.18). Of these, 13 patients were also reported to exhibit microcephaly and growth retardation, clinical features not normally associated with BPES. Recently, the microdeletion in one such patient was carefully mapped and was shown to encompass ATR, which localizes to the same region.18 It was therefore proposed that the non-BPES features observed in such patients might be due to haploinsufficiency for ATR.18
Hemizygous deletions on chromosome 17p also confer microcephaly and growth delay.19 PAHFAH1B1/Lis1 (MIM *601545) encodes a protein, lissencephaly 1 (Lis1) that functions in neuronal migration.20 Mutations in or heterozygous deletions of PAHFAH1B1/Lis1 alone cause isolated lissencephaly sequence (ILS), a disorder typified by reduced neuronal migration resulting in a “smooth brain” (lissencephaly).21 Larger deletions identified in some patients with ILS confer a more severe grade of lissencephaly associated with craniofacial abnormalities (ILS+). Even larger deletions extending from the PAHFAH1B1/Lis1 gene to the telomere are observed in Miller-Dieker lissencephaly syndrome (MDLS [MIM #247200]), a disorder characterized by the most severe grade of lissencephaly, with craniofacial abnormalities, microcephaly, and growth retardation. RPA1 (MIM *179835), the largest subunit of RPA, is heterozygously deleted in patients with MDLS and ILS+ but not ILS.19
Finally, Williams-Beuren syndrome (WBS [MIM #194050]) is caused by hemizygous deletion of chromosome 7q11.23, which results in the haploinsufficiency of multiple genes.22 WBS is characterized by facial dysmorphia, microcephaly, growth retardation, and supravalvular aortic stenosis (SVAS [MIM #185500]). Hemizygous deletion or mutations in ELN (MIM *130160) alone, the gene encoding elastin, a structural component of arteries, cause SVAS.23 Patients with WBS, in contrast, have larger deletions encompassing replication factor C2 (RFC2 [MIM *600404]), a subunit of replication factor C (RF-C).24 RF-C loads proliferating cell nuclear antigen (PCNA) onto chromatin during DNA replication, and four of its five subunits, Rfc2–5, form a complex with Rad17 that functions in ATR signaling.11,25–27
The association of severe microcephaly and growth retardation with heterozygous loss of genes encoding proteins involved in the ATR-signaling pathway prompted us to examine whether haploinsufficiency for these genes impacts the ATR-signaling response. We therefore examined cell lines obtained from these contiguous gene disorders for their ability to effect the ATR-dependent DNA-damage response. We employed sensitive assays capable of detecting defective ATR-pathway function that we had established elsewhere to examine SS cell lines. Strikingly, we observed defects in ATR signaling in all three disorders, demonstrating that ATR signaling is sensitive to haploinsufficiency.
Material and Methods
Cell Lines
Lymphoblastoid cell lines (LBLs) were cultured in RPMI 1640 with 15% fetal calf serum. GM02188 (wild type [WT]) and DK0064 (ATR-SS) have been described elsewhere.6 VD9396 (BPES-ATR+/−) LBLs were obtained from J.M.v.H.18 ILS, ILS+, and MDLS LBLs—DR00-063a1 (Con-MR), LP99-017 (ILS A), LP94-013 (ILS B), LP90-017 (ILS+ A), LP99-086 (ILS+ B), LP91-026 (ILS+ C), L95-059 (MDLS-A), LP92-005 (MDLS-B), LP90-006 (MDLS-C), and LP88-002 (MDLS-D)—have been described elsewhere and were provided by W.B.D.19 WBS and respective parental LBLs—GM14183 (WT-I), GM14182 (WBS-I), GM14295 (WT-II), and GM14297 (WBS-II)—were obtained from Coriell Cell Repository.
Antibodies
Antibodies were obtained as follows. α-ATR (N19), α-Lis1 (H-300), and α-β-tubulin (H235) were obtained from Autogen Bioclear; α-RPA1 (Ab-1), α-H2AX (DR-1016), and α-NBS1 (Ab-1) were obtained from Merck; α–γ-H2AX, α-pS10 Histone H3, and α-ATRIP were obtained from Upstate Technology; and α-pSer317-Chk1 was obtained from New England Biolabs.
Western Blotting and Chromatin Extraction
Western blotting was performed as described elsewhere.8 For γ-H2AX analysis, a chromatin extraction step was included. In brief, 1×107 cells were washed once in PBS and were resuspended in 100 μl hypotonic buffer (10 mM HEPES [pH 7.5], 5 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol, 10 mM NaF, 1 mM Na2VO3, 10 mM β-glycerolphosphate, 0.5% IPEGAL/Nonident P-40, and Protease Inhibitor Cocktail from Sigma). Lysates were incubated on ice for 15 min, were pelleted, and were washed twice (200 μl each) in hypotonic buffer. The pellet was treated with hypertonic buffer (hypotonic buffer with 0.5 M NaCl) and was incubated on ice for 15 min. After washing in hypertonic buffer, the chromatin pellets were resuspended in 100 μl of SDS-PAGE loading buffer (with 5% SDS, 10% β-mercaptoethanol) and were sonicated. Ten microliters of the chromatin fraction was separated on 17% SDS-PAGE.
G2/M Checkpoint Arrest
Cells were irradiated with 5 J/m2 UV-C, were immediately seeded into complete medium with 1.5 μM nocodazole, and were incubated for 24 h before being cytospun onto poly-D-lysine–coated slides and processed for immunofluorescence with α-pS10-Histone H3 and for counterstaining with 4',6-diamidino-2-phenylindole.
Nuclear Fragmentation (NF)
Cells were treated with 5 mM hydroxyurea (HU) in the presence of 1.5 μM nocodazole for 24 h and were processed as described elsewhere.6
Transfection
With use of Genejuice (Novagen), 3×105 cells/ml, in 3-ml quantities, were transiently transfected with 2 μg of pcDNA3-ATR, pcDNA3.1-RPA1, or pcDNA3.1-RFC2, according to the manufacturer's instructions, and were incubated for 24 h before NF processing. For complementation of the G2/M checkpoint, cells were incubated for 24 h and were retransfected for a further 24 h before processing.
RNA-Interference Transfection
The control WT LBL GM02188 was used for small-interfering RNA (siRNA) experiments. Cells were transfected once with 10 nM of respective siRNA duplex (GFPi, ATRi, Lis1i, or RPA1i), with use of SiPort NeoFX transfection reagent according to the manufacturer's instructions (Ambion). The oligonucleotides (sense) used were ATR 5′-AAC CUC CGU GAU GUU GCU UGA-3′, RPA1 5′-AAA CCA UCC ACG AAG CUU AUA GGC C-3′, Lis1 5′-UUG AUU UGG CCG UAC CAU ACG UAC C-3′, and a control oligonucleotide directed to GFP 5′-AAC ACU UGU CAC UAC UUU CTC.
Results
ATR Signaling Impaired in a BPES Cell Line Haploinsufficient for ATR
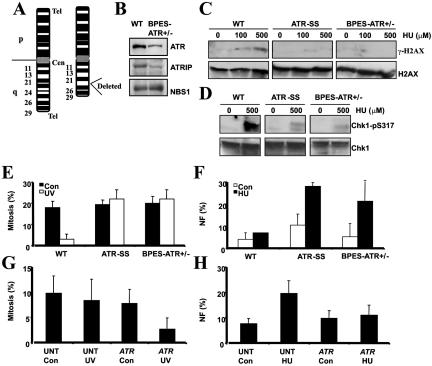
A previous study reported heterozygosity for ATR due to an interstitial deletion on 3q in a boy with BPES who displayed non-BPES clinical features that included mental retardation (MR), microcephaly, and growth delay.18 A representation of the deletion is shown in figure 1A. We obtained an LBL derived from this patient (BPES-ATR+/−) and, by immunoblotting, observed approximately twofold reduced ATR protein levels compared with those of a control LBL (fig. 1B). The level of ATRIP was also reduced approximately twofold, consistent with reports that ATR and ATRIP are coregulated.10 NBS1 served as a loading control and was expressed at similar levels in both lines (fig. 1B). An early step in the DNA damage response regulated by ATR is phosphorylation of the histone H2A variant, H2AX, on serine 139 (termed “γ-H2AX”), which can be detected by the immunoblotting of chromatin-bound proteins with α–γ-H2AX antibodies.28 After exposure to HU, an agent that causes replication-fork stalling, a strong γ-H2AX signal was observed in control LBLs, indicating activation of ATR-dependent damage-response signaling. However, in marked contrast, detectable γ-H2AX was not observed in the ATR-SS LBLs, nor in LBLs derived from the patient with BPES-ATR+/− (fig. 1C). Chk1 represents an important phosphorylation target of ATR that is required for the stability of arrested replication forks and cell-cycle arrest.29–33 HU-induced phosphorylation of Chk1 on serine 317, a known ATR target site, was also significantly diminished in LBLs from the patient with BPES-ATR+/−, similar to ATR-SS LBLs (fig. 1D).6 An important end point of ATR activation regulated by Chk1 is onset of G2/M checkpoint arrest, which serves to prevent cells harboring DNA damage from entering mitosis.6 To monitor G2/M checkpoint arrest, the percentage of mitotic cells was examined in untreated cells or 24 h postirradiation with UV (5 Jm−2). Control LBLs showed a marked reduction in mitotic cells due to arrest at the G2/M checkpoint, whereas ATR-SS and BPES-ATR+/− cells showed a mitotic index (MI) similar to that observed in the absence of UV treatment (fig. 1E). A further hallmark of impaired ATR signaling is the presence of cells with NF after treatment with HU.6 ATR-SS and BPES-ATR+/− LBLs showed markedly elevated levels of cells displaying NF after HU treatment, in contrast to those of control LBLs (fig. 1F). To verify that the failure to effect G2/M checkpoint arrest and that the damage-induced NF phenotype of BPES-ATR+/− cells were directly attributable to an impaired ATR response, the cells were transfected with ATR cDNA and were reexamined for these phenotypes. Significant UV-induced G2/M arrest (fig. 1G) and reduced NF (fig. 1H) were observed after transfection with ATR cDNA.
Figure 1. .
BPES-ATR+/− cells, which display an impaired ATR-dependent damage response. A, Chromosome 3 karyotype of the patient with BPES-ATR+/− showing the heterozygous deletion, del(3)(q23,q25). ATR localizes to 3q22-q24. Cen=centromere; Tel=telomere. B, WCE (100 μg) from WT and BPES-ATR+/− LBLs, analyzed by immunoblotting using α-ATR, ATRIP, and NBS1 antibodies. Reduced expression of ATR and ATRIP is seen in BPES-ATR+/− cells specifically. NBS1 served as a loading control and was expressed at normal levels. C, WT, ATR-SS, and BPES-ATR+/− LBLs, exposed to 100 or 500 μM HU for 1 h before chromatin fractionation. ATR-SS and BPES-ATR+/− cells display reduced γ-H2AX compared with WT cells. Blots were reprobed using α-H2AX to confirm loading. D, WT, ATR-SS, and BPES-ATR+/− LBLs, exposed to 500 μM HU for 1 h before extraction. ATR-SS and BPES-ATR+/− cells display reduced Chk1-pS317 formation compared with WT cells. Blots were reprobed using α-Chk1 to confirm loading. E, ATR-SS and BPES-ATR+/− LBLs showing defective UV-induced (5 J/m2) G2/M checkpoint arrest 24 h postirradiation. Arrest in WT LBLs is seen as a decrease in the MI after UV irradiation. F, ATR-SS and BPES-ATR+/− LBLs that, unlike WT cells, show increased NF after 24 h treatment with HU (5 mM). G, UV-induced G2/M defect in BPES-ATR+/− LBLs, complemented after transfection with ATR cDNA. BPES-ATR+/− cells, either untransfected (UNT) or transfected (ATR) with pc-DNA3-ATR, were UV irradiated (5 J/m2), and the MI was determined after 24 h. H, HU-induced NF in BPES-ATR+/−, complemented after transfection with ATR cDNA. BPES-ATR+/− LBLs either untransfected (UNT) or transfected (ATR) with pc-DNA3-ATR were untreated (Con=control) or treated with HU (5 mM) 24 h posttransfection. NF was analyzed 24 h posttreatment with HU.
Collectively, these results provide strong evidence that haploinsufficiency for ATR in the BPES-ATR+/− cell line confers an impaired response to DNA damage that is similar in magnitude to that observed in an ATR-SS cell line.
Cells Haploinsufficient for RPA1 Show Deficient ATR-Dependent DNA Damage–Response Signaling
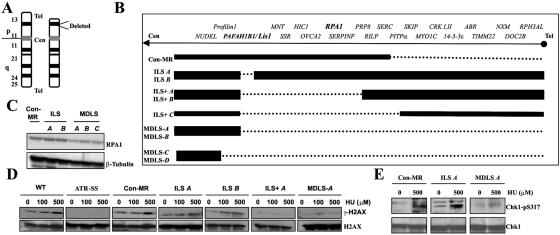
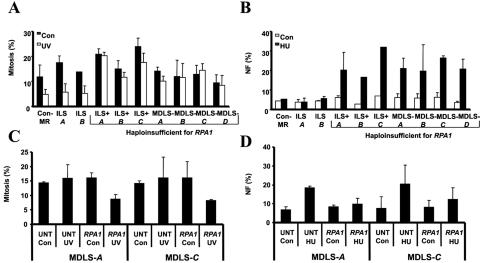
A representation of the deletion on chromosome 17p observed in MDLS is shown in figure 2A. The size of the heterozygous deletions on chromosome 17p in a panel of LBLs derived from patients with ILS, ILS+, and MDLS and from a control patient (Con-MR) are shown in figure 2B. ILS A and ILS B were derived from patients with mild ILS due to microdeletions involving Lis1 only. ILS+ A, ILS+ B, and ILS+ C were obtained from patients with more-severe ILS and craniofacial abnormalities. MDLS-A, -B, -C, and -D, which have deletions extending from Lis1 to the telomere, were derived from MDLS-affected patients with the severest grade of lissencephaly together with microcephaly and growth retardation. Con-MR, which has a heterozygous telomeric deletion that does not involve either RPA1 or Lis1, was derived from a patient with mild MR (fig. 2B). This patient does not exhibit MDLS, lissencephaly, microcephaly, or growth delay. The location of RPA1 is shown in figure 2B, demonstrating that this gene is deleted in some but not all LBLs in the panel. To examine whether haploinsufficiency for RPA1 correlated with impaired ATR-dependent damage-response signaling, we examined the response to DNA damage in this panel of LBLs. First, we examined RPA1 expression in whole-cell extracts (WCEs) derived from Con-MR, ILS A, and ILS B, compared with MDLS-A, -B, and -C. All three MDLS LBLs displayed reduced levels of RPA1 compared with those of Con-MR, ILS A, and ILS B LBLs, which is consistent with the deletion mapping (fig. 2B). Next, we examined HU-induced γ-H2AX formation in representative LBLs, either haploinsufficient for RPA1 or with both copies of the gene. Impaired HU-induced γ-H2AX was observed in the ATR-SS, ILS+ A, and MDLS A LBLs, the latter two of which exhibit haploinsufficiency for RPA1 (fig. 2B and 2D). Normal HU-induced γ-H2AX was observed in WT control LBLs and in those cell lines in which the deletion did not encompass RPA1 (Con-MR, ILS A, and ILS B) (fig. 2B and 2D). Similarly, MDLS-A LBLs also exhibited impaired HU-induced Chk1-pSer317 formation, unlike Con-MR and ILS A LBLs (fig. 2E). We also examined the ability to activate UV-induced G2/M checkpoint arrest and observed impaired arrest specifically in the seven LBLs with haploinsufficiency for RPA1 (figs. 2B and 3A). Furthermore, we examined HU-induced NF and observed elevated levels specifically in those LBLs from patients haploinsufficient for RPA1 (figs. 2B and 3B).
Figure 2. .
MDLS cells, which display an impaired ATR-dependent DNA damage response. A, Chromosome 17 karyotype of MDLS highlighting the heterozygous deletion at 17p13.3. B, Deletion mapping in the panel of LBLs from patients with ILS, ILS+, and MDLS. The dashed line indicates the heterozygously deleted region. Con-MR is a control LBL from a patient with mild MR who does not exhibit lissencephaly, microcephaly, growth retardation, or MDLS but has a hemizygous telomeric deletion that does not involve either RPA1 or PAFAH1B1/Lis1. ILS A and ILS B denote patients with low-grade ILS due to microdeletions involving PAFAH1B1/Lis1 only. ILS+A, ILS+B, and ILS+C denote patients with larger deletions, a more severe ILS, and additional craniofacial abnormalities, whereas MDLS-A, -B, -C, and -D denote patients with the largest deletions, who exhibit the most severe grade of lissencephaly along with microcephaly and growth retardation. The positions of PAFAH1B1/Lis1 and RPA1 are highlighted. C, Western-blot analysis of RPA1 expression from WCEs from Con-MR, ILS A, ILS B, MDLS-A, MDLS-B, and MDLS-C showing reduced expression of RPA1, specifically in the three MDLS cell lines. D, Defective HU-induced γ-H2AX formation, which segregates with RPA1 haploinsufficiency. Cells were treated as described for figure 1C. Defective γ-H2AX formation is seen in ATR-SS, ILS+ A, and MDLS-A cells, compared with the normal response in WT, Con-MR, ILS A, and ILS B cells. E, Impaired HU-induced Chk1-pS317, seen in MDLSA LBLs, compared with those of Con-MR and ILS A. Cells were treated with 500 μM HU for 1 h before extraction and were reprobed using α-Chk1 to confirm loading.
Figure 3. .
Haploinsufficiency of RPA1, which specifically segregates with a defective ATR-dependent DNA damage response. A, Defective ATR-dependent G2/M checkpoint arrest, which segregates with RPA1 haploinsufficiency. Con-MR, ILS A, and ILS B cells showing a reduction in MI (percentage of mitosis) at 24 h after UV irradiation (5 J/m2), indicating G2/M checkpoint arrest. ILS+ A, ILS+ B, ILS+ C, and MDLS-A, -B, -C, and -D cell lines failed to show a decrease in MI after UV treatment. B, Increased HU-induced NF segregating with RPA1 haploinsufficiency. No increase in HU-induced NF is seen in Con-MR, ILS A, or ILS B cells. In contrast, ILS+ A, ILS+ B, ILS+ C, and MDLS-A, -B, -C, and -D cells show elevated HU-induced NF. C, The ATR-dependent UV-induced G2/M defect in MDLS cells, complemented after transfection with RPA1 cDNA. MDLS-A and MDLS-C cells, either untransfected (“UNT”) or transfected (“RPA1”) with pc-DNA3-RPA1, were unirradiated (Con=control) or UV-irradiated (“UV”) (5 J/m2), and the MI was determined after 24 h. RPA1 cDNA specifically corrected the G2/M checkpoint defect of these cells, as seen by the reduced UV-induced MI after transfection (“RPA1 UV”), compared with untransfected irradiated cells (“UNT UV”). D, The increased HU-induced NF seen in MDLS, complemented after transfection with RPA1 cDNA. MDLS-A and -C, either untransfected (“UNT”), or transfected (“RPA1”) with pc-DNA3-RPA1 were untreated (“Con”) or treated with HU (5 mM) 24 h posttransfection. A reduction in HU-induced NF is specifically seen after transfection of MDLS LBLs with RPA1 cDNA (“RPA1 HU”), compared with the untransfected (“UNT HU”) cells.
To verify that the G2/M checkpoint and NF phenotypes were a consequence of haploinsufficiency for RPA1, these damage-response phenotypes were examined after transfection of two MDLS LBLs (MDLS-A and MDLS-C) with RPA1 cDNA. Expression of RPA1 cDNA resulted in recovery of G2/M checkpoint arrest and a diminished number of cells displaying HU-induced NF in both LBLs (fig. 3C and 3D).
Mildly Decreased Expression of RPA1 and ATR but Not Lis1 with Use of siRNA Affects ATR-Pathway Function after DNA Damage
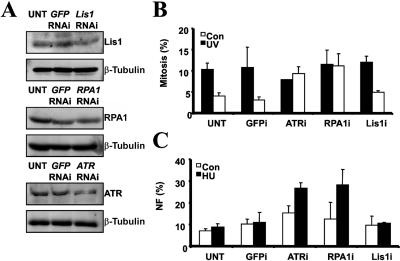
As an alternative way to determine whether the ATR-dependent DNA damage response might be sensitive to small perturbations in ATR or RPA1 expression, we used a single round of transfection with a low concentration (10 nM) of siRNA oligonucleotides to mildly reduce the expression of Lis1, RPA1, and ATR in the control LBLs. We obtained an approximately twofold reduction in Lis1, RPA1, and ATR expression in control LBLs (fig. 4A). This reduction in RPA1 and ATR protein levels conferred a failure to activate G2/M checkpoint arrest after UV irradiation (fig. 4B). In contrast, the clearly observable reduction in Lis1 expression after siRNA did not impact UV-induced G2/M checkpoint arrest (fig. 4B). Similarly, mild siRNA-mediated reduction in the expression of ATR and RPA1 caused elevated levels of NF after HU treatment, which was not observed after siRNA with use of oligonucleotides directed against Lis1 or GFP (fig. 4C).
Figure 4. .
Inefficient siRNA of ATR or RPA1, which impairs the ATR-dependent DNA damage response. A, WT LBLs transfected once with a low concentration (10 nM) of siRNA oligonucleotides for GFP, Lis1, RPA1, or ATR and analyzed for expression of Lis1, RPA1, and ATR by western blotting, using β-tubulin as a loading control, 24 h posttransfection. B, WT LBLs untransfected (UNT) or transfected with the indicated siRNA oligonucleotides, analyzed for UV-induced G2/M checkpoint arrest by monitoring MI. Transfection with siRNA oligonucleotides against ATR or RPA1 impaired G2/M arrest after UV. In contrast, after transfection with oligonucleotides against GFP and Lis1, an intact G2/M arrest was observed. C, WT LBLs transfected with the indicated siRNA oligonucleotides, analyzed for HU-induced NF. Transfection with siRNA oligonucleotides against ATR and RPA1 caused HU-induced NF, in contrast to the lack of impact of siRNA oligonucleotides against GFP and Lis1.
These results provide strong evidence suggesting that even a mild decrease in protein levels of either ATR or RPA1 impacts the ability of cells to mount a normal ATR-signaling response after exposure to DNA damage.
Impaired ATR-Dependent DNA Damage–Response Signaling in WBS: Correlation with Haploinsufficiency for RFC2
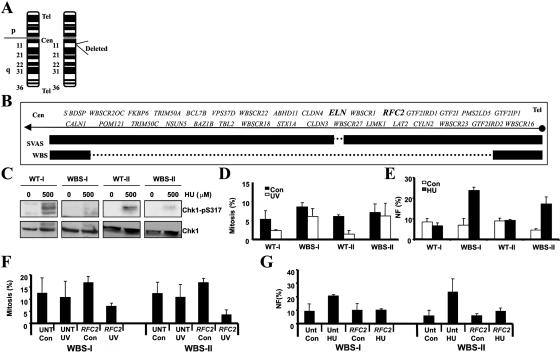
The heterozygous interstitial deletion on chromosome 7q11.23 that is seen in WBS is shown in figure 5A. The deletion encompasses multiple genes, including RFC2, a subunit of the Rad17/Rfc2-5 complex that plays a role in the ATR-dependent pathway (fig. 5B). Figure 5B also shows the microdeletion observed in a typical patient with SVAS that does not encompass RFC2 but includes ELN. Whereas a hypomorphic mutation in Saccharomyces cerevisiae RFC2 (rfc2-1) has been shown to impair normal cell-cycle checkpoint activation, a direct demonstration of a similar response in humans cells has not been described.34 To examine whether haploinsufficiency for RFC2 causes impaired ATR-dependent damage response, we examined two WBS LBLs (WBS-I and WBS-II) and LBLs derived from one clinically normal parent (WT-I and WT-II, respectively). Significantly, defective HU-induced Chk1-pS317 formation was also seen in WBS LBLs (WBS-I and WBS-II), compared with their clinically normal parental LBLs (WT-I and WT-II, respectively) (fig. 5C). Correspondingly, both WBS LBLs failed to induce any significant G2/M checkpoint arrest after exposure to UV, in contrast to the efficient checkpoint arrest observed in both parent lines (fig. 5D). Additionally, both WBS LBLs but neither parental line showed induction of NF after exposure to HU (fig. 5E). Elevated levels of G2/M checkpoint arrest (fig. 5F) and reduced HU-induced NF (fig. 5G) were observed in both WBS LBLs after transfection with RFC2 cDNA. Together, these data provide strong evidence that haploinsufficiency for RFC2 also confers an impaired ATR-dependent signaling response.
Figure 5. .
WBS cells, which show an impaired ATR-dependent DNA damage response. A, Chromosome 7 karyotype from WBS showing the location of the submicroscopic heterozygous interstitial deletion on chromosome 7q11.23. B, Deletion mapping of patients with WBS and SVAS showing the position of Elastin (ELN) and RFC2. The dashed line indicates the size of the heterozygous deletion. C, Impaired HU-induced Chk1-pS317, seen in WBS LBLs (WBS-I and WBS-II) compared with those of the clinically normal parent (WT-I and WT-II, respectively). Cells were treated with 500 μM HU for 1 h before extraction and were reprobed using α-Chk1 to confirm loading. D, WBS LBLs, which exhibit impaired UV-induced G2/M checkpoint arrest. The MI was determined 24 h after UV irradiation (5 J/m2). WBS-I and WBS-II LBLs show defective UV-induced G2/M arrest, unlike WT LBLs from their parents (WT-I and WT-II, respectively). E, Increased HU-induced NF, seen in WBS cell lines. LBLs were treated with HU (5 mM) and were examined for NF 24 h after treatment. Both WBS-I and WBS-II LBLs show increased HU-induced NF, unlike WT LBLs from their parents (WT-I and WT-II, respectively). F, UV-induced G2/M defect in WBS LBLs, complemented after transfection with RFC2 cDNA. WBS-I and WBS-II cells, either untransfected (“UNT”) or transfected (“RFC2”) with pc-DNA3-RFC2, were UV irradiated (“UV”) (5 J/m2), and the MI was determined after 24 h. A reduction in MI after UV irradiation is observed after transfection of WBS LBLs with RFC2 cDNA transfection (“RFC2 UV”) compared with the untransfected irradiated cells (“UNT UV”). G, The increased HU-induced NF seen in WBS LBLs, complemented after transfection with RFC2 cDNA. WBS-I and WBS-II, either untransfected (“UNT”) or transfected (“RFC2”) with pc-DNA3-RFC2, were treated with HU (5 mM) 24 h posttransfection. A reduction in HU-induced NF is seen after transfection of WBS LBLs with RFC2 cDNA (“RFC2 HU”), compared with the untransfected HU-treated (“UNT HU”) cells.
Discussion
Here, we examined cell lines derived from patients with BPES-ATR+/−, MDLS, and WBS, three distinct contiguous-gene-deletion disorders that manifest microcephaly and growth delay. In all cases, the heterozygous genomic deletions encompass a gene that is critical for the ATR-signaling response—namely, ATR itself (BPES-ATR+/−), RPA1 (MDLS), or RFC2 (WBS). Patient-derived cell lines haploinsufficient for any of these genes displayed an impaired response to DNA damage with use of assays that were selected to identify defective ATR signaling. Indeed, the defect was similar to that displayed by an SS cell line with a homozygous, hypomorphic mutation in ATR (ATR-SS).3,6 Our findings therefore provide strong evidence that haploinsufficiency for ATR, as well as additional components of the ATR-dependent signaling pathway, confers an impaired ability to respond to DNA damage or replication-fork stalling. Our assays were established to identify ATR-signaling defects in SS and employ relatively modest DNA damaging treatments, to allow the detection of potentially hypomorphic mutations.3,6 Indeed, we observed that, after more-dramatic treatments (e.g., high UV doses), the defect in ATR-SS cells is overridden, presumably because of the induction of a sufficient damage-response signal by the residual protein.3 This is distinct from other DNA damage–response assays, in which high doses are often used to overload the pathway and expose a repair defect.35 Although our assays are optimized to detect a subtle deficiency and may not completely reflect the role of ATR during development, they represent modest treatments that may not be entirely distinct from those occurring during cellular growth and development. Thus, the impact of haploinsufficiency appears to represent a unique phenotype of ATR signaling, in contrast with other DNA damage–response pathways.
Impaired ATR signaling is associated with microcephaly and growth delay in humans.3,7,8,36 The correlation of the presence of microcephaly and growth delay with impaired ATR-pathway dysfunction in the haploinsufficiency disorders presented here provides further evidence suggestive of a causal relationship. Since ATR-SS is a recessive disorder, ATR haploinsufficiency alone is unlikely to confer a clinical phenotype. Rather, the clinical features observed in these haploinsufficiency disorders are probably a consequence of combined haploinsufficiency for ATR-signaling genes and additional genes that may impact neuronal development and cell proliferation. Indeed, combined heterozygosity of Lis1 and 14-3-3ɛ has been shown to influence the severe clinical features of MDLS.37 Human brain size has increased dramatically during evolution, requiring enormous and rapid proliferation from a small number of precursor stem cells.38 Moreover, developing neurons incur high levels of oxidative DNA damage, placing a significant load on the damage-response pathways. Thus, the developing brain may have a high requirement for the ATR-signaling pathway, necessitating a diploid content of component proteins, particularly if further stress is imposed by haploinsufficiency of other genes. Mice, as models for haploinsufficiency disorders, have been used to investigate neuronal migration but may be limited in their application concerning microcephaly because of the relatively small size of murine brains compared with that of human brains.39,40
Finally, it should be noted that, to our knowledge, defective ATR-pathway function has not been described elsewhere for any of the contiguous-gene-deletion disorders investigated here (BPES-ATR+/−, MDLS, and WBS). Since increased life expectancy due to improved medical supervision is now a feature of conditions such as MDLS, a defective DNA damage response in this context may be relevant to potential tumor development.41 Furthermore, a defective DNA damage response can adversely affect treatment of malignancy by standard chemotherapeutic or bone marrow–transplantation regimens.42,43 It is unclear whether any of the conditions investigated here are tumor-predisposition conditions, although isolated reports exist of malignancy in patients with MDLS and particularly WBS.41,44–46 Interestingly, hypomorphic mutations in RFC subunits are associated with genetic instability in yeast, and a heterozygous missense mutation in RPA1 results in increased levels of lymphoid malignancy in mice.34,47,48 Defective ATR function has been described in various cancer types.49–51 Furthermore, it has been suggested that ATR may act as a tumor suppressor, when haploinsufficient, under certain circumstances.52
A recent study reported that copy-number variation of DNA sequences is a common genomic trait.53 Understanding the impact of haploinsufficiency is likely to be important for assessment of interindividual genetic variation, as well as the basis underlying haploinsufficiency disorders. In conclusion, we identify ATR signaling as a response sensitive to haploinsufficiency at the cellular level, with a provocative clinical link to microcephaly and growth delay. This provides novel insight into the impact of ATR-pathway haploinsufficiency and further strengthens the link between defective ATR-pathway function and the development of microcephaly and growth retardation in humans.
Acknowledgments
We thank Prof. Lehmann and Prof. Carr (Genome Damage and Stability Centre, University of Sussex) for critical reading of the manuscript. Thanks also to Gemma Alderton and Caroline Reis. We particularly acknowledge Dr. Hans Gille (Department of Clinical Genetics, Vrije Universiteit Medical Center) for making the BPES-ATR+/− LBL. The P.A.J. laboratory is supported by the UK Medical Research Council, the Human Frontiers Science Program, the UK Leukaemia Research Fund, the International Agency for Cancer Research, EU grant F16R-CT-2003-508842 (RiscRad), and 512113 (DNA Repair). M.O. is supported by the UK Leukaemia Research Fund, UK Medical Research Council, and Cancer Research UK.
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ATR, SS, Nijmegen breakage syndrome, Fanconi anemia, MCPH1, BPES, FOXL2, PAHFAH1B1/Lis1, MDLS, RPA1, WBS, SVAS, ELN, and RFC2)
References
- 1.Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15:2177–2196 10.1101/gad.914401 [DOI] [PubMed] [Google Scholar]
- 2.Shiloh Y (2001) ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev 11:71–77 10.1016/S0959-437X(00)00159-3 [DOI] [PubMed] [Google Scholar]
- 3.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA (2003) A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 33:497–501 10.1038/ng1129 [DOI] [PubMed] [Google Scholar]
- 4.O’Driscoll M, Jeggo PA (2003) Clinical impact of ATR checkpoint signalling failure in humans. Cell Cycle 2:194–195 [PubMed] [Google Scholar]
- 5.Seckel HPG (1960) Bird-headed dwarfs: studies in developmental anthropology including human proportions. Springer Karger, Basel, Switzerland [Google Scholar]
- 6.Alderton GK, Joenje H, Varon R, Borglum AD, Jeggo PA, O’Driscoll M (2004) Seckel syndrome exhibits cellular features demonstrating defects in the ATR signalling pathway. Hum Mol Genet 13:3127–3138 10.1093/hmg/ddh335 [DOI] [PubMed] [Google Scholar]
- 7.Stiff T, Reis C, Alderton GK, Woodbine L, O’Driscoll M, Jeggo PA (2005) Nbs1 is required for ATR-dependent phosphorylation events. EMBO J 24:199–208 10.1038/sj.emboj.7600504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alderton GK, Galbiati L, Griffith E, Surinya KH, Neitzel H, Jackson AP, Jeggo PA, O’Driscoll M (2006) Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol 8:725–733 10.1038/ncb1431 [DOI] [PubMed] [Google Scholar]
- 9.Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]
- 10.Cortez D, Guntuku S, Qin J, Elledge SJ (2001) ATR and ATRIP: partners in checkpoint signaling. Science 294:1713–1716 10.1126/science.1065521 [DOI] [PubMed] [Google Scholar]
- 11.Zou L, Liu D, Elledge SJ (2003) Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA 100:13827–13832 10.1073/pnas.2336100100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou L, Cortez D, Elledge SJ (2002) Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev 16:198–208 10.1101/gad.950302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 10.1016/S0168-9525(98)01555-8 [DOI] [PubMed] [Google Scholar]
- 14.Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, Fiegler H, Firth H, Sanlaville D, Winter R, Colleaux L, et al (2004) Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet 41:241–248 10.1136/jmg.2003.017731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page GP, George V, Go RC, Page PZ, Allison DB (2003) “Are we there yet?”: Deciding when one has demonstrated specific genetic causation in complex diseases and quantitative traits. Am J Hum Genet 73:711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crisponi L, Manila D, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, et al (2001) The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet 27:159–166 10.1038/84781 [DOI] [PubMed] [Google Scholar]
- 17.De Baere E, Dixon MJ, Small KW, Jabs EW, Leroy BP, Devriendt KG, Gillerot Y, Mortier G, Meire F, Van Maldergem L, et al (2001) Spectrum of FOXL2 gene mutations in blepharophimosis-ptosis-epicanthus inversus (BPES) families demonstrates a genotype-phenotype correlation. Hum Mol Genet 10:1591–1600 10.1093/hmg/10.15.1591 [DOI] [PubMed] [Google Scholar]
- 18.de Ru MH, Gille JJ, Nieuwint AW, Bijlsma JJ, van der Blij JB, van Hagen JM (2005) Interstitial deletion in 3q in a patient with blepharophimosis-ptosis-epicanthus inversus syndrome (BPES) and microcephaly, mild mental retardation and growth delay: clinical report and review of the literature. Am J Med Genetic A 137:81–87 10.1002/ajmg.a.30786 [DOI] [PubMed] [Google Scholar]
- 19.Cardoso C, Leventer RJ, Ward HL, Toyo-Oka K, Chung J, Gross A, Martin CL, Allanson J, Pilz DT, Olney AH, et al (2003) Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet 72:918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leventer RJ, Cardoso C, Ledbetter DH, Dobyns WB (2001) LIS1: from cortical malformation to essential protein of cellular dynamics. Trends Neurosci 24:489–492 10.1016/S0166-2236(00)01887-7 [DOI] [PubMed] [Google Scholar]
- 21.Cardoso C, Leventer RJ, Dowling JJ, Ward HL, Chung J, Petras KS, Roseberry JA, Weiss AM, Das S, Martin CL, et al (2002) Clinical and molecular basis of classical lissencephaly: mutations in the LIS1 gene (PAFAH1B1). Hum Mutat 19:4–15 10.1002/humu.10028 [DOI] [PubMed] [Google Scholar]
- 22.Tassabehji M (2003) Williams-Beuren syndrome: a challenge for genotype-phenotype correlations. Hum Mol Genet 12:R229–R237 10.1093/hmg/ddg299 [DOI] [PubMed] [Google Scholar]
- 23.Tassabehji M, Metcalfe K, Donnai D, Hurst J, Reardon W, Burch M, Read AP (1997) Elastin: genomic structure and point mutations in patients with supravalvular aortic stenosis. Hum Mol Genet 6:1029–1036 10.1093/hmg/6.7.1029 [DOI] [PubMed] [Google Scholar]
- 24.Wu YQ, Sutton VR, Nickerson E, Lupski JR, Potocki L, Korenberg JR, Greenberg F, Tassabehji M, Shaffer LG (1998) Delineation of the common critical region in Williams syndrome and clinical correlation of growth, heart defects, ethnicity, and parental origin. Am J Med Genet 78:82–89 [DOI] [PubMed] [Google Scholar]
- 25.Yao NY, Johnson A, Bowman GD, Kuriyan J, O’Donnell M (2006) Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J Biol Chem 281:17528–17539 10.1074/jbc.M601273200 [DOI] [PubMed] [Google Scholar]
- 26.Johnson A, Yao NY, Bowman GD, Kuriyan J, O’Donnell M (2006) The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem 281:35531–35543 10.1074/jbc.M606090200 [DOI] [PubMed] [Google Scholar]
- 27.Ellison V, Stillman B (2003) Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol 1:E33 10.1371/journal.pbio.0000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A (2004) H2AX: the histone guardian of the genome. DNA Repair 3:959–967 10.1016/j.dnarep.2004.03.024 [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Sanchez Y (2004) Chk1 in the DNA damage response: conserved roles from yeasts to mammals. DNA Repair 3:1025–1032 10.1016/j.dnarep.2004.03.003 [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al (2000) Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev 14:1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 31.Carr AM, Moudjou M, Bentley NJ, Hagan IM (1995) The chk1 pathway is required to prevent mitosis following cell-cycle arrest at “start.” Curr Biol 5:1179–1190 [DOI] [PubMed] [Google Scholar]
- 32.Petermann E, Caldecott KW (2006) Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle 5:2203–2209 [DOI] [PubMed] [Google Scholar]
- 33.Zachos G, Rainey M, Gillespie D (2003) Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J 22:713–723 10.1093/emboj/cdg060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noskov VN, Araki H, Sugino A (1998) The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol Cell Biol 18:4914–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Girard P-M, Riballo E, Begg A, Waugh A, Jeggo PA (2002) Nbs1 promotes ATM dependent phosphorylation events including those required for G1/S arrest. Oncogene 21:4191–4199 10.1038/sj.onc.1205596 [DOI] [PubMed] [Google Scholar]
- 36.O’Driscoll M, Jeggo PA (2006) The role of double-strand break repair-insights from human genetics. Nat Rev Genet 7:45–54 10.1038/nrg1746 [DOI] [PubMed] [Google Scholar]
- 37.Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns WB, Ledbetter D, et al (2003) 14-3-3ɛ is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet 34:274–285 10.1038/ng1169 [DOI] [PubMed] [Google Scholar]
- 38.Cox J, Jackson AP, Bond J, Woods CG (2006) What primary microcephaly can tell us about brain growth. Trends Mol Med 12:358–366 10.1016/j.molmed.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 39.Yingling J, Toyo-oka K, Wynshaw-Boris A (2003) Miller-Dieker syndrome: analysis of a human contiguous gene syndrome in the mouse. Am J Hum Genet 73:475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheen VL, Ferland JR, Harney RM, Hill S, Neal J, Banham AH, Brown P, Chenn A, Corbo J, Hecht J, et al (2006) Impaired proliferation and migration in human Miller-Dieker neural precursors. Ann Neurol 60:137–144 10.1002/ana.20843 [DOI] [PubMed] [Google Scholar]
- 41.Shizuyo U, Masaru K, Shigekazu K, Michihiko W (2006) Gallbladder cancer in a patient with Miller-Dieker syndrome. Acta Paediatrica 95:113–114 [DOI] [PubMed] [Google Scholar]
- 42.Bakhshi S, Cerosaletti KM, Concannon P, Bawle EV, Fontanesi J, Gatti RA, Bhambhani K (2003) Medulloblastoma with adverse reaction to radiation therapy in Nijmegen breakage syndrome. J Pediatr Hematol Oncol 25:248–251 10.1097/00043426-200303000-00013 [DOI] [PubMed] [Google Scholar]
- 43.Rogers PB, Plowman PN, Harris SJ, Arlett CF (2000) Four radiation hypersensitivity cases and their implications for clinical radiotherapy. Radiother Oncol 57:143–154 10.1016/S0167-8140(00)00249-8 [DOI] [PubMed] [Google Scholar]
- 44.Amenta S, Moschovi M, Sofocleous CH, Kostaridou S, Mavrou A, Fryssira H (2004) Non-Hodgkin lymphoma in a child with Williams syndrome. Cancer Genet Cytogenet 154:86–88 10.1016/j.cancergencyto.2004.02.004 [DOI] [PubMed] [Google Scholar]
- 45.Thornburg C, Roulston D, Castle V (2005 ) Burkitt lymphoma and Williams syndrome: a model for children with a multisystem disorder and malignancy. J Pediatr Hematol Oncol 27:109–111 10.1097/01.mph.0000153444.43816.ea [DOI] [PubMed] [Google Scholar]
- 46.Semmekrot BA, Rotteveel JJ, Bakker-Niezen SH, Logt F (1985–1986) Occurrence of an astrocytoma in a patient with Williams syndrome. Pediatr Neurosci 12:188–191 [DOI] [PubMed] [Google Scholar]
- 47.Kim H-S, Brill SJ (2001) Rfc4 Interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol Cell Biol 21:3725–3737 10.1128/MCB.21.11.3725-3737.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R, Carrion DV, Chin L, Kucherlapati R, Kolodner RD, et al (2005) Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat Genet 37:750–755 10.1038/ng1587 [DOI] [PubMed] [Google Scholar]
- 49.Lewis KA, Mullany S, Thomas B, Chien J, Loewen R, Shridhar V, Cliby WA (2005) Heterozygous ATR mutations in mismatch repair-deficient cancer cells have functional significance. Cancer Res 65:7091–7095 10.1158/0008-5472.CAN-05-1019 [DOI] [PubMed] [Google Scholar]
- 50.Liu A, Takakuwa T, Fujita S, Ham MF, Luo WJ, Daibata M, Aozasa K (2005) Alterations of DNA damage-response genes ATM and ATR in pyothorax-associated lymphoma. Lab Invest 85:436–446 10.1038/labinvest.3700235 [DOI] [PubMed] [Google Scholar]
- 51.Dierov J, Dierova R, Carroll M (2004) BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S phase checkpoint. Cancer Cell 5:275–285 10.1016/S1535-6108(04)00056-X [DOI] [PubMed] [Google Scholar]
- 52.Fang Y, Tsao CC, Goodman BK, Furumai R, Tirado CA, Abraham RT, Wang XF (2004) ATR functions as a gene dosage-dependent tumor suppressor on a mismatch repair-deficient background. EMBO J 23:3164–3174 10.1038/sj.emboj.7600315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Redon R, Ishikawa S, Fitch KR, Feuk LP, Andrews G, Fiegler D, Shapero H, Carson MH, Chen AR, Cho W, et al (2006) Global variation in copy number in the human genome. Nature 444:444–454 10.1038/nature05329 [DOI] [PMC free article] [PubMed] [Google Scholar]