Abstract
Mutational heterogeneity represents a significant barrier to development of therapies for many dominantly inherited diseases. For example, >100 mutations in the rhodopsin gene (RHO) have been identified in patients with retinitis pigmentosa (RP). The development of therapies for dominant disorders that correct the primary genetic lesion and overcome mutational heterogeneity is challenging. Hence, therapeutics comprising two elements—gene suppression in conjunction with gene replacement—have been investigated. Suppression is targeted to a site independent of the mutation; therefore, both mutant and wild-type alleles are suppressed. In parallel with suppression, a codon-modified replacement gene refractory to suppression is provided. Both in vitro and in vivo validation of suppression and replacement for RHO-linked RP has been undertaken in the current study. RNA interference (RNAi) has been used to achieve ∼90% in vivo suppression of RHO in photoreceptors, with use of adeno-associated virus (AAV) for delivery. Demonstration that codon-modifed RHO genes express functional wild-type protein has been explored transgenically, together with in vivo expression of AAV-delivered RHO-replacement genes in the presence of targeting RNAi molecules. Observation of potential therapeutic benefit from AAV-delivered suppression and replacement therapies has been obtained in Pro23His mice. Results provide the first in vivo indication that suppression and replacement can provide a therapeutic solution for dominantly inherited disorders such as RHO-linked RP and can be employed to circumvent mutational heterogeneity.
Elucidation of the molecular pathogenesis of inherited disorders has highlighted the presence of substantial levels of genetic heterogeneity (OMIM database). One of the most genetically heterogeneous conditions defined to date is retinitis pigmentosa (RP [MIM +180380]), which involves photoreceptor-cell degeneration and affects ∼1 in 3,000 people.1 More than 40 causative genes have been implicated in RP (Retinal Information Network database). Although it is timely to explore gene therapies for RP, inter- and intragenic heterogeneity represent significant barriers to therapeutic development. For example, >100 mutations in the human rhodopsin gene (RHO [GenBank accession number NM_000539.2]), which encodes the photosensitive pigment in rod photoreceptors, have been identified in autosomal dominantly inherited RP (adRP). Notably, the mutational heterogeneity in RHO-linked RP is mirrored in many other dominantly inherited conditions (OMIM database). Development of therapies for each individual mutation would be technically difficult to achieve and not economically viable; thus, a therapeutic approach that circumvents mutational diversity would be of great value. Elsewhere, we described a therapeutic strategy for dominant genetic diseases, termed “mutation-independent suppression and replacement.”2 This approach involves suppression of both mutant and wild-type alleles of a gene causative of a dominant disorder.2–4 Suppressors are designed to be complementary in nucleotide sequence to a site within a target transcript that is independent of the disease-causing mutation(s). Since both mutant and wild-type alleles are suppressed, suppression is undertaken in conjunction with the provision of codon-modified replacement gene(s) refractory to suppression, thereby providing the wild-type protein. Such an approach overcomes the need to design specific suppression agents targeting each mutation in a given gene, which enables correction of a genetic defect in a mutation-independent manner. However, the approach involves two components—suppression and replacement—and therefore is, in principle, more complex than replacement-only therapies for recessive disorders. For this reason, no in vivo demonstration of the approach has been achieved to date.
Exploiting the degeneracy of the genetic code to facilitate therapeutic development has been suggested for dominantly inherited conditions with the hallmark of mutational heterogeneity.1–4 More specifically, in relation to retinopathies, the approach has been proposed elsewhere for RHO- and peripherin-linked adRP.2,3,5 Studies have been undertaken, using ribozymes, that focused on the suppression component of the approach.6 RNA interference (RNAi)–mediated suppression with synthesized small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs) in conjunction with codon-modified replacement genes were explored for mouse rhodopsin7 and peripherin.8 Additionally, mutation-specific suppression of RHO was attempted, employing either ribozymes or RNAi.9,10 However, suppression targeted to specific mutation(s) limits flexibility in suppressor design, thereby reducing the probability of identifying potent molecules.
In parallel with the design of therapeutic strategies for dominant retinopathies, vectors for delivery of nucleotide-based therapies to photoreceptors have been evaluated. In this regard, a hybrid recombinant adeno-associated virus (AAV) with serotype 2 inverted terminal repeats (ITRs) and the serotype 5 capsid (AAV2/5) was shown to infect photoreceptors preferentially in a variety of species, including mouse, rat, dog, and macaque.11–14 Efficient transduction of photoreceptors by AAV2/5, the safety profile of the virus in the CNS, and the proposed use of AAV2 in human clinical trials15 have encouraged the exploration of AAV2/5 for the therapeutic approach outlined in the current study.
Key elements required for mutation-independent suppression and replacement for dominantly inherited disorders, with human RHO as an example, are presented in the current study. RNAi has been employed as a means of potent suppression of RHO expression, as evaluated in cell culture, organotypic retinal explants, and in vivo, with use of AAV-mediated delivery. Furthermore, the resistance of transcripts from codon-modified replacement genes to RNAi-mediated suppression, the generation of a transgenic mouse providing evidence of the functional equivalence of RHO replacement genes, and potential benefits of AAV-delivered therapies in mouse models of RP are presented. Results suggest that suppression and replacement can provide a therapeutic solution for dominantly inherited disorders such as adRP and can be employed to circumvent mutational heterogeneity.
Material and Methods
siRNA and shRNA
RHO-targeting siRNAs siB (target-position nucleotide [nt] 256–277), siBB (nt 254–274), siC (nt 270–292), and siCC (nt 274–294) were designed according to the method of Elbashir et al.16 siQ1 and siQ2, also targeting RHO (nt 650–670 and nt 671–694, respectively), were designed using the HiPerformance siRNA design algorithm (Qiagen). siRNAs—an siRNA targeting enhanced green fluorescent protein (EGFP [Entrez Gene accession number U57608]), siEGFP (nt 256–277), and a nontargeting siRNA (siNT, 5′-UUCUCCGAACGUGUCACGU-3′)—were synthesized by Qiagen. siRNA target sequences differed by at least 4 nt from any sequences in mouse and human databases (BLAST 2.2.617). siBB, siQ1, and siNT were initially cloned downstream of the H1 promoter, to generate shRNAs,18 and subsequently in pEGFP-1 (BD Biosciences), to generate shBB-EGFP, shQ1-EGFP, and shNT-EGFP (fig. 1A).
Figure 1. .
RHO-suppression and -replacement constructs. A, Representation of RHO suppressor-EGFP construct shBB-EGFP. (shQ1-EGFP and shNT-EGFP have the same format.) shRNAs were expressed from the H1 promoter and EGFP from the CMV immediate early promoter. The SV40 polyadenylation signal was located at the 3′ end of the EGFP gene. B, Two-component suppression and replacement construct shBB-rBB. (shQ1-rQ1 and shNT-rBB have the same format.) Suppressors were expressed from the H1 promoter and replacement RHO cDNAs from RhoP. Polyadenylation signals of the RHO gene were included in the 1,829-bp fragment. For tissue culture and retinal explant experiments, these constructs were maintained in pEGFP-1 (A) or a CMV-promoterless derivative of pcDNA-3.1 (B) and for in vivo experiments in the AAV vector. Restriction-enzyme sites used for cloning are indicated. Promoters were separated by spacer-DNA fragments. Numbers indicate molecular sizes (bp), and arrows indicate the direction of transcription. HGH int=human growth hormone intron.
Replacement RHO Genes
Replacement (r) RHO cDNAs rBB, rCC, and rQ1, with degenerative nucleotides altered at siBB, siCC, and siQ1 target sites, respectively, were generated by primer-directed mutagenesis and were cloned into pcDNA3.1 (Invitrogen). The cytomegalovirus (CMV) promoter was replaced with either the human ubiquitin C promoter (pUB6/V5-His [Invitrogen]) or a 1.7-kb fragment of the mouse Rho promoter (RhoP). We thank Prof. W. Baehr, University of Utah, for the original RHO cDNA construct. Altered nucleotides, underlined in target sites of replacement genes, are as follows: rBB (5′-ATAAATTTTTTGACCCTGTAT-3′), rCC (5′-CTGTATGTGACGGTG-3′), and rQ1 (5′-TGTAGCTGCGGTATAGATTAT-3′).
Cell Culture, Mouse Retinal Explants, and Fluorescence-Activated Cell Sorting (FACS)
CMV promoter–driven wild-type RHO or ubiquitin promoter–driven replacement constructs rBB, rCC, or rQ1 and siRNAs, siB, siBB, siC, siCC, siQ1, siQ2, siNT, or siEGFP were cotransfected into HeLa cells (ATCC accession number CCL-2) as described in the work of Millington-Ward et al.19 Twenty-four hours posttransfection, RNA or cytoplasmic protein was isolated as described elsewhere.8 Mouse retinal explants were prepared; were electroporated with shBB-EGFP, shQ1-EGFP, or shNT-EGFP; and were maintained as described. Subsequently, retinas were trypsin dissociated, and retinal cells expressing EGFP were identified and sorted by FACS.8
Real-Time RT-PCRs
RHO mRNA expression levels were assessed by real-time RT-PCR on a 7300 Real Time PCR System (Applied Biosystems) with use of a QuantiTect SYBR Green RT-PCR kit (Qiagen) and with β-actin as an endogenous control, with RHO primers (forward, 5′-CTTTCCTGATCTGCTGGGTG-3′; reverse, 5′-GGCAAAGAACGCTGGGATG-3′) and β-actin primers (forward, 5′-TCACCCACACTGTGCCCATCTACGA-3′; reverse, 5′-CAGCGGAACCGCTCATTGCCAATGG-3′). Typically, real-time RT-PCRs were performed twice in triplicate.
AAV
Recombinant AAV2/5 viruses were generated using a helper virus–free system.20 Expression cassettes were cloned into pAAV-MCS (Stratagene), between the inverted terminal repeats of AAV2. The resulting constructs were transfected into human embryonic kidney (HEK)–293 cells (ATCC [accession number CRL-1573]) with pRep2/Cap521 and pHelper (Stratagene), at a ratio of 1:1:2. Fifty 150-mm plates of confluent cells were transfected (50 μg DNA per plate) with polyethylenimine.22 Forty-eight hours posttransfection, crude viral lysates were cleared23 and purified by CsCl2-gradient centrifugation. AAV-containing fractions were dialyzed against PBS. Genomic titres—that is, viral particles (vp/ml)—were determined by quantitative real-time PCR.24 The generated AAVs contained the shBB-EGFP and shNT-EGFP constructs (AAV-shBB-EGFP and AAV-shNT-EGFP, respectively) (fig. 1A); RHO-suppression and -replacement constructs shBB-rBB, shQ1-rQ1, and shNT-rBB (AAV-shBB-rBB, AAV-shQ1-rQ1, and AAV-shNT-rBB, respectively) (fig. 1B); or a CMV promoter–driven EGFP gene (AAV-EGFP).
Animals
Transgenic mice NHR+/− Rho−/−, Pro23His+/− Rho+/−, and RHO-M+/− Rho−/− (see below) and Rho−/− were used in this study.25 Rho−/− mice have a severe degenerative retinopathy. The mice fail to elaborate rod photoreceptor outer segments and show no rhodopsin immunostaining. At age 12 wk, the rod electroretinogram (ERG) is not recordable, and the photoreceptor layer comprises a single row of nuclei, probably representing cone cells.26 NHR and Pro23His mice were first described by Olsson et al.27 NHR mice on a Rho−/− background express a human transgene and display a wild-type phenotype.26 Histology of retinas from 10-d-old Pro23His mice on a Rho−/− background showed marked photoreceptor degeneration. ERG analyses in 10-d-old mice showed reduced a- and b-wave amplitudes. Only a single row of photoreceptor cell nuclei remained in the outer nuclear layer (ONL). A transgenic mouse model (RHO-M) was generated, which expresses a replacement human RHO gene designed to avoid suppression by short complementary oligonucleotides targeting a specific site (codons 59–63) of the RHO mRNA. With use of PCR-directed mutagenesis, the original sequence of CTCTACGTCACCGTC was replaced by CTGTATGTGACGGTG (altered nucleotides underlined), both encoding the same amino acid sequence of LYVTV. The replacement RHO gene was placed under the control of a 3.8-kb fragment of the mouse RhoP. Intron 9 of the HPRT gene was cloned downstream of the RHO polyadenylation signal. The construct was microinjected into male pronuclei of fertilized eggs, to generate transgenic progeny. Four founder animals, identified by Southern blotting, yielded four individual transgenic mouse lines; one of these was evaluated in this study. All mice were maintained under specific pathogen-free housing conditions.
Subretinal AAV Injection
Subretinal injections were performed in strict compliance with the European Communities Regulations 2002 and 2005 (Cruelty to Animals Act) and the Association for Research in Vision and Ophthalmology statement for the use of animals in ophthalmic and vision research. Mice were anesthetized by intraperitoneal injection of medetomidine and ketamine (10 and 750 μg/10 g body weight, respectively). Pupils were dilated with 1% cyclopentolate and 2.5% phenylephrine, and, under local analgesia (Amethocaine), a small puncture was made in the sclera. A 34-gauge blunt-ended microneedle attached to a 10-μl syringe (Hamilton) was inserted through the puncture, and 1–3 μl of 10 vp/ml13–14 AAV in PBS was administered to the subretinal space, and a retinal detachment was induced. Following subretinal injection, a reversing agent (100 μg/10 g body weight [Atipamezole Hydrochloride]) was delivered by intraperitoneal injection. Body temperature was maintained using a homeothermic heating device. Newborn mice were prepared for subretinal injection with the method described by Matsuda and Cepko.27
ELISA, Immunocytochemistry, and Microscopy
Rhodopsin ELISAs were performed in triplicate as described elsewhere.8 The rhodopsin primary antibody, kindly provided by Prof. R. S. Molday, University of British Colombia, Vancouver, was used in a 1:500 dilution. Rhodopsin immunocytochemistry, fluorescent microscopy, epon embedding, and semithin sectioning were performed as described elsewhere.7,28 ONL thickness was evaluated in semithin resin-embedded sections, which were taken from the central meridian of the eye at 50-mm intervals. Approximately 40 measurements of ONL thickness per section from suppression and replacement eyes and from control eyes were taken using analysis 5.0 (Olympus Biosystems) on an HBO 100 Axioplan 2 microscope (Carl Zeiss). ONL measurements for treated and control eyes were grouped and ordered by thickness, and, subsequently, the highest and lowest 15% of values were analyzed.
RNase Protection Assay
RNA probes specific to Rho (5′-ACGGGCUCUUCGUAGACAGAGAC-3′), RHO (5′-GCGUACCACACCCGUCGCAUUGG-3′), or shBB (5′-GUAGAGCGUGAGGAAGUUGAUG-3′) were obtained from Sigma-Proligo. Probes and Decade size marker (Ambion) were 5′-end labeled with P32-γATP (Amersham GE Healthcare) with use of the mirVana Probe and Marker kit according to the manufacturer’s protocol (Ambion). RNase protection assays were performed using the RPA III Ribonuclease Protection Assay kit and the manufacturer’s protocol (Ambion). One picomole of labeled probe was hybridized to 3 μg of mouse retinal RNA. Samples were separated on 15% denaturing polyacrylamide gels.
ERG
Animals were dark adapted overnight and were prepared under dim red light. Pupils were dilated with 1% cyclopentalate and 2.5% phenylephrine. Animals were anesthetized with ketamine and xylazine (16 and 1.6 μg/10 g body weight, respectively) injected intraperitoneally. Standardized flashes of light were presented to the mouse in a Ganzfeld bowl. ERG responses from both eyes were recorded simultaneously by means of contact lens electrodes (Medical Workshop), with use of 1% amethocaine as topical anesthesia. Reference and ground electrodes were positioned subcutaneously, ∼1 mm from the temporal canthus and anterior to the tail, respectively. Responses were analyzed using a RetiScan RetiPort electrophysiology unit (Roland Consulting). The protocol was based on that approved by the International Clinical Standards Committee for human ERG. Rod-isolated responses were recorded using a dim white flash (−25 dB maximal intensity, where maximal flash intensity was 3 candelas/m2/s) presented in the dark-adapted state. Maximal combined rod-cone response to the maximal intensity flash was then recorded. After a 10-min light adaptation to a background illumination of 30 candelas/m2, cone-isolated responses were recorded to the maximal intensity flash, presented initially as a single flash and subsequently as 10-Hz flickers. a-Waves were measured from the baseline to the trough and b-waves from the baseline (in the case of rod-isolated responses) or from the a-wave to the trough.
Statistical Analysis
Data sets for a given construct were pooled and averaged, and SD values were calculated. Statistical significance of differences between control and target data sets were determined by either Student’s two-tailed t test or analysis of variance (ANOVA) with the least-significant-difference post hoc test (Data Desk 6.1 [Data Descriptions]); differences with P<.05 were considered statistically significant.
Results
siRNA-Mediated Suppression and Replacement of Human Rhodopsin in HeLa Cells
RNAi-mediated suppression of RHO was initially evaluated in HeLa cells. siRNAs targeting RHO were cotransfected with a CMV promoter–driven wild-type RHO. Transfections were performed three times in quadruplicate. Real-time RT-PCRs, performed on RNAs extracted from transfected cells 24 h posttransfection, demonstrated up to 87% suppression (P<.01) (fig. 2A). siRNAs siBB, siCC, and siQ1 were selected for further analysis. Similar levels of rhodopsin protein suppression were quantified by ELISA (up to 88%; P<.01) (fig. 2A) and were demonstrated by immunocytochemistry 24 h posttransfection (fig. 2B). Subsequently, replacement RHO constructs rBB, rCC, and rQ1 were generated, incorporating nucleotide changes at degenerate positions over the target sites for siRNAs siBB, siQ1, and siCC. Transfections were performed three times in quadruplicate in HeLa cells. Replacement RHO constructs were not suppressed by corresponding siRNAs—for example, rBB by siBB (fig. 3). However, significant levels of suppression were obtained with other noncorresponding siRNAs—for example, siQ1 suppressed rBB and rCC (fig. 3).
Figure 2. .
RHO suppression in HeLa cells. HeLa cells were transiently cotransfected three times in triplicate with wild-type RHO and RHO-targeting siRNAs (siB, siBB, siC, siCC, siQ1, or siQ2) or control siRNAs (siEGFP or siNT). After transfection, RHO mRNA and protein levels were evaluated by real-time RT-PCR (A), ELISA (A), and Alexa Fluor 568–labeled immunocytochemistry (B). Cell nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI). Error bars represent SD values.
Figure 3. .
Replacement RHO suppression in HeLa cells. Replacement RHO sequences were generated with altered degenerate nucleotides at siRNA target sites. HeLa cells were transiently cotransfected three times in triplicate with a replacement RHO-expression vector (rBB, rCC, or rQ1) and a RHO-targeting siRNA (siBB, siCC, or siQ1) or a nontargeting siRNA (siNT). Replacement RHO mRNA levels were evaluated by real-time RT-PCR. Error bars represent SD values.
shRNA-Mediated Suppression of Human Rhodopsin in Retinal Explants
To provide long-term RHO suppression, siBB and siQ1 were cloned as shRNAs into an EGFP-expressing vector (shBB-EGFP and shQ1-EGFP, respectively) (fig. 1A). Plasmids were electroporated into retinal explants from newborn NHR+/− Rho−/− mice. NHR+/− mice express a wild-type human RHO gene and display a wild-type phenotype.25 Cells from retinal explants (n=6) were dissociated 2 wk postelectroporation, and EGFP-positive cells were isolated by FACS (fig. 4A). Real-time RT-PCR was undertaken on RNA extracted from EGFP-positive FACS-isolated cells, and results obtained in explants mirrored those found in HeLa cells. RHO suppression of >85% was achieved (P<.001) (fig. 4B).
Figure 4. .
RHO suppression in retinal explants. Mouse retinas (n=6), dissected from newborn NHR+/− Rho−/− pups, were electroporated with a construct coexpressing an shRNA targeting RHO or a nontargeting shRNA and EGFP (shBB-EGFP, shQ1-EGFP, or shNT-EGFP). Negative control explants were not electroporated. After 2 wk, organotypic cultures were dissociated with trypsin and were analyzed by FACS. Red and blue dots represent gated and ungated populations of dissociated explants, respectively. Scatterplots of forward- (FSC) versus side-scatter (SSC) and histograms of EGFP fluorescence of the gated population of nonelectroporated (A1 and A2, EGFP-negative) and electroporated (A3 and A4, EGFP-positive) retinas are given. The bar chart indicates RHO mRNA levels in retinal explant cells expressing sNT-EGFP, sBB-EGFP, and sQ1-EGFP, quantified by real-time RT-PCR. Error bars represent SD values.
AAV-shRNA–Mediated Suppression of Human RHO in Vivo
It is clear that long-term expression of therapies will be required for a progressive retinopathy such as adRP. To achieve long-term suppression in vivo, shBB-EGFP and the nontargeting shNT-EGFP were engineered into AAV vectors (AAV-shBB-EGFP and AAV-shNT-EGFP). The EGFP gene enabled viral transduction to be monitored. Three microliters of AAV-shBB-EGFP (2×1012 vp/ml) or AAV-shNT-EGFP (3×1012 vp/ml) were subretinally injected into adult NHR+/− Rho−/− mice. Two weeks postinjection, two animals were sacrificed, and expression of the 21-nt shBB was shown in two retinas with use of RNase protection (fig. 5A). Retinas were dissociated, and EGFP-positive cells were collected by FACS. RNAi-mediated suppression of RHO, as evaluated by real-time RT-PCR 2 wk postinjection (n=6), was ∼90% (P<.001) in AAV-shBB-EGFP–transduced photoreceptor cells (fig. 5B). Four retinas were dissociated, and significant suppression of rhodopsin protein expression was demonstrated in vivo in EGFP-positive transduced cells by immunocytochemistry (fig. 5C).
Figure 5. .
RHO suppression in photoreceptor cells in vivo. Adult transgenic NHR+/− Rho−/− mice were subretinally injected with 3 μl of 2×1012 vp/ml AAV coexpressing a RHO-targeting or nontargeting shRNA and EGFP (AAV-shBB-EGFP or AAV-shNT-EGFP). Retinas were analyzed 2 wk postinjection. Expression of the 21-nt shRNA BB, detected by RNase protection in two transduced retinas, is depicted in lanes L1 and L2 (A). RHO RNA probes were labeled with P32-γATP, and protected RNA was separated on 15% denaturing acrylamide gels (A). In lane M, size markers indicate 10, 20, and 30 nt. Bars represent RHO mRNA levels in FACS-sorted cells from dissociated retinas (n=6) transduced with either AAV-shBB-EGFP or AAV-shNT-EGFP (B). Suppression levels were determined by real-time RT-PCR. Error bars represent SD values. Rhodopsin immunocytochemistry (Cy3 labeled) and EGFP protein expression in cells from dissociated retinas, transduced with either AAV-shBB-EGFP (arrows) or AAV-shNT-EGFP (arrow heads), are depicted (C). Cell nuclei were counterstained with DAPI.
Functional Rescue of Rho−/− Mice with a Codon-Modified RHO-Replacement Transgene
A transgenic mouse expressing a sequence-modified RHO gene was generated (RHO-M). The 3.8-kb mouse rhodopsin promoter used to drive expression of the transgene was also used to drive expression of EGFP in the plasmid pEGFP-1 (Rho-EGFP). A significant level of expression was demonstrated by Rho-EGFP, and it was confined to photoreceptor cells when newborn CD1 mouse retinal explants were electroporated with this construct (data not shown). RHO-M+/− Rho−/− mice were evaluated at age 2 mo, for rescue of the retinal pathology present in Rho−/− mice, by histology (fig. 6A–6C) and ERG (fig. 6D).
Figure 6. .
Retinal histology and ERG analysis of RHO-M mouse. Two-month-old Rho+/+ (wild type), Rho+/−, NHR+/− Rho−/−, and RHO-M+/− Rho−/− mice were analyzed by retinal histology and ERG (n=8). A–C, Rhodopsin immunocytochemistry (Cy3) showing similar rod outer segment (ROS) labeling in Rho+/+, NHR+/− Rho−/−, and RHO-M+/− Rho−/− retinas, respectively. Nuclear layers were stained with DAPI. INL=inner nuclear layer; GCL=ganglion cell layer. D, Representative rod-isolated ERG responses. Note that there is natural variation in b-wave amplitude. The range with use of our protocol in wild-type animals is 349–720 μv (mean 485) and, in Rho+/− animals, is 270–700 μv (mean 447).
Rhodopsin immunolabeling in rod outer segments and the thickness of ONLs were similar in wild-type Rho+/+ (fig. 6A), NHR+/− Rho−/− (fig. 6B) and RHO-M+/− Rho−/− (fig. 6C) mice. Additionally, ERG responses were similar in wild-type Rho+/+, Rho+/−, NHR+/− Rho, −/−, and RHO-M+/− Rho−/− mice. ERG b-waves of rod-isolated responses of 500–700 μV were observed in mice of all genotypes (fig. 6D). The amplitudes and timings of the combined rod and cone responses to the maximal intensity flash presented in the dark-adapted state, as well as the light-adapted cone-isolated responses both to single flash and 10-Hz flickers, were equivalent in all the genotypes examined (data not shown). These results validate the use of the degeneracy of the genetic code for engineering codon-modified human RHO genes that can provide functional human rhodopsin protein.
AAV-Delivered Suppression and Replacement of Human RHO in Vivo
Since shBB and shQ1 were established as potent suppressors and rBB and rQ1 as refractory to their corresponding suppressors, shBB-rBB and shQ1-rQ1 were cloned into AAV vectors, and viruses containing both elements of the therapeutics were generated (AAV-shBB-rBB and AAV-shQ1-rQ1). Three microliters of AAV-shBB-rBB was subretinally injected into adult wild-type Rho+/+ mice (n=12), and replacement RHO mRNA expression was confirmed by RT-PCR and RNase protection with RNA extracted 10 d postinjection (data not shown). To demonstrate that AAV-delivered rBB is translated into protein, 2 μl of a 1:1 mix of AAV-shBB-rBB and AAV-EGFP was subretinally injected into 10-d-old Rho−/− mice (n=6). Two weeks postinjection, rhodopsin and EGFP protein expression were determined using fluorescent microscopy. Marked rhodopsin expression, overlapping with EGFP, was observed in transduced areas (fig. 7).
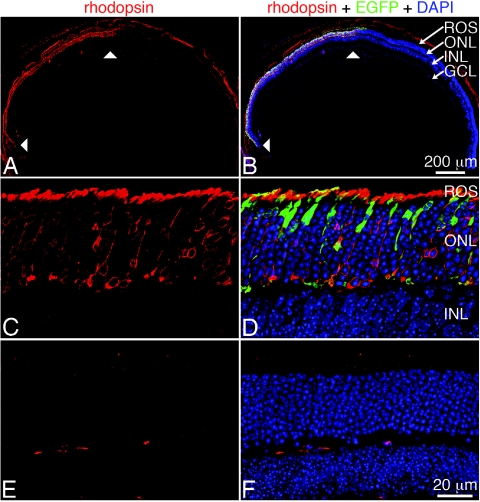
Figure 7. .
Expression of replacement RHO in vivo. Ten-day-old Rho−/− mice were subretinally injected with a 1:1 mixture of 2 μl of 2×1012 vp/ml of two AAV vectors: AAV-EGFP and AAV-shBB-rBB. Rhodopsin, EGFP protein, and nuclei were detected by Cy3-labeled immunocytochemistry, native fluorescence, and nuclear DAPI staining, respectively. Low-magnification images show a cross section of a whole injected eye, with arrowheads indicating the transduced area (A and B). High-magnification laser-scanning micrographs show transduced (C and D) and nontransduced (E and F) areas. INL=inner nuclear layer; GCL=ganglion cell layer; ROS=rod outer segments.
Subsequently, 1 μl of AAV-shBB-rBB or AAV-shQ1-rQ1 was subretinally injected into newborn Pro23His+/− Rho+/− mice (n=10) that presented with a retinal degeneration resulting in complete loss of photoreceptors by age 2 wk. In all animals, one eye was injected with therapeutic virus (either AAV-shBB-rBB or AAV-shQ1-rQ1) and the other with a control virus (AAV-EGFP). The early onset and rapid nature of the retinopathy in young Pro23His pups precluded use of ERG as a readout for benefit. However, at age 10 d, retinal histology was evaluated in semithin resin-embedded sections cut at ∼50-μm intervals throughout the central meridian of the eye (n=10). From each section ∼40 measurements of ONL thickness were taken. Since only a part of the retina is transduced by a single subretinal injection of AAV (particularly in newborn pups), to identify the transduced area, ONL measurements were ordered by thickness, and the highest and lowest 15% of values were grouped for analysis. Lowest values represent thinnest ONL readings, most likely corresponding to peripheral areas of the retina and thus not in close proximity to injection sites. Highest values represent thickest ONL readings, most likely corresponding to central areas of the retina and thus in closer proximity to injection sites. Significant differences in ONL thickness between AAV-shBB-rBB– and AAV-EGFP–treated eyes were observed. The ONL of treated eyes was found to be ∼33% (P<.001) thicker than control-injected counterparts for the highest value groupings (fig. 8A–8C). In the lowest value groupings, a difference of ∼10% was observed (fig. 8A). These data provide evidence at the histological level that AAV2/5-delivered RNAi, in conjunction with provision of a codon-modified replacement gene, can beneficially modulate the retinopathy in Pro23His+/− Rho−/− mice.
Figure 8. .
Histology of AAV-transduced Pro23His retinas. Newborn Pro23His+/− Rho+/− mice were subretinally injected with 1 μl of 2×1012 vp/ml AAV-shBB-rBB or AAV-EGFP (n=6). Ten days posttransduction, eyes were processed for semithin sectioning and were stained with toluidine blue. Approximately 40 measurements of ONL thickness (μm) in three layers per eye were taken. A, ONL thickness (bars) of the central meridian of the eye, of the lowest and highest 15% of values (P<.01). B and C, Representative images of AAV-shBB-rBB– and AAV-EGFP (control)–injected sections corresponding to highest ONL thickness values. Yellow arrows indicate ONL thickness. INL=inner nuclear layer; GCL=ganglion cell layer. Error bars represent SD values.
Discussion
Central to the design of the study is the exploration of therapies that overcome the significant mutational heterogeneity in dominant disorders such as RHO-linked adRP. The degeneracy of the genetic code facilitates such an approach by enabling RNAi-mediated suppression in conjunction with use of sequence-modified replacement genes whose transcripts are RNAi resistant (fig. 3). Such an approach has been proposed but as yet not validated in vivo for a number of other dominantly inherited conditions. For example, Millington-Ward et al.19 have considered suppression and replacement for type I collagen-linked osteogenesis imperfecta type I (MIM #166200) in mesenchymal progenitor stem cells. Xia et al.29 proposed that suppression and replacement represents a valuable therapeutic strategy for copper zinc superoxide dimutase (SOD1)–linked amyotrophic lateral sclerosis (MIM *147450) and evaluated the approach in HEK-293 cells. Similarly, Kim and Rossi4 described the use of suppression and replacement, employing expression of an EGFP-reporter gene in vitro to demonstrate the principle. Additionally, whereas RNAi-mediated suppression of mutant alleles has been shown to provide therapeutic benefit for spinocerebellar ataxia (MIM #183090)30 and Huntington disease (MIM +143100),31 it still has to be established whether therapies for these disorders will require inclusion of replacement genes. Suppression and replacement may also prove necessary as a therapeutic strategy for such disorders. More generally, elucidation of the molecular etiologies of genetic conditions over the past 2 decades has served to highlight the enormous mutational heterogeneity in many disorders and hence the need for therapeutic strategies such as the suppression and replacement presented in this work.
In the current study, substantial RNAi-based suppression of human RHO has been achieved in HeLa cells (figs. 2 and 3), organotypic retinal explants (fig. 4), and in vivo subsequent to subretinal injection of AAV vectors (fig. 5). Observation of >88% RHO suppression in AAV-shRNA–treated retinas represents the first demonstration of potent suppression of a gene in photoreceptors after viral delivery of shRNAs. In addition, the generation of a transgenic mouse (RHO-M) expressing a codon-modified human RHO-replacement gene has provided evidence in vivo of the functionality of codon-modified replacement RHO genes. Results obtained from both ERG and histology in RHO-M+/− Rho−/− transgenic mice carrying this replacement RHO gene were equivalent to those observed in wild-type mice (Rho+/+), Rho +/− mice, and NHR+/− Rho−/− mice carrying a wild-type human RHO transgene (fig. 6).
An objective of the current study was in vivo demonstration of suppression and replacement. Hence, shRNAs targeting RHO together with codon-modified RHO replacement genes have been engineered into AAV vectors. Expression of RNAi-resistant replacement genes in the presence of shRNAs was demonstrated in mice subretinally injected with AAV vectors with use of real-time RT-PCR, RNase protection assays, and immunocytochemistry (fig. 7). In addition, evidence suggesting therapeutic benefit after administration of AAV vectors incorporating suppression and replacement components has been obtained in Pro23His+/− Rho+/− mice expressing a mutant human RHO allele and presenting with a rapid retinal degeneration. Retinas from eyes injected with AAV-shBB-rBB or AAV-shQ1-rQ1 showed preservation of the ONL when compared with those injected with control virus—a 33% difference in ONL thickness was observed between treated and control eyes in an animal model simulating human RHO-linked RP (fig. 8).
Suppression and replacement, although providing significant advantages in terms of condensing mutational heterogeneity, is challenging, since the approach involves 2-nt–based components within a single therapeutic. Ensuring sufficient RNAi-mediated suppression in conjunction with appropriate levels of expression of the RNAi-resistant replacement gene will clearly be important. However, in the current study, significant strides toward this end were achieved—notably, ∼90% in vivo suppression of RHO with use of AAV-delivered RNAi, demonstration that a codon-modifed RHO gene expresses functional wild-type protein, in vivo expression of AAV-delivered RHO replacement genes in the presence of targeting RNAi molecules, and observation of potential therapeutic benefit in Pro23His mice. In summary, the study represents the first in vivo indication that suppression and replacement as a strategy can provide therapeutic benefit and can overcome the mutational heterogeneity associated with RHO-linked RP, a significant barrier to therapeutic development for this and many other dominantly inherited genetic conditions. Our results, in conjunction with accumulating evidence that AAV represents a safe and efficient means of delivering molecular therapies to photoreceptors,11 suggest that AAV-delivered suppression and replacement may well provide future therapies for patients with RHO-linked RP, as well as for other disorders.
Acknowledgments
We thank the vector core of the University Hospital of Nantes, supported by the Association Française contre le Myopathies, for providing rAAV2/5-EGFP. We also thank Prof. Ted Dryja (Harvard Medical School, Massachusetts Eye and Ear Infirmary, Boston) for providing the Pro23His mouse, Dr. Alfonso Blanco Fernandez (Flow Cytometry Core Facility in University College, Dublin) for assisting with FACS analysis, and Ms. Orla Hanrahan (School of Biochemistry and Immunology, Trinity College, Dublin) for helping with confocal microscopy. The research was supported by grant awards from Science Foundation Ireland, Enterprise Ireland, the 6th Framework Programme of the European Union (RETNET MRTN-CT-2003-504003), and Fighting Blindness Ireland.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- ATCC, http://www.atcc.org/ (for HeLa cells [accession number CCL-2]) and HEK-293 cells [accession number CRL-1573])
- BLAST, http://www.ncbi.nlm.nih.gov/blast
- Entrez Gene, http://www.ncbi.nlm.nih.gov/entrez/ (for EGFP [accession number U57608])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for RHO [accession number NM_000539.2])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RP, type I collagen-linked osteogenesis imperfecta type I, SOD1-linked amyotrophic lateral sclerosis, spinocerebellar ataxia, and Huntington disease)
- Retinal Information Network database, http://www.sph.uth.tmc.edu/Retnet/
References
- 1.Farrar GJ, Kenna PF, Humphries P (2002) On the genetics of retinitis pigmentosa and on mutation-independent approaches to therapeutic intervention. EMBO J 21:857–864 10.1093/emboj/21.5.857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Millington-Ward S, O’Neill B, Tuohy G, Al-Jandal N, Kiang AS, Kenna PF, Palfi A, Hayden P, Mansergh F, Kennan A, et al (1997) Strategems in vitro for gene therapies directed to dominant mutations. Hum Mol Genet 6:1415–1426 10.1093/hmg/6.9.1415 [DOI] [PubMed] [Google Scholar]
- 3.Hauswirth WW, Lewin AS (2000) Ribozyme uses in retinal gene therapy. Prog Retin Eye Res 19:689–710 10.1016/S1350-9462(00)00007-0 [DOI] [PubMed] [Google Scholar]
- 4.Kim DH, Rossi JJ (2003) Coupling of RNAi-mediated target downregulation with gene replacement. Antisense Nucleic Acid Drug Dev 13:151–155 10.1089/108729003768247619 [DOI] [PubMed] [Google Scholar]
- 5.Cashman SM, Binkley EA, Kumar-Singh R (2005) Towards mutation independent silencing of genes involved in retinal degeneration by RNA interference. Gene Ther 12:1223–1228 10.1038/sj.gt.3302512 [DOI] [PubMed] [Google Scholar]
- 6.Gorbatyuk M, Justilien V, Liu J, Hauswirth WW, Lewin AS (2007) Preservation of photoreceptor morphology and function in P23H rats using an allele independent ribozyme. Exp Eye Res 84:44–52 10.1016/j.exer.2006.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiang AS, Palfi A, Ader M, Kenna PF, Millington-Ward S, Clark G, Kennan A, O’Reilly M, Tam LC, Aherne A, et al (2005) Toward a gene therapy for dominant disease: validation of an RNA interference based mutation-independent approach. Mol Ther 12:555–561 10.1016/j.ymthe.2005.03.028 [DOI] [PubMed] [Google Scholar]
- 8.Palfi A, Ader M, Kiang AS, Millington-Ward S, Clark G, O’Reilly M, McMahon HP, Kenna PF, Humphries P, Farrar GJ (2006) RNAi-based suppression and replacement of rds-peripherin in retinal organotypic culture. Hum Mutat 27:260–268 10.1002/humu.20287 [DOI] [PubMed] [Google Scholar]
- 9.Lewin AS, Drenser KA, Hauswirth WW, Nishikawa S, Yasumura D, Flannery JG, LaVail MM (1998) Ribozyme rescue of photoreceptor cells in a transgenic rat model of autosomal dominant retinitis pigmentosa. Nat Med 4:967–971 (erratum 4:1081) 10.1038/nm0898-967 [DOI] [PubMed] [Google Scholar]
- 10.Tessitore A, Parisi F, Denti MA, Allocca M, Di Vicino U, Domenici L, Bozzoni I, Auricchio A (2006) Preferential silencing of a common dominant rhodopsin mutation does not inhibit retinal degeneration in a transgenic model Mol Ther 14:692–699 [DOI] [PubMed] [Google Scholar]
- 11.Rolling F (2004) Recombinant AAV-mediated gene transfer to the retina: gene therapy perspectives. Gene Ther Suppl 11:S26–S32 10.1038/sj.gt.3302366 [DOI] [PubMed] [Google Scholar]
- 12.Dinculescu A, Glushakova L, Min SH, Hauswirth WW (2005) Adeno-associated virus-vectored gene therapy for retinal disease. Hum Gene Ther 16:649–663 10.1089/hum.2005.16.649 [DOI] [PubMed] [Google Scholar]
- 13.Allocca M, Tessitore A, Cotugno G, Auricchio A (2006) AAV-mediated gene transfer for retinal diseases. Expert Opin Biol Ther 6:1279–1294 10.1517/14712598.6.12.1279 [DOI] [PubMed] [Google Scholar]
- 14.Stieger K, Le Meur G, Lasne F, Weber M, Deschamps JY, Nivard D, Mendes-Madeira A, Provost N, Martin L, Moullier P, et al (2006) Long-term doxycycline-regulated transgene expression in the retina of nonhuman primates following subretinal injection of recombinant AAV vectors. Mol Ther 13:967–975 10.1016/j.ymthe.2005.12.001 [DOI] [PubMed] [Google Scholar]
- 15.Bennett J (2004) Gene therapy for Leber congenital amaurosis Novartis Found Symp 255:195–202 [PubMed] [Google Scholar]
- 16.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498 10.1038/35078107 [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- 19.Millington-Ward S, McMahon HP, Allen D, Tuohy G, Kiang AS, Palfi A, Kenna PF, Humphries P, Farrar GJ (2004) RNAi of COL1A1 in mesenchymal progenitor cells. Eur J Hum Genet 12:864–866 10.1038/sj.ejhg.5201230 [DOI] [PubMed] [Google Scholar]
- 20.Xiao X, Li J, Samulski RJ (1998) Production of high-titer recombinant adenoassociated virus vectors in the absence of helper adenovirus. J Virol 72:2224–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildinger M, Auricchio A, Gao G, Wang L, Chirmule N, Wilson JM (2001) Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J Virol 75:6199–6203 10.1128/JVI.75.13.6199-6203.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed SE, Staley EM, Mayginnes JP, Pintel DJ, Tullis GE (2006) Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J Virol Methods 138:85–98 10.1016/j.jviromet.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 23.Auricchio A, Hildinger M, O’Connor E, Gao GP, Wilson JM (2001) Isolation of highly infectious and pure adeno-associated virus type 2 vectors with a single-step gravity-flow column. Hum Gene Ther 12:71–76 10.1089/104303401450988 [DOI] [PubMed] [Google Scholar]
- 24.Rohr UP, Wulf MA, Stahn S, Steidl U, Haas R, Kronenwett R (2002) Fast and reliable titration of recombinant adeno-associated virus type-2 using quantitative real-time PCR. J Virol Methods 106:81–88 10.1016/S0166-0934(02)00138-6 [DOI] [PubMed] [Google Scholar]
- 25.Humphries MM, Rancourt D, Farrar GJ, Kenna P, Hazel M, Bush RA, Sieving PA, Sheils DM, McNally N, Creighton P, et al (1997) Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat Genet 15:216–219 10.1038/ng0297-216 [DOI] [PubMed] [Google Scholar]
- 26.McNally N, Kenna P, Humphries MM, Hobson AH, Khan NW, Bush RA, Sieving PA, Humphries P, Farrar GJ (1999) Structural and functional rescue of murine rod photoreceptors by human rhodopsin transgene. Hum Mol Genet 8:1309–1312 10.1093/hmg/8.7.1309 [DOI] [PubMed] [Google Scholar]
- 27.Olsson JE, Gordon JW, Pawlyk BS, Roof D, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP (1992) Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron 9:815–830 10.1016/0896-6273(92)90236-7 [DOI] [PubMed] [Google Scholar]
- 28.Matsuda T, Cepko CL (2004) Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA 101:16–22 10.1073/pnas.2235688100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia XG, Zhou H, Zhou S, Yu Y, Wu R, Xu Z J (2005) An RNAi strategy for treatment of amyotrophic lateral sclerosis caused by mutant Cu,Zn superoxide dismutase. J Neurochem 92:362–367 (erratum 92:1554) 10.1111/j.1471-4159.2004.02860.x [DOI] [PubMed] [Google Scholar]
- 30.Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL (2004) RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med 10:816–820 10.1038/nm1076 [DOI] [PubMed] [Google Scholar]
- 31.Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL (2005) RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci USA 102:5820–5825 10.1073/pnas.0501507102 [DOI] [PMC free article] [PubMed] [Google Scholar]