Abstract
“Autosomal dominant retinitis pigmentosa” (adRP) refers to a genetically heterogeneous group of retinal dystrophies, in which 54% of all cases can be attributed to 17 disease loci. Here, we describe the localization and identification of the photoreceptor cell-specific nuclear receptor gene NR2E3 as a novel disease locus and gene for adRP. A heterozygous mutation c.166G→A (p.Gly56Arg) was identified in the first zinc finger of NR2E3 in a large Belgian family affected with adRP. Overall, this missense mutation was found in 3 families affected with adRP among 87 unrelated families with potentially dominant retinal dystrophies (3.4%), of which 47 were affected with RP (6.4%). Interestingly, affected members of these families display a novel recognizable NR2E3-related clinical subtype of adRP. Other mutations of NR2E3 have previously been shown to cause autosomal recessive enhanced S-cone syndrome, a specific retinal phenotype. We propose a different pathogenetic mechanism for these distinct dominant and recessive phenotypes, which may be attributed to the dual key role of NR2E3 in the regulation of photoreceptor-specific genes during rod development and maintenance.
Retinitis pigmentosa (RP [MIM 268000]), a group of retinal dystrophies, is clinically characterized by early-onset night blindness, followed by progressive constriction of the peripheral visual field and, eventually, loss of central vision in a majority of cases.1 RP is a genetically heterogeneous monogenic disorder, with 17 autosomal dominant, 25 autosomal recessive and 6 X-linked recessive loci (RetNet, the Retinal Information Network).
Autosomal dominant RP (adRP) accounts for 20%–40% of all RP cases. Until now, only 54% of the adRP cases could be attributed to 17 known loci, of which 15 genes are known. Overall, both founder mutations as well as allelic heterogeneity are seen.2 Most of the adRP disease genes are either expressed preferentially in the photoreceptors or the retinal pigment epithelium and encode diverse proteins involved in the phototransduction cascade, retinoid cycle, photoreceptor structure, or transcription (RetNet).
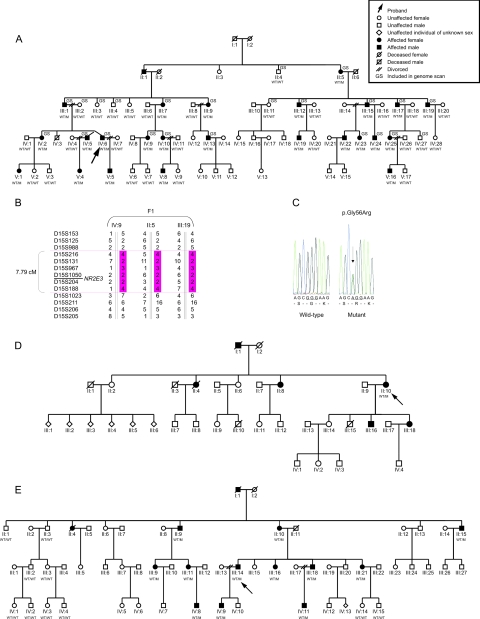
In this study, a four-generation Belgian family (F1) was ascertained in which adRP segregates in 25 individuals (fig. 1A). After informed consent was given, genomic DNA was obtained from 24 affected and 26 unaffected individuals. As a first step, 15 adRP loci, known at the onset of the study, were excluded, implicating the involvement of a novel adRP locus. Subsequently, a genomewide screen was performed on 23 individuals (ABI Prism Linkage Mapping Set, MD-10 v2.5 [Applied Biosystems]) (fig. 1A). Two-point LOD scores between the disease phenotype and each of the markers were calculated (SuperLink v1.4, EasyLINKAGE Plus v4.01beta),3 under the assumption of a dominant mode of inheritance with a penetrance of 98%, a phenocopy rate of 0.1%, and a disease-allele frequency of 0.0001. Significant linkage was obtained for two markers on chromosome 15, of which D15S131 yielded the higher LOD score (5.10 at θ=0.00) (data not shown). Segregation analysis of 10 additional flanking microsatellite markers in all available family members revealed critical recombination events between D15S988 and D15S216 at the proximal end, and between D15S188 and D15S1023 at the distal end. This defined a candidate region of 7.79 cM on 15q22–15q25, containing 172 genes (fig. 1B and table 1). Two of these—the Bardet-Biedl syndrome-4 gene (BBS4 [MIM 600374]) and the photoreceptor cell-specific nuclear receptor gene NR2E3—have a retinal expression pattern and are involved in retinal biology and disease. Mutations in BBS4 are known to cause Bardet-Biedl syndrome, an autosomal recessive disease characterized by pigmentary retinopathy and other systemic features.4 Mutations in the NR2E3 gene have been described elsewhere as causing enhanced S-cone syndrome (ESCS), an autosomal recessive retinopathy.5 Moreover, NR2E3 is a key player in the retinal transcriptional network, with a dual regulatory role in the terminal differentiation and in the maintenance of the rod phenotype.6,7
Figure 1. .
A, Pedigree of F1, in which NR2E3 mutation p.Gly56Arg was found. WT = wild type; M = mutation p.Gly56Arg. B, Haplotypes of critical recombinants of F1 used to narrow candidate region. C, Sequence electropherogram of heterozygous substitution c.166G→A (p.Gly56Arg) versus wild type. D and E, Pedigrees of F2 and F3, respectively.
Table 1. .
Two-Point LOD Scores for Microsatellites in the Candidate Region at 15q22–15q25[Note]
| LOD at θ = |
||||||||||
| Marker | Genetic Distancea | .00 | .01 | .05 | .10 | .20 | .30 | .40 | Zmax | θmax |
| D15S153 | 62.40 | 5.43 | 6.49 | 6.92 | 6.65 | 5.50 | 3.91 | 1.99 | 6.92 | .05 |
| D15S988 | 66.90 | .93 | 1.67 | 2.09 | 2.09 | 1.75 | 1.22 | .55 | 2.11 | .07 |
| D15S216 | 70.73 | 7.99 | 7.86 | 7.33 | 6.63 | 5.13 | 3.47 | 1.66 | 7.99 | .00 |
| D15S131 | 71.28 | 8.59 | 8.47 | 7.96 | 7.29 | 5.77 | 4.04 | 2.05 | 8.59 | .00 |
| D15S1050 | 71.82 | 9.71 | 9.55 | 8.90 | 8.05 | 6.21 | 4.18 | 1.97 | 9.71 | .00 |
| D15S204 | 71.82 | 9.24 | 9.09 | 8.48 | 7.69 | 5.97 | 4.05 | 1.91 | 9.24 | .00 |
| D15S188 | 71.82 | 7.41 | 7.29 | 6.83 | 6.23 | 4.92 | 3.46 | 1.82 | 7.41 | .00 |
| D15S1023 | 74.69 | 3.44 | 5.18 | 6.21 | 6.21 | 5.30 | 3.83 | 1.97 | 6.28 | .07 |
| D15S205 | 78.92 | −0.46 | 1.67 | 3.33 | 3.77 | 3.50 | 2.60 | 1.33 | 3.80 | .12 |
Note.— The markers D15S153, D15S131, and D15S205 originate from the ABI Prism Linkage Mapping Set (MD-10 v2.5, Applied Biosystems). Marker D15S131 generated the highest LOD score in a total genome scan (data not shown). Recombination events between D15S988 and D15S216 and between D15S188 and D15S1023 defined a candidate region of 7.79 cM. The maximum LOD score (Zmax) was obtained for D15S1050, as indicated in bold. The NR2E3 gene is located between D15S1050 and D15S204. LOD score calculations were performed on all available family members.
According to the Marshfield map (NCBI Map Viewer).
Hence, mutation screening of NR2E3 was performed in the proband of F1, revealing a novel heterozygous transition c.166G→A in exon 2 of the NR2E3 gene, which results in the missense mutation p.Gly56Arg (figs. 1C and 2A) (Ensembl reference sequence ENSP00000317199). This mutation cosegregated with disease. Subsequently, NR2E3 was screened in a panel of 86 unrelated probands, mainly of Belgian origin, with the following potentially dominant degenerative retinal diseases: rod-cone dystrophy (58 probands) including RP (46 probands), cone-rod dystrophy (22 probands), and cone dystrophy (6 probands). This led to the identification of the same sequence change c.166G→A in two additional probands with adRP (one French, one Belgian) (families F2 and F3) (fig. 1D and 1E). In F2, no additional family members were available for segregation analysis. The autosomal dominant inheritance is evident from the pedigree, however (fig. 1D). Segregation analysis of c.166G→A was performed in 12 additional affected and 10 unaffected individuals from F3 (fig. 1E). Finally, several known polymorphisms and novel changes were identified in the test cohort and in a control group (table 2).
Figure 2. .
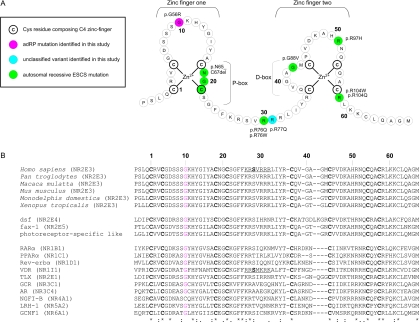
A, Theoretical model of the two C4 zinc fingers located in the DBD of NR2E3. The first zinc finger contains the P-box, in which two or three exposed residues are responsible for half-site sequence recognition. The second zinc finger harbors a dimerization interface for DBDs (D-box) and is involved in recognition of half-site spacing and mutual orientation. A consensus numbering system, starting from the first Cys of the first zinc finger, is used for comparison with other NRs in panel B. B, top rows, Alignment of the human protein sequence of NR2E3 with five orthologs. Middle rows, Three nonhuman NRs belonging to the same subfamily as NR2E3 (dsf, D. melanogaster; fax-1, C. elegans; and photoreceptor-specific like, D. melanogaster). Bottom rows, Human NRs belonging to one of six NR subfamilies: (1) Thyroid hormone receptor–like (RARα, PPARα, Rev-erbα and VDR); (2) HNF4-like (TLX); (3) Estrogen receptor–like (GR and AR); (4) Nerve Growth factor IB receptor–like (NGFI-B); (5) Fushi tarazu-F1–like (LRH-1); and (6) Germ cell nuclear factor–like (GCNF1). The official NR nomenclature is given in parentheses.8 Cys residues composing the C4 zinc-finger motif are indicated in bold. Gly residues in other NRs, corresponding to Gly56 in NR2E3, are indicated in red. Gly56 is conserved in almost all members of the human NR superfamily and in different orthologs, suggesting a functional constraint. Underlined sequences indicate a nuclear localization signal (NLS) in VDR and the corresponding putative NLS in NR2E3. The amino acid Ser, in bold, represents Ser74 and Ser51 in NR2E3 and VDR, respectively (details provided in table 3).
Table 2. .
Overview of Novel and Known Sequence Changes in NR2E3 Identified in the Present Study[Note]
| No. of Probands with Change |
||||||||
| Nucleotide Change |
Amino Acid Change | RP | CRD | RCD | Exon or Intron | Nature of Variant | Control Allele Frequency | Reference |
| c.166G→A | p.Gly56Arg | 3 | 0 | 0 | Exon 2 | M | 0 | Present study |
| c.230G→A | p.Arg77Gln | 0 | 0 | 0 | Exon 2 | UV | 1/380 | Present study |
| c.419A→G | p.Glu140Gly | 2 | 1 | 1 | Exon 4 | P | 7/380 | Ensembl |
| c.488T→C | p.Met163Thr | 2 | 1 | 1 | Exon 4 | P | 7/380 | Ensembl |
| c.119–27del16 | − | 1 | 0 | 0 | Intron 1 | P | 1/380 | Haider et al.5 |
| c.119–27dup16 | − | 1 | 0 | 0 | Intron 1 | P | 0 | Present study |
| c.119–28T→C | − | 4 | 1 | 1 | Intron 1 | P | 9/380 | Ensembl |
Note.— CRD, cone-rod dystrophy; RCD, rod-cone dystrophy; M, mutation; UV, unclassified variant; P, polymorphism. A minus sign (−) indicates no predicted effect. Numbering of NR2E3 mutations was performed following HGVS nomenclature guidelines (Ensembl reference sequence ENSP00000317199, with A of ATG=+1). Of the variants, p.Glu140Gly, p.Met163Thr, and the intronic change c.119-28T→C are known polymorphisms (Ensembl). One unaffected control person was heterozygous for the novel change c.230G→A (p.Arg77Gln). Interestingly, this novel missense change is flanking residue Arg76, which has been shown to be mutated in ESCS (p.Arg76Gln and p.Arg76Trp) (fig. 2A). Although NR2E3 mutations are considered relatively rare in the general population (frequency estimated to be <1 in 500),9 this change may represent a recessive NR2E3 mutation rather than a rare polymorphism. No other potentially pathogenic NR2E3 variant was found in this individual. The novel intronic sequence changes found in patients and controls are predicted not to affect normal splicing. They are considered polymorphisms.
The pathogenic nature of p.Gly56Arg is supported by the following arguments: (1) cosegregation with disease in the large adRP-affected families F1 and F3; (2) absence in 380 control chromosomes; (3) conservation in several orthologs and paralogs of the nuclear receptor (NR) superfamily (fig. 2B); (4) Polyphen and SIFT predictions, as well as Grantham score calculation, suggesting that p.Gly56Arg may affect protein structure or function; (5) ESEfinder and RESCUE-ESE results predicting the introduction of novel exonic splicing enhancers; and (6) NetPhos 2.0 and NetPhosK 1.0 predictions indicating the introduction of novel (protein kinase C-specific) phosphorylation sites, possibly affecting both function and nuclear localization of NR2E3 (table 3).10,11
Table 3. .
Evaluation of Pathogenic Potential of Missense Mutations in the DBD of NR2E3: p.Gly56Arg in Comparison to p.Arg76Gln, p.Arg76Trp, and p.Arg77Gln[Note]
| Mutation | Polyphen | SIFT | Grantham Score |
ESEfinder (exon 2) |
RESCUE-ESE (exon 2) |
NetGene 2 (intron 1–exon 2–intron 2) |
BDGP (intron 1–exon 2–intron 2) |
SpliceSiteFinder (intron 1–exon 2–intron 2) |
NetPhos 2.0 | NetPhosK 1.0 |
| p.Gly56Arg | Probably damaging | Affects protein function | 125 | One novel ESE | Two novel ESEs | One novel acceptor splice site | − | − | One novel Ser-phosphorylation site | Two novel Ser-phosphorylation sites |
| p.Arg77Gln | Possibly damaging | Affects protein function | 43 | − | One novel ESE | − | − | − | Loss of one Ser-phosphorylation site | − |
| p.Arg76Gln | Probably damaging | Affects protein function | 43 | Two novel ESEs, disruption of one ESE | − | − | − | − | Loss of one Ser-phosphorylation site | − |
| p.Arg76Trp | Probably damaging | Affects protein function | 101 | Disruption of two ESEs | − | Loss of two acceptor splice sites | − | − | Loss of one Ser-phosphorylation site | − |
Note.— Ser, serine. A minus sign (−) indicates no predicted effect. Overall, Polyphen and SIFT predictions, together with the Grantham score, sustain a pathogenic effect of p.Gly56Arg. Moreover, predictions of the phosphorylation site pattern are even more supportive. Phosphorylation is an important regulatory mechanism of nuclear receptors. The novel Ser phosphorylation sites predicted for p.Gly56Arg are in contrast with the predicted loss of one Ser phosphorylation site at position 74 for NR2E3 proteins with the recessive ESCS mutations p.Arg76Gln, p.Arg76Trp, and p.Arg77Gln, respectively. Interestingly, the Ser74 residue in NR2E3 corresponds to Ser51 in the VDR, in which phosphorylation not only influences the activity of the VDR but also regulates an NLS.10,11 On the basis of the alignment of NR2E3 and VDR, one could hypothesize that the corresponding sequence in NR2E3 might also contain an NLS (fig. 2B). The recessive NR2E3 mutations p.Arg76Gln, p.Arg76Trp, and p.Arg77Gln are located in this putative NLS and are predicted to disrupt the Ser74 phosphorylation site, which suggests an influence on phosphorylation, transcriptional activity, and nuclear localization (fig. 2B). This is supported by observations in corresponding mutated residues in the VDR.10,11 In contrast, for p.Gly56Arg, the predicted novel phosphorylation sites might compete with Ser74. In this way, it may be that both p.Gly56Arg and the recessive ESCS mutations p.Arg76Gln, p.Arg76Trp, and p.Arg77Gln have an important, although distinct, effect on NR2E3 activation, as well as on nuclear localization.
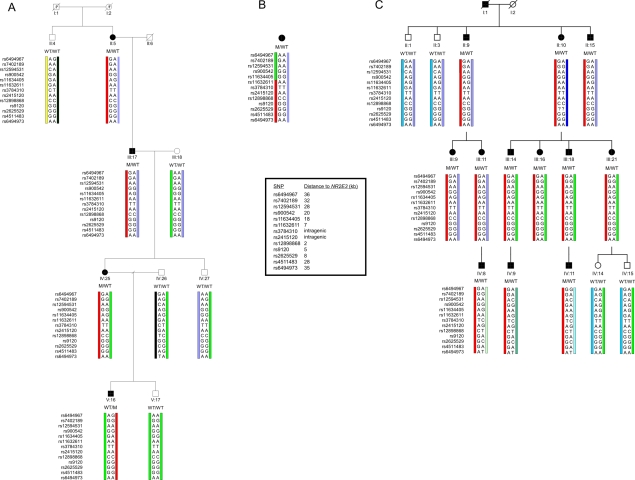
Two of the three adRP-affected families in which p.Gly56Arg was found are of Belgian descent (F1 and F3), and one is French (F2). To assess the origin of p.Gly56Arg, haplotype analysis was performed using 2 intragenic SNPs, 11 flanking SNPs that are located in a region of 70 kb around NR2E3, and 1 anonymous microsatellite (at 50 kb downstream of NR2E3) (HapMap, Ensembl, UCSC Genome Browser, Primer3). Genotyping of these markers was performed in core families of F1 and F3 and in the proband of F2. Interestingly, all affected individuals analyzed in these three families harbored the same intragenic alleles and SNPs up to a distance of 7 kb upstream and 35 kb downstream of NR2E3, respectively. Overall, the maximal common region between the three families covers ∼75 kb, suggesting that an ancient founder mutation cannot be excluded (fig. 3). The identification of p.Gly56Arg in 3 unrelated pedigrees with adRP among 47 families with RP (i.e., 6.4%) may indicate the potential importance of this mutation in a significant fraction of cases of adRP, at least in families of European descent. Further studies in larger cohorts are required to support this hypothesis.
Figure 3. .
Haplotype analysis with intragenic and closely flanking SNPs. Genotyping in a core family of F1 (A), the proband of F2 (B), and a core family of F3 (C), performed using 2 intragenic SNPs and 11 flanking SNPs that are located in a region of 70 kb around the 7.7-kb NR2E3 gene (HapMap, Ensembl). In F1 and F3, the red bar represents the common disease haplotype. In F2, this disease haplotype is partially shared from SNP rs11632611 (located 7 kb upstream of NR2E3) to SNP rs6494973 (35 kb downstream of NR2E3), as represented by the red bar. The green bar represents the distinct haplotype block (ranging from SNP rs11634405 at 18 kb to SNP rs6494967 at 36 kb both upstream of NR2E3). Genotyping of an anonymous microsatellite at 50 kb downstream of NR2E3 (UCSC Genome Browser, Primer3) allowed delineation of a maximal common region of ∼75 kb (upstream rs11634405 at 18 kb – NR2E3 7.7 kb – downstream microsatellite at 50 kb) (data not shown). WT, wild type; M, mutant. Filled bars represent affected individuals. Pedigree numbers are represented below the symbols and are in agreement with the numbering of figure 1.
Until now, NR2E3 mutations have been associated only with autosomal recessive ESCS and related recessive conditions,12 whereas the mutation identified in this study was found in families with a clinical diagnosis of adRP. In ESCS, a gain of function of S-cones and a maldevelopment and/or degeneration of rods lead to an increased sensitivity to blue light, night blindness, and central visual loss.5 Retinal pigmentation is of the nummular type, typically located immediately beyond the temporal vascular arcades and nasal to the optic disc. Electroretinography (ERG) in ESCS is characterized by severely reduced or absent rod-mediated responses to a weak light stimulus and by near-identical responses to a high-intensity stimulus in both dark- and light-adapted conditions, caused by supernormal S-cone–mediated responses.13,14 Classic adRP is an autosomal dominant rod-cone dystrophy with progressive degeneration of rods and subsequent involvement of cones, further characterized by progressive intraretinal pigment migration, generally of the spicular type. Although the ERG may be initially normal, rod responses progressively decline over time, with cone-mediated responses suffering the same fate with some delay in time.1
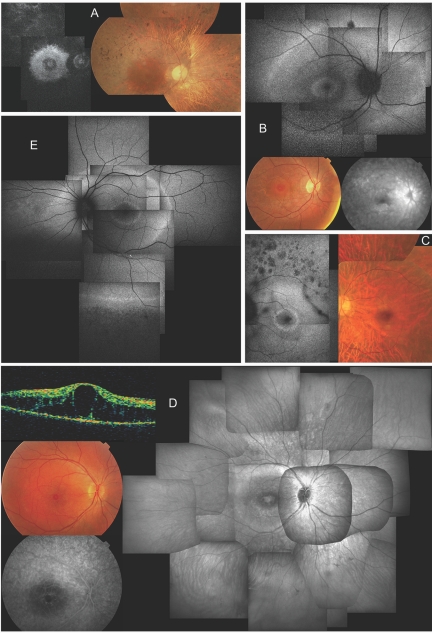
Ten affected members of F1 and three affected members of F2 and F3 underwent an extensive ophthalmological evaluation (clinical details are provided in tables 4 and 5). Interestingly, the phenotype found in F1, F2, and F3 corresponds to classic adRP but also shares some specific characteristics with ESCS. Patients showed a decline of cone function relatively late, at a time when rod function was already undetectable (fig. 4). In addition, the phenotype is characterized by three concentric rings of hyperautofluorescence: around the fovea, along the vascular arcades, and in the far periphery. Notably, the two more central rings are located in the same areas as those affected in ESCS.14 Moreover, the rather limited intraretinal pigmentation seen in the later stages, is of the mixed nummular and spicular type. In patients without any pigmentation, nummular areas of hypoautofluorescence around the vascular arcades are reminiscent of the pigment clumps observed in ESCS (fig. 5). As is often seen in adRP, the phenotype in F1 is rather variable from one patient to another (fig. 5 and tables 4 and 5).
Table 4. .
Clinical Characteristics of Several Affected Individuals in Families F1, F2, and F3[Note]
| Patient | Age (years) |
Sex | Best-Corrected Visual Acuity and Refraction RE | Best-Corrected Visual Acuity and Refraction LE | Night Blindness | Photophobia | First Symptom | Age at First Symptom (years) |
Cataract | Cystoid Macular Degeneration (Both Eyes) |
| Fam 1 III:1 | 76 | M | 5/10 w/o corr | LP (perforating trauma) | Yes | Night blindness | SCP | No | ||
| Fam 1 IV:2 | 50 | F | 9/10 w/ −1.0 D | 9/10 w/ −1.0 D | Yes | No | Night blindness | 15 | No | No |
| Fam 1 IV:6 | 45 | M | 3/10 w/ +3.0 D (−1,75 D) 20° | 5/10 w/ +1.0 D (−1.45 D) 145° | Yes | Yes | Night blindness | 7 | SCP | Yes |
| Fam 1 V:1 | 24 | F | 10/10 w/ −0.5 D | 10/10 w/o corr | Yes | No | Night blindness | 15 | No | No |
| Fam 1 III:12 | 64 | M | 5/10 w/ +0.75 D (−0.75 D) 10° | 7/10 w/ +2.25 D (−0.75 D) 180° | Yes | Yes | Night blindness | 10 | SCP | No |
| Fam 1 IV:19 | 29 | M | CF at 2 m (anisometropic amblyopia) | 10/10 w/o corr | Yes | No | Night blindness | 10 | No | No |
| Fam 1 III:15 | 62 | M | 2/10 w/ plano (−1.25 D) 175° | 5/10 w/ −2.5 D (−1.25 D) 175° | Yes | No | Night blindness | 10 | SPC and corticonuclear | No |
| Fam 1 IV:23 | 31 | F | 10/10 w/ −2.75 D (−2.75 D) 10° | 10/10 w/ −3.50 D (−2.25 D) 167° | Yes | No | Night blindness | 15 | No | No |
| Fam 1 IV:24 | 16 | M | 10/10 w/o corr | 10/10 w/o corr | Yes | No | Night blindness | 13 | No | No |
| Fam 1 IV:25 | 36 | F | 6/10 w/ +0.5 D | 7/10 w/ +0.5 D | Yes | Yes | Night blindness | 15 | No | No |
| Fam 1 V:16 | 10 | M | 10/10 w/ +4.25 D (−0.5 D) 180° | 10/10 w/ +4.0 D (−0.5 D) 130° | Yes | Yes | Night blindness | 5 | No | No |
| Fam 2 II:10 | 55 | F | 3/10 w/ −1.0 D (−0.75 D) 170° | 5/10 w/o corr | Yes | No | Night blindness | 30 | Nuclear cataract + | No |
| Fam 3 III:14 | 43 | M | 6/10 w/ +1.25 D | 1/20 w/ +0.5 D | Yes | No | Night blindness | 15 | SCP | No |
| Fam 3 IV:9 | 16 | M | 2/10 w/ −2.0 D | 4/10 w/ −2.5 D | Yes | No | Night blindness | 12 | Mild SCP | Yes |
Note.— Fam, family; w/, with correction; w/o corr, without correction; CF, counting fingers; LP, light perception; D, diopters; SCP, subcapsularis posterior; RE, right eye; LE, left eye; BE, both eyes; R/G, red/green; B/Y, blue/yellow; ERG, electroretinography.
Table 5. .
Clinical Characteristics of Several Affected Individuals in Families F1, F2, and F3[Note]
| Patient | Color Vision | Goldmann Visual Fields | ISCEV Standard-Pattern ERG (Both Eyes) | ISCEV Standard Full-Field Flash ERG (Both Eyes) | 1/ Fundus 2/ Infrared, autofluorescence and red-free imaging (Both eyes unless otherwise indicated) |
| Fam 1 III:1 | Not performed | Concentric constriction to <5° | Not performed | Absent rod- and cone-mediated responses | 1/ Pigmentary retinopathy (no further details known); 2/ Not performed |
| Fam 1 IV:2 | Normal | Mild superior indentation peripheral limits RE; normal LE | Normal | Rod-cone dystrophy; rod-mediated responses w/ amplitudes 1/5 of normal; cone-mediated responses w/ amplitudes 1/2 of normal | 1/ Outer retinal atrophy limited to the temporal and inferior periphery w/ intraretinal pigmentation of a mixed nummular and spicular type; mild attenuation of retinal vasculature; fovea normal; 2/ Mild bull’s eye maculopathy on infrared and red-free; small number of hypo- and hyperautofluorescent flecks inferior to temporal vascular arcade |
| Fam 1 IV:6 | Moderate mixed R/G and B/Y defect | Concentric constriction to <10° | Not performed | Absent rod- and cone-mediated responses | 1/ Outer retinal atrophy more pronounced around vascular arcades w/ intraretinal pigmentation of mixed nummular and spicular types; mild attenuation of retinal vasculature; RPE alterations in fovea; RE: synchisis scintillans; 2/ Not performed |
| Fam 1 V:1 | Normal | Normal peripheral limits; relative concentric pericentral scotoma BE | 1/2 of normal amplitudes | Rod-cone dystrophy; rod-mediated responses w/ amplitudes 1/5 of normal; cone-mediated responses w/ amplitudes 1/2 of normal | 1/ Outer retinal atrophy more pronounced in two concentric rings around vascular arcades and around fovea as bull’s eye maculopathy w/ only very few areas of intraretinal pigmentation limited to a few nummular hyperpigmentations in periphery; moderate attenuation of retinal vasculature; 2/ Double concentric hyperautofluorescent rings with one located just inside vascular arcades and another around fovea |
| Fam 1 III:12 | Mild mixed R/G and B/Y defect | Concentric constriction to <10° | Absent | Absent rod- and cone-mediated responses | 1/ Outer retinal atrophy more pronounced in two concentric rings around vascular arcades and around fovea as bull’s eye maculopathy w/ considerable intraretinal pigmentation of mixed nummular and spicular types; severe attenuation of retinal vasculature; fovea fairly normal; |
| 2/ Diffuse nummular areas of hypo-autofluorescence around vascular arcades; double concentric hyperautofluorescent rings with one located inside vascular arcades and another around fovea | |||||
| Fam 1 IV:19 | Mild mixed R/G and B/Y defect LE | Generalized sensitivity loss w/ pericentral scotoma; normal peripheral limits | Absent RE; 1/2 of normal amplitudes LE | Rod-cone dystrophy | 1/ Outer retinal atrophy more pronounced around vascular arcades w/ intraretinal pigmentation limited to a few nummular hyperpigmentations in periphery (RE>LE); mild attenuation of retinal vasculature; fovea normal; |
| Rod-mediated responses w/ residual amplitudes; delayed cone-mediated responses w/ amplitudes1/2 of normal | 2/ Diffuse nummular areas of hypo-autofluorescence around vascular arcades; triple concentric hyperautofluorescent rings with one located around fovea and another just inside vascular arcades, with a third in periphery | ||||
| Fam 1 III:15 | Mild mixed R/G and B/Y defect | Concentric constriction to <10° | Residual | Absent rod- and cone-mediated responses | 1/ Outer retinal atrophy more pronounced in two concentric rings around vascular arcades and around fovea as bull’s eye maculopathy w/ considerable intraretinal pigmentation of mixed nummular and spicular types; severe attenuation of retinal vasculature; fovea fairly normal; |
| 2/ Diffuse nummular areas of hypo-autofluorescence around vascular arcades; double concentric hyperautofluorescent rings with one located inside vascular arcades and another around fovea | |||||
| Fam 1 IV:23 | Normal | Generalized sensitivity loss w/ pericentral scotoma; normal peripheral limits | Not performed | Absent | 1/ Outer retinal atrophy in midperiphery, most pronounced around vascular arcades; no intraretinal pigmentation; moderate attenuation of retinal vasculature; fovea normal; 2/ Diffuse large, nummular areas of hypoautofluorescence around vascular arcades; concentric hyperautofluorescent ring around fovea |
| Fam 1 IV:24 | Normal | Pericentral scotoma; normal peripheral limits | 2/3 of normal amplitudes | Rod-cone dystrophy; rod-mediated responses w/ residual amplitudes; cone-mediated responses w/ amplitudes 2/3 of normal | 1/ Outer retinal atrophy more pronounced in two concentric rings around vascular arcades and around fovea as bull’s eye maculopathy w/ no intraretinal pigmentation; moderate attenuation of vasculature; 2/ Double concentric hyperautofluorescent rings with one located just inside vascular arcades and another around fovea |
| Fam 1 IV:25 | Normal | Normal peripheral limits; relative concentric pericentral scotoma BE | Residual | Rod (-cone) dystrophy; rod-mediated responses w/ residual amplitudes; cone-mediated responses w/ amplitudes 1/6 of normal | 1/ Outer retinal atrophy more pronounced in two concentric rings around vascular arcades and around fovea as bull’s eye maculopathy w/ only very few areas of intraretinal pigmentation limited to a few nummular hyperpigmentations in inferior periphery; moderate attenuation of retinal vasculature; some very fine intraretinal white punctate flecks in midperiphery along vascular arcades; 2/ Double concentric hyperautofluorescent rings with one located just inside vascular arcades and another around fovea |
| Fam 1 V:16 | Normal | Normal | Normal | Rod-cone dystrophy; rod-mediated responses w/ amplitudes 1/2 of normal; cone-mediated responses w/ amplitudes normal | 1/ Discrete outer retinal atrophy in inferior periphery; some very fine intraretinal white punctate flecks in midperiphery; no intraretinal pigmentation 2/ Discrete mottling of outer retina on infrared imaging; some very fine intraretinal white punctate flecks in midperiphery, rest normal |
| Fam 2 II:10 | Not performed | Concentric constriction to <10° | Absent | Absent rod- and cone-mediated responses | 1/ Extensive outer retinal atrophy more pronounced in two concentric rings around vascular arcades and around fovea as bull’s eye maculopathy w/ patches of total atrophy and moderate intraretinal pigmentation of mixed nummular and spicular types; severe attenuation of retinal vasculature; central fovea fairly normal; 2/ Double concentric hyperautofluorescent rings with one located just inside vascular arcades and another around fovea |
| Fam 3 III:14 | R/G Defect | Mild constriction of peripheral limits; considerable loss of pericentral sensitivity BE | 1/3 of normal amplitudes RE; absent LE | Rod-cone dystrophy; absent rod-mediated responses; delayed cone-mediated responses w/ amplitudes 1/4 of normal | 1/ Outer retinal atrophy w/ retinal oedema more pronounced in two concentric rings around vascular arcades and around fovea as bull’s eye maculopathy; some very fine intraretinal white punctate flecks around fovea and in midperiphery; considerable intraretinal pigmentation of mixed nummular and spicular types in periphery; severe attenuation of retinal vasculature; large nummular areas of chorioretinal atrophy temporal to the macula 2/ Discrete mottling of outer retina on infrared imaging; some very fine intraretinal white punctate flecks in midperiphery, |
| Fam 3 IV:9 | Not performed | Mild constriction of peripheral limits; relative concentric pericentral scotoma BE | 2/3 of normal amplitudes | Rod-cone dystrophy; absent rod-mediated responses; cone-mediated responses w/ amplitudes 1/2 of normal | 1/ Outer retinal atrophy more pronounced around vascular arcades w/ intraretinal pigmentation limited to a few nummular hyperpigmentations in periphery (RE>LE); mild attenuation of retinal vasculature; cystoid degeneration of fovea; 2/ Discrete double concentric hyperautofluorescent rings with one located around vascular arcades and another around fovea |
Note.— Fam, family; M, male; F, female; w/, with correction; w/o corr, without correction; CF, counting fingers; LP, light perception; D, diopters; SCP, subcapsularis posterior; RE, right eye; LE, left eye; BE, both eyes; R/G, red/green; B/Y, blue/yellow; ERG, electroretinography.
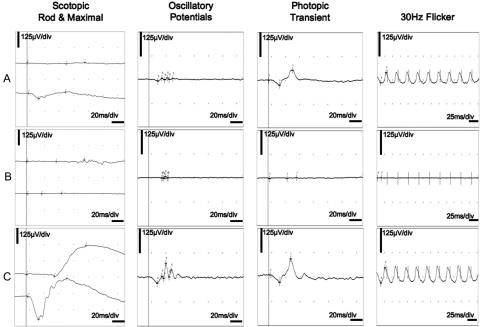
Figure 4. .
Full-field flash ERG. A, ERG traces of patient IV:24 of F1 at age 13 years. Scotopic (dark-adapted) rod-specific responses are completely absent, whereas scotopic maximal combined rod-cone responses are derived only from cones; amplitudes of photopic (light-adapted) transient and 30-Hz flicker cone-specific responses are moderately reduced, with limited latency delays; aspects of scotopic maximal combined rod-cone responses and photopic cone-specific responses to a transient flash are not identical. B, ERG traces of patient III:15 of F1 at age 62 years. All responses are absent, illustrating that gross retinal function is unmeasurable. C, Normal traces, for comparison. All ERGs are according to standards of International Society for Clinical Electrophysiology of Vision (ISCEV). RE, right eye; LE, left eye; μV/div, microvolts per division.
Figure 5. .
Clinical characteristics of affected members of F1 and F3, revealing a novel NR2E3-related clinical subtype of adRP. A, Late-stage phenotype in patient III:15 of F1 at age 62 years; composite autofluorescence (AF) (left) and composite fundus (right) of right eye (RE). Note small, relatively well-preserved macular area on fundus picture—although bull’s-eye pattern of hyperautofluorescence can be seen on AF in that area, with two hyperfluorescent rings that have virtually merged into one; also note severe narrowing of retinal vasculature. B, Early-stage phenotype in patient IV:24 of F1 at age 13 years; composite of AF (top) and fundus (bottom left) and late-phase fluorescein angiography (FA) of posterior pole of RE. Note the presence of two hyperautofluorescent rings on AF: one in bull’s-eye pattern around the fovea, another in midperiphery beyond temporal vascular arcades and nasal to optic disc (OD). Also note diffuse chorioretinal atrophy in areas corresponding to hyperautofluorescent rings visible on fundus and angiography picture. The small darker spot above the superior temporal vascular arcade on AF corresponds to a small retinal hemorrhage unrelated to RP phenotype. Discrete narrowing of retinal vasculature is also shown. C, Midstage phenotype in patient IV:23 of F1 at age 31 years. Composite AF (left) and composite fundus pictures (right) of posterior pole of left eye (LE) shows a bull’s-eye pattern with one small, discrete, intensely hyperautofluorescent ring around the fovea and a second, more diffusely hyperautofluorescent ring within area of temporal vascular arcades. Also note nummular areas of hypoautofluorescence above temporal vascular arcade, corresponding to areas of outer retinal atrophy; a dark string-like opacity superotemporal between vascular arcades which corresponds to a floater; marked attenuation of retinal vasculature; choroidal vasculature shining through due to diffuse chorioretinal atrophy, without intraretinal pigmentation; and a good-quality fovea. D, Midstage phenotype in patient IV:9 of F3. Clockwise from right, composite infrared image (IR) of fundus of RE, late phase FA and fundus picture of posterior pole, and optical coherence tomography (OCT) of central macular area of RE at age 16 years. Note cystic degeneration of fovea on IR, fundus picture, and OCT, without progressive fluorescein diffusion (leakage), unlike cystic macular edema, on FA, but identical to cystic degeneration seen in ESCS. IR further shows two whiter concentric rings in bull’s-eye pattern around the fovea, with the larger ring located just inside the temporal vascular arcades; also note sparse spicular pigmentation in inferior periphery and mild attenuation of retinal vasculature. E, Midstage phenotype in patient IV:19 of F1 at age 29 years; composite AF of RE. Note three concentric rings around posterior pole: one in a bull’s-eye pattern around fovea, a second at level of temporal vascular arcades and nasal to OD, and a third in periphery (only depicted inferiorly); hypoautofluorescence due to outer retinal degeneration beyond the peripheral hyperautofluorescent ring; and moderate vascular attenuation.
Importantly, the phenotypic findings observed in F1, F2, and F3 represent a novel recognizable clinical subtype of adRP. Further screening of additional families may provide more evidence of a robust genotype-phenotype correlation. Genotype-phenotype correlations become more important in directing molecular testing in adRP, because of extreme locus heterogeneity. Our findings add to a slowly growing list of phenotypically recognizable features relating to underlying adRP genes, such as nonpenetrance (PRPF31 mutations)15 and clumped pigment deposits (NRL mutations).16
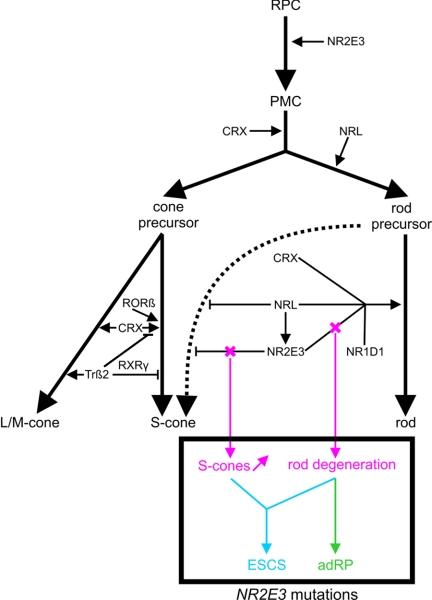
In addition to phenotypic differences, the RP phenotype described here and ESCS have a different inheritance pattern. This is comparable with inheritance of conditions caused by mutations in CRX and NRL, encoding two other key players of the retinal transcriptional network (RetNet) (fig. 6). Moreover, individuals with adRP resulting from a particular NRL mutation (p.Ser50Thr)16 have features reminiscent of the NR2E3-related adRP phenotype found in our families: mixed spicular-nummular intraretinal pigmentation and hyperautofluorescence surrounding the fovea in younger individuals. In addition, recessive mutations in both NR2E3 and NRL also result in comparable phenotypes.14,24 Indeed, such similarities are not surprising, because NRL activates transcription of NR2E3 (fig. 6).
Figure 6. .
Role of retinal TFs during photoreceptor development. Several TFs are required to determine a specific photoreceptor cell type (rod or cone). First, multipotent retinal progenitor cells (RPC) pass through different stages of competence, finally becoming postmitotic cells (PMC). The expression of CRX commits a population of PMCs toward photoreceptor lineage.17 A pool of these are directed to cone fate. It is presumed that cone precursors follow an S-cone default pathway (influenced by CRX and RORβ18,19) and that subsequent L- and M-cone induction is mediated by additional TFs. One of these is Trβ2, inducing M-opsin expression but repressing S-opsin expression, probably as a heterodimer with the nuclear receptor RXRγ.20,21 Later in development, another pool of PMCs will acquire the rod fate by NRL expression. NRL activates transcription of NR2E3, having a positive synergistic influence with CRX, NR1D1, and NRL on the transcription of rod-specific genes.6,7 Moreover, NR2E3 contributes to rodlike morphology.17 Importantly, rod precursors are relatively “plastic,” because they are still capable of developing S-cones. NRL and especially NR2E3 are able to repress transcription of cone-specific genes, to stabilize the rod fate before further development of rodlike characteristics.7,19,22 This dual function of NR2E3, involving both transrepression of cone-specific genes and transactivation of rod-specific genes, may underlie molecular pathogenesis of distinct NR2E3-related retinal dystrophies, as illustrated in this study. We hypothesize that mutations affecting both functions lead to an increased density of S-cones and a degeneration of rods, as seen in ESCS, whereas mutations affecting only or predominantly the transactivation function result in a predominant degeneration of rods, as seen in adRP. Recent studies have suggested that temporal differences exist between those NR2E3 functions. NR2E3 expression has been shown in mitotic as well as postmitotic cells of the developing retina.23,35 Moreover, NR2E3 expression was also seen in both rods and cones of the mature retina. It is presumed that in postmitotic rods NR2E3 influences rod cell differentiation, whereas, in late mitotic progenitors, its primary function is to suppress the cone-generation program.35
In general, adRP and ESCS may have a different pathogenesis that is mainly related to the pathophysiology of the patients' S-cone photoreceptors. This is reflected by their typical ERG patterns, which reveal a difference in S-cone response.1,13 We presume this may be attributed to the dual regulatory role of NR2E3 and its relation with other key players of the retinal transcriptional network. During rod development, NR2E3 has two distinct functions: (1) it suppresses the transcription of cone-specific genes and (2) it enhances transcriptional activation of rod-specific genes in concert with other transcription factors (TFs) such as NR1D1, CRX, and NRL (fig. 6).6,7,17,22 Because of this dual function, we hypothesize that the predominant degeneration of rods in adRP in this study results from a loss of activation of rod-specific genes, whereas the increased S-cone density and the degeneration of rods in ESCS are due to a derepression of cone-specific genes and to a loss of activation of rod-specific genes, respectively (fig. 6).7,17 Since S-cone function is not enhanced in adRP, this would imply that p.Gly56Arg, either exclusively or predominantly, affects the transactivation function of NR2E3.
We hypothesize that the specific location of NR2E3 mutations may be an important factor in determining their functional effect. NR2E3 is part of the NR superfamily, in which the DNA-binding and ligand-binding domains (DBD and LBD, respectively) are most conserved (fig. 2A).25 ESCS-causing missense mutations in the LBD of NR2E3 have been shown to lead to derepression of cone-specific genes, whereas missense mutations in the DBD in ESCS, affect both transactivation of rhodopsin and derepression of cone-specific genes.7,22
On the basis of alignment of NR2E3 with other NRs, p.Gly56Arg is most likely to be located in the DBD (fig. 2B), more specifically in a region between the second and third cysteine of the first zinc finger of NR2E3, where no mutations have yet been identified (fig. 2A). The potential functional importance of Gly56 is sustained by the effect of natural missense mutations affecting corresponding residues in other NRs, namely p.Gly30Asp in the vitamin D receptor (VDR [MIM 601769])26,27 and both p.Gly568Trp and p.Gly568Val in the androgen receptor (AR [MIM 313700]) (fig. 2B).28,29 They cause autosomal recessive hypocalcemic 1.25-dihydroxyvitamin D-resistant rickets (HVDRR [MIM 277440]) and X-linked recessive partial androgen insensitivity syndrome (AIS [MIM 300068]), respectively. Their occurrence suggests that this Gly residue is prone to mutations, which is in agreement with the conclusions of this study.
VDRs with p.Gly30Asp display normal ligand binding but show low affinity for DNA and are transcriptionally inactive.26,27,30 Weak DNA binding and reduced transactivation is also seen with mutation p.Gly568Val in the AR, but some normal transrepression functions are still maintained.31 This observation, together with an impaired coactivator binding, suggests that, besides DNA binding, cross-talk with other proteins may be an essential function of this Gly residue.32 Involvement in specific protein-protein interactions is also supported by reporter gene assays with p.Gly568Trp in the AR, showing that the Gly568 residue is probably more important for transrepression than for transactivation.33 In addition, the Gly→Val mutation has been studied in the mouse glucocorticoid receptor (GR [MIM 138040]), where it induces alterations in affinity and/or transcriptional activation in a promotor-dependent manner.34
Thus, from these studies, it can be deduced that the corresponding Gly56 in NR2E3 may play an important role in DNA binding, promotor-specific transcriptional activation and/or repression, or specific protein-protein interactions. The absence of potential NR2E3 consensus target sequences in the minimal rhodopsin promoter6 suggests a mechanism of action independent of DNA binding, however, and consequently suggests a major role for protein-protein interactions. We hypothesize that NR2E3 mutations causing ESCS affect protein interactions involved in transrepression and/or transactivation, whereas NR2E3 mutations causing adRP predominantly affect those that lead to transactivation of rod-specific genes. More specifically, the Gly56 residue may play an important role in determining interactions between the DBD of NR2E3 and proteins favoring the transactivation function.
TFs that interact with NR2E3 during transactivation of rod-specific genes are CRX and NR1D1 (fig. 6). They form a multiprotein complex with NRL in the retina (fig. 6).6,7 A recent in vivo study showed that NR2E3 can suppress expression of cone genes and activate expression of rhodopsin autonomously but also that it requires coexpression of NRL during the activation of other rod-specific genes (fig. 6).17
In summary, p.Gly56Arg may predominantly influence the terminal differentiation and maintenance of rods through disruption of the transactivation function of NR2E3—but not, or only to a minimal extent, of the transrepression function—possibly through loss of interactions with CRX, NRL, or other cofactors. Although it is not yet clear whether this mutation leads to loss of function, gain of function, or a dominant-negative effect, any change in transactivation function of NR2E3 may lead to an imbalance in the dosage-sensitive retinal transcription network, with subsequent rod degeneration, as is seen in adRP (fig. 6).
In conclusion, this study provides evidence for a potentially important role of NR2E3 in the causation of adRP. We propose a different pathogenetic mechanism for the distinct dominant and recessive NR2E3-related phenotypes that may be attributed to the dual role of NR2E3 in the regulation of photoreceptor-specific genes. In addition, a novel recognizable clinical subtype of adRP is identified, pointing to NR2E3 mutations and thus serving as a clinical marker improving targeted molecular genetic testing in adRP. Further molecular screening of NR2E3 and phenotypic characterization of a larger cohort of families with adRP will be performed to corroborate our findings.
Acknowledgments
F.C. is a doctoral student (Fund for Scientific Research–FWO) (1.1.387.07.N.00). K.D.B. and E.D.B. are funded by the FWO. This study is supported by FWO grants 1.2.843.07.N.01, 1.5.244.05 (EDB), and OZP 3G004306 (E.D.B. and B.P.L.). J.H. is funded by the Institute for Promotion of Innovation by Science and Technology- Flanders (IWT). The authors are grateful to Sarah De Jaegere for her technical assistance. The authors also gratefully acknowledge the patients participating in this study.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Berkeley Drosophila Genome Project (BDGP), http://www.fruitfly.org/seq_tools/splice.html
- Ensembl Genome Browser, http://www.ensembl.org/index.html
- ESEfinder, http://rulai.cshl.edu/tools/ESE/
- International HapMap Project, http://www.hapmap.org/
- NCBI Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/static/MVstart.html
- NetGene2, http://www.cbs.dtu.dk/services/NetGene2/
- NetPhos 2.0 Server, http://www.cbs.dtu.dk/services/NetPhos/
- NetPhosK 1.0 Server, http://www.cbs.dtu.dk/services/NetPhosK/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RP, BBS4, VDR, AR, HVDRR, AIS, and GR)
- Polymorphism Phenotyping (PolyPhen), http://genetics.bwh.harvard.edu/pph/
- Primer3, http://biotools.umassmed.edu/bioapps/primer3_www.cgi
- RESCUE-ESE, http://genes.mit.edu/burgelab/rescue-ese/
- RetNet, http://www.sph.uth.tmc.edu/Retnet/
- Sorting Intolerant From Tolerant (SIFT), http://blocks.fhcrc.org/sift/SIFT.html
- SpliceSiteFinder, http://www.genet.sickkids.on.ca/~ali/splicesitefinder.html
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Bird AC (1995) Retinal photoreceptor dystrophies LI. Am J Ophthalmol 119:543–562 [DOI] [PubMed] [Google Scholar]
- 2.Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA, Garcia CA, Ruiz RS, Blanton SH, Northrup H, et al (2006) Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci 47:3052–3064 10.1167/iovs.05-1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindner TH, Hoffmann K (2005) easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics 21:405–407 10.1093/bioinformatics/bti009 [DOI] [PubMed] [Google Scholar]
- 4.Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, et al (2001) Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet 28:188–191 10.1038/88925 [DOI] [PubMed] [Google Scholar]
- 5.Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, et al (2000) Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet 24:127–131 10.1038/72777 [DOI] [PubMed] [Google Scholar]
- 6.Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A (2004) Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet 13:1563–1575 10.1093/hmg/ddh173 [DOI] [PubMed] [Google Scholar]
- 7.Peng GH, Ahmad O, Ahmad F, Liu J, Chen S (2005) The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet 14:747–764 10.1093/hmg/ddi070 [DOI] [PubMed] [Google Scholar]
- 8.Nuclear Receptors Nomenclature Committee (1999) A unified nomenclature system for the nuclear receptor superfamily. Cell 97:161–163 10.1016/S0092-8674(00)80726-6 [DOI] [PubMed] [Google Scholar]
- 9.Miano MG, Jacobson SG, Carothers A, Hanson I, Teague P, Lovell J, Cideciyan AV, Haider N, Stone EM, Sheffield VC, et al (2000) Pitfalls in homozygosity mapping. Am J Hum Genet 67:1348–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh JC, Jurutka PW, Nakajima S, Galligan MA, Haussler CA, Shimizu Y, Shimizu N, Whitfield GK, Haussler MR (1993) Phosphorylation of the human vitamin D receptor by protein kinase C: biochemical and functional evaluation of the serine 51 recognition site. J Biol Chem 268:15118–15126 [PubMed] [Google Scholar]
- 11.Hsieh JC, Shimizu Y, Minoshima S, Shimizu N, Haussler CA, Jurutka PW, Haussler MR (1998) Novel nuclear localization signal between the two DNA-binding zinc fingers in the human vitamin D receptor. J Cell Biochem 70:94–109 [DOI] [PubMed] [Google Scholar]
- 12.Sharon D, Sandberg MA, Caruso RC, Berson EL, Dryja TP (2003) Shared mutations in NR2E3 in enhanced S-cone syndrome, Goldmann-Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch Ophthalmol 121:1316–1323 10.1001/archopht.121.9.1316 [DOI] [PubMed] [Google Scholar]
- 13.Jacobson SG, Marmor MF, Kemp CM, Knighton RW (1990) SWS (blue) cone hypersensitivity in a newly identified retinal degeneration. Invest Ophthalmol Vis Sci 31:827–838 [PubMed] [Google Scholar]
- 14.Marmor MF, Jacobson SG, Foerster MH, Kellner U, Weleber RG (1990) Diagnostic clinical findings of a new syndrome with night blindness, maculopathy, and enhanced S cone sensitivity. Am J Ophthalmol 110:124–134 [DOI] [PubMed] [Google Scholar]
- 15.Rivolta C, McGee TL, Rio Frio T, Jensen RV, Berson EL, Dryja TP (2006) Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat 27:644–653 10.1002/humu.20325 [DOI] [PubMed] [Google Scholar]
- 16.Bessant DA, Holder GE, Fitzke FW, Payne AM, Bhattacharya SS, Bird AC (2003) Phenotype of retinitis pigmentosa associated with the Ser50Thr mutation in the NRL gene. Arch Ophthalmol 121:793–802 10.1001/archopht.121.6.793 [DOI] [PubMed] [Google Scholar]
- 17.Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A (2006) In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum Mol Genet 15:2588–2602 10.1093/hmg/ddl185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivas M, Ng L, Liu H, Jia L, Forrest D (2006) Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor beta. Mol Endocrinol 20:1728–1741 10.1210/me.2005-0505 [DOI] [PubMed] [Google Scholar]
- 19.Oh EC, Khan N, Novelli E, Khanna H, Strettoi E, Swaroop A (2007) Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci USA 104:1679–1684 10.1073/pnas.0605934104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagi Y, Takezawa S, Kato S (2002) Distinct functions of photoreceptor cell-specific nuclear receptor, thyroid hormone receptor β2 and CRX in one photoreceptor development. Invest Ophthalmol Vis Sci 43:3489–3494 [PubMed] [Google Scholar]
- 21.Roberts MR, Hendrickson A, McGuire CR, Reh TA (2005) Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci 46:2897–2904 10.1167/iovs.05-0093 [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Rattner A, Nathans J (2005) The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci 25:118–129 10.1523/JNEUROSCI.3571-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takezawa S, Yokoyama A, Okada M, Fujiki R, Iriyama A, Yanagi Y, Ito H, Takada I, Kishimoto M, Miyajima A, et al (2007) A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J 26:764–774 10.1038/sj.emboj.7601548 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Nishiguchi KM, Friedman JS, Sandberg MA, Swaroop A, Berson EL, Dryja TP (2004) Recessive NRL mutations in patients with clumped pigmentary retinal degeneration and relative preservation of blue cone function. Proc Natl Acad Sci U S A 101:17819–17824 10.1073/pnas.0408183101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renaud JP, Moras D (2000) Structural studies on nuclear receptors. Cell Mol Life Sci 57:1748–1769 10.1007/PL00000656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes MR, Malloy PJ, Kieback DG, Kesterson RA, Pike JW, Feldman D, O’Malley BW (1988) Point mutations in the human vitamin D receptor gene associated with hypocalcemic rickets. Science 242:1702–1705 10.1126/science.2849209 [DOI] [PubMed] [Google Scholar]
- 27.Malloy PJ, Hochberg Z, Pike JW, Feldman D (1989) Abnormal binding of vitamin D receptors to deoxyribonucleic acid in a kindred with vitamin D-dependent rickets, type II. J Clin Endocrinol Metab 68:263–269 [DOI] [PubMed] [Google Scholar]
- 28.Lobaccaro JM, Belon C, Lumbroso S, Olewniczack G, Carre-Pigeon F, Job JC, Chaussain JL, Toublanc JE, Sultan C (1994) Molecular prenatal diagnosis of partial androgen insensitivity syndrome based on the Hind III polymorphism of the androgen receptor gene. Clin Endocrinol (Oxford) 40:297–302 [DOI] [PubMed] [Google Scholar]
- 29.Allera A, Herbst MA, Griffin JE, Wilson JD, Schweikert HU, McPhaul MJ (1995) Mutations of the androgen receptor coding sequence are infrequent in patients with isolated hypospadias. J Clin Endocrinol Metab 80:2697–2699 10.1210/jc.80.9.2697 [DOI] [PubMed] [Google Scholar]
- 30.Sone T, Scott RA, Hughes MR, Malloy PJ, Feldman D, O’Malley BW, Pike JW (1989) Mutant vitamin D receptors which confer hereditary resistance to 1,25-dihydroxyvitamin D3 in humans are transcriptionally inactive in vitro. J Biol Chem 264:20230–20234 [PubMed] [Google Scholar]
- 31.Aarnisalo P, Santti H, Poukka H, Palvimo JJ, Janne OA (1999) Transcription activating and repressing functions of the androgen receptor are differentially influenced by mutations in the deoxyribonucleic acid-binding domain. Endocrinology 140:3097–3105 10.1210/en.140.7.3097 [DOI] [PubMed] [Google Scholar]
- 32.Poukka H, Aarnisalo P, Santti H, Janne OA, Palvimo JJ (2000) Coregulator small nuclear RING finger protein (SNURF) enhances Sp1- and steroid receptor-mediated transcription by different mechanisms. J Biol Chem 275:571–579 10.1074/jbc.275.1.571 [DOI] [PubMed] [Google Scholar]
- 33.Lobaccaro JM, Poujol N, Terouanne B, Georget V, Fabre S, Lumbroso S, Sultan C (1999) Transcriptional interferences between normal or mutant androgen receptors and the activator protein 1—dissection of the androgen receptor functional domains. Endocrinology 140:350–357 10.1210/en.140.1.350 [DOI] [PubMed] [Google Scholar]
- 34.Necela BM, Cidlowski JA (2004) A single amino acid change in the first zinc finger of the DNA binding domain of the glucocorticoid receptor regulates differential promoter selectivity. J Biol Chem 279:39279–39288 10.1074/jbc.M405489200 [DOI] [PubMed] [Google Scholar]
- 35.Haider NB, Demarco P, Nystuen AM, Huang X, Smith RS, McCall MA, Naggert JK, Nishina PM (2006) The transcription factor Nr2e3 functions in retinal progenitors to suppress cone cell generation. Vis Neurosci 23:917–929 10.1017/S095252380623027X [DOI] [PubMed] [Google Scholar]