Abstract
The acyl-CoA dehydrogenases are a family of multimeric flavoenzymes that catalyze the α,β-dehydrogenation of acyl-CoA esters in fatty acid β-oxidation and amino acid catabolism. Genetic defects have been identified in most of the acyl-CoA dehydrogenases in humans. Acyl-CoA dehydrogenase 9 (ACAD9) is a recently identified acyl-CoA dehydrogenase that demonstrates maximum activity with unsaturated long-chain acyl-CoAs. We now report three cases of ACAD9 deficiency. Patient 1 was a 14-year-old, previously healthy boy who died of a Reye-like episode and cerebellar stroke triggered by a mild viral illness and ingestion of aspirin. Patient 2 was a 10-year-old girl who first presented at age 4 mo with recurrent episodes of acute liver dysfunction and hypoglycemia, with otherwise minor illnesses. Patient 3 was a 4.5-year-old girl who died of cardiomyopathy and whose sibling also died of cardiomyopathy at age 21 mo. Mild chronic neurologic dysfunction was reported in all three patients. Defects in ACAD9 mRNA were identified in the first two patients, and all patients manifested marked defects in ACAD9 protein. Despite a significant overlap of substrate specificity, it appears that ACAD9 and very-long-chain acyl-CoA dehydrogenase are unable to compensate for each other in patients with either deficiency. Studies of the tissue distribution and gene regulation of ACAD9 and very-long-chain acyl-CoA dehydrogenase identify the presence of two independently regulated functional pathways for long-chain fat metabolism, indicating that these two enzymes are likely to be involved in different physiological functions.
The acyl-CoA dehydrogenases (ACADs) are a group of mitochondrial flavoenzymes that catalyze the α,β-dehydrogenation of acyl-CoA esters, using the electron transfer flavoprotein (ETF) as their physiologic electron acceptor. There are at least nine known members of the ACAD gene family active in branched-chain amino acid catabolism or fatty acid oxidation. All of the ACADs are nuclear-encoded proteins that must first be translated in the cytoplasm, then imported into mitochondria and processed into a mature form.
Deficiencies of the ACADs are among the most common inborn errors of metabolism. Seven different ACAD genetic defects have been described in the past.1 ACAD-deficient patients typically accumulate intermediate metabolites with profiles that reflect the substrate specificity of the deficient enzyme. However, of the three long-chain–specific ACADs, inherited defects have been identified only in very-long-chain ACAD (VLCAD) (ACADVL [MIM 609575]). Patients present with recurrent episodes of hypoglycemia with or without rhabdomyolysis, sudden unexplained death, and cardiomyopathy.1 Recently, tandem mass spectrometry has been used to detect characteristic metabolites in blood spots collected from newborns, allowing diagnosis of VLCAD deficiency before symptoms develop.2 Long-chain ACAD (LCAD) was characterized about a decade before VLCAD was discovered3; however, its physiologic function remains a mystery. Acyl-CoA dehydrogenase 9 (ACAD9) has only recently been recognized.4,5 Substrate-utilization studies have shown that ACAD9 is active against both saturated and unsaturated long-chain acyl-CoA substrates, with optimum activity toward the latter.4 In this article, we report three cases of ACAD9 deficiency presenting with episodic liver dysfunction during otherwise mild illnesses or cardiomyopathy, along with chronic neurologic dysfunction. In spite of the high degree of homology between ACAD9 and VLCAD, it is clear that their physiological functions do not complement each other when either is deficient.6 Rather, our studies of the enzymatic function, tissue distribution, and gene expression of these two enzymes identify the presence of two independently regulated functional pathways for long-chain fat metabolism and expand the potential role for fatty acid oxidation in intermediary metabolism.
Methods
Overexpression and Purification of Long-Chain ACADs
Full-length cDNAs corresponding to the mature protein sequence of ACAD9, VLCAD, and LCAD were cloned individually into the Escherichia coli expression vector pET-21a(+) (Novagen) and were overexpressed, and the proteins were purified as described elsewhere.4,7 The purity of each enzyme was estimated, by SDS-PAGE, to be >95%.
Quantitative Real-Time RT-PCR
Reverse transcription was performed using a panel of total RNA pooled from 20 human tissues or total RNA pooled from human hippocampus (purchased from BD Biosciences Clontech), with 1 μg of RNA and Powerscript reverse transcriptase (BD Biosciences Clontech), in accordance with the manufacturer’s instructions. cDNA corresponding to 20 ng of total RNA was used for real-time RT-PCR on an ABI 7900HT Sequence Detection System (PE Applied Biosystems), with the use of heat-activated TaqDNA polymerase (BD Biosciences Clontech). Specific probes and primers (sequences available on request) for VLCAD, ACAD9, LCAD, medium-chain ACAD (MCAD), and short-chain ACAD (SCAD) were designed to target near the coding sequence for the catalytic domain of each enzyme. Probes for the housekeeping gene glucuronidase, beta (GUSB) and the 18S ribosomal RNA (rRNA) gene were used as controls. All the probes and primers were synthesized by PE Applied Biosystems (Assay on Demand). PCR assays were performed in 96-well optical plates, in accordance with the manufacturer’s recommendations. The amplification efficiency of primer and probe sets was tested using different dilutions of total RNA, and all were similar (close to 1). For quantitative analysis of the data, CT (threshold-cycle number) values were normalized to those of GUSB or 18S rRNA, with use of the ΔΔCT method.8
cDNA Amplification and Sequencing
Total RNA was extracted from patient fibroblasts or frozen postmortem liver with the use of TRIzol reagent (Invitrogen) in accordance with the manufacturer’s instructions. Reverse transcription was performed using 1 μg of total RNA and Powerscript reverse transcriptase, with the use of random hexamers as primers. The ACAD9 coding region was then amplified with the Yieldace PCR kit (Stratagene), with primers as follows: 5′-AGACGTGTGTGTGTCCCTGCGGCGCTA-3′ and 5′-GAGTAACAGTCATCCAGCAACGGGCCATGGGTA-3′. Various primers at intron/exon junctions of ACAD9 were also used, and their sequences are available on request. The PCR products were purified using the PCR Cleanup kit (Qiagen) and were sequenced directly by the Genomics and Proteomics Core Facility at the University of Pittsburgh.
Immunoblotting and Immunocompetition Assay
Fifty micrograms of total protein from human fibroblast or liver extracts and 150 μg of total protein from muscle lysates were separated on a 12% SDS polyacrylamide gel, were transferred to nitrocellulose, and were immunostained with the indicated antibodies, by use of our previously published methods.4 Immunocompetition assays were performed similar to routine immunobloting, except that purified ACAD9 protein was added to the antibody incubation solution. The ratio between purified ACAD9 protein and affinity-purified ACAD9 antibody was 1:1 (mole:mole).
ETF Fluorescence-Reduction Assay for ACAD Activity
ACAD activity in cultured skin fibroblasts and frozen liver obtained 6–10 h postmortem from patient 1 and an age-matched control who died of trauma (Brain and Tissue Bank for Developmental Disorders, Baltimore) was assayed using the sensitive and specific anaerobic ETF fluorescence-reduction assay with the indicated substrates at 5 μM final concentration, as described elsewhere.9 Mitochondrial matrix and membrane fractions were prepared and assayed in triplicate, with use of palmitoyl (C16:0)-CoA as substrate, as described elsewhere.10 Five normal liver samples were used as controls. Saturated and monounsaturated straight-chain acyl-CoA substrates—hexanoyl (C6:0)–CoA, octanoyl (C8:0)–CoA, decanoyl (C10:0)–CoA, dodecanoyl (C12:0)–CoA, tetradecanoyl (C14:0)–CoA, C16:0–CoA, palmitoleoyl (C16:1)–CoA, stearoyl (C18:0)–CoA, octadecenoyl (C18:1)–CoA, octadecynoyl (C18:2)–CoA, and arachidoyl (C20:0)–CoA—were purchased from Sigma. Arachidonoyl (C20:4)–CoA and docosahexaenoyl (C22:6)–CoA were purchased from Avanti Polar Lipids. Branched-chain and some polyunsaturated-chain acyl-CoA substrates included the CoA ester of 3α, 7α, l2α-trihydroxy-5β-cholestan-26-oic acid (THC-CoA); S, 2-methyl-hexadecanoyl-CoA (S, 2-methyl C16-CoA); S, 2-methyl-pentadecanoyl-CoA (S, 2-methyl C15-CoA); S, 4, 8, 12 trimethyl-tridecanoyl-CoA (S, 4, 8, 12, trimethyl C13-CoA); eicosapentaenoyl (C20:5)-CoA (kindly provided by Dr. Paul P. Veldhoven [Leuven University, Belgium]), and S, 2,6-dimethylheptanoyl-CoA (S, dimethyl C7-CoA) (kindly provided by Dr. R. J. A. Wanders [University of Amsterdam]). Cytokine stimulation of cultured human fibroblasts was performed as described elsewhere.11 ACAD activity was assayed in cultures before and after stimulation.
Immunoinactivation of Purified ACAD9
A rabbit polyclonal antiserum was raised to ACAD9 and was purified through a Staphylococcus aureus protein A column (Pierce), in accordance with the manufacturer’s instructions. For immunoinactivation assays, 1 μg of purified antibody was added per 10 μg of liver lysate protein. The mixture was incubated at 4°C overnight, with gentle mixing. The next morning, protein A–bound resin was added to the mixture and was incubated at 4°C for an additional hour, and bound antibody complex was precipitated by centrifugation. The supernatant was assayed for ACAD activity.
Immunohistochemical and Immunofluorescent Staining of Fixed Tissues and Skin Fibroblast
Formalin-fixed, paraffin-embedded sections of cerebellum, muscle, and liver from patient 1 were provided by the Department of Pathology, Children’s Hospital of Alabama. Sections from the same tissues from a 14-year-old boy who died of sudden cardiac failure were selected from the files of the Department of Pathology, Children’s Hospital of Pittsburgh. Five-micrometer sections were cut at regular intervals and were mounted on glass slides. Immunohistochemical staining was performed as described elsewhere, with minor modification, with use of ACAD9, VLCAD, LCAD, and MCAD antibodies at dilutions of 1:200–300.12 Staining was performed using the avidin–biotinylated peroxidase complex (ABC) method (Vector Laboratories), was visualized by peroxidase substrate and 3,3-diaminobenzidine (DAB), and was counterstained with hematoxylin.
Human cerebellum sections were reacted with 1:200 dilution goat antiserum to the high-affinity aspartate/glutamate transporter (EAAT4 [Molecular Probes]) and rabbit anti-ACAD9 antibodies and were visualized with Alexa Fluor 680–conjugated donkey anti-goat and IRDye 800CW–conjugated donkey anti-rabbit immunoglobulin G (IgG) (H+L [Rockland Immunochemicals]) at a 1:5,000 dilution. The immune signals were detected with the LI-COR Odyssey (42-μm resolution and 1-mm offset at the highest quality capture settings), in accordance with the manufacturer’s instructions. For human liver sections, goat anti-human alpha-1-antitrypsin (α-1-AT [Fisher Scientific]) was used as a control antibody. For human lung sections, mouse anti-human MUC1 antibody (kindly provided by Dr. Joseph Pilewski, University of Pittsburgh) was used as a control antibody. Human fibroblasts were fixed with 4% formaldehyde on coverslips and were incubated with primary antibody and fluorescent-labeled secondary antibodies, in accordance with the manufacturer’s recommendation, and were then visualized as described above.
Promoter Analysis
ACAD9 DNA fragments were amplified by PCR from control and patient genomic DNA, starting at coding nucleotide position +50 and containing increasing lengths of 5′ untranslated sequence to nucleotide positions −264, −380, and −590. Fragments were cloned upstream of the firefly luciferase reporter gene in the promoterless pGL3-Basic plasmid (Promega) and were confirmed by DNA sequencing. Transfection experiments in HepG2 cells were performed as described elsewhere.13 A constitutive reporter vector was cotransfected as a control. Cells were lysed 48 h after transfection, and 20 μl of cell lysate was used for a luciferase reporter assay with a Dual Luciferase Assay kit (Promega), in accordance with the manufacturer’s instructions. Light intensity was measured by a Monolight 2010 luminometer for 10 s after manual reagent injection. All transfections were performed in triplicate, and average relative luciferase activities were determined.13
Clinical Presentation
Patient 1
Case report
A 14-year-old boy presented with a Reye-like episode triggered by ingestion of aspirin during a minor viral illness. Initial laboratory evaluation revealed a markedly elevated plasma ammonia concentration (>700 μmol/liter; reference range 9–33 μmol/liter) and elevated results from liver function tests: aspartate aminotransferase (AST) was 3,355 U/liter (reference range 15–40 U/liter), alanine aminotransferase was 2,488 U/liter (reference range 10–45 U/liter), and prothrombin time was 23.6 s (reference range 10.6–12.6 s). Plasma glucose findings were normal. His serum salicylate level was 23.2 mg/dL (therapeutic range 2.8–20 mg/dL). Results of additional laboratory evaluation were remarkable for a serum lactate level of 10.8 mmol/dL (reference range 0.7–2.1 mmol/liter), lactate dehydrogenase level of 15,394 U/liter (reference range 360–730 U/liter), creatine kinase level of 2,824 U/liter (reference range 30–150 U/liter), and creatine kinase, muscle, and brain subunit levels of 9.5 ng/ml (reference range 0.0–5.0 ng/ml). Results from multiple viral studies were negative. Acute hemodialysis was instituted, and, after 3 d in the hospital, his ammonia level had decreased to 158 mmol/liter. In spite of this, the child remained unresponsive, and a head CT scan showed diffuse cerebral edema with tonsillar herniation. Neurologic examination confirmed brain death, and support was discontinued.
Autopsy report
Findings on autopsy included diffuse hepatic microvesicular steatosis and some macrovesicular steatosis, which were interpreted as being consistent with Reye-like syndrome. Brain findings were notable for generalized edema with diffuse ventricular compression, acute left tonsillar herniation, and diffuse multifocal acute damage in the hippocampus. In addition, some abnormalities consistent with nonacute changes were seen. These included a subacute right cerebellar hemispheric infarct measuring ∼2.5×3.5 cm and a moderate-to-severe reduction in the number of neurons in neocortex, cerebellum, and hippocampus compared with age-matched controls, even in areas not affected by acute damage. Liver and muscle samples were rapidly frozen for additional biochemical evaluation. Activities of electron transfer–chain complexes were normal in muscle (Center for Inherited Disorders of Energy Metabolism, Cleveland), and no deletions, duplications, or depletion or point mutations were detected in mtDNA isolated from patient muscle and liver (Horizon Molecular Medicine, Atlanta).
Patient 2
Case report
The patient is a 10-year-old girl who initially presented with fulminant liver failure at age 4 mo. Her blood glucose level at that time was lower than the detectable range, and the AST was >100,000 U/liter. She had a rapid and effective response to intravenous glucose therapy, and results from her prothrombin time and liver function tests were back to near normal by the time she was discharged. A head CT scan showed a small area of low attenuation in the right inferior basal ganglia, interpreted as representing either prominent perivascular space or old lacunar infarct. Results from extensive viral studies were all negative. Findings from mtDNA mutation and deletion screening from blood showed no abnormality. She continued to have recurrent episodes of hepatocellular dysfunction with hypoglycemia, usually triggered by viral infections, as summarized in table 1. The acute episodes have become less severe as she has aged. She has also been noted to have a persistently low platelet count (hovering near 120×109/liter) suggestive of chronic thrombocytopenia.
Table 1. .
Liver Function Tests from Patient 2 during Episodes of Mild Viral Illness
| Age | Alkaline Phosphatasea (U/liter) |
ASTb (U/liter) |
Total/Direct Bilirubinc (mg/dl) |
Prothrombin Timed (s) |
| 17 mo | 1,250 | 17,150 | 3.1/2.3 | 98.1 |
| 18 mo | 637 | 18,400 | 4.3/3.1 | 81.7 |
| 3 1/2 years | 684 | 13,720 | 3.0/1.8 | 35.2 |
| 5 10/12 years | 1,031 | 5,730 | 3.7/2.6 | 19.6 |
| 6 11/12 years | Normal | 652 | Normal | Normal |
| 9 years | Normal | 162 | Normal | Normal |
Reference range: 367–805 U/liter.
Reference range: 20–40 U/liter.
Reference range: total, 0.1–0.9 mg/dl; direct, 0.0–0.3 mg/dl.
Reference range: 8.4–12.0 s.
Patient 3
Case report
The patient was a 4.5-year-old girl with cardiomyopathy and dilated left ventricle, whose family history included a sibling who died of cardiomyopathy at age 22 mo. During an acute illness at age 18 mo, she was first given a diagnosis of severe left ventricular dysfunction. Hepatomegaly was also present. Her initial blood glucose level was <20 mg/dl, and her plasma carnitine level was 67 μM (reference range 25–79 μM), with a free carnitine level of 16.3 μM (reference range 21–68 μM) and a markedly elevated esterified fraction of 50.7 μM (reference range 2–7 μM). A defect in mitochondrial fatty acid oxidation was suspected, but a specific diagnosis was not made. She subsequently developed a pattern of recurrent rhabdomyolysis with intercurrent illness, with total creatine phosphokinase level during acute episodes usually rising to 11,000–13,000 U/liter (reference range 0–200 U/liter). On carnitine supplementation, she had persistent neurologic deficits with abnormal gait and clumsiness, muscle weakness, and hepatomegaly with acute episodes. She died of congestive heart failure at age 4.5 years. Autopsy revealed a dilated cardiomyopathy with prominent liver necrosis.
Metabolic Evaluation of Patients
Measurement of urine organic acids in patient 1 showed grossly elevated lactate and ketones with dicarboxylic and hydroxydicarboxylic acids. Unsaturated species and 3-hydroxysebacic acid were the most prominent compounds reported. The acylcarnitine profile from a postmortem liver extract from this patient showed elevated long-chain saturated and unsaturated species (Center for Inherited Disorders of Energy Metabolism, Cleveland) (table 2). This pattern was not characteristic of any known disorder of mitochondrial β-oxidation but was strikingly similar to the substrate-utilization profile of ACAD9; most notably, C18:1 and C18:2 carnitine species, corresponding to the optimal substrates of ACAD9, were markedly elevated.4,5 The acylcarnitine profile from postmortem muscle extract from the patient exhibited a similar pattern of metabolites (not shown). Additionally, several short branched-chain species were present in a nonspecific pattern typical of Reye syndrome.14 Blood acylcarnitine analysis was not performed before death, and no postmortem blood, urine, or other bodily fluid samples were available for study. To assess overall mitochondrial β-oxidation, patient fibroblasts were analyzed for production of acylcarnitine species after incubation with tritium-labeled palmitic acid (C16; 16-2H3-palmitic acid) and deuterium-labeled oleic acid (C18:1; 9, 10-2H2-cis-9-C18:1) by the Institute of Metabolic Disease, Baylor University Medical Center, in accordance with their standard clinical testing protocols. No abnormalities were seen, except for a somewhat mild suppression of the whole metabolite spectrum.
Table 2. .
Acylcarnitine Species in Patient 1 Liver and Muscle
| Carbon Chain | Liver (Reference Rangea) | Muscle (Reference Rangea) |
| C12 | .58 (.16–.23) | 6.5 (.07–.81) |
| C14 | 1.47 (0) | 20.25 (.0–.92) |
| C16 | 5.38 (.79–1.54) | 62.00 (.30–5.32) |
| C16:1 | 3.07 (0–.13) | 32.00 (.00–2.50) |
| C18 | 5.86 (.5–1.9) | 41.00 (.24–3.09) |
| C18:1 | 14.6 (.23–1.67) | 139.50 (.64–35.59) |
| C18:2 | 3.84 (.34–1.8) | 62.75 (.00–15.31) |
Laboratory reference range.
Blood glucose levels in both patients 2 and 3 were below the detectable range at their initial presentation. No further episodes of hypoglycemia were reported in patient 3, whereas a rapid drop in blood glucose level was reported during two subsequent acute episodes in patient 2 through age 2 years. Urine organic acid profiles of both patients 2 and 3 during acute illness showed hypoketotic dicarboxylic aciduria with prominent unsaturated species, as well as 3-hydroxyadipic, 3-hydroxysuberic, and 3-hydroxysebacic acids. Urine acylglycine levels were normal in both patients during acute illness, excluding most of the known fatty acid oxidation disorders. The total plasma carnitine level in patient 2 fell during illness but recovered to a normal level without supplementation, and her plasma acylcarnitine profile was normal. Interestingly, the urine acylcarnitine profile of both patients 2 and 3 was notable for elevation of acetylcarnitine in the face of normal or reduced ketones. Fatty acid oxidation studies of fibroblasts from patient 2 showed reduced oxidation of myristate (16.1 pmol/min/mg protein; reference range [±SD] 26.5±3.0) and palmitate (33.4 pmol/min/mg protein; reference range 62.7±16.1), in agreement with the presence of a defect in long-chain fatty acid oxidation, but findings from assays of long-chain 3-hydroxyacyl-CoA dehydrogenase (LCHAD), long-chain 3-ketothiolase, short-chain 3-hydroxyacyl-CoA dehydrogenase, short-chain 3-ketothiolase, and carnitine palmitoyl transferases 1 and 2 were normal (reported by Dr. Michael Bennett, Children’s Hospital of Philadelphia). In addition, DNA sequencing of the entire coding region of short-chain 3-ketothiolase, 3-hydroxymethylglutaryl-CoA synthase, VLCAD, MCAD, LCAD, and selected exons of long-chain 3-hydroxyacyl-CoA dehydrogenase from patient 2 failed to identify a mutation (reported by Dr. Arnold Strauss, Vanderbilt School of Medicine). No frozen blood or tissue samples were available from patient 3 for study.
Results
Biochemical Comparison of Purified Recombinant ACAD9, VLCAD, and LCAD
All three patients had biochemical findings suggestive of an unknown long-chain fat metabolism defect. To further pursue this finding, we first sought to better characterize the enzymatic properties and tissue distribution of the known long-chain ACADs. VLCAD, ACAD9, and LCAD have all been purified from tissue or recombinant sources, but reports on their enzymatic function have varied in technique, making direct comparison of their catalytic properties difficult. To facilitate this comparison, we overexpressed the mature forms of each in an E. coli system and purified them to >95% homogeneity. The specific activities of purified recombinant VLCAD, ACAD9, and LCAD with palmitoyl-CoA as substrate were 13 U/mg protein, 1.2 U/mg protein, and 0.9 U/mg protein, respectively, as measured by the ETF fluorescence-reduction assay. All three purified enzymes had similar specificity for the saturated and unsaturated substrates used predominantly for energy generation, such as C16:0-CoA, C16:1-CoA, C18:0-CoA, and C18:1-CoA (fig. 1a). In contrast, ACAD9 differed significantly from VLCAD in the use of polyunsaturated substrates not primarily used for energy, especially C22:6-CoA, and LCAD is the only one of three enzymes that uses long- and branched-chain substrates, including THC-CoA (fig. 1a).
Figure 1. .
Comparison of activity, expression, and tissue localization of VLCAD, ACAD9, and LCAD. The specific activity of VLCAD, ACAD9, and LCAD, as measured with palmitoyl-CoA, was 13, 1.2, and 0.9 U/mg, respectively (a). All substrates used are described in the “Methods” section. b and c, Expression of mRNA of ACAD9 and LCAD (b) and VLCAD, MCAD, and SCAD (c), measured in 21 human tissues by real-time RT-PCR and compared with the relative level of expression in muscle. The ratio of expression of VLCAD, ACAD9, LCAD, MCAD, and SCAD in muscle to the housekeeping gene GUSB was 34, 1.0, 0.4, 4.9, and 17.1, respectively. d, Immunoinactivation of ACAD activity in human liver, muscle, and brain, measured with C18:0-CoA as substrate. VLCAD antibodies inactivated 32%, 100%, and 0% of the activity measured in the mitochondrial membrane fraction of human liver, muscle, and brain, respectively, whereas ACAD9 antibodies inactivated 41%, 0%, and 100% in these same tissues.
Tissue Expression of Long-Chain ACADs
Quantitative real-time PCR was used to measure the expression of each long-chain ACAD gene, along with MCAD and SCAD, in 21 adult and fetal human tissues. The expression of two housekeeping genes (GUSB and the gene for 18S rRNA) was used as an internal control. Figure 1b and 1c show expression of ACADs normalized to GUSB; the pattern was unchanged when 18S rRNA expression was used as control instead (not shown). In muscle, a tissue with a high rate of β-oxidation, all of the ACADs tested were highly expressed. The expression levels of VLCAD, MCAD, and SCAD were 34, 4.9, and 17 times that of GUSB, respectively, whereas the expression levels of ACAD9 and LCAD were nearer to that of GUSB, suggesting that expression of the long-chain ACADs may be differentially regulated. It has been reported that significant levels of VLCAD nonsense transcripts exist in cells and in most human tissues—for example, transcripts with retention of intron 10 that introduces a premature stop codon into the message.15 Such a high level of nonsense VLCAD message may be associated with post-transcriptional regulatory functions.16 The expression level of each ACAD in each tissue is compared with its expression level in muscle, and all are shown in figure 1b and 1c. VLCAD, MCAD, and SCAD were the most highly expressed in muscle and heart (fig. 1c), tissues that use long-chain fatty acids for 85% of their energy, which is consistent with the function of these enzymes in energy homeostasis.1 Whereas the expression pattern of ACAD9 overlapped VLCAD in muscle and heart, ACAD9 was the only long-chain ACAD mainly expressed in brain (fig. 1b). LCAD was the only long-chain ACAD mainly expressed in kidney, lung, thyroid gland, and prostate gland (fig. 1b). These tissues, along with human brain, have high levels of lipid recycling and synthesis. LCAD had higher expression in fetal than adult brain, a pattern opposite of ACAD9 (fig. 1b).
Immunohistochemical staining in human muscle, brain, and lung revealed that the protein distribution of each of the enzymes followed their gene expression patterns (fig. 2 and table 3). VLCAD staining was much stronger than ACAD9 (fig. 2a and 2b) and LCAD (not shown) staining in muscle, and the distribution of its staining was different than that of ACAD9. The VLCAD staining pattern was unevenly distributed, with the greatest signal in slow-twitch muscle fibers (fig. 2a). In contrast, ACAD9 was predominantly localized perivascularly (fig. 2b). Immunohistochemical staining of normal human brain sections with ACAD9 antibody revealed a primary neuronal location for ACAD9 (fig. 2d–2i). This included most neurons of all five layers of frontal cortex (fig. 2d) and various neurons in hippocampus, including all the cornus ammonis regions and the dentate gyrus (fig. 2e and 2f). ACAD9 staining was particularly strong in the small granular neurons of the dentate gyrus, and the neurons of the CA4 region (fig. 2f) gave a much stronger signal than those in the CA1 region (fig. 2e). In human cerebellum, ACAD9 decorated Purkinje neurons and dentate nucleus. It also was present in neuronal dendrites in the granular layer, very likely in granular neurons (fig. 2c, 2g, and 2i). Thus, the distribution of ACAD9 is distinct for specific subgroups of neurons. The staining pattern of ACAD9 in cerebellum cortex is a diffuse punctate pattern extending to neuronal dendrites, which is characteristic for mitochondrial staining (fig. 2c). Staining of this region with a VLCAD antibody gave no signal (not shown), consistent with previous literature reports.12 The distribution of long-chain β-oxidation enzymes, including VLCAD, ACAD9, LCAD, and LCHAD, is summarized in table 3.
Figure 2. .
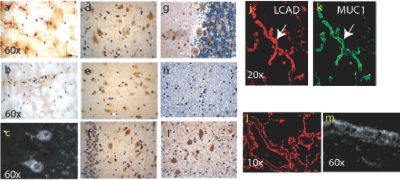
Immunohistochemical and immunofluorescent staining of normal human brain and muscle sections with ACAD9 antisera and human lung sections with LCAD antisera. a and b, Immunohistochemical staining of human muscle with VLCAD (a) and ACAD9 (b) antibody visualized with DAB (brown), counterstained with a blue nuclear stain. c, Diffuse and punctate pattern of ACAD9 staining, extending to neuronal dendrites of Purkinje cells. The nucleus of Purkinje and granular neurons does not stain with ACAD9 antibodies visualized with fluorescent secondary antibody (white). d–i, Normal human brain section immunostained with ACAD9 antibody and visualized with DAB (brown). All sections are counterstained with a nuclear stain (blue). d, Neurons of human neocortex. e, Neurons in the CA1 region of normal human hippocampus showing relatively weak ACAD9 staining. f, Neurons in the CA4 region and granular neurons of hippocampus. g, Cerebellar cortex, including Purkinje neurons, their dendrites, and neurons in the granular layer, showing relatively strong ACAD9 staining. h, Absence of ACAD9 staining in cerebellar white matter. i, Strong ACAD9 staining in dentate nucleus in the deep white matter of cerebellum. j and k, Normal human lung sections stained with LCAD (j) or the marker protein MUC1 (k) (a membrane protein, specific to human alveolar septae) antibody and visualized by fluorescent secondary antibodies, LCAD (j, red) and MUC1 ( k, green), colocalizing to type II alveolar epithelial cells (white arrow), as described in the “Methods” section. l and m, LCAD antibodies specifically staining bronchial epithelium and the alveolar septae of normal human lung tissue (l). At high magnification, the staining of LCAD in bronchial epithelium (m) shows a diffused punctate pattern consistent with mitochondrial staining.
Table 3. .
Labeling of Brain Sections with Antibodies to ACAD9, LCHAD, LCAD, and VLCAD
| Cerebral Region | ACAD9 | LCHAD | LCAD | VLCAD |
| Frontal cortex: | ||||
| Pyramidal cells | + | + | + | − |
| Endothelial cellsa | − | + | − | + |
| Hippocampus: | ||||
| CA1 | + | + | + | − |
| CA4 | +++ | + | + | − |
| Granular layer | ++ | + | + | − |
| Cerebellum: | ||||
| Purkinje | +++ | + | − | − |
| Granular layer | +++ | +++ | − | − |
| Dentate nucleus | +++ | ++ | +++ | − |
ACAD9-specific staining was not seen in the endothelial cells in frontal cortex; however, it was present in the endothelial cells in the deep white matter of cerebellum.
Enzymatic and immunoinactivation studies confirmed that long-chain ACAD activity in human cerebellum cortex was predominately ACAD9 (figs. 1d and 3). In contrast, ACAD9 and VLCAD activities were equally represented in liver, whereas nearly all of the long-chain ACAD activity in muscle was from VLCAD (fig. 1d). LCAD antigen was the only one of the three long-chain ACADs identified in lung, where it colocalized with a lung epithelial cell marker (MUC1 protein) in bronchial epithelium and throughout the alveolar septae (fig. 2j-2m). It was especially abundant in the alveolar type II epithelial cells responsible for synthesizing surfactant (fig. 2j and 2k).17,18
Figure 3. .
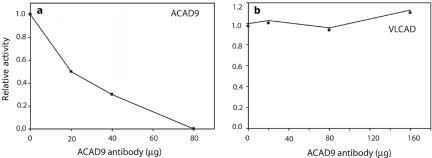
Immunoinactivation of purified ACAD9 protein. Serial dilutions of purified rabbit anti-ACAD9 IgG were used to inactivate 1.5 μg of purified recombinant ACAD9 or VLCAD. Reactions were performed in the presence of 0.5 mg/ml of BSA and 5% glycerol. After incubation with antibody, reactions were precipitated with S. aureus protein A–conjugated Sepharose beads, and the remaining ACAD activity was measured with the ETF fluorescence-reduction assay. The data plotted represent the average of duplicate measurements. a, Twenty to 80 μg of ACAD9 antibody incubated with purified ACAD9. The Y-axis shows the relative activity compared with enzyme without antibody treatment. b, Up to 160 μg of ACAD9 antibody incubated with purified VLCAD protein and plotted as was that in panel a.
ACAD9 Promoter Analysis and Functional Studies
The distinctive tissue-distribution pattern of expression of the long-chain ACADs led us to further study the regulatory factors for the ACAD9 gene. The computer programs PROSCAN and TFSEARCH were used to evaluate the 5′ region of the ACAD9 gene (2 kb of sequence covering the 5′ UTR of the ACAD9 gene, exon 1, intron 1, and exon 2) for possible promoter sequences, including major transcription-factor binding sites. The minimal promoter region predicted for the ACAD9 gene (starting at position −264) is shown in figure 4a. The transcriptional start sites 1 and 2 (TSS1 and TSS2) were inferred from the consensus of the 5′ end of EST tags in the dbEST database, using the BLAST tool in combination with PCR analysis. TSS2 appears to be a major transcriptional start site, since it matches most of the 5′-end EST tags; however, there are also a number of 5′-end EST sequences with TSS1 as the start site. PCR primers close to either TSS site efficiently amplified full-length ACAD9 coding cDNA. Three eukaryotic expression vectors were constructed to test the ability of the sequences around this region to act as a promoter (fig. 4c). Varying lengths of 5′ UTRs of the ACAD9 gene were cloned upstream of a promoterless luciferase reporter gene, and the level of expression was compared with that of a constitutive reporter gene on a control vector after cotransfection into the human hepatoma cell line HepG2. ACAD9 sequences including nucleotides −590 to +50 (p1A9) and −264 to +50 (p3A9) both led to high levels of expression of the reporter gene, whereas a fragment from −380 to +50 (p2A9) gave very low expression. The growth of all cultures after transfection was similar. This confirms that the predicted minimal promoter region contains the major regulatory elements for expression of ACAD9.
Figure 4. .
Promoter function of the ACAD9 gene and its putative regulatory pathway. a, Sequence of the human ACAD9 gene promoter region, as predicted by computational analysis. The transcriptional start sites (TSS1 and TSS2) were inferred from the consensus of the 5′ end of EST tags in dbEST in combination with PCR analysis. b, Two conserved nucleotide blocks that match the consensus sequences for NRF-1 and CRE transcription-factor binding sites. The dotted line indicates the invariant nucleotides in the recognition sites among different species. The NRF-1 and CRE sites are duplicated in all primates. c, A series of fragments containing increasing lengths of 5′ UTR from the ACAD9 gene, inserted upstream of a firefly luciferase reporter gene (Luc) in the promoterless pGL3-basic vector. The TAAG insertion identified in the patient is shown as a vertical bar in exon 1. d, The promoter-luciferase fusion constructs, transfected into HepG2 cells along with an internal control. The luciferase activity is presented as the mean and SD from at least three independent experiments, as described in the “Methods” section. The 4-bp insertion was introduced in plasmid p3A9 (e), and the activities were measured as were those in panel d.
Phylogenetic alignment of the ACAD9 promoter region revealed two evolutionarily highly conserved regions, each with duplicated sequences homologous to nuclear respiratory factor 1 (NRF-1) and cAMP-responsive element transcription-factor binding sites (CREB) (fig. 4b). Scanning of 5 kb of the 5′ UTR of both human and mouse ACAD9 genes, with use of the online software NHR SCAN, failed to identify a putative direct repeat site 1 (DR1) binding site for the peroxisomal proliferation-activated receptors (PPARs). In contrast, multiple DR1 sites are present in the promoter region of the VLCAD gene and in all other known human and mouse ACAD genes. Thus, it appears that expression of the ACAD9 gene is independent of PPARα but, rather, is controlled by NRF-1 and CREB, important regulators of the genes involved in mitochondrial biogenesis and those for most respiratory-chain enzyme subunits.19,20
Stimulation of various human cell lines with cytokines has been shown to down-regulate retinoid X receptor (RXR)/PPAR–dependent genes and to up-regulate NRF-1–controlled genes.21 Addition of interferon-α (INF-α) or a combination of both INF-α and tumor necrosis factor-α (TNF-α) to cultured fibroblasts led to significant reduction of ACAD activity measured with C18-CoA activity in cells from patient 2 but not in those from control cells (table 4). In contrast, activity measured with C12-CoA, metabolized primarily by MCAD (the expression of which is regulated by multiple independent elements besides RXR/PPAR22,23), was unaltered with cytokine treatment. Thus, defective long-chain ACAD activity can be detected in ACAD9-deficient patient (patient 2) fibroblast on cytokine simulation, since down-regulation of VLCAD in patient 2 cells was apparently not compensated by induction of ACAD9 activity, as it was in controls.
Table 4. .
ETF Fluorescence Reduction Assay of Patient 2 and Control Fibroblast Lysates after Cytokine Treatment
| Fibroblast Specific Activitya(mU/mg) |
||||
| Control |
Patient 2 |
|||
| Cytokines | C12 | C18 | C12 | C18 |
| Untreated | 1.42 | 1.20 | 1.59 | 1.66 |
| INFα | 1.43 | 1.18 | 2.14 | NDb |
| INFα+TNFα | 1.68 | 1.13 | 2.01 | NDb |
All the activity data are averages of measurements from two independent experiments.
ND = not detectable.
Two ACAD9 mRNA and Protein Isoforms Exist in Cells
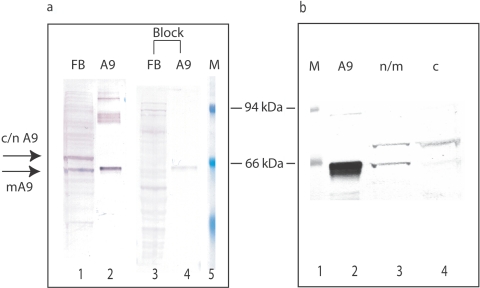
Subcellular localization studies of ACAD9 protein in human skin fibroblasts revealed that it was present not only in mitochondria but also in the cytoplasm and nucleus, suggesting the presence of so-called moonlighting functions for this protein24 (figs. 5c–5e and 6). This was also seen in hepatocytes (data not shown) but not in most of the neurons in the cerebellum (fig. 2c). We were able to confirm the existence of two protein forms in fibroblast and liver lysate that differed by ∼10 kDa in molecular mass (figs. 5a, 5b, and 6). One form corresponds to the previously recognized mitochondrial enzyme (64.8 kDa). The other form corresponds in size to the predicted protein product encoded by an alternative mRNA species identified in the public databases that appears to be generated by alternative splicing of ACAD9 message (ACAD9 variant a in fig. 7a) and eliminates the mitochondrial targeting signal. Subcellular fractionation experiments with fibroblast lysate showed that the 76-kDa protein was present in both cytoplasm and nuclear (c/n) fractions (fig. 6b). Both ACAD9 protein species react specifically with our ACAD9 antibody, as shown by immunocompetition experiments (fig. 6a).
Figure 5. .
Subcellular location of ACAD9 protein. a and b, Western blot of fibroblast and liver lysate with anti-human ACAD9 antibodies. One major band (M) in control samples (a, lanes 2 and 4; b, lane 2) co-migrated with purified recombinant mitochondrial ACAD9 (a, lane 1; b, lane 1), and another was larger (c/n). Both forms were missing or reduced in patient samples (a, lanes 3 and 5; b, lane 3). VLCAD-deficient fibroblasts (V) (a, lane 2) and normal human fibroblast (N) (b, lane 2) or postmortem liver (N) (a, lane 4) served as controls. c–e, Immunofluorescent staining of normal fibroblasts with ACAD9 (c, green) and mitochondrial ATP synthesase (mATPase) (d, red) antibodies, visualized by fluorescent secondary antibodies. The merged image (e) shows that the two proteins colocalize in part (yellow). A green ACAD9 signal is seen in the nucleus (white arrow). Panels f–h and i–k are the same as panels c–e but use fibroblasts from patients 2 and 1 (P2 and P1), respectively. l–n, Immunofluorescent staining of postmortem liver from patient 3 (P3) and a control (C) with antibodies against ACAD9 (l, green) or liver-specific α1-AT (m, red), visualized by fluorescent secondary antibodies. The merged signal is shown in panel n. ACAD9 (αA9) (lanes 1 and 2) staining is deficient in patient 3 (lane 2), but MCAD (αM) (lane 3) and VLCAD (αV) (lane 4) staining are normal. o–t, Immunofluorescent staining of postmortem cerebellar cortex of patient 1 (P1) (o–q) and control (r–t) with antibodies to the cerebellar cortex marker EAAT4 (o and r, red) or ACAD9 (p and s, green); the merged signal is shown in panels q and t, in pseudocolored yellow. ACAD9 is prominent in control (s) but is absent in patient cerebellum (p). A white arrow indicates ACAD9-specific staining of dentate nucleus in deep white matter of the normal control.
Figure 6. .
Immunocompetition and subcellular fractionation of ACAD9 antigen in human fibroblast lysate. a, Immunocompetition of ACAD9 antigen in human fibroblast lysate by purified ACAD9 protein. Fifty micrograms of normal human fibroblast lysate or 40 ng of purified ACAD9 protein were separated by SDS-PAGE and were transferred to a PVDF membrane. The membrane was then incubated with ACAD9 antibodies (affinity purified) with and without an equal amount (mole/mole) of purified ACAD9 protein. Lanes 1 and 2 show the reaction of fibroblast extract and purified ACAD9 with ACAD9 antibody. Lanes 3 and 4 show the same as do lanes 1 and 2, except that the ACAD9 signals on the membrane are diminished when purified ACAD9 protein is included in the antibody-incubation mixture. Arrows point to two ACAD9 protein species with different molecular masses. The band marked “mA9” corresponds in size to purified mature mitochondrial ACAD9, whereas the other ACAD9 species (c/n A9) is ∼10 kDa bigger than mA9 and is of a size consistent with the predicted cytosolic/nuclear form of ACAD9. Lane 5, Molecular mass markers. The two marked bands are β-galactosidase (94 kDa) and albumin (66 kDa). b, Subcellular distribution of ACAD9 proteins in normal fibroblast extract. Fibroblasts were lysed by repeated cycles of freezing and thawing, and the cytoplasmic fraction (lane 4) was separated from the nuclear/mitochondrial fraction (lane 3) by centrifugation. Lane 2, Purified ACAD9. Protein markers in lane 1 are the same as those in panel a.
Figure 7. .
Molecular analysis of ACAD9-deficient patients. a, ACAD9 consisting of 22 exons and generating two predominant molecular species through alternative splicing. The schematic drawing illustrates that the basic ACAD domain structure of both translated proteins is conserved (ACDN, ACDM, and ACDC); however, in one, a nuclear targeting signal (N) is substituted for the mitochondrial targeting signal (M). This variant message also contains a longer exon 1, giving a longer N-terminus (U) and a shorter exon 18 along with additional exons 19–22. b and c, Amplification of ACAD9 sequences from control (C), patient 1 (P1), and patient 2 (P2) mRNA. PCRs with control fibroblast cDNA as template readily amplified full-length ACAD9 coding sequences (b, lane 6; c, lane 2 ) and overlapping subfragments (b, lane 2, exons 1–7; b, lane 4, exons 6–18). Full-length message (exons 1–18) could not be amplified from mRNA either from patient 1 (b, lane 7) or patient 2 (c, lane 3). Shorter-than-expected fragments were amplified from patient 1 cDNA (b, lane 3, exons 1–7; lane 5, exons 6–18). d, Sequence of ACAD9 UTR from genomic DNA and cDNA. Sequencing chromatograms of genomic DNA from patient 1 (left panel) identified a dual pattern due to a 4-bp insertion. Patient cDNA in that region showed only the normal sequence, without the insertion (right panel). e, Diagrams of ACAD9 species amplified from cDNA from patient 1. One consisted of exon 1 joined to exon 6 (α), whereas the other consisted of exon 11 jointed to exon 18 (β).
The ACAD9 variant a (GenBank accession numbers CR613592 and BX415793) (aAugust05 [AceView]) mRNA differs from variant b (GenBank accession number NM_014049) (bAugust05 [AceView]) by the presence of an alternative exon 1a at the 5′ terminus, one alternative exon 18 resulting from differential splicing, and four additional exons (19–22) in the 3′ UTR (fig. 7a). At least 33 human ESTs containing exon 1a exist in dbEST. However, this region is conserved only in primates and not in other mammals. Alternative exon 1 could be amplified by PCR from human fibroblast cDNA, substantiating its existence in vivo (fig. 7b, lane 2). A number of human ESTs in the public database also support the existence of the alternative exons in the 3′ UTR in some transcripts; however, the combination of exons and the alternative splicing of exon 18 is variable. Therefore, the exact sequence of the 3′ UTR of ACAD9 variant a remains unclear. This may explain the absence of the second species when full-length ACAD9 is amplified from cDNA (fig. 7b, lane 6, and 7c, lane 2), since the 3′ amplification primer likely sits in a region that varies between variants a and b. Analysis with PSORT II predicts that variant a most likely localizes to the cytoplasm (43%) or nucleus (30%). A putative nuclear targeting sequence (PRERKEV) is encoded by alternative exon 1a and replaces the mitochondrial targeting sequence in variant b. However, the translation start codon predicted for this transcript is a CTG instead of an ATG, a codon rarely used in mammals. Further investigation of the alternative ACAD9 protein is necessary to understand its molecular origin. Interestingly, intron 21 and exon 22 of ACAD9 variant a overlap an unrelated gene, KIAA1257, on the opposite DNA strand. Thus, it is possible that expression of ACAD9 variant a can also be regulated by the expression of the KIAA1257 gene.
ACAD9 Activity and Antigen Is Reduced or Deficient in Patient Cells
All of the apparent ACAD9-deficient patients exhibited marked abnormalities in ACAD9 protein (fig. 5). In contrast, VLCAD and MCAD antigen was comparable to control in all patient samples (fig. 5l–5n, lanes 3 and 4 for patient 3; data not shown for patient 1 and 2). LCAD antigen was minimal in both patient and control fibroblast and liver and was normal in patient muscle (not shown). Patient 3 lacked all identifiable ACAD9 antigen (fig. 5l–5n, lane 2). Although a minimal amount of ACAD9 antigen was detected in samples from patients 1 (fig. 5a, 5i–5k, and 5o–5q) and 2 (fig. 5b and 5f–5h), it was not likely to be enzymatically active, since none was appropriately targeted to mitochondria (fig. 5f–5k). Instead, the residual ACAD9 protein in patient 1 was predominantly cytoplasmic. Most detectable ACAD9 antigen in patient 2 localized to the perinuclear region, with a much lower signal in cytoplasm.
VLCAD has been demonstrated elsewhere to account for nearly all of the activity toward palmitoyl-CoA in crude fibroblast extracts.25 In keeping with this finding, both patients 1 and 2 had normal long-chain activity in fibroblasts, whereas a known VLCAD-deficient cell line had no detectable activity (tables 4 and 5). In contrast, VLCAD and ACAD9 were present in equal proportions in liver mitochondrial membrane preparations (fig. 1d). Consistent with this, VLCAD-deficient patients and VLCAD-null mice have previously been shown to have a 50% reduction in palmitoyl-CoA activity in liver.25,26 A frozen liver sample available from patient 1 had 46% ACAD activity measured with palmitoyl-CoA as substrate, as compared with a control sample (table 5), and reduction of C16 activity in mitochondrial membrane fraction of patient liver was >70% (not shown), but sample size did not permit additional immunoinactivation experiments. However, fibroblasts from patient 1 had normal activity measured with palmitoyl-CoA as substrate, indicating normal VLCAD activity. Thus, the reduction in activity in patient liver must be due to loss of ACAD9. No liver tissue from patient 2 was available for assay.
Table 5. .
ETF Fluorescence Reduction Assay of Patient 1 and Control Samples
| Sample | Specific Activity (mU/mg) |
| Liver: | |
| Control | 8.6±1.6a |
| Patient | 4.7 |
| VLCAD deficient | 5.9b |
| Fibroblasts: | |
| Control | 2.4±0.3a |
| Patient | 2.2 |
| VLCAD deficient | NDc |
The SD is derived from the average activities of five normal human liver or cultured skin fibroblast samples.
Literature-reported value.25
ND = not detectable.
Abnormalities of ACAD9 in the Brain of Patient 1
The autopsy report of the brain of patient 1 described mild, apparently chronic abnormalities, with a significantly reduced number of neurons in the gray matter of hippocampus, neocortex, and cerebellum, but it was not clear if this was due to a developmental or degenerative defect. This distribution matches that of the expression pattern of ACAD9 protein in the CNS. Our immunofluorescence staining of ACAD9-specific antigen in patient cerebellum cortex tissue confirmed that ACAD9 protein was almost absent (fig. 5o–5t). We therefore performed glial fibrillary acid protein (GFAP) staining (a marker of gliosis) of various brain sections from patient postmortem brain, including neocortex, hippocampus, and cerebellum. GFAP staining in patient gray matter was similar to that in control samples and does not support the presence of chronic degeneration (not shown). Interestingly, a mild increase of leukocyte common antigen (LCA) staining was seen in perivascular cells around the blood vessels in the white matter of the centrum semiovale (fig. 8a and 8b). LCA staining was, however, normal in sections of the gray matter and leptomeninges from neocortex. Morphologically, the staining cells were a mixture of macrophages (including some hemosiderin laden) and lymphocytes. Similar inflammatory cell infiltration was seen in the white matter of brain stem from the patient but not in hippocampus or cerebellum, where the damage from his acute clinical course was the most severe. Such inflammatory cell cuffing is not normal in the brain of a 14-year-old.
Figure 8. .
Mild perivascular chronic inflammatory cell cuffing in postmortem patient brain. Scattered perivascular lymphocytes (arrows) cuff the blood vessels in the brain stem of patient 1. a, Hematoxylin and eosin stain (640× magnification) and the neocortical white matter. b, LCA immunohistochemistry stain (640× magnification). This inflammatory cell infiltrate is also accompanied by some hemosiderin-laden macrophages, seen only in the deep white matter of the centrum semiovale and the brain stem but not in gray matter or leptomeninges where the acute damage was most severe. Neither of these findings appears to be related to the child’s acute demise; rather, they appear to be chronic. These changes are reminiscent of, though not as severe as, those reported in X-linked adrenoleukodystrophy (MIM 300100).
Molecular Analysis of ACAD9 mRNA and Genomic DNA
Full-sized ACAD9 message could easily be amplified from control liver and fibroblast mRNA but not from cDNA made from fibroblast mRNA from either patient 1 or 2 (fig. 7b and 7c) or liver mRNA from patient 1. When primers corresponding to subsections of the ACAD9 message were used for amplification, two dominant species were identified in cDNA from patient 1 that were smaller than the expected size for normal message (fig. 7b). Direct sequencing of each fragment revealed that one was missing sequences corresponding to exons 2–5, whereas the other was missing exon 12–17 sequences (fig. 7e). Additional minor species were also occasionally seen in repeat experiments, all of which also contained absent exons (not shown). Misspliced RNA species were also amplified from cDNA made from patient 2 fibroblast mRNA, with a deletion of exon 3 found in the most prominent species. Only paraffin-fixed tissue was available from patient 3, and attempts to amplify DNA from it were unsuccessful.
All 18 exons, the intron-exon boundaries, and 650 bp of 5′ UTR and untranscribed region of the ACAD9 gene were amplified and sequenced from genomic DNA from patients 1 and 2. A 4-bp insertion was found 44 bp upstream of the first ATG in one allele from patient 1 (fig. 7d, left). Only a minimal signal corresponding to this insertion was visible when fragments amplified from cDNA made from patient liver mRNA were directly sequenced, suggesting a transcriptional defect in this allele (fig. 7d, right). Introduction of this 4-bp insertion into a p3A9 promoter construct led to 75% reduction of expression of a reporter gene (fig. 4e). This insertion is not found in dbSNP, and screening by PCR of genomic DNA from 244 alleles from ethnically matched controls identified only one containing the 4-bp TAAG insertion. There is only one report of this insertion in >330 independent ACAD9 cDNA sequence entries in the human EST database (BI 911835.1), and it appears in a sample made from pooled human leukocytes from an unknown number of individuals. In addition, the insertion is reported in the Hela cell line (DA 567380).
Although a number of intronic sequence variants were identified near intron-exon boundaries in both patients 1 and 2 (table 6), none affect canonical splicing sequences, nor did Southern blotting reveal a large deletion in either patient’s ACAD9 gene (not shown). Thus, the origin of the apparently misspliced transcripts in mRNA from patients 1 and 2 remains undetermined. Variations in exon sequences were also found in both patients. 379A→C (exon 4) and 1476C→T (exon 14) are common variants in dbSNP. Both the 787T→C (exon 7) and g.40508G→A (exon 22) are also present in dbSNP, but their population frequency has not been determined. None of the sequence variants in exons 4, 7, or 14 cause amino acid changes, and exon 22 is in the 3′ UTR. The sequence variant g.34927C→T in exon 20 of patient 1 is not present in dbSNP. However, since exon 20 is also in the 3′ UTR, the effect of this private sequence change remains unclear. Parental DNA was not available for study from either family.
Table 6. .
Summary of DNA Sequence Variations Found in Patients 1 and 2
| Patient and Allele |
INS4TAAG g.−44 Promoter |
20IVS3C→T | 379A→C Exon 4 |
787T→C Exon 7 |
7IVS9G→A | 1476C→T Exon 14 |
g.34927C→Ta Exon 20 |
g.40508G→A Exon 22 |
| Patient 1: | ||||||||
| Allele 1 | + | − | + or − | +b | + or − | + | + or − | + or − |
| Allele 2 | − | − | + or − | − | + or − | + | + or − | + or − |
| Patient 2: | ||||||||
| Allele 1 | − | + or − | + | − | − | + | − | − |
| Allele 2 | − | + or − | + | − | − | + | − | − |
Novel mutation, not in dbSNP.
The 787T→C variation was not seen in the cDNA sequence, indicating it is on the same allele with the INS4TAAG mutation.
Discussion
This report presents the first cases of ACAD9 deficiency. Although the patients have different phenotypes, the spectrum of findings is similar to that seen in other known long-chain fatty acid β-oxidation defects, including VLCAD deficiency.1 The presentation of a Reye-like episode after aspirin ingestion during a viral illness has most frequently been reported in LCHAD-deficient (HADHA [MIM 600890] and MCAD-deficient (ACADM [MIM 607008] patients,27,28 but ACAD9 deficiency should now be considered if other β-oxidation defects are not identified. Cardiomyopathy is common in both VLCAD and LCHAD deficiencies1 and was present in the pair of ACAD9-deficient sibs described here. Fulminant liver failure was the most dramatic aspect of the case of patient 2 and is not typical of other β-oxidation disorders in the absence of Reye syndrome. The incidence of cerebral strokes in children and young adults is ∼2.5:100,000.29 Whereas a number of metabolic disorders are associated with stroke or strokelike episodes, they are rarely vascular in nature in disorders of mitochondrial energy metabolism.29 Patient 1 in this report had pathologic evidence of a bona fide cerebellar vascular accident. On the basis of histologic criteria, it is likely that the stroke dated to the beginning of the patient’s acute illness. It is not clear whether the stroke was directly related to his ACAD9 deficiency or was secondary to his liver disease and coagulopathy, though the latter was mild. Of note, a remote infarct close to the brain-stem region also was identified by magnetic resonance imaging scan in patient 2. Thus, ACAD9 deficiency should be considered in unexplained cerebrovascular stroke.
To better understand the differences in the physiological functions of the long-chain ACADs, we compared the substrate specificity and tissue-distribution pattern of ACAD9, VLCAD, and LCAD. ACAD9 and LCAD were found to be distinct from the other known ACADs involved in energy production, including VLCAD, implicating them in other metabolic pathways. C22:6-CoA was a preferred substrate for ACAD9 but not for VLCAD. However, neither enzyme had significant activity with C20:5-CoA as substrate (not shown), indicating that ACAD9 shortens C22:6-CoA to C20:5-CoA through a single cycle of β-oxidation. LCAD was the only one of the three to have significant activity with a variety of long, branched-chain substrates, including THC-CoA, an intermediate in an alternative mitochondrial pathway for free bile acid synthesis implicated in regulation of the cellular metabolic rate (fig. 1a).30 The ability of ACAD9 to use the CoA esters of polyunsaturated fatty acids such as C22:6 is consistent with the finding that it is the predominant long-chain ACAD in human cerebellum, a rich source of polyunsaturated fatty acids. LCAD is the only long-chain ACAD mainly expressed in human lung (in type 2 pneumocytes), thyroid, prostate gland, and kidney. One possible role of LCAD in the lung is in surfactant metabolism, since dipalmitoylphosphatidylcholine is the most abundant long-chain lipid in surfactant, synthesized predominantly in type 2 pneumocytes. The functional difference among the three longchain ACADs and the identification of distinct VLCAD- and ACAD9-deficient patients indicates that these enzymes cannot compensate for each other in vivo.
The overlap in substrate use by the long-chain ACADs presents potential challenges in distinguishing deficient patients on the basis of metabolic profiles, since deficient patients may accumulate similar intermediate metabolites from long-chain fatty acid β-oxidation. Moreover, many of these long-chain intermediates are known to be toxic and may lead to similar clinical presentation, such as cardiomyopathy, as seen in our patient 3 as well as in patients with other long-chain fatty acid oxidation defects.31,32 Differences in tissue distribution of ACAD9 and VLCAD provide a method to distinguish ACAD9 or VLCAD deficiency by direct enzymatic assay of tissue samples, though this would require an invasive procedure.
One of the major functions of mitochondrial β-oxidation is in maintenance of energy homeostasis. Loss of this function in patients with genetic defects in β-oxidation triggers fasting or stress-induced metabolic crises that can usually be treated by providing patients with an alternative energy source.1 Another major function of mitochondrial β-oxidation is to recycle carbons from a variety of long-chain fatty acids for lipid synthesis.6 Little is known about the effect of disruption of this process on the pathogenesis of symptoms in defects of β-oxidation, and it may, in part, explain the features of these disorders that respond poorly to treatment with alternative energy sources such as neurological dysfunction and acute respiratory distress syndrome.33,34 These symptoms are not reported in patients with deficiencies in VLCAD or MCAD but are seen in patients with deficiencies in LCHAD and mitochondrial trifunctional protein (MTP [MIM 609015]).33,34
ACAD9 is the only ACAD identified to date that is highly expressed in human brain and is mostly localized to a subpopulation of neurons that are coincident with LCHAD/MTP (table 2), most notably in the cerebellum. The role of mitochondrial β-oxidation of long-chain fatty acid in the human CNS has long been controversial. Neuropathological problems are uncommon in patients with defects of fatty acid oxidation (except for LCHAD/MTP deficiency) and, when present, are usually considered secondary to recurrent episodes of hypoglycemia. However, >50% of patients with a deficiency of LCHAD/MTP manifest neurological symptoms with demonstrable neuropathology, including demyelination in peripheral neurons.35–37 Although LCHAD/MTP has been found to be highly expressed in various neurons, its function remains unclear, which is largely due to the common perception that long-chain ACADs are not present in the CNS to provide substrate to the MTP.12 The finding that ACAD9 is, in fact, expressed in the CNS in humans and the chronic neuronal abnormalities seen in the ACAD9-deficient patients strongly support a previously unappreciated but important role for mitochondrial long-chain fatty acid β-oxidation in human neuronal tissue. Since brain does not rely on mitochondrial β-oxidation of long-chain fatty acids for energy, this pathway must have alternative functions in the CNS. One possible role for ACAD9 is in the recycling of complex neural lipids and production of intermediate metabolites for synthetic pathways. It is unusual that both patients 2 and 3 chronically secreted large amounts of acetylcarnitine in their urine. However, this supports the presence of a defect in lipid remodeling in these patients, with wasting of acetylcarnitine induced by reduced channeling of medium-chain substrates to lipid synthesis from a pathway involving ACAD9.
Alternative splicing of pre-mRNAs is widely used by higher eukaryotes to generate different protein isoforms in specific cell or tissue types and is especially prominent in humans in brain, liver, and testis.38 Between one-third and two-thirds of all human genes are estimated to undergo alternative splicing, and the disruption of this process has been implicated in several human diseases.39,40 Our studies have identified two ACAD9 protein isoforms corresponding in size to the predicted products of two ACAD9 messages generated by alternative splicing. Database analysis and subcellular fractionation indicate that each isoform localizes to a different subcellular location on the basis of the targeting signals present in each (figs. 5a–5e, 6a, and 6b). Interestingly, patients 1 and 2 show abnormalities in the cellular distribution of ACAD9 protein (fig. 5a and 5b), consistent with the possibility that disruption of alternative splicing is occurring.
The 4-bp insertion in the promoter region of one allele of patient 1 clearly impairs transcription of this allele in vivo and in vitro and constitutes a pathologic mutation. However, the complexity of the ACAD9 gene and its transcripts hindered our ability to elucidate the molecular defect in the three remaining mutant ACAD9 alleles available for examination. Regardless, cDNA amplification experiments unequivocally demonstrated that both patients 1 and 2 lacked normal full-length ACAD9 message in liver and/or fibroblasts. The presence of multiple abnormal species in patient mRNA, with apparent deletions of coding exons, obviously implicates a defect in RNA splicing. The identification of only one (and sometimes no) mutations in clear-cut genetic disorders is a relatively common phenomenon and has been reported in as many as 5%–10% of cases of enzymatically proven MCAD and VLCAD deficiency,41–43 whereas, in the case of metabolic enzymes that are primarily active in brain—such as GTP cyclohydrolase—no mutations have been reported in >40% of the patients with deficient enzymatic activity.11 In these patients, as in ours, the assumption is that other intronic or untranscribed sequence changes are responsible for the deficiency.39
The ACAD9 gene has a unique 5′ UTR compared with the other ACAD genes. The RXR heterodimer pathway PPARα/RXR regulates expression of most of the enzymes of β-oxidation involved in energy homeostasis (including VLCAD) and is a major drug target for treating common diseases, such as metabolic syndrome.44 Rapid use of carbons from fatty acids for synthesis of certain complex lipids through β-oxidation is known to be critical for the host defense system during acute phase response to infection or cytokine stimulation and includes up-regulation of enzymes involved in synthesis and degradation of lipids such as glycosphingolipids and ceramides.45 The RXR-associated pathway is largely inactive under these conditions, and the β-oxidation enzymes that are regulated by PPARα/RXR are down-regulated, including VLCAD.46 However, the NRF-1–associated regulatory pathway is activated in response to stimulation of cytokines or stress.21 Control of expression of the ACAD9 gene by NRF-1 suggests that it could be regulated opposite of other ACAD genes under conditions of cellular stress or stimulation of cytokines. In fact, cytokine stimulation of ACAD9-deficient and normal fibroblasts supports this (table 4). Participation of ACAD9 in a pathway that is rapidly induced by cytokines suggests a unique role for long-chain fatty acid β-oxidation in acute phase response upon infection.
In summary, we present the first report, to our knowledge, of genetic defects in ACAD9. The dramatic nature of the clinical presentations seen in these patients makes it imperative to learn the frequency of this disorder in individuals with unexplained liver failure, cerebellar stroke, and cardiomyopathy of unknown etiology and to characterize the full spectrum of symptoms in this disease. Additional study of this enzyme is likely to provide us with new insights into the pathogenesis of disorders of fat metabolism in humans.
Acknowledgments
We thank Paul P. Van Veldhoven and R. J. A. Wanders for providing the branched-chain and polyunsaturated-chain acyl-CoA substrates. We also thank J. J. Kim for providing purified human LCAD. We thank Joseph Pilewski and David Schowalter, respectively, for providing MUC1 and VLCAD antibodies. We thank Joseph Latoche for technical help. We also thank Dr. Ronald Jaffe for providing the control human tissues. This work was supported by National Institutes of Health (NIH) Public Health Service grant R01 DK45482 (to J.V.), by the Pennsylvania State Tobacco Settlement Fund (to J.V.), and as a pilot project of NIH grant U54 DK078377 (J.V.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- AceView, http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/
- BLAST, http://www.ncbi.nlm.nih.gov/blast/
- dbEST, http://www.ncbi.nlm.nih.gov/dbEST/
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for full-length human ACAD9 cDNA sequence from mRNA variant b [accession number NM_014049] and partial human ACAD9 cDNA sequence from mRNA variant a [accession numbers CR613592 and BX415793])
- NHR SCAN, http://mordor.cgb.ki.se/cgi-bin/NHR-scan/nhr_scan.cgi
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ACADVL, HADHA, ACADM, MTP, and X-linked adrenoleukodystrophy)
- PROSCAN, http://bimas.dcrt.nih.gov/molbio/proscan/function.html
- PSORT II (K. Nakai), http://psort.nibb.ac.jp
- TFSEARCH, http://www.cbrc.jp/research/db/TFSEARCH.html
References
- 1.Vockley J, Whiteman DA (2002) Defects of mitochondrial β-oxidation: a growing group of disorders. Neuromuscul Disord 12:235–246 10.1016/S0960-8966(01)00308-X [DOI] [PubMed] [Google Scholar]
- 2.Spiekerkoetter U, Sun B, Zytkovicz T, Wanders R, Strauss AW, Wendel U (2003) MS/MS-based newborn and family screening detects asymptomatic patients with very-long-chain acyl-CoA dehydrogenase deficiency. J Pediatr 143:335–342 10.1067/S0022-3476(03)00292-0 [DOI] [PubMed] [Google Scholar]
- 3.Furuta S, Miyazawa S, Hashimoto T (1981) Purification and properties of rat liver acyl-CoA dehydrogenases and electron transfer flavoprotein. J Biochem (Tokyo) 90:1739–1750 [DOI] [PubMed] [Google Scholar]
- 4.Ensenauer R, He M, Willard JM, Goetzman ES, Corydon TJ, Vandahl BB, Mohsen AW, Isaya G, Vockley J (2005) Human acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial beta-oxidation of unsaturated fatty acids. J Biol Chem 280:32309–32316 10.1074/jbc.M504460200 [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Zhang W, Zou D, Chen G, Wan T, Zhang M, Cao X (2002) Cloning and functional characterization of ACAD-9, a novel member of human acyl-CoA dehydrogenase family. Biochem Biophys Res Commun 297:1033–1042 10.1016/S0006-291X(02)02336-7 [DOI] [PubMed] [Google Scholar]
- 6.Cunnane SC, Ryan MA, Nadeau CR, Bazinet RP, Musa-Veloso K, McCloy U (2003) Why is carbon from some polyunsaturates extensively recycled into lipid synthesis? Lipids 38:477–484 10.1007/s11745-003-1087-8 [DOI] [PubMed] [Google Scholar]
- 7.Aoyama T, Souri M, Ushikubo S, Kamijo T, Yamaguchi S, Kelley RI, Rhead WJ, Uetake K, Tanaka K, Hashimoto T (1995) Purification of human very-long-chain acyl-coenzyme A dehydrogenase and characterization of its deficiency in seven patients. J Clin Invest 95:2465–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 9.Vockley J, Parimoo B, Tanaka K (1992) Identification of the molecular defects responsible for the various genotypes of isovaleric acidemia. Prog Clin Biol Res 375:533–540 [PubMed] [Google Scholar]
- 10.Battaile KP, McBurney M, Van Veldhoven PP, Vockley J (1998) Human long chain, very long chain and medium chain acyl-Coa dehydrogenases are specific for the S-enantiomer of 2-methylpentadecanoyl-CoA. Biochim Biophys Acta 1390:333–338 [DOI] [PubMed] [Google Scholar]
- 11.Bonafe L, Thony B, Leimbacher W, Kierat L, Blau N (2001) Diagnosis of dopa-responsive dystonia and other tetrahydrobiopterin disorders by the study of biopterin metabolism in fibroblasts. Clin Chem 47:477–485 [PubMed] [Google Scholar]
- 12.Tyni T, Paetau A, Strauss AW, Middleton B, Kivela T (2004) Mitochondrial fatty acid β-oxidation in the human eye and brain: implications for the retinopathy of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency. Pediatr Res 56:744–750 10.1203/01.PDR.0000141967.52759.83 [DOI] [PubMed] [Google Scholar]
- 13.Friday RP, Pietropaolo SL, Profozich J, Trucco M, Pietropaolo M (2003) Alternative core promoters regulate tissue-specific transcription from the autoimmune diabetes-related ICA1 (ICA69) gene locus. J Biol Chem 278:853–863 10.1074/jbc.M210175200 [DOI] [PubMed] [Google Scholar]
- 14.Corkey BE HD, Glennon MC, Kelley RI, Coates PM, Kilpatrick L, Stanley CA (1988 ) Relationship between unusual hepatic acyl coenzyme A profiles and the pathogenesis of Reye syndrome. J Clin Invest 82:782–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andresen BS, Olpin S, Poorthuis B, Scholte HR, Vianey-Saban C, Wanders R, Ijlst L, Morris A, Pourfarzam M, Bartlett K, et al (1999) Clear correlation of genotype with disease phenotype in very-long-chain acyl-CoA dehydrogenase deficiency. Am J Hum Genet 64:479–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehwinkel J, Raes J, Izaurralde E (2006) Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem Sci 31:639–646 10.1016/j.tibs.2006.09.005 [DOI] [PubMed] [Google Scholar]
- 17.Mason RJ (2006) Biology of alveolar type II cells. Respirology 11:S12–S15 10.1111/j.1440-1843.2006.00800.x [DOI] [PubMed] [Google Scholar]
- 18.Young SL, Fram EK, Larson E, Wright JR (1993) Recycling of surfactant lipid and apoprotein-A studied by electron microscopic autoradiography. Am J Physiol 265:L19–L26 [DOI] [PubMed] [Google Scholar]
- 19.Kelly DP, Scarpulla RC (2004) Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev 18:357–368 10.1101/gad.1177604 [DOI] [PubMed] [Google Scholar]
- 20.Scarpulla RC (2002) Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta 1576:1–14 [DOI] [PubMed] [Google Scholar]
- 21.Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM (2001) Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol Cell 8:971–982 10.1016/S1097-2765(01)00390-2 [DOI] [PubMed] [Google Scholar]
- 22.Huss JM, Kopp RP, Kelly DP (2002) Peroxisome proliferator-activated receptor coactivator-1α (PGC-1α) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. Identification of novel leucine-rich interaction motif within PGC-1α. J Biol Chem 277:40265–40274 10.1074/jbc.M206324200 [DOI] [PubMed] [Google Scholar]
- 23.Djouadi F, Weinheimer CJ, Saffitz JE, Pitchford C, Bastin J, Gonzalez FJ, Kelly DP (1998) A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator-activated receptor α-deficient mice. J Clin Invest 102:1083–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sriram G, Martinez JA, McCabe ER, Liao JC, Dipple KM (2005) Single-gene disorders: what role could moonlighting enzymes play? Am J Hum Genet 76:911–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathur A, Sims HF, Gopalakrishnan D, Gibson B, Rinaldo P, Vockley J, Hug G, Strauss AW (1999) Molecular heterogeneity in very-long-chain acyl-CoA dehydrogenase deficiency causing pediatric cardiomyopathy and sudden death. Circulation 99:1337–1343 [DOI] [PubMed] [Google Scholar]
- 26.Cox KB, Hamm DA, Millington DS, Matern D, Vockley J, Rinaldo P, Pinkert CA, Rhead WJ, Lindsey JR, Wood PA (2001) Gestational, pathologic and biochemical differences between very long-chain acyl-CoA dehydrogenase deficiency and long-chain acyl-CoA dehydrogenase deficiency in the mouse. Hum Mol Genet 10:2069–2077 10.1093/hmg/10.19.2069 [DOI] [PubMed] [Google Scholar]
- 27.Tyni T, Pihko H (1999) Long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency. Acta Paediatr 88:237–245 10.1080/08035259950169954 [DOI] [PubMed] [Google Scholar]
- 28.Bzduch V, Behulova D, Lehnert W, Fabriciova K, Kozak L, Salingova A, Hrabincova E, Benedekova M (2001) Metabolic cause of Reye-like syndrome. Bratisl Lek Listy 102:427–429 [PubMed] [Google Scholar]
- 29.Vallee L, Fontaine M, Nuyts JP, Ricart G, Krivosic I, Divry P, Vianey-Saban C, Lhermitte M, Vamecq J (1994) Stroke, hemiparesis and deficient mitochondrial β-oxidation. Eur J Pediatr 153:598–603 10.1007/BF02190669 [DOI] [PubMed] [Google Scholar]
- 30.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, et al (2006) Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439:484–489 10.1038/nature04330 [DOI] [PubMed] [Google Scholar]
- 31.Olpin SE, Clark S, Andresen BS, Bischoff C, Olsen RK, Gregersen N, Chakrapani A, Downing M, Manning NJ, Sharrard M, et al (2005) Biochemical, clinical and molecular findings in LCHAD and general mitochondrial trifunctional protein deficiency. J Inherit Metab Dis 28:533–544 10.1007/s10545-005-0533-8 [DOI] [PubMed] [Google Scholar]
- 32.Gilbert-Barness E (2004) Review: metabolic cardiomyopathy and conduction system defects in children. Ann Clin Lab Sci 34:15–34 [PubMed] [Google Scholar]
- 33.Lundy CT, Shield JP, Kvittingen EA, Vinorum OJ, Trimble ER, Morris AA (2003) Acute respiratory distress syndrome in long-chain 3-hydroxyacyl-CoA dehydrogenase and mitochondrial trifunctional protein deficiencies. J Inherit Metab Dis 26:537–541 10.1023/A:1025995813914 [DOI] [PubMed] [Google Scholar]
- 34.Das AM, Illsinger S, Lucke T, Hartmann H, Ruiter JP, Steuerwald U, Waterham HR, Duran M, Wanders RJ (2006) Isolated mitochondrial long-chain ketoacyl-CoA thiolase deficiency resulting from mutations in the HADHB gene. Clin Chem 52:530–534 10.1373/clinchem.2005.062000 [DOI] [PubMed] [Google Scholar]
- 35.Tein I, Donner EJ, Hale DE, Murphy EG (1995) Clinical and neurophysiologic response of myopathy and neuropathy in long-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency to oral prednisone. Pediatr Neurol 12:68–76 10.1016/0887-8994(94)00109-F [DOI] [PubMed] [Google Scholar]
- 36.Bertini E, Dionisivici C, Garavaglia B, Burlina AB, Sabatelli M, Rimoldi M, Bartuli A, Sabetta G, Didonato S (1992) Peripheral sensory-motor polyneuropathy, pigmentary retinopathy, and fatal cardiomyopathy in long-chain 3-hydroxy-acyl-CoA dehydrogenase deficiency. Eur J Pediatr 151:121–126 10.1007/BF01958956 [DOI] [PubMed] [Google Scholar]
- 37.Tyni T, Rapola J, Paetau A, Palotie A, Pihko H (1997) Pathology of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency caused by the G1528C mutation. Pediatr Pathol Lab Med 17:427–447 10.1080/107710497174732 [DOI] [PubMed] [Google Scholar]
- 38.Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al (2006) Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol 2:e4 10.1371/journal.pcbi.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faustino NA, Cooper TA (2003) Pre-mRNA splicing and human disease. Genes Dev 17:419–437 10.1101/gad.1048803 [DOI] [PubMed] [Google Scholar]
- 40.Yeo G, Holste D, Kreiman G, Burge CB (2004) Variation in alternative splicing across human tissues. Genome Biol 5:R74 10.1186/gb-2004-5-10-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liebig M, Schymik I, Mueller M, Wendel U, Mayatepek E, Ruiter J, Strauss AW, Wanders RJ, Spiekerkoetter U (2006) Neonatal screening for very long-chain acyl-coA dehydrogenase deficiency: enzymatic and molecular evaluation of neonates with elevated C14:1-carnitine levels. Pediatrics 118:1065–1069 10.1542/peds.2006-0666 [DOI] [PubMed] [Google Scholar]
- 42.Carpenter K, Wiley V, Sim KG, Heath D, Wilcken B (2001) Evaluation of newborn screening for medium chain acyl-CoA dehydrogenase deficiency in 275 000 babies. Arch Dis Child Fetal Neonatal Ed 85:F105–F109 10.1136/fn.85.2.F105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andresen B, Dobrowolski S, O’Reilly L, Muenzer J, McCandless S, Frazier DM, Udvari S, Bross P, Knudsen I, Banas R, et al (2001) Medium-chain acyl-coA dehydrogenase (MCAD) mutations identified by MS/MS-based prospective screening of newborns differ from those observed in patients with clinical symptoms: identification and characterization of a new, prevalent mutation that results in mild MCAD deficiency. Am J Hum Genet 68:1408–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shulman AI, Mangelsdorf DJ (2005) Retinoid X receptor heterodimers in the metabolic syndrome. N Engl J Med 353:604–615 10.1056/NEJMra043590 [DOI] [PubMed] [Google Scholar]
- 45.Khovidhunkit W, Kim MS, Memon RA, Shigenaga JK, Moser AH, Feingold KR, Grunfeld C (2004) Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J Lipid Res 45:1169–1196 10.1194/jlr.R300019-JLR200 [DOI] [PubMed] [Google Scholar]
- 46.Yoo JY, Desiderio S (2003) Innate and acquired immunity intersect in a global view of the acute-phase response. Proc Natl Acad Sci USA 100:1157–1162 10.1073/pnas.0336385100 [DOI] [PMC free article] [PubMed] [Google Scholar]