Abstract
Listeria monocytogenes is a significant food-borne pathogen that is capable of adhering to and producing biofilms on processing equipment, making it difficult to eliminate from meat-processing environments and allowing potential contamination of ready-to-eat (RTE) products. We devised a fluorescence-based microplate method for screening isolates of L. monocytogenes for the ability to adhere to abiotic surfaces. Strains of L. monocytogenes were incubated for 2 days at 30°C in 96-well microplates, and the plates were washed in a plate washer. The retained cells were incubated for 15 min at 25°C with 5,6-carboxyfluorescein diacetate and washed again, and then the fluorescence was read with a plate reader. Several enzymatic treatments (protease, lipase, and cellulase) were effective in releasing adherent cells from the microplates, and this process was used for quantitation on microbiological media. Strongly adherent strains of L. monocytogenes were identified that had 15,000-fold-higher levels of fluorescence and 100,000-fold-higher plate counts in attachment assays than weakly adherent strains. Strongly adherent strains of L. monocytogenes adhered equally well to four different substrates (glass, plastic, rubber, and stainless steel); showed high-level attachment on microplates at 10, 20, 30, and 40°C; and showed significant differences from weakly adherent strains when examined by scanning electron microscopy. A greater incidence of strong adherence was observed for strains isolated from RTE meats than for those isolated from environmental surfaces. Analysis of surface adherence among Listeria isolates from processing environments may provide a better understanding of the molecular mechanisms involved in attachment and suggest solutions to eliminate them from food-processing environments.
Listeria monocytogenes is a psychrotrophic bacterium that is pathogenic to humans and animals. Its presence in feces or on the hides of food production animals facilitates its entry into meat slaughter areas, onto carcasses, and subsequently onto raw meat products. The presence of L. monocytogenes on incoming raw meat ingredients is a continuous source of contamination for facilities manufacturing ready-to-eat (RTE) meats, making it difficult to eliminate from meat-processing environments. Contamination problems with L. monocytogenes have resulted in numerous recalls and outbreaks that have been addressed by the U.S. Department of Agriculture (USDA) Food Safety Inspection Service with various notices, directives, and regulatory actions regarding control of L. monocytogenes. In the United States, there are estimated to be about 2,500 cases of listeriosis per year, with 20 to 40% mortality in large outbreaks, and although the incidence of listeriosis has decreased in recent years, it remains an important health risk (5, 20). L. monocytogenes poses such a formidable problem for the RTE meat industry that both the USDA and the Food and Drug Administration (FDA) have established zero tolerance for its presence in RTE foods. The ability to attach to abiotic surfaces in meat-processing environments can only exacerbate problems associated with control of L. monocytogenes.
Many bacteria are known to attach to abiotic and biotic surfaces by various means, including ionic charges (29), hydrophobic attraction (11), and “biochemical appendages,” such as pili (27), fimbriae (8), flagella (30), and specific proteins (18) and extracellular polysaccharides (4). Depending on the microorganism, initial attachment may lead to more highly developed “biofilms,” which can be considered three-dimensional communities showing structured environments involving channels and nutrient flow (4). L. monocytogenes is well known for its ability to form biofilms and to establish harborages on food-processing equipment (stainless steel, plastic, and rubber surfaces), making its eradication even more difficult, which may allow the contamination of RTE food products. Wong (34) found that not only could L. monocytogenes adhere to stainless steel and rubber, but under favorable conditions, it could multiply on stainless steel. Bacteria within biofilms are considered sessile and metabolically different than planktonic bacteria. Biofilms are self-regulating, and as they grow, individual cells or parts of the biofilm may break off, and these pieces may subsequently colonize new substrates or pass contaminating bacterial cells onto food products. Another feature of being buried within biofilms is that the bacteria are often more resistant to sanitizers and removal strategies (6, 16).
Several methods have been developed in an attempt to quantify the number of cells attached to surfaces or associated with biofilms. Some investigators have used crystal violet to stain biofilm cells and absorbance readings to estimate cell numbers (24). Crystal violet does not differentiate between live and dead cells or between cells and extracellular polymers, and different strains may produce varying levels of extracellular polysaccharides. This variability in staining may be complicated by variability in removing excess stain with alcohol. Narisawa et al. (23) modified the crystal violet method by using biofilms in microplates and extracting the crystal violet with alcohol, which was then transferred to new plates and quantified. Other investigators have used acridine orange fluorescence to visualize and enumerate biofilm cells (13); however, this method also stains live and dead cells, and the viability of the live cells is compromised by the stain.
The purpose of this study was to develop a convenient fluorescence assay to screen and identify the adherence characteristics of strains of L. monocytogenes that were isolated from raw and processed RTE meats and from RTE meat-processing environments. Adherence may be considered the first stage of biofilm formation and, at the very least, a sanitation nightmare for meat-processing facilities. Using a microplate assay system, plate washer, and plate reader, we devised and implemented an in situ method of detecting cells adhering to microplates by using 5,6-carboxyfluorescein diacetate (5,6-CFDA), after which cells remain viable for subsequent analyses (15).
MATERIALS AND METHODS
Bacterial cultures and growth conditions.
Initial attachment and detachment assays were developed using four strains of L. monocytogenes (Scott A-2, serotype 4b; V7-2, serotype 1/2a; retail hot dog isolate 39-2; and ground beef isolate 383-2). The bacterial strains were cultured by transferring 100 μl of thawed frozen culture suspension into 9 ml of brain heart infusion (BHI) broth (Difco; Becton-Dickinson, Franklin Lakes, NJ), incubating it overnight (18 to 24 h) at 30°C, and subculturing the bacteria twice before use. Frozen culture stocks were prepared by centrifuging 9 ml of culture, resuspending the pellet in 2 ml of sterile BHI broth (containing 10% glycerol), and storing it at −76°C. Colony enumeration was performed on general-purpose agar for 24 h at 37°C (tryptic soy agar; Difco). Additional strains of L. monocytogenes were obtained from our culture collection and contained strains isolated from retail frankfurters (31), raw meats (9, 26), and RTE meat-processing facilities (26) (Table 1).
TABLE 1.
Bacterial strains used in this study
| L. monocytogenes strain | Source | Reference |
|---|---|---|
| Scott A-2 | Rifamycin- and streptomycin-resistant derivative of Scott A | 22 |
| V7-2 | Rifamycin- and streptomycin-resistant derivative of V-7 | 22 |
| 39-2 | Retail frankfurter isolate; rifamycin- and streptomycin-resistant derivative of CW39 | 22 |
| 383-2 | Retail ground beef isolate; rifamycin- and streptomycin-resistant derivative of PMM383 | 22 |
| 99-5 | Ground beef isolate; retail store no. 8.2 | 9 |
| 99-15 | Ground beef isolate; retail store no. 16 | 9 |
| 99-25 | Ground beef isolate; commercial processor no. 3 | 9 |
| 99-38 | Ground beef isolate; retail store no. 8.4 | 9 |
| 99-52 | Ground beef isolate; retail store no. 11 | 9 |
| 99-56 | Ground beef isolate; retail store no. 21 | 9 |
| 99-60 | Ground beef isolate; retail store no. 23 | 9 |
| CW34 | Retail frankfurter isolate | 31 |
| CW35 | Retail frankfurter isolate | 31 |
| CW50 | Retail frankfurter isolate | 31 |
| CW52 | Retail frankfurter isolate | 31 |
| CW62 | Retail frankfurter isolate | 31 |
| CW72 | Retail frankfurter isolate | 31 |
| CW73 | Retail frankfurter isolate | 31 |
| CW77 | Retail frankfurter isolate | 31 |
| G1-36 | Environmental isolate, RTE processing facility B | 26 |
| G1-40 | Environmental isolate, RTE processing facility A | 26 |
| G1-73 | Environmental isolate, RTE processing facility C | 26 |
| G1-108 | Environmental isolate, RTE processing facility B | 26 |
| G1-112 | Environmental isolate, RTE processing facility B | 26 |
| G1-120 | Environmental isolate, RTE processing facility B | 26 |
| G1-122 | Environmental isolate, RTE processing facility B | 26 |
| G1-126 | Environmental isolate, RTE processing facility A | 26 |
| G1-148 | Environmental isolate, RTE processing facility C | 26 |
| G1-158 | Environmental isolate, RTE processing facility B | 26 |
| G1-168-4 | Environmental isolate, RTE processing facility C | 26 |
| G2-2 | Environmental isolate, RTE processing facility C | 26 |
| G2-6 | Environmental isolate, RTE processing facility C | 26 |
| G2-25 | Environmental isolate, RTE processing facility C | 26 |
| G2-54-3 | Environmental isolate, RTE processing facility C | 26 |
| G2-55-4 | Environmental isolate, RTE processing facility C | 26 |
| G2-75-1 | Environmental isolate, RTE processing facility C | 26 |
| G2-96 | Environmental isolate, RTE processing facility C | 26 |
| G7-4 | Environmental isolate, RTE processing facility C | 26 |
| G7-5 | Environmental isolate, RTE processing facility C | 26 |
| J46 | Environmental isolate, RTE processing facility C | 26 |
| J52 | Environmental isolate, RTE processing facility C | 26 |
| J53 | Environmental isolate, RTE processing facility C | 26 |
| J204 | Environmental isolate, RTE processing facility C | 26 |
| J207 | Environmental isolate, RTE processing facility C | 26 |
| J212 | Environmental isolate, RTE processing facility C | 26 |
| J220 | Environmental isolate, RTE processing facility C | 26 |
| J232 | Environmental isolate, RTE processing facility B | 26 |
| J233-1 | Environmental isolate, RTE processing facility B | 26 |
| SM1 | Retail ground beef isolate | 21 |
| SM2 | Retail ground beef isolate | 21 |
| SM3 | Retail ground beef isolate | 21 |
| SM4 | Retail ground pork isolate | 21 |
| SM5 | Retail ground turkey isolate | 21 |
Fluorescent microplate assay for surface attachment.
A method for microplate incubation of various strains was devised and compiled partly from similar procedures and conditions found in the literature, as well as our own modifications (i.e., washing and addition of fresh medium). Strains to be tested were subcultured overnight in BHI broth held at 30°C. The overnight culture was diluted 105-fold (i.e., from ∼109 CFU/ml to ∼104 CFU/ml) in fresh BHI broth, and 200 μl was transferred to designated wells of a 96-well black microwell plate with a clear lid (Nunc, Denmark). The edge of the plate was wrapped in Parafilm to prevent evaporation, and the plate was incubated at 30°C for 24 h (the temperature was chosen to be the same as the culture incubation temperature). After incubation, the microplate was washed three times with Tris buffer (pH 7.4; 0.05 M) in a Biotec Elx405 Magna plate washer (Ipswich, Suffolk, United Kingdom) with 96 pairs of needles (one for aspiration; another for dispensing) to remove loosely adhered cells (1). The plate washer was sanitized with 200 ppm sodium hypochlorite (pH 6.5) after each use. The washing was followed by the addition of 200 μl of fresh (sterile) BHI broth to each experimental well, and the plate was again wrapped in Parafilm, incubated at 30°C, and washed three times with Tris buffer (pH 7.4; 0.05 M) after another 24 h. After the final washing, 200 μl of 5,6-CFDA (Molecular Probes/Invitrogen, Carlsbad, CA) fluorescent substrate solution was added. The 5,6-CFDA fluorescent substrate working stock was prepared by adding 10 μl of a 2% 5,6-CFDA solution in dimethyl sulfoxide to 1 ml of cold Tris buffer (pH 7.4; 0.05 M). Following incubation with the 5,6-CFDA substrate, the plates were washed three times with Tris buffer (pH 7.4; 0.05 M) in the plate washer, and the medium was replaced with 200 μl of the same medium. The plate was then read from above or below in a Tecan GENios fluorescent-plate reader (Phenix Research Products, Hayward, CA) using a fixed signal gain of 75% with excitation at 485 nm and detection at 535 nm.
Optimization of cell detection with CFDA.
Various substrate incubation times (15, 30, 45, 60, and 90 min) and temperatures (25, 30, and 37°C) with CFDA substrate were examined to determine the optimal conditions for an effective fluorescence response. The fluorescence signal obtained with the mixed-isomer substrate (5,6-CFDA) was also compared to that obtained with the single-isomer substrate (5-CFDA; Molecular Probes/Invitrogen). The temperature chosen for substrate incubation was also examined at shorter time intervals (5, 10, 15, 30, 45, and 60 min). We also examined fluorescence detection of attached cells after 1 day of attachment (at 30°C) versus replacement of planktonic cells with fresh sterile BHI medium and continued incubation; this cycle was examined after 1, 2, and 3 medium replacements. Comparisons were also made of fluorescence signals obtained from different-color microplates (untreated black, clear, and white; Phenix Research Products) and between top and bottom (clear) sides from which different plates could be read (i.e., plates read from the bottom had clear bottoms, while those of the same color that were read from the top had solid bottoms).
Quantification of cell attachment by enzymatic detachment.
We examined the use of various enzymes to cause the release of attached cells for the purpose of subsequent enumeration. Various proteases, including pronase E, trypsin, papain, pepsin, and thermolysin (Sigma-Aldrich, St. Louis, MO) (all constituted in Tris buffer [pH 7.4; 0.05 M] at 1,000 U/ml), as well as BAX protease (Qualicon), were tested for the ability to release adherent cells from microplates. BAX protease was used according to the manufacturer's directions (12.5 μl per ml Tris buffer, pH 7.4, 0.05 M; specific enzyme and concentration unknown). The effects of lipoprotein lipase B, lipase, alpha amylase, and cellulase (VWR) were also examined for detachment of adherent cells. Each enzyme (except BAX protease) was used at 100 enzyme units (U) per 200-μl microwell plate assay.
Nonproteolytic enzymes were tested with RediPlate 96 EnzChek (Invitrogen), an enzymatic assay in microplate format to test for metallo-, serine, acid, and sulfhydryl protease activities, which we used to ensure the absence of protease contamination in the nonproteolytic enzyme preparations mentioned above. The assay was performed according to the manufacturers' directions, generating a green fluorescent signal upon hydrolysis, and was read in the Tecan GENios plate reader with excitation at 485 nm and detection at 535 nm.
A “detachment assay” was run on attached cells using the 48-h microplate assay described above. After 48 h of incubation, the microplates were washed twice with Tris buffer (pH 7.4) using the automated plate washer, followed by a final rinse with either Tris buffer, pH 7.4, 0.05 M (i.e., controls), or Tris buffer containing 100 U of enzyme per 200 μl (i.e., enzyme-treated samples). After incubation at 37°C for 1 h, the liquid in the wells was harvested and plated for microbial enumeration of detached cells. All plating was done on tryptic soy agar plates incubated at 37°C for 48 h. After the detachment assays, the microplates were washed with the automated plate washer and subjected to the 5,6-CFDA-based fluorescence assays for comparison of the fluorescent signals of attached cells (control wells without added enzyme) and detached cells (wells treated with enzyme), as well as comparison with microbial-cell counts recovered from both control and enzyme treatments.
Planktonic cells in BHI broth culture were also treated with enzyme to determine if the enzyme(s) affected cell viability and, therefore, the integrity of our bacterial plate counts recovered after enzymatic detachment. Overnight 9-ml cultures of the four test strains of L. monocytogenes described above were centrifuged at 4,500 × g for 30 min in a Sorvall RC5 Plus centrifuge at 5°C; the supernatant broth was discarded, and the cell pellets were resuspended in 9 ml of Tris buffer (0.05 M; pH 7.4). Eight hundred-microliter samples of the resuspended cells were placed into an Eppendorf tube, along with 200 μl of enzyme/Tris buffer (pH 7.4) so that the final concentration of enzyme was 100 U per 200 μl (as would be used in microplates with attached cells). A control was used for each enzyme, consisting of cells resuspended in buffer without enzyme. After 1 h at 37°C, appropriate dilutions were made of both controls and enzyme-treated planktonic cells using 0.1% buffered peptone water (BPW), which were plated on tryptic soy agar, followed by 48 h of incubation at 30°C before enumeration.
In another assay, fluorescence readings were also obtained with cells in suspension for strains designated “strongly adherent” or “weakly adherent” in order to determine if differences observed in microplate fluorescence assays were perhaps attributable to the abilities of the strains to take up and/or hydrolyze the fluorescence substrate. The 5,6-CFDA-derived fluorescence was obtained by using equivalent numbers of planktonic cells from liquid culture (BHI broth), which were centrifuged and resuspended in 0.1% BPW as described above and then incubated with 5,6-CFDA substrate in Eppendorf tubes for the same time and at the same temperature used in microplate assays. The cells were pelleted in a microcentrifuge (Eppendorf model 5417C; 8000 × g) to remove residual fluorescence substrate, resuspended with 0.1% BPW, and quickly placed in microplates for fluorescence readings in the GENios plate reader. The plate counts of the cell suspensions used for the plate readings were also recorded to ensure that equivalent numbers of cells were used in the assays.
Fluorescence microscopy.
Fluorescence microscopy was conducted with cultures in a modified attachment assay using untreated eight-compartment CultureSlides (Falcon; Becton-Dickinson, Bedford, MA), which were polystyrene chambers fixed onto glass slides with the intention that, after culturing, the liquid would be removed, the chambers would be washed and disassembled, and the bottom surface of the chamber would be a microscope slide useful for microscopic observation and comparison of the eight bottom surfaces. Overnight cultures of select strains of L. monocytogenes were diluted 105-fold (i.e., ∼104 CFU/ml) in fresh, sterile BHI broth, and 200 μl of the resulting dilution was placed into chambers on the culture slides. The cultures were incubated under the same conditions as in the microplate assay (48 h; 30°C), rinsed by manual pipette aspiration using Tris buffer (pH 7.4; 0.05 M), and incubated with CFDA-based substrate as previously described. The chambers were removed using the manufacturer's tool, and the bottom slides were examined by fluorescence microscopy using a Nikon Eclipse E400 fluorescence microscope (excitation at 450 to 490 nm; detection at 500 nm) using a BA 515 B-2A filter and outfitted with a digital camera attachment.
SEM.
Scanning electron microscopy (SEM) images were obtained by comparison of eight strains of L. monocytogenes selected from the results of our microplate fluorescence assays and CultureSlide microscopic assays. We selected four strains that demonstrated high-level fluorescence in our attachment assay in comparison with four strains that gave low-level fluorescence. The cultures were grown in the presence of glass microscope coverslips placed in a sterile 24-well microplate (Falcon) with 500 μl of culture at ∼104 CFU/ml in fresh BHI broth and incubated overnight at 30°C. As with our microplate attachment assay, the cells were removed and the wells/coverslips were washed three times with Tris buffer (pH 7.4; 0.05 M) and replaced with 500 μl of fresh BHI broth for further incubation. After a total of 48 h, the coverslips were transferred to new wells and again washed three times with Tris buffer (pH 7.4; 0.05 M) for transfer to the Electron Microscopy Core Facility at Oklahoma State University (SEM analysis was performed by Terry Colberg).
Attachment to different substrates.
In another detachment assay, comparisons were made of cell counts recovered from similar-size pieces of stainless steel, rubber, glass, and plastic (polypropylene) using four strongly adherent strains (L. monocytogenes CW50, CW62, CW77, and 99-38) and two weakly adherent strains (L. monocytogenes CW34 and CW35). Attachment assays were performed in 24-well microplates in a manner similar to that performed in the 96-well plates (i.e., 2-day incubation at 30°C). The pieces of substrate were then moved to clean wells for manual rinses before treatment with BAX protease for recovery and plating.
Effects of incubation temperature on attachment.
We examined attachment using our 2-day microplate assay with incubation at 10, 20, 30, and 40°C to determine if attachment was affected by temperature (i.e., temperature-regulated gene expression), as these extremes of temperature can be encountered at various points within food-processing facilities where L. monocytogenes may be found as an environmental contaminant.
Experimental design and statistical analysis.
All trials were carried out in triplicate, and data are presented as the means. Standard deviations were obtained for the multiple replications and are represented by error bars. Statistical analysis was performed for multiple comparisons of the means and standard deviations obtained for different treatments. Analysis of variance was performed using the Holm-Sidak test for pairwise multiple comparisons to determine significant differences (P < 0.05) using the software program SigmaStat 3.1 (SPSS Inc., Chicago, IL).
RESULTS AND DISCUSSION
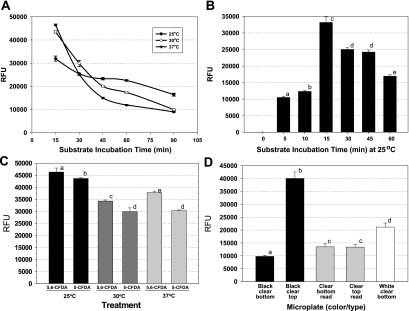
Microplates offer a convenient format for testing a wide variety of strains (10, 14), especially since this plate format has been integrated with plate washers and plate readers, which we utilized to complement our microplate fluorescence attachment assay. We used 5,6-CFDA because of its historical use with flow cytometry and as a general indicator of cellular activity (15). We initially examined several features that could influence biologically derived fluorescence signals, including the substrate, substrate incubation temperature and time, and type of microplate. CFDA-based assays provided excellent correlation and linearity (r2 = 0.9979) when cell populations of L. monocytogenes Scott A-2 (104 to 109 CFU/ml) were serially diluted and tested for fluorescence with the 5,6-CFDA substrate (data not shown). L. monocytogenes Scott A-2 was incubated in microplate wells at 30°C for attachment and then examined at various substrate incubation temperatures for uptake and response with the 5,6-CFDA substrate (Fig. 1A). The results showed that the highest fluorescence levels were obtained in 15 min at each of the substrate incubation temperatures examined, with all decreasing significantly after 15 min (Fig. 1A). Since the rate of decrease of fluorescence was least when the cells were incubated at 25°C, we chose that substrate incubation temperature for the remainder of the study. We also examined shorter substrate incubation periods at 25°C and found that a 15-min incubation period provided higher fluorescence levels from attached cells than did either shorter or longer substrate incubation periods (Fig. 1B); however, the lower levels of fluorescence at shorter incubation times may be due to a minimum time necessary for the substrate to enter the cell and become hydrolyzed to the fluorescent by-product. The subsequent decreasing fluorescence levels may likewise be due to metabolic quenching or leakage from the attached cells, since leakage of the fluorescent derivative outside the attached cells would still be removed by microplate washing immediately after the substrate incubation period (Fig. 1B). However, the carboxy-diacetate modification (i.e., 5,6-CFDA) is supposed to reduce cytoplasmic leakage of the hydrolyzed carboxyfluorescein product relative to traditional fluorescein due to the presence of negative charges at cytoplasmic pH levels (15). We also examined a single-isomer substrate (5-CFDA) in comparison with 5,6-CFDA and found no enhancement of signal performance with L. monocytogenes Scott A (Fig. 1C). Other fluorescein-based substrates that may also prove beneficial in such applications are the succinimidyl ester and acetoxymethyl modifications; however, the costs of these substrates were, respectively, 7- and 20-fold more than that of 5,6-CFDA, and they were not considered further. One other aspect of assay optimization was the testing of several different microplate formats, including white and black plates (with clear or solid bottoms), as well as clear microplates. We obtained the best signals using solid black microplates read from above (Fig. 1D).
FIG. 1.
Optimization of the 5,6-CFDA assay for Listeria attachment. (A) Fluorescences of planktonic cells of L. monocytogenes Scott A-2 at different substrate incubation times at 25, 30, and 37°C. (B) Examination of fluorescence after different substrate incubation times at 25°C. (C) Comparison of fluorescences obtained with mixed and single isomers of CFDA incubated for 15 min at 25, 30, and 37°C. (D) Examination of fluorescence signals obtained from adherence of L. monocytogenes Scott A-2 with black, clear, and white microplates when read from the top or the bottom. All data are presented as the means of triplicate replications with standard deviations; means with different lowercase letters are significantly different (P < 0.05). RFU, relative fluorescence units.
Our finalized microplate fluorescence assay consisted of using black microplates with a 2-day incubation/attachment period at 30°C. After the first day, planktonic cells were removed, the plates were washed with a microplate washer, and the medium was replaced with sterile medium for continued incubation for a second day (i.e., only those cells that were attached would contribute to continued growth). After the second and final day of attachment, planktonic cells were again removed, and the plates were washed with buffer using the plate washer, followed by the addition of 5,6-CFDA substrate solution and incubation at 25°C for 15 min. After the substrate incubation period, the plates were washed again, the medium was replaced with buffer, and the plates were read on a plate reader.
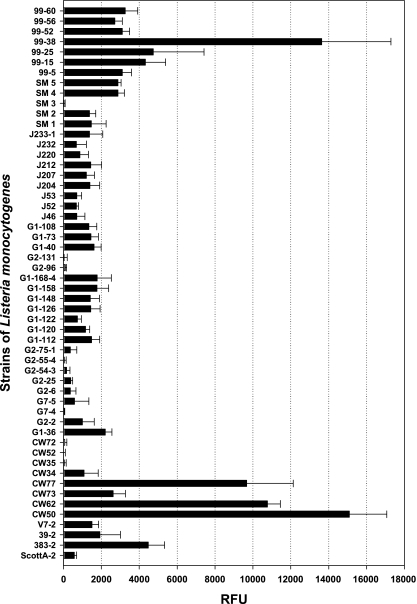
Using this modified procedure, we screened more than 50 strains of L. monocytogenes isolated from RTE meat-processing facilities, raw retail meats, and RTE meats for the ability to adhere in our attachment assays (Table 1 and Fig. 2). Of the strains tested, a >15,000-fold difference in fluorescence signal was obtained between various strains, suggesting that some may have demonstrated greater levels of attachment than others (Fig. 2). It is interesting that a rather high percentage of strains isolated from raw or processed meats (Fig. 2, 99 SM, and CW series) were moderately to highly adherent whereas general environmental isolates recovered from RTE meat-processing facilities (Fig. 2, J and G series) were mostly weakly adherent isolates. This may simply reflect the fact that the meat products may have been the recipient of strongly adherent strains from food contact surfaces that were selectively retained from the general environmental biota and subsequently transferred to the raw or processed meats. This further emphasizes not only the need to ensure the elimination of contamination in crevices on food contact surfaces, but also the need to reduce the burden of contamination in the general food-processing environment from which these contaminants were likely derived.
FIG. 2.
Microplate fluorescence attachment assay of various strains of L. monocytogenes from retail ground beef (99 series strains) and ground pork and turkey samples (G and SM series), from environmental surfaces in commercial processing plants making RTE meats (J series), and isolated from retail frankfurters (CW series). RFU, relative fluorescence units. Signals were obtained using a fixed manual gain of 75%. The data bars are presented as the mean of triplicate replications, and the error bars represent the standard deviations of the mean.
Based on our microplate fluorescence assays, strains were tentatively differentiated as strong versus weakly adherent and confirmed in head-to-head testing on the same plate (Table 2). Although we considered higher levels of fluorescence to correspond to higher levels of attachment, one possible explanation for the variations in signals from our microplate fluorescence assays may also have been that different strains were able to take up and hydrolyze the substrate better than others. In that case, the fluorescence signals may have merely represented strain differences in biochemical handling of the substrate rather than differences in attachment. We therefore examined the fluorescence of the same level of planktonic cells in suspension to determine if there were strain differences that correlated with what was observed in microplate attachment assays. When planktonic cells of the weakly adherent strains were treated with substrate, we obtained levels of fluorescence equivalent to or higher than those of strains considered strongly adherent in microplate attachment assays (Table 2). Considering that the planktonic fluorescence assay was performed with an equivalent numbers of cells for each of the strains tested (Table 2), we were satisfied that the attachment assay was representative of the relative adherence levels of the various strains.
TABLE 2.
Comparison of the 5,6-CFDA fluorescence assays for strains of L. monocytogenes as attached or planktonic cells
| L. monocytogenes strain | Microplate fluorescence attachment assay
|
Planktonic-cell fluorescence assay
|
Planktonic-cell plate count assay
|
|||
|---|---|---|---|---|---|---|
| RFUa | SDb | RFU | SD | Log CFU/ml | SD | |
| CW50 | 14,000 | 3,700 a | 20,000 | 2,000 a | 9.1 | 0.14 a |
| CW62 | 13,000 | 210 ab | 26,000 | 2,800 a | 9.2 | 0.17 a |
| CW73 | 11,000 | 1,900 ab | 22,000 | 4,200 a | 9.3 | 0.12 a |
| CW77 | 9,700 | 1,800 b | 21,000 | 4,000 a | 9.3 | 0.05 a |
| CW34 | −7 | 31 c | 31,000 | 2,000 a | 9.3 | 0.10 a |
| CW35 | 900 | 430 c | 23,000 | 4,600 a | 9.2 | 0.16 a |
| CW72 | 1,100 | 220 c | 23,000 | 6,800 a | 9.2 | 0.11 a |
| CW52 | 280 | 8 c | NTc | NT | NT | NT |
RFU, relative fluorescence units; values represent “net” values (with background subtracted).
Means within an assay (column) followed by the same letter are not significantly different (P > 0.05).
NT, not tested.
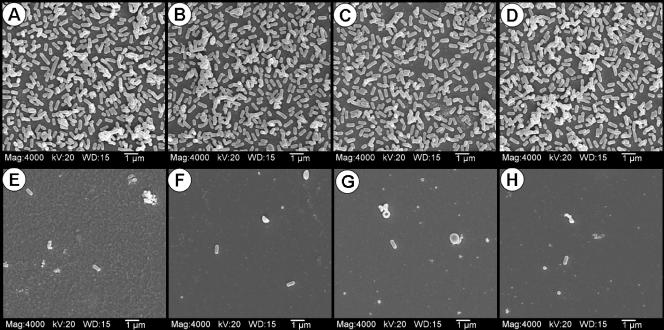
In order to confirm adherence by more quantitative means, we compared eight strains of L. monocytogenes (four strongly and four weakly fluorescing strains from attachment assays) for the ability to attach in head-to-head comparisons when tested under the same conditions using microscope slide chambers. After incubation for attachment and substrate uptake, the chambers were removed, and the slides were examined by both light and fluorescence microscopy. The microscopy results confirmed that cells from strains that yielded strong fluorescence signals were present in higher numbers on the slides than those from strains giving weak fluorescence signals in attachment assays (data not shown). The same strains were again incubated under identical conditions in microplates with glass chips that were washed five times with buffer before being submitted for SEM analysis. The strains that were chosen from our attachment assays for high fluorescence signals and shown to have high levels of attachment by light and fluorescence microscopy were also found to be strongly adhering by SEM analysis (Fig. 3A to D). The same strains that showed consistently low levels of fluorescence in the attachment and microscopic assays also showed low levels of attachment by SEM analysis relative to the more highly adhering strains (Fig. 3E to H). The SEM photographs demonstrate a visually striking comparison of the weakly versus strongly adherent strains. It is very likely that the strong adherence possessed by these strains may play a role in their persistence in plant environments. It is interesting that, unlike raw ground meats, in which Listeria contaminants may be present due to acquisition either from processing equipment/surfaces or from original carcass biota during slaughter, the presence of L. monocytogenes on RTE meats can mainly be attributed to acquisition from food contact surfaces after processing (i.e., cooking), and our RTE isolates demonstrated a high incidence of strong adherence characteristics (Fig. 2, CW series).
FIG. 3.
SEM images of various strongly and weakly adherent strains of L. monocytogenes screened using the microplate attachment assay with glass chips. The strains of L. monocytogenes are as follows: top row, CW50 (A), CW62 (B), CW77 (C), and 99-38 (D); bottom row, CW34 (E), CW35 (F), CW52 (G), and SM3 (H). Mag, magnification; WD, working distance (mm).
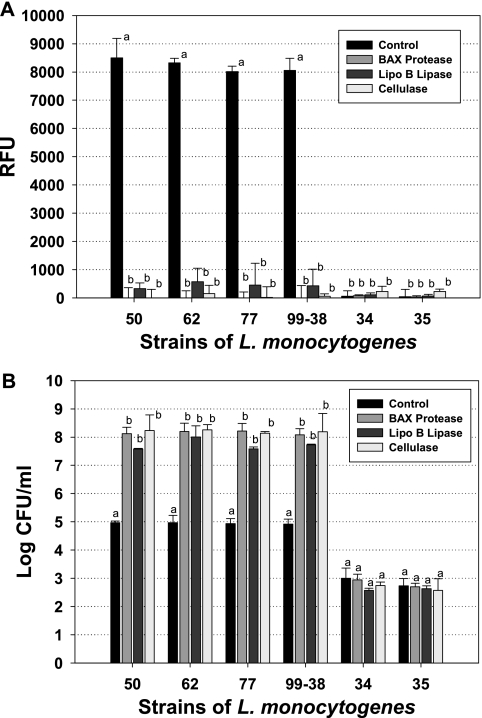
In efforts to quantify the numbers of cells attached to surfaces, investigators have previously used methods such as scraping or swabbing cells to determine their relative populations (17, 19). Proteolytic enzymes have also been proposed for use in removing bacteria trapped as parts of biofilms on prosthetic devices (28) and food-processing equipment (25). In a related modification, we used proteases to help quantify the levels of attachment by proteolytic release (or “detachment”) from microplate well surfaces. In order to rely on plate counts from “detachment” assays, we had to ensure that neither the substrate incubation nor enzymatic treatments would have any adverse effects on the viability of the treated cells; otherwise, the counts would not be representative of what was previously attached. We found little or no effect on cell viability after treatment for as long as 90 min with our 5,6-CFDA substrate solution or after extended treatments with the various proteases, lipase, or cellulase used (data not shown). We further tested and found several nonproteolytic enzyme preparations (alpha-amylase and lipase) to contain considerable proteolytic activity when tested with the EnzChek assay (data not shown). Because of their lack of protease activity with the EnzChek assay, we compared the effects of lipoprotein B lipase and cellulase with that of BAX protease prior to our microplate fluorescence assay and for quantitation of L. monocytogenes after detachment from microplates (Fig. 4A). When control wells for strongly adhering strains were treated with buffer instead of enzyme, we obtained typical high-level fluorescence signals when performing our microplate assay, although little or no signal was obtained with controls for several weakly adhering strains also included in the assay (Fig. 4A). When wells containing strongly attaching strains were treated with BAX protease or cellulase, we obtained complete loss of fluorescence and nearly complete loss with lipoprotein B lipase (Fig. 4A). The data suggest that substrates for the three types of enzymes may be involved in attachment by L. monocytogenes or possibly that the cellular constituent may be embedded in the peptidoglycan layer, which contains protein, carbohydrate, and lipid moieties that can all be acted upon by the enzymes tested. When we examined the abilities of the same enzymes to detach attached Listeria cells, the data complemented those obtained with fluorescence in that BAX protease and cellulase gave the highest recovered plate counts while those obtained with lipoprotein B lipase treatment were slightly lower for all four strongly adhering strains (Fig. 4B). In this series of assays, all attached wells were washed five times prior to final treatment of control wells (i.e., with buffer) or test wells (with enzyme) to obtain samples for plating (Fig. 4B). For the strongly adhering strains (L. monocytogenes CW50, CW62, CW77, and 99-38), the data show that >3-log-unit-lower levels were recovered when they were treated with buffer than when they were treated with enzymes, indicating that only about 0.1% of what was attached came off in the buffer wash (Fig. 4B). However, the weakly adhering strains (L. monocytogenes CW34 and CW35) showed approximately 5-log-unit-lower levels of attached cells than the strongly adhering strains, and the controls showed comparable levels of release with buffer treatment and with enzymatic detachment (Fig. 4B). The differences in recovery of cells after buffer versus after enzyme treatments are further representative of their relative levels of attachment.
FIG. 4.
Enzymatic detachment of attached cells of various strains of L. monocytogenes (CW50, CW62, CW77, 99-38, CW34, and CW35) using BAX protease, lipoprotein (Lipo) B lipase, or cellulase. (A) Effect of enzyme treatment on fluorescence signals of attached cells in comparison to buffer treatment (controls) using the fluorescence microplate assay. (B) Microbial enumeration of detached cells after buffer (control) or enzyme treatment. For assays of the same strain, means with the same letter are not significantly different from each other; means with different letters are significantly different (P < 0.05). The error bars indicate standard deviations. RFU, relative fluorescence units.
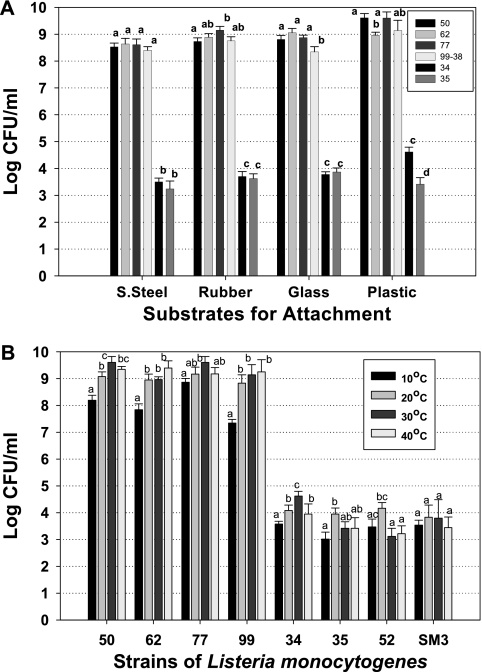
The same strains were also tested for attachment to each of four types of surfaces (glass, plastic, stainless steel, and rubber) as determined by detachment recovery after 2 days of incubation on the same-size pieces of material. Similar to what we observed with microplate wells, attachment of the strongly adherent strains was approximately 5 log units higher than what was observed for the weakly adherent strains (Fig. 5A). One possible cellular constituent that may contribute to attachment is flagella, which have been associated with adherence to surfaces. Our attachment incubation temperature could have straddled the temperature limits for expression (as a possible explanation of why some strains did not show strong attachment). Although expression of flagella for L. monocytogenes is generally considered to be down-regulated above 25°C (12, 30, 32, 33), Bigot et al. (3) found that 20 of 100 clinical isolates of L. monocytogenes they examined expressed flagella and motility at 37°C. We therefore incubated cells at two temperatures above (30°C and 40°C) and two temperatures below (10°C and 20°C) this level to see if any differences were observed that would indicate temperature-dependent attachment characteristics. The cell levels recovered from detachment assays did not show enhanced adherence at lower temperatures that would suggest a contribution of flagella among our strains (Fig. 5B). This may be due to the fact that culture and adherence assay temperatures were 30°C, and therefore, we likely selected for adherent strains of L. monocytogenes that either did not express flagella or expressed them at this temperature. Furthermore, the contribution of flagella to adherence has generally been only a 1-log-unit (or less) enhancement out of 4 to 6 log CFU of total cell adherence in studies where attachment due to temperature regulation of flagellar expression or flagellar mutant versus wild-type strains was examined (3, 7, 30). It is also important to note that all of these temperatures are likely to be found in different areas of meat-processing facilities, where L. monocytogenes can be troublesome as an environmental contaminant, either during steam sanitation in large processing facilities when temperature control has been stopped to prevent fogging or in unrefrigerated side rooms of smaller processors, where carts and other equipment may be stored. It is important to note that the strongly adherent strains were still attached at levels far greater than the weakly adherent strains, even at 10°C. Jeong and Frank (17) previously noted that L. monocytogenes can develop biofilms at 10°C, and this was further analyzed at 8, 20, and 37°C for L. monocytogenes strain LO28 by Chavant et al. (7). In our study, the level of cells recovered from microplates incubated at 10°C was less than those observed for the other three temperatures and likely represents the drastically reduced growth rate at 10°C compared to higher temperatures (Fig. 5B).
FIG. 5.
Attachment characteristics of strongly and weakly adhering strains of L. monocytogenes. (A) Microbial plating of strains detached with BAX protease after attachment to various substrates. S. Steel, stainless steel. (B) Microbial plating of cells detached after attachment with BAX protease for 48 h (20, 30, and 40°C) or 96 h (10°C) at various incubation temperatures for attachment. The strains of L. monocytogenes used were CW52, CW62, CW77, 99-38, CW34, CW35, CW50, and SM3. For a given substrate (A) or strain (B), means sharing the same letter are not significantly different (P > 0.05), whereas those with different letters are significantly different (P < 0.05). The error bars indicate standard deviations.
These data present a practical and important distinction among strains isolated from raw or processed meats and from meat-processing environments based on the ability to adhere to surfaces. Meat and poultry processors cannot predetermine the adherence traits of strains that may enter their plants on raw meat ingredients. Strongly adhering strains, as shown in Fig. 3A to D, may prove more difficult to remove from processing plants, provide a greater likelihood of persistence and subsequent food contamination, and perhaps more readily promote the initiation of long-lasting biofilms on processing equipment and environmental surfaces than those that adhere weakly (Fig. 3E to H). Such strains are able to adhere strongly irrespective of the type of surface or the temperature (Fig. 5). The prospect of viable L. monocytogenes on environmental or food contact surfaces has significant consequences and can result in the manufacture of Listeria-contaminated RTE meats that may lead to consumer illness and death, product recalls, reduced confidence, and/or loss of retail customers and increased USDA Food Safety Inspection Service regulatory actions. Although we did not identify whether attachment occurs constitutively with planktonic cells, from expression of adherence traits during active growth, or when triggered after initial surface adherence, any of these possibilities would be important to food plant sanitation. The data presented here further emphasize the importance of plant sanitation and of microbial interventions that eradicate L. monocytogenes from food products themselves should they become contaminated. Although we used enzymatic detachment as a means of quantifying strain attachment, this approach may conceivably be useful as part of a sanitizing regimen, similar to the use of proteases in laundry detergents to eradicate protein-based stains (2).
CFDA-based fluorescence has proven useful in applications such as flow cytometry assays (15). In this work, we combined 5,6-CFDA-based fluorescence with a microplate format to develop an easy attachment assay to evaluate the adherence characteristics of individual strains of L. monocytogenes isolated from meat and meat plant environments. The identification of strains of L. monocytogenes with such different adherence characteristics is significant for practical consumer food safety considerations. Since attachment is also the first step in initiating cellular infection, it would be interesting to see in future research if strong adherence to abiotic surfaces correlates with enhanced cellular attachment in tissue culture assays.
Acknowledgments
This research was partially funded by the Oklahoma Agricultural Experiment Station, Oklahoma State University, Stillwater (HATCH project no. 2335).
We thank Pornpimon Pimonpan for providing some early analyses at the start of this project.
The manuscript was approved by the Oklahoma Experiment Station.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.An, Y. H., and R. J. Friedman. 1997. Laboratory methods for studies of bacteria adhesion. J. Microbiol. Methods 30:141-152. [Google Scholar]
- 2.Banik, R. M., and M. Prakash. 2004. Laundry detergent compatibility of the alkaline protease from Bacillus cereus. Microbiol. Res. 159:135-140. [DOI] [PubMed] [Google Scholar]
- 3.Bigot, A., H. Pagniez, E. Botton, C. Frehel, I. Dubail, C. Jacquet, A. Charbit, and C. Raynaud. 2005. Role of FliF and FliI of Listeria monocytogenes in flagellar assembly and pathogenicity. Infect. Immun. 73:5530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branda, S. S., A. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2007. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 sites, United States, 2006. Morb. Mortal. Wkly. Rep. 56:336-339. [PubMed] [Google Scholar]
- 6.Chae, M. S., and H. Schraft. 2000. Comparative evaluation of adhesion and biofilm formation of different Listeria monocytogenes strains. Int. J. Food Microbiol. 62:103-111. [DOI] [PubMed] [Google Scholar]
- 7.Chavant, P., B. Martinie, T. Meylheuc, M.-N. Bellon-Fontaine, and M. Hebraud. 2002. Listeria monocytogenes LO28: surface physiochemical properties and ability to form biofilms at different temperatures and growth rates. Appl. Environ. Microbiol. 68:728-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Q., H. Wu, and P. M. Fives-Taylor. 2004. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol. Microbiol. 53:843-856. [DOI] [PubMed] [Google Scholar]
- 9.Davidson, C., S. S. Reilly, E. Harp, S. E. Gilliland, and P. M. Muriana. 1999. Incidence of Esherichia coli, Listeria monocytogenes, Campylobacter spp., and Salmonella spp. in ground beef and on beef carcasses in Oklahoma, abstr. 79C-17. Ann. Meet. Inst. Food Technol., Chicago, IL, 24 to 28 July 1999.
- 10.Djordjevic, D., M. Wiedmann, and L. A. McLandsborough. 2000. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68:2950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donlan, R. M. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dons, L., O. F. Rasmussen, and J. E. Olsen. 1992. Cloning and characterization of a gene encoding flagellin of Listeria monocytogenes. Mol. Microbiol. 6:2919-2929. [DOI] [PubMed] [Google Scholar]
- 13.Fessia, S. L., and M. J. Griffin. 1991. A method for assaying biofilm capacity on polyurethane-coated slides. Perit. Dial. Int. 11:144-146. [PubMed] [Google Scholar]
- 14.Harvey, J., K. P. Keenan, and A. Gilmour. 2007. Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiol. 24:380-392. [DOI] [PubMed] [Google Scholar]
- 15.Hoefel, D., W. L. Grooby, P. T. Monis, S. Andrews, and C. P. Saint. 2003. A comparative study of carboxyfluorescein diacetate and carboxyfluorescein diacetate succinimidyl ester as indicators of bacterial activity. J. Microbiol. Methods 52:379-388. [DOI] [PubMed] [Google Scholar]
- 16.Hood, S. K., and E. A. Zottola. 1995. Biofilms in food processing. Food Control 6:9-18. [Google Scholar]
- 17.Jeong, D. K., and J. F. Frank. 1994. Growth of Listeria monocytogenes at 10°C in biofilms with microorganisms isolated from meat and dairy processing environments. J. Food Prot. 57:576-586. [DOI] [PubMed] [Google Scholar]
- 18.Latasa, C., C. Solano, J. R. Penadés, and I. Lasa. 2006. Biofilm-associated proteins. C. R. Biol. 329:849-857. [DOI] [PubMed] [Google Scholar]
- 19.Marion, K., J. Freney, G. James, E. Bergeron, F. N. R. Renaud, and J. W. Costerton. 2006. Using an efficient biofilm detaching agent: an essential step for the improvement of endoscope reprocessing protocols. J. Hosp. Infect. 64:136-142. [DOI] [PubMed] [Google Scholar]
- 20.Mead, P. S., L. Slutsker, V. Diez, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitra, S., and P. M. Muriana. 2004. Detection of Listeria monocytogenes from meats using fluorescent-based universal realtime PCR primers, abstr. 99A-36. Ann. Meet. Inst. Food Technol., Las Vegas, NV, 12 to 16 July 2004.
- 22.Muriana, P. M., W. Quimby, C. Davidson, and J. Grooms. 2002. Post-package pasteurization of RTE deli meats by submersion heating for reduction of Listeria monocytogenes. J. Food Prot. 65:963-969. [DOI] [PubMed] [Google Scholar]
- 23.Narisawa, N., S. Furukawa, H. Ogihara, and M. Yamasaki. 2005. Estimation of the biofilm formation of Escherichia coli K-12 by the cell number. J. Biosci. Bioeng. 99:78-80. [DOI] [PubMed] [Google Scholar]
- 24.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 25.Oulahal-Lagsir, N., A. Martial-Gros, M. Bonneau, and L. J. Blum. 2003. “Escherichia coli-milk” biofilm removal from stainless steel surfaces: synergism between ultrasonic waves and enzymes. Biofouling 19:159-168. [DOI] [PubMed] [Google Scholar]
- 26.Quimby, W., J. Grooms, and P. M. Muriana. 2001. PFGE genomic subtyping of Listeria spp. from cattle holding, slaughter, and processing facilities, abstr. 59F-33. Ann. Meet. Inst. Food Technol., New Orleans, LA, 20 to 24 June 2001.
- 27.Scott, J. R., and D. Zahner. 2006. Pili with strong attachments: Gram-positive bacteria do it differently. Mol. Microbiol. 62:320-330. [DOI] [PubMed] [Google Scholar]
- 28.Selan, L., F. Berlutti, C. Passariello, M. R. Comodi-Ballanti, and M. C. Thaller. 1993. Proteolytic enzymes: a new treatment strategy for prosthetic infections. Antimicrob. Agents Chemother. 37:2618-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Merode, A. E. J., H. C. van der Mei, H. J. Busscher, K. Waar, and B. P. Krom. 2006. Enterococcus faecalis strains show culture heterogeneity in cell surface charge. Microbiology 152:807-814. [DOI] [PubMed] [Google Scholar]
- 30.Vatanyoopaisarn, S., A. Nazli, C. E. R. Dodd, C. E. D. Rees, and W. M. Waites. 2000. Effect of flagella on initial attachment of Listeria monocytogenes to stainless steel. Appl. Environ. Microbiol. 66:860-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, C., and P. M. Muriana. 1994. Incidence of Listeria monocytogenes in packages of retail franks. J. Food Prot. 57:382-386. [DOI] [PubMed] [Google Scholar]
- 32.Way, S. S., L. J. Thompson, J. E. Lopes, A. M. Hajjar, T. R. Kollmann, N. E. Freitag, and C. B. Wilson. 2004. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell Microbiol. 6:235-242. [DOI] [PubMed] [Google Scholar]
- 33.Williams, T., B. Joseph, D. Beier, W. Goebel, and M. Kuhn. 2005. Response regulator DegU of Listeria monocytogenes regulates the expression of flagella-specific genes. FEMS Microbiol. Lett. 252:287-298. [DOI] [PubMed] [Google Scholar]
- 34.Wong, A. C. 1998. Biofilms in food processing environments. J. Dairy Sci. 81:2765-2770. [DOI] [PubMed] [Google Scholar]