Abstract
Genome-wide analysis of the wine yeast strain Saccharomyces cerevisiae PYCC4072 identified 36 genes highly expressed under conditions of low or absent nitrogen in comparison with a nitrogen-replete condition. Reverse transcription-PCR analysis for four of these transcripts with this strain and its validation with another wine yeast strain underlines the usefulness of these signature genes for predicting nitrogen deficiency and therefore the diagnosis of wine stuck/sluggish fermentations.
Nitrogen deficiency has been associated with major problems encountered in contemporary wine making, especially those related to slow (sluggish) and incomplete (stuck) fermentations (2, 3, 10, 11, 19). Under wine-making conditions, initial low levels of nitrogen act by limiting growth and biomass, resulting in a reduced fermentation rate (14, 23). Until completing alcoholic fermentation, the fermenting juice is at risk of spoilage due to oxidation and microbial activity, which can reduce the quality and thus the commercial value of the final product. A few systematic studies have been done to identify changes in gene expression that take place in response to nitrogen deficiency under enological conditions. From these studies, some genes, such as CAR1 (5) and ACA1 (9), were indicated as being more strongly expressed with nitrogen limitation.
Genome-wide expression analysis has also emerged as a powerful tool that can be used for identification of signature genes that behave in a similar fashion at a particular time point or under particular conditions (13). It has been successfully used in the identification of predictive biomarkers for clinical diagnosis (17, 20, 22). An analogous approach has been recently used with Saccharomyces cerevisiae for identifying CO2-responsive genes (1) and macronutrient (4, 21) and micronutrient (8) limitation under laboratory growth conditions. In a previous study, we used a genome-wide approach integrating the different situations of nitrogen supply: (i) with enough nitrogen to complete sugar fermentation, (ii) with nitrogen-limiting fermentation, and (iii) with addition of nitrogen to the nitrogen-deficient fermentation (15). In our previous study (15), samples for DNA macroarrays were taken from 11 points corresponding to different stages of the three fermentations, combining low and/or high concentrations of glucose, nitrogen, and ethanol (Table 1). The public data set, submitted to the GEO data repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE5842 (15), contains hybridization values for all time point replicates. Normalized final values are available from our website (http://scsie.uv.es/chipsdna/).
TABLE 1.
Fermentation parameters evaluated during experiments carried out with synthetic grape juice medium with 20% glucose and different initial nitrogen concentrationsa
| Expt | Time (h) | Glucose concn (g liter−1) | Nitrogen concn (mg liter−1) |
|---|---|---|---|
| CF | 0 | 197.8 ± 2.4 | 261.2 ± 6.2 |
| 24 | 183.8 ± 0.6 | 178.1 ± 6.4 | |
| 48 | 136.1 ± 0.3 | 2.4 ± 2.2 | |
| 96 | 24.7 ± 4.0 | 0.0 ± 0.0 | |
| LN | 0 | 197.6 ± 2.0 | 66.5 ± 2.2 |
| 24 | 189.1 ± 0.7 | 2.3 ± 0.2 | |
| 48 | 171.5 ± 5.7 | 0.0 ± 0.0 | |
| 80 | 154.1 ± 4.4 | 0.0 ± 0.0 | |
| 96 | 148.3 ± 4.3 | 0.0 ± 0.0 | |
| 144 | 137.7 ± 3.7 | 0.0 ± 0.0 | |
| RF | 72 | 164.9 ± 4.2 | 196.4 ± 1.4 |
| 80 | 148.2 ± 1.7 | 136.5 ± 3.9 | |
| 96 | 116.6 ± 0.3 | 0.8 ± 1.4 | |
| 144 | 24.9 ± 10.8 | 0.0 ± 0.0 |
CF, control fermentation (267 mg N liter−1); LN, low-nitrogen fermentation (66 mg N liter−1); RF, refed fermentation (66 + 200 mg N liter−1 supplied at 72 h as diammonium phosphate). Time points previously selected for macroarray analysis are in bold. Values represent means ± standard deviations for three independent experiments.
In the present study, the main goal was to identify genes that showed robust changes in their expression levels specifically associated with nitrogen deficiency and that could be potential candidates as biomarkers for predicting sluggish or stuck fermentations. For this purpose we used the DNA macroarray data obtained with the wine yeast strain S. cerevisiae PYCC4072 and compared gene expression levels between the nitrogen-replete condition and various defined situations of low nitrogen and N starvation, irrespective of the glucose availability, ethanol production, or other variations than can occur during vinification. Based on the data presented in Table 1, two time points, CF48 (control fermentation at 48 h) and LN24 (low-nitrogen fermentation at 24 h), were selected for the low-nitrogen conditions and five time points, CF96, LN48, LN80, LN96, and LN144, were chosen for the N starvation conditions. Pairwise comparisons were done using sample CF24 as the reference, since at that stage nitrogen was still abundant (178 mg liter−1). To estimate significantly differentially expressed genes in pairwise comparisons, a z-test was applied, and false-discovery-rate analysis was the method used for false-positive error correction.
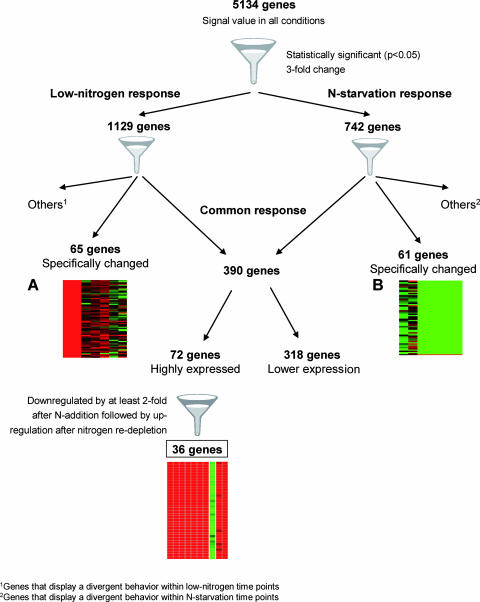
To select genes that displayed a consistent change in expression in the different nitrogen situations, the expression profiles were filtered as schematically presented in Fig. 1. First, only those genes with signal values in all time points selected above were considered (5,134 genes). Second, two selection criteria were cumulatively applied: the expression level had to change (i) significantly (P < 0.05) and (ii) by at least threefold compared to that of the reference sample under low-nitrogen and/or N starvation conditions. Genes displaying opposite changes in expression behavior within each of the two conditions were discarded. Genes were grouped into three sets: (i) those with significant changes at both time points only with a low extracellular nitrogen level (LN24 and CF48) (low-nitrogen response); (ii) those that showed significant changes in expression only between CF96, LN48, LN80, LN96, and LN144 and the reference cultures (CF24) (N starvation response); and (iii) those that displayed significant changes in expression under both low-nitrogen and N starvation conditions (common response).
FIG. 1.
Overview of gene selection criteria. Transcript profiles of genes that specifically respond to low nitrogen (A) and to N starvation (B) under alcoholic fermentation conditions; results, log2 expression ratios obtained by dividing the experimental results by the reference sample results, are represented with a green-to-red color scale. Each expression diagram shows, from top to bottom, relative expression levels of each set of genes and, from left to right, comparisons between LN24, CF48, CF96, LN48, LN80, LN96, and LN144 and the reference cultures, CF24. Down-regulated genes are green, whereas up-regulated genes are red.
Sixty-five genes were identified that specifically changed in response to low nitrogen, and all were up-regulated (see Fig. S1 in the supplemental material), while 61 genes were specifically reset in response to nitrogen starvation. In this last set, only CUP1-2, involved in resistance to high concentrations of copper, had higher expression under the nitrogen-deprived conditions (see Fig. S1 in the supplemental material). Indeed, this gene has already been reported to be one of the highly expressed genes in the late stationary phase during alcoholic fermentation (24). A total of 390 genes were found to be significantly affected under both low-nitrogen and N starvation conditions. Among those genes, 72 genes had consistently higher expression (see Table S2 in the supplemental material) while 318 had lower expression (see Table S3 in the supplemental material) under all nitrogen deprivation conditions relative to the reference situation. Results for these genes were compared with those for the environmental stress response (ESR) family genes obtained by evaluating the transcriptional responses with a wide range of stress stimuli, including nitrogen depletion (6). It was found that 27 of the 72 up-regulated genes (P value, 1 × 10−18) and 128 of the 318 down-regulated genes (P value, 3 × 10−53) are among the ESR genes, indicating that the yeast cell response to nitrogen restriction under vinification conditions involves the ESR.
To identify potential candidate biomarker genes for predicting nitrogen shortage during alcoholic fermentation, analyses were restricted to the 72 up-regulated genes, and their responses were examined after nitrogen refeeding. Only those genes whose expression decreased by at least twofold after nitrogen addition (RF80 [refed fermentation at 80 h]/LN48) and increased again at least threefold when nitrogen became depleted (RF96/RF80 and RF144/RF80) were considered. Thirty-six genes passed all rather stringent criteria applied, suggesting that they could be promising candidates for predicting nitrogen deficiency during alcoholic fermentation (Table 2). The remaining genes could be involved in nonspecific responses to nitrogen limitation and associated with a more general response, namely, those responding to other intracellular and/or environmental changes that occur under nitrogen deficiency conditions. Further research is required to clarify this aspect. Several interesting aspects of this set of candidate genes should be highlighted (Table 2). Half of these genes have been identified as being ESR upregulated (6), and thus, their usefulness for the specific diagnosis of the nitrogen deficiency response could be limited. A large number of genes (50%) have no known molecular function. Determining their physiological role could give further insights into their contributions to the yeast response to nitrogen deficiency. Twelve genes are among the 127 genes induced in cells in stationary-phase cultures (P value, 3 × 10−12). Two of them, HBT1 and FMP45, are considered essential for long-term survival in stationary phase (12). Genes whose expression is known to be under glucose repression, such as IDP2, XYL2, and the transcription factor CAT8, involved in the derepression of a number of genes during the diauxic shift (7), are part of this restricted group of genes, despite the high glucose levels present at all time points. Furthermore, 14 genes of our signature group were found to be specifically more highly expressed under carbon limitation (4, 21). This result supports the previous suggestion that the yeast cell responses to nitrogen limitation/starvation observed in this study have similarities to the yeast cell response to glucose limitation (15). In addition, two other transcription factors, XBP1, whose transcriptional activation has been considered a key response of yeast cells to nitrogen limitation (16), and CUP2, involved in copper-responsive transcriptional regulation of the metallothionein genes CUP1-1 and CUP1-2, are part of the candidate genes identified in the current study. The high mRNA levels of CUP1-2 observed under N starvation indicate that maintenance of copper homeostasis in yeast cells could be decisive for yeast cell survival under such conditions. Experimental identification of the set of genes that are regulated by each one of these transcription factors, either by transcriptional profiling of knockout mutants or by overexpression of the various factors, will be necessary to access their functions in the regulation of nitrogen availability responses. Finally, seven of the proposed candidate genes (P value, 1 × 10−8) were formerly included in the top 50 open reading frames induced by ammonium starvation (26). The finding of an overlap between the current set of data, corresponding to a wine yeast strain grown under batch fermentation conditions, and that from Wu et al. (26), obtained with a laboratory S. cerevisiae strain in chemostat cultures, supports the accuracy of the approach considered herein. Despite the overlap shown in Table 2, it is worth mentioning that this study introduced seven genes for which no previous relationship had been established with ESR (6), ammonium starvation (26), nitrogen and carbon limitation (4, 21), or stationary phase (12) but which are induced under all of the conditions of nitrogen limitation and starvation considered in this work.
TABLE 2.
Thirty-six signature genes identified in this work as potential candidates for predicting nitrogen deficiency under wine-making conditions and their overlapping with other reported conditionsa
| ORF name | Gene name | Association with:
|
Reference(s) | ||||
|---|---|---|---|---|---|---|---|
| Ammonium starvation | Nitrogen limitation | Carbon limitation | ESR | Stationary phase | |||
| YNL270c | ALP1 | x | 4, 21 | ||||
| YMR280c | CAT8 | x | 4, 21 | ||||
| YGL166w | CUP2 | ||||||
| YOR180c | DCI1 | x | 4, 21 | ||||
| YKR076w | ECM4 | x | x | x | 4, 6, 21 | ||
| YPL222w | FMP40 | ||||||
| YDL222c | FMP45 | x | x | 6, 12 | |||
| YMR250w | GAD1 | x | x | x | 6, 12, 21 | ||
| YDL223c | HBT1 | x | x | 4, 12, 21 | |||
| YOR391c | HSP33 | x | 26 | ||||
| YLR174w | IDP2 | x | x | x | 4, 12, 21, 26 | ||
| YML128c | MSC1 | x | x | x | x | 4, 6, 12, 21, 26 | |
| YPL134c | ODC1 | x | x | x | 4, 12, 21, 26 | ||
| YDR313c | PIB1 | ||||||
| YDL204w | RTN2 | x | x | x | x | 4, 6, 12, 21, 26 | |
| YIL113w | SDP1 | x | 6 | ||||
| YDR238c | SEC26 | ||||||
| YMR175w | SIP18 | x | x | 4, 12, 21 | |||
| YGL208w | SIP2 | x | x | 6, 21 | |||
| YGR248w | SOL4 | x | 6 | ||||
| YBL106c | SRO77 | ||||||
| YLR178c | TFS1 | x | 6 | ||||
| YBR006w | UGA2 | x | 6 | ||||
| YIL101c | XBP1 | x | x | 6, 12 | |||
| YLR070c | XYL2 | ||||||
| YCR061w | YCR061w | x | 6 | ||||
| YLR272c | YCS4 | ||||||
| YDL218w | YDL218w | x | x | 12, 26 | |||
| YDR271c | YDR271c | ||||||
| YGR043c | YGR043c | x | x | x | 4, 6, 12, 21 | ||
| YLR312c | YLR312c | x | x | x | x | 6, 12, 21, 26 | |
| YMR090w | YMR090w | x | x | 6, 21 | |||
| YMR206w | YMR206w | x | 4 | ||||
| YNL115c | YNL115c | x | 6 | ||||
| YNL194c | YNL194c | x | x | 4, 6, 21 | |||
| YNL195c | YNL195c | x | x | 4, 6, 21 | |||
See included references. x, gene expression is induced.
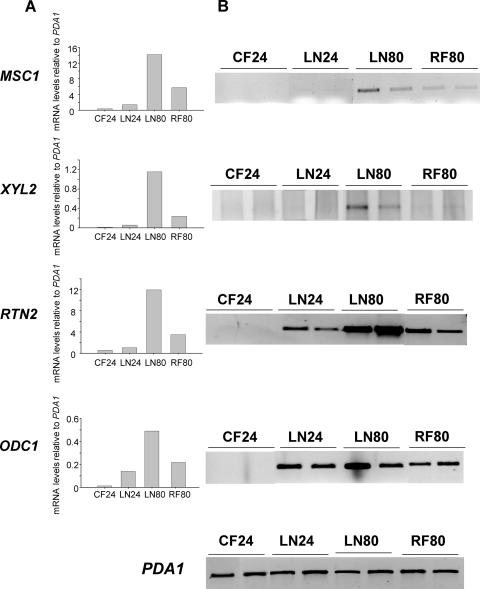
In order to validate the differential expression of candidate genes obtained by macroarray analysis, semiquantitative reverse transcription-PCR (RT-PCR) was performed using the same RNA from the original experiments with S. cerevisiae PYCC4072 according to the protocol described by Zuzuarregui et al. (27). Table 3 includes the sequences of the oligonucleotides used in these amplification reactions, the number of cycles, and the hybridization temperature. The PDA1 gene was used for normalization of the data, since it shows a constitutive expression in batch and chemostat cultures in the presence of various carbon sources (25). Four up-regulated genes (MSC1, XYL2, RTN2, and ODC1) were selected from the candidate gene list for this purpose (Fig. 2). The expression levels of RTN2 and ODC1 in both RT-PCR and macroarray analysis showed an increase under nitrogen limitation and especially starvation. Also, for the MSC1 and XYL2 genes, the pattern obtained by RT-PCR supported the macroarray results; the mRNA levels were almost undetectable in the reference sample (CF24), and the highest mRNA abundance was shown at time LN80, under starvation conditions, decreasing after nitrogen addition to a greater degree in the case of XYL2. RTN2 and MSC1 have been described as part of the ESR (6). On the other hand, these two genes together with ODC1 were included in the top 50 open reading frames induced by ammonium starvation in S. cerevisiae grown in chemostat culture (21). The validation of these genes reinforces the consistency of the results obtained in this study and shows that some relationship can be found between growth under vinification and laboratory conditions. Besides, this analysis allows the addition of several genes, including XYL2, to the list of those previously proposed to be induced by nitrogen deficiency (4, 21).
TABLE 3.
Gene-specific primers for semiquantitative RT-PCR assays
| Gene | Primer 1 | Primer 2 | Temp (°C) | No. of cycles |
|---|---|---|---|---|
| MSC1 | TAATGCGGTTTCCGCAT | TAGCTCGTCCTTGCTTT | 50 | 25 |
| XYL2 | GCCCTCAATGATCGCTTGA | TGACTTAACTACACAAGA | 45 | 22 |
| RTN2 | TATTGCCATTGGCCTT | CAAACCAACCGCATTGTT | 50 | 22 |
| ODC1 | TATACCAGTTCACAGCC | AATCCATGACGTTCGTG | 55 | 22 |
| PDA1 | GCTTCATTCAAACGCCAACC | TCCCTAGAGGCAAAACCTTG | 45 | 22 |
FIG. 2.
Validation of macroarray data obtained with S. cerevisiae PYCC4072 by semiquantitative RT-PCR. Panel A shows a histogram corresponding to the results obtained with macroarray analysis, relative to PDA1 results. Panel B contains the result of the semiquantitative RT-PCR analysis carried out with samples from two independent cultures.
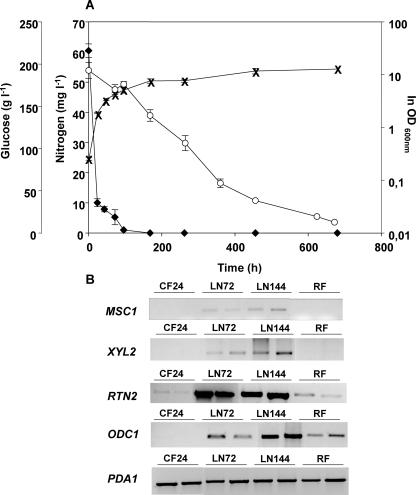
To assess whether the results obtained for the wine yeast strain S. cerevisiae PYCC4072 could be extrapolated to other wine strains and other fermentation conditions, we studied the transcriptional response of the same four genes with another commercial wine yeast strain. The experiments using S. cerevisiae strain ICV16 (Fermicru primeur, DSM) were performed with synthetic grape juice (18) containing 300 mg liter−1 (MS300) or 60 mg liter−1 (MS60) of assimilable nitrogen as a mixture of ammonium and amino acids in a proportion of 2:3, respectively, following the experimental details described by Jiménez-Martí et al. (9). Nitrogen was added (240 mg liter−1 of ammonium) to the MS60 experiment 72 h after inoculation, when total assimilable nitrogen was almost completely consumed (5.25 mg liter−1 of nitrogen remained in the extracellular medium at this time point). In these experiments, active dry yeasts were rehydrated in water according to the manufacturer's instructions and used for inoculation to a final count of 5 × 106 cells ml−1, as determined by total cell counts. The results shown in Fig. 3 further confirmed that the selected genes are also applicable to other wine strains irrespective of growth medium composition and fermentation conditions.
FIG. 3.
Analysis of some of the candidate genes in S. cerevisiae strain ICV 16 (Fermicru primeur, DSM). Panel A shows the changes in extracellular nitrogen concentrations (⧫) and yeast cell growth (×) versus glucose consumption (○) during fermentation with an initial assimilable-nitrogen concentration of 60 mg liter−1. Data presented are representative of at least three independent experiments. Panel B contains the results obtained by RT-PCR. CF24, sample from control fermentation carried out with 300 mg N/liter at 24 h after inoculation, when the amount of assimilable nitrogen remaining in the must was 260 mg liter−1 (9); LN72 and LN144, samples obtained at 72 and 144 h, respectively, from the nitrogen-limiting fermentation; RF, sample obtained 9 h after addition of nitrogen to the limiting vinification. OD, optical density.
It is worth mentioning that in order to get a reasonable number of marker genes for this signature group, we have focused only on those with higher levels of expression under nitrogen deficiency conditions. A similar approach could be followed for the down-regulated genes. The presence in this group of several MET genes, essential for the assimilatory reduction pathway of sulfate to sulfide, is of great interest, and further studies are being carried out to understand their relevance in the wine-making process.
A designed DNA chip incorporating some of the genes identified in this study can be developed to assist the winemaker in assessing the nitrogen status of the fermenting grape juice. The ultimate goal is that such a chip can be used to predict premature fermentation arrest due to a nitrogen shortage and to customize treatment strategies. Nevertheless, it should be emphasized that these observations must be viewed as preliminary, and expansion to other yeast strains is required before a specialized DNA chip for predictive diagnosis of this problem can be designed.
Supplementary Material
Acknowledgments
This work was carried out in the framework of project PTDC/AGR-ALI/71460/2006 (grant to A.M.-F.). This work was also supported by grants AGL2005-00508 and BMC2003-07072-C03-02 from the Spanish Ministerio de Educación y Ciencia to M.O. and to J.E.P.-O., respectively. Elena Jiménez-Martí is an FPI Fellow for the Spanish Government.
We thank R. N. Bennett for English revision of the manuscript.
Footnotes
Published ahead of print on 29 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aguilera, J., T. Petit, J. H. de Winde, and J. T. Pronk. 2005. Physiological and genome-wide transcriptional responses of Saccharomyces cerevisiae to high carbon dioxide concentrations. FEMS Yeast Res. 5:579-593. [DOI] [PubMed] [Google Scholar]
- 2.Bisson, L. F. 1991. Influence of nitrogen on yeast and fermentation of grapes, p. 78-89. In J. M. Rantz (ed.), Proceedings of the International Symposium on Nitrogen in Grapes and Wine. American Society for Enology and Viticulture, Davis, CA.
- 3.Bisson, L. F. 1999. Stuck and sluggish fermentations. Am. J. Enol. Vitic. 50:107-119. [Google Scholar]
- 4.Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265-3274. [DOI] [PubMed] [Google Scholar]
- 5.Carrasco, P., J. E. Pérez-Ortín, and M. del Olmo. 2003. Arginase activity is a useful marker of nitrogen limitation during alcoholic fermentations. Syst. Appl. Microbiol. 26:471-479. [DOI] [PubMed] [Google Scholar]
- 6.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedges, D., M. Proft, and K. D. Entian. 1995. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15:1915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins, V. J., P. J. Rogers, and I. W. Dawes. 2003. Application of genome-wide expression analysis to identify molecular markers useful in monitoring industrial fermentations. Appl. Environ. Microbiol. 69:7535-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiménez-Martí, E., A. Aranda, A. Mendes-Ferreira, A. Mendes-Faia, and M. del Olmo. 2007. The nature of the nitrogen source added to nitrogen depleted vinifications conducted by a Saccharomyces cerevisiae strain in synthetic must affects gene expression and the levels of several volatile compounds. Antonie Leeuwenhoek 92:61-75. [DOI] [PubMed] [Google Scholar]
- 10.Kunkee, R. E. 1991. Relationship between nitrogen content in must and sluggish fermentation, p. 148-155. In J. M. Rantz (ed.), Proceedings of the International Symposium on Nitrogen in Grapes and Wine. American Society for Enology and Viticulture, Davis, CA.
- 11.Manginot, C., J. L. Roustan, and J. M. Sablayrolles. 1998. Nitrogen demand of different yeast strains during alcoholic fermentation. Importance of the stationary phase. Enzyme Microb. Technol. 23:511-517. [Google Scholar]
- 12.Martinez, M. J., S. Roy, A. B. Archuletta, P. D. Wentzell, S. S. Anna-Arriola, A. L. Rodriguez, A. D. Aragon, G. A. Quinones, C. Allen, and M. Werner-Washburne. 2004. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol. Biol. Cell 15:5295-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martoglio, A. M., J. W. Miskin, S. K. Smith, and D. J. MacKay. 2002. A decomposition model to track gene expression signatures: preview on observer-independent classification of ovarian cancer. Bioinformatics 18:1617-1624. [DOI] [PubMed] [Google Scholar]
- 14.Mendes-Ferreira, A., A. Mendes-Faia, and C. Leão. 2004. Growth and fermentation patterns of Saccharomyces cerevisiae under different ammonium concentrations and its implications in winemaking industry. J. Appl. Microbiol. 97:540-545. [DOI] [PubMed] [Google Scholar]
- 15.Mendes-Ferreira, A., M. del Olmo, J. Garcia-Martinez, E. Jiménez-Martí, A. Mendes-Faia, J. Perez-Ortin, and C. Leão. 2007. Transcriptional response of Saccharomyces cerevisiae to different nitrogen concentrations during alcoholic fermentation. Appl. Environ. Microbiol. 73:3049-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miled, C., C. Mann, and G. Faye. 2001. Xbp1-mediated repression of CLB gene expression contributes to the modifications of yeast cell morphology and cell cycle seen during nitrogen-limited growth. Mol. Cell. Biol. 21:3714-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modena, P., E. Lualdi, F. Facchinetti, J. Veltman, J. F. Reid, S. Minardi, I. Janssen, F. Giangaspero, M. Forni, G. Finocchiaro, L. Genitori, F. Giordano, R. Riccardi, E. F. Schoenmakers, M. Massimino, and G. Sozzi. 2006. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J. Clin. Oncol. 24:5223-5233. [DOI] [PubMed] [Google Scholar]
- 18.Riou, C., J. M. Nicaud, P. Barre, and C. Gaillardin. 1997. Stationary-phase gene expression in Saccharomyces cerevisiae during wine fermentation. Yeast 13:903-915. [DOI] [PubMed] [Google Scholar]
- 19.Salmon, J. M. 1989. Effect of sugar transportation inactivation in Saccharomyces cerevisiae on sluggish and stuck enological fermentations. Appl. Environ. Microbiol. 55:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorlie, T., C. M. Perou, R. Tibshirani, T. Aas, S. Geisler, H. Johnsen, T. Hastie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, T. Thorsen, H. Quist, J. C. Matese, P. O. Brown, D. Botstein, P. Eystein Lonning, and A. L. Borresen-Dale. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 98:10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai, S. L., V. M. Boer, P. Daran-Lapujade, M. C. Walsh, J. H. de Winde, J. M. Daran, and J. T. Pronk. 2005. Two-dimensional transcriptome analysis in chemostat cultures. Combinatorial effects of oxygen availability and macronutrient limitation in Saccharomyces cerevisiae. J. Biol. Chem. 280:437-447. [DOI] [PubMed] [Google Scholar]
- 22.van de Vijver, M., Y. He, L. van't Veer, H. Dai, A. Hart, D. Voskuil, G. Schreiber, J. Peterse, C. Roberts, M. Marton, M. Parrish, D. Atsma, A. Witteveen, A. Glas, L. Delahaye, T. van der Velde, H. Bartelink, S. Rodenhuis, E. Rutgers, S. Friend, and R. Bernards. 2002. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347:1999-2009. [DOI] [PubMed] [Google Scholar]
- 23.Varela, C., F. Pizarro, and E. Agosin. 2004. Biomass content governs fermentation rate in nitrogen-deficient wine musts. Appl. Environ. Microbiol. 70:3392-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varela, C., J. Cárdenas, F. Melo, and E. Agosin. 2005. Quantitative analysis of wine yeast gene expression profiles under winemaking conditions. Yeast 22:369-383. [DOI] [PubMed] [Google Scholar]
- 25.Wenzel, T. J., M. A. Luttik, J. A. van den Berg, and H. Y. de Steensma. 1993. Regulation of the PDA1 gene encoding the E1 alpha subunit of the pyruvate dehydrogenase complex from Saccharomyces cerevisiae. Eur. J. Biochem. 218:405-411. [DOI] [PubMed] [Google Scholar]
- 26.Wu, J., N. Zhang, A. Hayes, K. Panoutsopoulou, and S. G. Oliver. 2004. Global analysis of nutrient control of gene expression in Saccharomyces cerevisiae during growth and starvation. Proc. Natl. Acad. Sci. USA 101:3148-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuzuarregui, A., L. Monteoliva, C. Gil, and M. del Olmo. 2006. Transcriptomic and proteomic approach for understanding the molecular basis of adaptation of Saccharomyces cerevisiae to wine fermentation. Appl. Environ. Microbiol. 72:836-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.