Abstract
Wild-type Bacillus subtilis ferments 20 g/liter glucose in 48 h, producing lactate and butanediol, but not ethanol or acetate. To construct an ethanologenic B. subtilis strain, homologous recombination was used to disrupt the native lactate dehydrogenase (LDH) gene (ldh) by chromosomal insertion of the Zymomonas mobilis pyruvate decarboxylase gene (pdc) and alcohol dehydrogenase II gene (adhB) under the control of the ldh native promoter. The values of the intracellular PDC and ADHII enzymatic activities of the engineered B. subtilis BS35 strain were similar to those found in an ethanologenic Escherichia coli strain. BS35 produced ethanol and butanediol; however, the cell growth and glucose consumption rates were reduced by 70 and 65%, respectively, in comparison to those in the progenitor strain. To eliminate butanediol production, the acetolactate synthase gene (alsS) was inactivated. In the BS36 strain (BS35 ΔalsS), ethanol production was enhanced, with a high yield (89% of the theoretical); however, the cell growth and glucose consumption rates remained low. Interestingly, kinetic characterization of LDH from B. subtilis showed that it is able to oxidize NADH and NADPH. The expression of the transhydrogenase encoded by udhA from E. coli allowed a partial recovery of the cell growth rate and an early onset of ethanol production. Beyond pyruvate-to-lactate conversion and NADH oxidation, an additional key physiological role of LDH for glucose consumption under fermentative conditions is suggested. Long-term cultivation showed that 8.9 g/liter of ethanol can be obtained using strain BS37 (BS35 ΔalsS udhA+). As far as we know, this is the highest ethanol titer and yield reported with a B. subtilis strain.
Ethanol production from renewable resources, such as biomass, is a promising alternative to compete with and eventually replace nonrenewable fossil fuels; it has considerable advantages in terms of sustainability, lower greenhouse gas emissions, and cost reduction (25). Currently, fuel ethanol is produced by using Saccharomyces cerevisiae that ferments sucrose from sugarcane or glucose from hydrolyzed cornstarch. Nonetheless, large amounts of sugars are available in plant biomass. This biomass is a complex mixture of carbohydrate polymers, mainly cellulose and hemicellulose. Cellulose utilization requires a previous enzymatic breakdown to release glucose; however, the use of cellulases raises the cost of ethanol production (18, 22, 25).
Gram-positive bacteria have several traits, such as the capacity to withstand relatively low pH, high temperature, high sugar, salt and ethanol concentrations, and various other harsh conditions, which could be used to develop an advanced biocatalyst and improve the commercial competitiveness of fuel ethanol production (9, 17). Several groups have tried to develop ethanologenic gram-positive bacteria, but with limited success (2, 15, 19, 21, 27, 32).
Different chimerical pet operons have been constructed by employing pdc and adhB genes from Zymomonas mobilis (gram-negative bacterium) or by using the pdc gene from Sarcina ventriculy (gram-positive bacterium) for expression in monocopy or multicopy vectors in gram-positive bacteria. In some of these cases, functional pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH) enzymes have been detected, although the ethanol yields have been low or nonexistent (19, 21, 27, 32).
Bacillus subtilis has a generally-recognized-as-safe status (29), metabolizes an extensive range of sugars (31), and can efficiently synthesize a wide variety of proteinases and transport them out of the cell using secretion systems. This capacity can potentially be used to export cellulases, which could break down plant wastes, releasing cellobiose and glucose. Hence, ethanologenic B. subtilis strains engineered in the future might consume both carbohydrates to produce ethanol. This design could help in reducing fuel ethanol production cost. However, under fermentative conditions, B. subtilis produces lactate, acetate, butanediol, and traces of ethanol from glucose, amino acids, and pyruvate (Fig. 1) (7). Lactate is produced by the reduction of pyruvate, a reaction catalyzed by lactate dehydrogenase (LDH), with the simultaneous oxidation of one molecule of NADH per molecule of pyruvate reduced (7). In this study, the heterofermentative metabolism of B. subtilis was modified to obtain a gram-positive strain that produces ethanol as the main fermentation product. By inactivating the lactate dehydrogenase gene (ldh) via chromosomal integration of the Z. mobilis pdc and adhB genes, also eliminating butanediol synthesis, and by inserting the E. coli udhA transhydrogenase in the alsS locus, the resulting recombinant B. subtilis strain produced ethanol as the sole fermentation product.
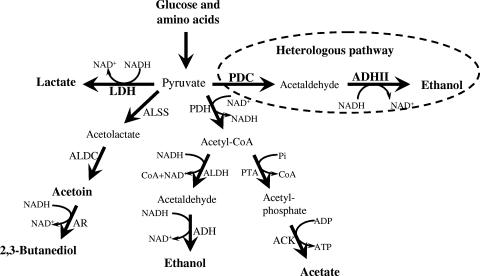
FIG. 1.
Bacillus subtilis fermentation pathways. The heterologous pathway used in this study is highlighted with a dashed-line ellipse. PDC and ADHII are from Z. mobilis. ALDC, acetolactate decarboxylase; AR, acetoin reductase; PDH, pyruvate dehydrogenase; CoA, coenzyme A; ALDH, acetaldehyde dehydrogenase; PTA, phosphotransacetylase; ACK, acetate kinase. (Modified from references 7 and 26 with permission.)
LDH inactivation caused a B. subtilis growth defect in spite of complementation with a stoichiometrically equivalent redox pathway (PDC-ADHII). We found that LDH oxidizes NADH to NAD+ and NADPH to NADP+. The apparent Km value for both cofactors is comparable to Km values of LDH for NADH of other gram-positive bacteria (14). Hence, an additional key physiological role of LDH for glucose consumption under fermentative conditions is suggested. The results presented in this work clearly indicate that this key physiological role of LDH for glucose consumption under fermentative conditions should be considered in the development of new ethanologenic B. subtilis strains.
MATERIALS AND METHODS
Strains and plasmids.
The B. subtilis strains and plasmids used in this study are listed in Table 1, and the oligonucleotides in Table 2. To avoid the Trp requirement, competent B. subtilis WB700 cells (35) were transformed by using chromosomal DNA from a Trp+ B. subtilis BSR1 strain (20), recombinants were selected in mineral medium containing glucose, and the resulting prototrophic strain was designated CH1 (Table 1). B. subtilis CH1 was used to construct ethanologenic B. subtillis strains. During strain and plasmid construction, strains were grown in Luria-Bertani (LB) agar (23) containing appropriate antibiotics. The final antibiotic concentrations were as follows: 5 μg/ml chloramphenicol (Cm), 5 μg/ml erythromycin (Ery), 5 μg/ml lincomycin (Lin), 100 μg/ml spectinomycin (Spt), 10 μg/ml tetracycline (Tc), 50 μg/ml kanamycin (Km), and 100 μg/ml ampicillin (Ap).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| B. subtilis WB700 | B. subtilis 168 trpC2 ΔnprE ΔaprE Δepr Δbpf Δmpr ΔnprB ΔvrpE Eryr Linr | 35 |
| B. subtilis CH1 | WB700 prototroph | This study |
| B. subtilis CH1 ΔalsS mutant | CH1 als::Spt | This study |
| B. subtilis BS35 | CH1 ldh::pdc-adhB Cmr | This study |
| B. subtilis BS36 | CH1 ldh::pdc-adhB Cmrals::Spt | This study |
| B. subtilis BS37 | CH1 ldh::pdc-adhB Cmrals::udhA_Spt | This study |
| Plasmids | ||
| pUC19 | pMB1 ori Apr | BioLabs |
| pCR-Blunt II-TOPO | pBR322 ori KmrccdB selection | Invitrogen |
| pLOI276 | pUC19 carrying pdcZm | 5 |
| pLOI284 | pUC19 carrying adhBZm | 6 |
| IpTOPO-pdc | pCR carrying pdcZm | This study |
| pTOPO-adh-term | pCR carrying adhBZm-cryIIIAt | This study |
| pTOPO-pdc-adh-term | pCR carrying pdcZm, adhBZm, and Cmr | This study |
| pSG-PLK | pBR322 ori Apr | 20 |
| pSG-operon | pSG-PLK carrying pdcZm, adhBZm, and Cmr | This study |
| pUC-ldh | pUC19 carrying ldhBs | This study |
| pUC-ldh::operonCm | pUC19 carrying ldhBs::(pdcZmadhBZm) | This study |
| pTOPO-alsS | pCR carrying alsS | This study |
| pCm::Spt | pIC177 carrying Cmr::Sptr | 30 |
| pTOPO-alsS::Spt | pCR carrying alsS::Sptr | This study |
| pTOPO-alsS::udhA_Spt | pCR carrying alsS::(udhAEc Sptr) | This study |
Zm, Z. mobilis; Bs, B. subtilis; Ec, E. coli.
TABLE 2.
Oligonucleotides used in this study
| Primer | Target gene | Sequence (5′→3′) |
|---|---|---|
| 1 | Z. mobilis pdc | GTC GGC CGG CCA AGG AGG AGT AAG CAA TGA GTT ATA CTG TCG G |
| 2 | Z. mobilis pdc | CTT GGC GCG CCT TAC GGC TGT TGG CGG GCA GC |
| 3 | Z. mobilis adhB | TAA GGC GCG CCA AGG AGG GTA TAG CTA TGG CTT CTT CAA CTT TTT ATA TTC |
| 4 | Z. mobilis adhB | TCC GGT CAA TTG GAG TGA TGT CCG TTT TCC TGT TTT GAA ATT AG |
| 5 | B. thuringiensis cryIIIAt | AAA AGA ATT CAA AAA AAA ACG GAC ATC ACT CCG GTC AAT TGG AGT G |
| 6 | B. subtilis ldh | CCG CGG ATC CAA GGA GGG ATG ATT AAT GAT GAA CAA ACA TGT A |
| 7 | B. subtilis ldh | GCC CGA ATT CAC TCT AAA GTT GCG GTT AGT TGA C |
| 8 | B. subtilis alsS | GTG TCA CAC ATG TAA TTG GCA TTC C |
| 9 | B. subtilis alsS | GGC TGA GCA CTT AAA TGT TGC TTT C |
| 10 | E. coli udhA | GCC GGT CGA CAA GGA GGA CCC TAC CAT GCC ACA TTC C |
| 11 | E. coli udhA | GCC GGT CGA CTT AAA ACA GGC GGT TTA AAC CG |
Construction of ethanologenic B. subtilis strains. (i) Genetic procedures.
Standard procedures were used for plasmid preparation, restriction enzyme digestions, ligations, transformations, and agarose gel electrophoresis (23). E. coli XL1-Blue was used as the host for plasmid constructions, and cells were grown in LB medium. B. subtilis was transformed by using the natural competence method (8). Chromosomal integrations and deletions were confirmed by using appropriate antibiotic markers and PCR analysis and by the examination of fermentation products.
(ii) Inactivation of ldh and chromosomal integration of Z. mobilis pdc and adhB genes.
Specific primers were designed to amplify Z. mobilis pdc and adhB genes (Table 2). The primer design included the B. subtilis Shine-Dalgarno consensus sequence and also the conserved distance between this motif and the start codon of each gene. The Z. mobilis pdc gene was amplified from vector pLOI276, using primers 1 and 2. The 1.7-kb PCR-amplified segment was cloned into pCR-TOPO, obtaining the vector IpTOPO-pdc.
The Z. mobilis adhB gene, followed by the terminator of the Bacillus thuringiensis cryIIIA gene (cryIIIAt), was amplified (34). In the first PCR, using primers 3 and 4, the adhB gene was amplified from vector pLOI284, including half of cryIIIAt; in the second PCR, using primers 3 and 5, the adhB gene, followed by the complete terminator, was amplified; this PCR segment was cloned into pCR-TOPO, generating pTOPO-adh-term.
The pdc gene was purified as a 1.7-kbp EcoRV-AscI fragment from vector IpTOPO-pdc. This fragment was ligated into the blunt-treated BamHI and AscI sites upstream from the adhB gene in pTOPO-adh-term, obtaining the vector pTOPO-pdc-adh-term. The pdc and adhB genes were purified as a 2.88-kbp EcoRI-NaeI fragment from vector pTOPO-pdc-adh-term. This fragment was ligated into the EcoRI-HincII sites of vector pSG-PLK (20), obtaining the vector pSG-operon. This plasmid contained the pdc and adhB genes followed by the transcription terminator cryIIIAt and the chloramphenicol-resistant cassette (cat).
The ldh gene of B. subtilis was amplified by PCR, using primers 6 and 7, and cloned into the HincII site of plasmid pUC19 to construct pUC-ldh.
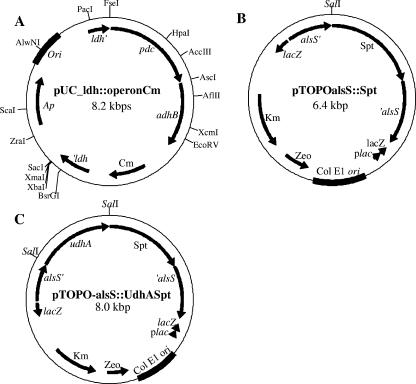
The promoterless pdc and adhB genes, cryIIIAt, and the cat gene were purified as a 4.5-kbp blunt-treated AccI fragment from vector pSG-operon and ligated into the EcoRV site of the ldh gene in pUC-ldh; the resulting plasmid was pUC-ldh::operonCm (Fig. 2A).
FIG. 2.
Vectors constructed to inactivate (A) the B. subtilis ldh gene (ldhBs) by chromosomal integration of the Z. mobilis pdc gene (pdcZm) and adhBZm, (B) alsSBs by chromosomal integration of the Spt cassette, and (C) alsSBs by chromosomal integration of E. coli udhA and the Spt cassette.
Competent B. subtilis CH1 cells were transformed by using the suicide vector pUC-ldh::operonCm; thereby, pdc and adhB genes were integrated by double-crossover homologous recombination in the ldh locus into the B. subtilis CH1 genome (8) under the control of the ldh promoter. B. subtilis recombinants were selected by Cmr and identified as being defective in lactate production under fermentative conditions. The resulting strain was named BS35.
(iii) alsS cloning and inactivation.
The alsS gene was amplified by PCR from the B. subtilis genome using primers 8 and 9. This segment was cloned into vector pCR-TOPO in order to construct pTOPO-alsS. A 1.2-kbp segment containing the Spt cassette was cut out by double digestion with BamHI-HindIII from vector pCm::Spt (30). After the ends were blunt-treated, the Spt cassette was ligated into the SspI site of the alsS gene of pTOPO-alsS, generating the pTOPO-alsS::Spt plasmid (Fig. 2B). The inactivation of the alsS gene, by the insertion of the Spt cassette contained in this plasmid, was transferred to the alsS locus of B. subtilis BS35 by a double homologous recombination event. alsS mutants were selected by Sptr and identified for the absence of 2,3-butanediol production under fermentative conditions. This new strain was designated BS36.
The CH1 alsS mutant strain was constructed to evaluate the apparent Km of LDH in the cellular extracts of the B. subtilis strain. Competent cells of strain CH1 were transformed with plasmid pTOPO-alsS::Spt. alsS mutants were selected by Sptr and identified for the absence of 2,3-butanediol production under fermentative conditions.
(iv) Inactivation of alsS by chromosomal integration of E. coli transhydrogenase gene (udhA).
The primer design to amplify the E. coli udhA gene included the same translational elements used in the amplification of the pdc and adhB genes. The PCR product containing the udhA gene, amplified from the E. coli JM101 genome using primers 10 and 11, was ligated into the unique SalI site of pTOPO-alsS::Spt. The resulting plasmid was pTOPO-alsS::UdhA_Spt (Fig. 2C). Competent B. subtilis BS35 cells were transformed with this plasmid; the udhA gene was thereby integrated by double homologous recombination in the alsS locus into the B. subtilis BS35 chromosome (8), allowing the expression of udhA under the control of the alsS promoter. Transformants were selected on LB-Spt agar plates. B. subtilis BS37 (BS35 ΔalsS udhA+) was confirmed by PCR amplification of the udhA gene from the alsS mutant's genome and identified as being defective in lactate and butanediol production under fermentative conditions.
Inoculum preparation and fermentation conditions.
LB-glucose 0.2% agar plates were inoculated with cells stored at −70°C in glycerol and incubated overnight at 37°C. These cells were used to inoculate 500-ml flasks containing 150 ml of LB broth with 20 g/liter glucose. The flasks were incubated overnight at 37°C and 100 rpm. Cells from the flasks were centrifuged and used as inocula at an initial optical density at 600 nm (OD600) of 0.1. Cultures were performed by duplication in minifleakers (3) containing 200 ml of LB supplemented with 20 g/liter glucose. The working conditions were 35°C, 100 rpm, and pH 7 (controlled by the automatic addition of 2 N KOH). The growth was monitored by measuring the OD600 with a spectrophotometer (Lambda 11; Perkin Elmer, Pomona, CA). Samples were periodically taken and centrifuged; supernatants were used for analytical determinations. Cells with an OD600 adjusted to 1 were collected at 48 h elapsed time of fermentation, centrifuged, and utilized for enzymatic activity assays.
Analytical methods.
Biomass measured by the OD600 was converted to dry cellular weight (DCW) using a standard curve (1 OD600 = 0.35 g DCW/liter). The PDC and ADHII enzymatic activities were assayed at 30°C following protocols previously described (5, 6). The LDH activity was assayed using pyruvate, NADH, and the buffers used for the PDC assay. The apparent Km was determined in cellular extracts of B. subtilis CH1 ΔalsS, from cells collected at 48 h elapsed time of fermentation. B. subtilis BS35 and BS36 were included as negative controls. For the apparent Km determination, different concentrations of NADH (0 to 0.15 mM) and NADPH (0 to 1.3 mM) were tested, with a constant pyruvate concentration (5.0 mM), employing the same buffers used for the PDC assay, at 30°C.
The protein concentration was determined by using a commercial Bradford reagent according to the manufacturer's instructions (Bio-Rad Laboratories, Inc., United States). The glucose, pyruvate, lactate, and butanediol levels were measured by high-pressure liquid chromatography and the ethanol levels by gas chromatography as previously reported (13).
RESULTS
B. subtilis produces lactate and butanediol under fermentative conditions.
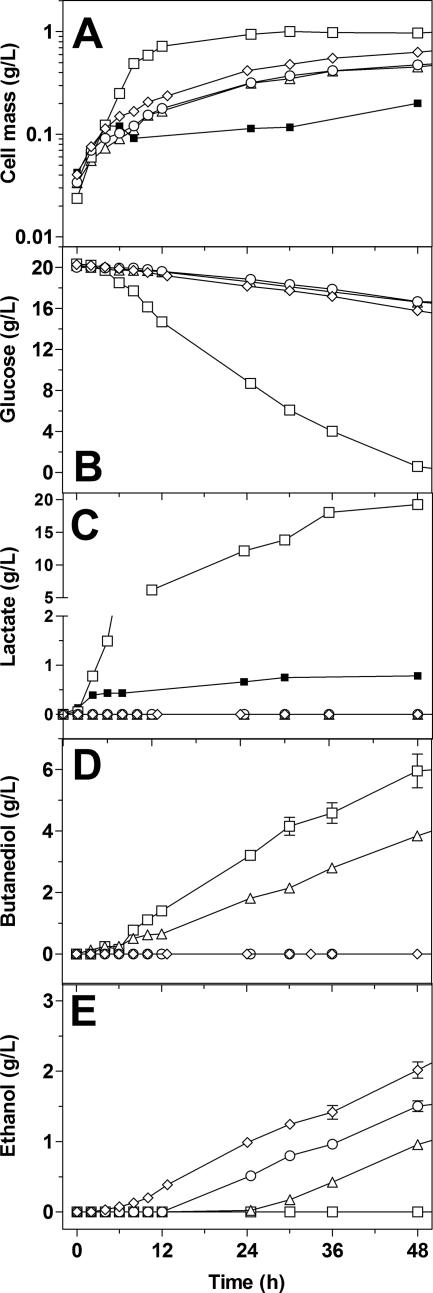
B. subtilis under anaerobic conditions metabolizes glucose to different products, depending on the culture conditions, culture media, and terminal electron acceptor (7, 10, 26). The baseline performance of the prototrophic strain CH1 was characterized under nonaerated conditions, using LB supplemented with 20 g/liter glucose (Fig. 3). During the first 10 h of the culture, B. subtilis CH1 grew exponentially, generating 0.6 g/liter of cell mass, producing mainly lactate (4.7 g/liter) and 2,3-butanediol (1 g/liter). Table 3 shows a summary of the kinetic parameters determined for these cultures.
FIG. 3.
Characterization of B. subtilis strains under fermentative conditions in LB medium containing glucose (20 g/liter). Cell mass formation (A), glucose consumption (B), lactate (C), butanediol (D), and ethanol (E) production of B. subtilis strains CH1 (□), BS35 (▵), BS36 (○), and BS37 (⋄). B. subtilis CH1 (▪) was included as a control, using LB medium without glucose. The error bars represent the variations of duplicated experiments.
TABLE 3.
Kinetic parameters of B. subtilis CH1 and its transformants under fermentative growth in LB broth with 20 g glucose/litera
| Strain | μ (h−1 ± SD)b |
q glucosec (g glucose/g DCW/h ± SD)
|
q ethanol (g ethanol/g DCW/h ± SD)d | q lactate (g lactate/g DCW/h ± SD)d | q butanediol (g butanediol/g DCW/h ± SD)d | |
|---|---|---|---|---|---|---|
| Exponential phase | Stationary phase | |||||
| CH1 | 0.35 ± 0.014 | 1.86 ± 0.03 | 0.44 ± 0.002 | 0 | 0.43 ± 0.005 | 0.15 ± 0.000 |
| BS35 | 0.11 ± 0.015 | 0.66 ± 0.01 | 0.26 ± 0.010 | 0.08 ± 0.002 | 0 | 0.27 ± 0.016 |
| BS36 | 0.09 ± 0.007 | 0.44 ± 0.02 | 0.23 ± 0.006 | 0.12 ± 0.002 | 0 | 0 |
| BS37 | 0.11 ± 0.014 | 0.70 ± 0.03 | 0.23 ± 0.010 | 0.11 ± 0.009 | 0 | 0 |
q, specific rate.
Specific growth rate.
Specific glucose consumption rate.
Specific rates of ethanol, lactate, and butanediol production during the stationary phase.
During the stationary phase, a large amount of lactate (14.5 g/liter) was accumulated (Fig. 3C and Table 3). The specific glucose consumption rate diminished 75% in comparison with the rate in the exponential phase, and glucose was depleted at 48 h. A total of 5 g/liter of 2,3-butanediol was formed. Ethanol, acetate, succinate, formate, and acetoin were not detected during the culture.
B. subtilis CH1 was included as a control, using LB medium without glucose. The CH1 cells grew exponentially, generating only 0.11 g/liter of cell mass and 0.8 g/liter of lactate (Fig. 3).
B. subtilis containing pdc and adhB genes produced ethanol, but ldh knockout reduced growth rate and glucose consumption.
Based on the high lactate productivity observed during the exponential and stationary growth phases, the ldh promoter was considered a good candidate to be used for driving heterologous gene expression under nonaerated conditions. Therefore, this promoter was selected to drive pdc and adhB expression in the B. subtilis chromosome.
Characterization of strain BS35 (CH1 ldh::pdc-adhB) under nonaerated conditions showed that the heterologous genes pdc and adhB are expressed, leading to the synthesis of functional enzymes. Table 4 shows a summary of the PDC, ADHII, and LDH enzymatic activities determined in strains CH1 and BS35. Interestingly, interruption of the ldh gene impaired the growth capacity of strain BS35 (Fig. 3A), diminishing its growth rate by 70% in comparison with that of strain CH1, possibly as a consequence of a 65% decrease in the specific glucose consumption rate (Fig. 3B and Table 3). In comparison with the titer and specific rate of butanediol production in strain CH1, the final titer and the specific rate of butanediol production were slightly reduced in strain BS35, but lactate production was completely abolished (Table 3 and Fig. 3C and D). Even though the growth and glucose consumption rates in strain BS35 were impaired, it produced ethanol during the stationary phase (Fig. 3E), with a specific productivity of 0.08 g ethanol/g DCW/h (Table 3). Interestingly, ethanol accumulation in the culture broth was detected until 24 h elapsed time of fermentation, and 0.95 g/liter was obtained at 48 h.
TABLE 4.
Specific enzymatic activities of PDC, ADH, and LDH in B. subtilis strainsa
| Strain | LDH (IU/mg protein ± SD) | PDC (IU/mg protein ± SD) | ADHII (IU/mg protein ± SD) |
|---|---|---|---|
| B. subtilis CH1 | 7.36 ± 0.02 | 0.00 | 0.03 ± 0.02 |
| B. subtilis BS35 | 0.02 ± 0.01 | 4.21 ± 0.03 | 3.19 ± 0.04 |
Samples were collected at 48 h of nonaerated culture. Activities were replicated four times.
Butanediol synthesis suppression increased ethanol production capacity.
Acetoin synthesis from pyruvate involves two steps, catalyzed by acetolactate synthase (ALSS) and acetolactate decarboxylase (Fig. 1). Butanediol is formed from acetoin reduction (7). Thus, ALSS directly competes with PDC for pyruvate in strain BS35. Hence, to increase pyruvate availability, the alsS gene was interrupted as described in Materials and Methods. Insertional inactivation of alsS totally abolished butanediol production (Fig. 3D) and allowed the onset of ethanol production at 12 h of culture time (Fig. 3E). The specific ethanol production rate in strain BS36 increased 50% in comparison with the rate in strain BS35 (Table 3). Analysis of glucose consumption and ethanol production in the stationary phase (from 12 to 48 h) revealed that 1.5 g/liter of ethanol was produced from 3 g/liter of glucose consumed. This value corresponds to 98% of the maximum theoretical yield (0.51 g ethanol/g glucose). In comparison with the rates in strain CH1, the growth and glucose consumption rates in BS36 remained impaired.
B. subtilis l-LDH can use both NADH and NADPH as cofactors.
The heterologous ethanol pathway in strain BS35 is equivalent to the native lactate pathway in strain CH1 in terms of NADH reoxidation (Fig. 1 and Table 4). Nevertheless, as shown above, the growth and glucose consumption rates were impaired when the homologous lactate pathway was replaced with the heterologous ethanol pathway.
To determine the LDH cofactor specificity, we evaluated if NADPH could be a substrate of B. subtilis LDH. Enzymatic assays clearly showed that both NADH and NADPH are substrates for LDH (Table 5). A control test was also performed using BS35 and BS36 cellular extracts, showing that in the absence of LDH, no NADPH reduction can be detected (Table 5).
TABLE 5.
Determination of specific enzymatic activities of LDH of B. subtilis strains towards two different cofactorsa
| Strain | Cofactor | Sp act (IU/mg protein ± SD) |
|---|---|---|
| BS35 | NADPH | 0.01 ± 0.01 |
| BS36 | NADPH | 0.02 ± 0.005 |
| CH1 | NADPH | 0.68 ± 0.04 |
| CH1 | NADH | 7.36 ± 0.02 |
Activities were replicated four times. Samples were collected at 48 h of nonaerated culture.
Furthermore, to verify that only the LDH activity was responsible for NADPH oxidation, the apparent Km (Michaelis-Menten affinity constant) was determined in cellular extracts from a B. subtilis ldh+ ΔalsS strain (CH1 alsS mutant strain) (Table 1). The apparent Km values of LDH were 0.013 mM for NADH and 0.288 mM for NADPH.
The expression of a transhydrogenase improved growth and reduced the onset time for ethanol production.
The E. coli udhA gene encodes a soluble pyridine nucleotide transhydrogenase (4). This enzyme catalyzes the reversible transfer of reducing equivalents between NADP+ and NAD+ pools according to the following equation: NADPH + NAD+ ⇔ NAP+ + NADH.
Neither any putative transhydrogenase-encoding gene nor any enzymatic system(s) that could fulfill a similar function has been identified in the B. subtilis genome. To evaluate if LDH could play a role in maintaining NADPH/NADP+ equilibrium, the udhA gene from E. coli was integrated into the B. subtilis BS35 chromosome to disrupt the native alsS gene and, at the same time, be under the control of the alsS promoter (see Materials and Methods for details). Under anaerobic conditions, the alsS promoter is induced during the exponential phase and its expression is maintained during the stationary phase (7). A 22% improvement in the growth rate and a 59% increase in the glucose consumption rate during the exponential phase were observed (Fig. 3 and Table 3); as expected, butanediol production was abolished. Ethanol production started at 6 h of culture, and 2 g/liter of ethanol was produced at 48 h; 4.1 g/liter of glucose was consumed during this time, indicating a high conversion yield of glucose into ethanol (96% of the theoretical maximum).
B. subtilis can produce lactate and butanediol from rich medium.
The results with the progenitor strain CH1 indicated that higher yields were obtained in comparison with the amount of glucose consumed, considering that the theoretical maximum yields of lactate and butanediol from glucose are 1.0 g lactate/g glucose and 0.5 g butanediol/g glucose (Fig. 3). In addition, some lactate was produced by strain CH1 in glucose-free LB medium (Fig. 3). These results indicated that components from the rich medium were used to build up cell mass and synthesize fermentation products. Furthermore, LB medium supplemented with glucose allowed the generation of reducing power by strain CH1, to produce large amounts of lactate and butanediol. On one hand, BS35 produced 0.95 g ethanol/liter and 3.8 g butanediol/liter. After the inactivation of ALSS in strain BS36, no butanediol but only 1.5 g/liter of ethanol was produced at 48 h; only 0.65 g/liter more of ethanol was produced than in strain BS35, and a larger amount could be expected. Based on carbon, 2 mol of pyruvate is used to produce 1 mol of butanediol, but just 1 mol of NADH is needed. In comparison, 2 mol of pyruvate and NADH is required to form 2 mol of ethanol. Thereby, in terms of reducing power, although a greater availability of carbon precursors to produce ethanol probably exists in BS36, the reducing power must be limiting. These results suggest that BS35 produced butanediol from carbon sources obtained from rich medium components with a lesser reducing power than glucose.
In addition, ldh inactivation reduced cell mass formation, but small amounts of the glucose consumed were efficiently converted into ethanol in the BS36 and BS37 strains. In these cases, it is suggested that components from the LB were only used for cell mass formation. BS37 was also cultivated in LB without glucose, and biomass formation was only 0.19 g/liter (similar to that found for the first 12 h of culture with glucose); no ethanol was produced in this condition (data not shown). These results confirm that components from rich medium were used to build up cell mass, and that actual ethanol obtained with the BS36 and BS37 strains was synthesized from the glucose consumed.
Exogenous pyruvate was channeled to ethanol, improving the growth rate.
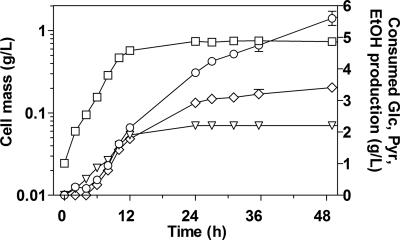
B. subtilis has a poor capacity to grow anaerobically in glucose-mineral medium (26). However, when glucose-mineral medium is supplemented with pyruvate or amino acids, B. subtilis can grow anaerobically, producing lactate, acetate, and butanediol (7, 26). Consequently, to test if pyruvate availability or the PDC- and ADHII-specific activities limit BS37's anaerobic growth in rich medium (LB), LB-glucose medium was supplemented with 2 g/liter of pyruvate (Fig. 4). It was observed that glucose and pyruvate were cometabolized during the exponential growth phase. During this phase, pyruvate and glucose were consumed at the same specific rate. The growth rate of BS37 increased from 0.11 h−1 (Table 3) to 0.27 h−1 when pyruvate was added (Fig. 3 and 4). Pyruvate was exhausted at 12 h, coincidental with the end of the exponential growth phase.
FIG. 4.
Characterization of B. subtilis BS37 in LB supplemented with glucose (Glc; 20 g/liter) and pyruvate (Pyr; 2 g/liter) under fermentative conditions. Cell mass (□), consumed glucose (○), consumed pyruvate (▿), and ethanol (EtOH) production (⋄). The error bars represent the variations of duplicated experiments.
The onset of ethanol production was at 6 h of culture time; the ethanol production rate was 0.9 g ethanol/g DCW/h when glucose and pyruvate were consumed simultaneously. This rate is similar to the ethanol production rates of ethanologenic organisms like E. coli KO11 and yeast (13, 33). In molar terms, the sum of the glucose and pyruvate consumption rates was similar to the ethanol production rate (19 versus 22 mmol ethanol/g DCW/h), indicating that the PDC and ADHII levels in BS37 do not limit ethanol production; therefore, the expression level of these enzymes was not the reason for glucose consumption and growth impairment. After pyruvate depletion, 3.5 g/liter of glucose was consumed and 1.6 g/liter of ethanol was produced (i.e., 90% of the theoretical yield).
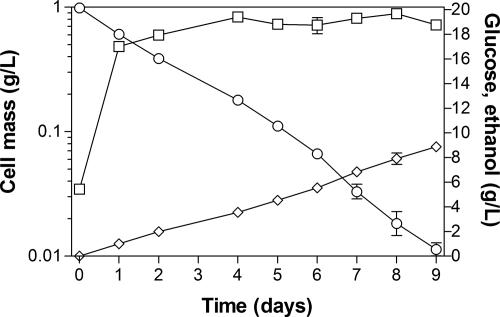
In long-term cultivation, B. subtilis BS37 produced 8.9 g/liter ethanol from glucose.
The growth and formation of products from B. subtilis BS37 (ldh::pdc_adhA ΔalsS udhA+) were characterized in LB broth with 20 g/liter glucose under nonaerated conditions in long-term cultivation (Fig. 5). During the first 4 days, strain BS37 produced ethanol (3.6 g/liter) from glucose (7.5 g/liter) with a high ethanol yield (95% of the theoretical). Although the specific rate of ethanol production was low, it was constant during the 9 days of batch culture. Hence, B. subtilis BS37 produced 8.9 g/liter of ethanol from 20.13 g/liter of glucose, despite the ethanol yield diminishing (87% of the theoretical) during the last days, which could be an effect of ethanol evaporation. The ethanol formation during the 9 days indicates that PDC and ADHII were functional during this long period.
FIG. 5.
Long-term fermentation of B. subtilis BS37 in LB supplemented with glucose (20 g/liter). Cell mass (□), glucose consumption (○), and ethanol production (⋄). The error bars represent the variations of duplicated experiments.
DISCUSSION
Several groups have tried to develop ethanologenic gram-positive bacteria by engineering lactic acid bacteria, Corynebacterium, and Bacillus species. Nevertheless, little or no ethanol has been produced by these recombinant microorganisms (2, 15, 19, 21, 27, 32).
Barbosa and Ingram (2) tried to express the Z. mobilis pdc and adhB genes by using a multicopy vector in B. subtilis. Using Western blot analysis, they observed that both proteins were synthesized in B. subtilis, although no enzymatic activity or ethanol production was reported (2). Talarico et al. (32) constructed different chimeric operons using pdc genes from gram-positive and gram-negative bacteria and from Saccharomyces cerevisiae, to be expressed in multicopy vectors in Bacillus megaterium. They reported different PDC activity levels for each source, indicating the importance of differences in codon usage between gram-positive and gram-negative bacteria. Despite high levels of PDC and ADH in the recombinant B. megaterium strain, in vivo ethanol production was null, and an in vitro assay was necessary to prove ethanol production from pyruvate (32).
LDH plays a complex role in B. subtilis fermentative metabolism.
It has been reported that ldh inactivation reduced the rate of growth of B. subtilis under fermentative conditions (7). In the present study, with the substitution of the combined PDC and ADH activities for LDH activity, we tried to maintain the NADH/NAD+ redox balance; however, the glucose consumption and growth rates of the B. subtilis ldh mutant were impaired. Two hypotheses could explain this phenomenon.
(i) The PDC and ADHII activity levels might not be sufficient to substitute for the previous LDH activity; thus, the NADH/NAD+ redox balance in this strain might be limited. Nevertheless, the Z. mobilis PDC and ADHII enzymes were found to be functional in B. subtilis, with enzymatic activity levels similar to those observed in ethanologenic E. coli (28). The addition of pyruvate led to an increase of 32% in the glucose consumption rate. Furthermore, it was also shown that the activities of these enzymes were sufficient to produce ethanol at a rate similar to that reported for ethanologenic E. coli KO11 and yeast (0.9 g ethanol/g DCW/h) (13, 33). This fact demonstrates the potential of ethanol production in BS37 and eliminates the possibility that the PDC and ADHII levels were not sufficient to support the anaerobic growth of B. subtilis.
(ii) B. subtilis LDH might contribute to maintaining NADPH/NADP+ equilibrium under fermentative conditions. It has been reported that B. subtilis LDH oxidizes NADH to NAD+ (14, 36). Our data proved that B. subtilis LDH can use both NADH and NADPH as cofactors, although the apparent Km of LDH showed a preferential binding of NADH over NADPH. The Km value for NADH was similar to that reported by Yoshida (36). The Km values of LDH for NADH from different gram-positive bacteria range from 0.001 to 0.22 mM. LDH from B. subtilis has one of the largest reported activities and highest affinities for NADH (14). The Km value for NADPH of B. subtilis LDH was similar to the Km values for NADH of LDH from Lactobacillus acidophilus and Lactobacillus jensenii (14). Thus, the LDH from B. subtilis has a significant affinity for NADPH, whose influence in the NADPH/NADP+ equilibrium must be decisive in the fermentative metabolism of B. subtilis.
The transhydrogenase's enzymatic activity (4) partially restored the observed diminished growth rate, supporting the hypothesis that LDH might have an important physiological and regulatory role related to the NADPH/NADP+ balance. In this sense, the cofactor-level imbalance provoked by LDH inactivation might affect the metabolism of B. subtilis, diminishing glucose consumption in the ethanologenic B. subtilis—a role that the ethanologenic pathway might not fulfill.
LDH inactivation in Lactobacillus has also been reported to have negative effects on the glucose consumption rate and cellular growth (1, 12, 21) and on peptidoglycan precursor synthesis (11). Based on the results of these previous works and those presented in this study, it can be speculated that the LDH role in some gram-positive bacteria is as complex as it is in B. subtilis; it sustains not only the pyruvate transformation to lactate and NADH+ oxidation but also, apparently, the NADPH oxidation. Hence, the development of an ethanologenic microorganism derived from a gram-positive bacteria, such as B. subtilis, might require the use of other enzymatic activities, such as those of a transhydrogenase, or the directed evolution of protein to obtain new ADHs that might fulfill the complex role of LDH in the metabolism of gram-positive bacteria, or metabolic strategies to reduce the generation of NADPH in glucose catabolism.
High ethanol production from glucose in B. subtilis.
This study reports the metabolic engineering of B. subtilis for the production of ethanol as the main fermentation product. The strategy was based on the replacement of the LDH activity by the PDC and ADHII activities. The resulting strain produced ethanol, while the parental strain did not produce this compound under the same growth conditions. To ensure the efficient expression of these genes to transcriptional and translational levels in this study, the use of a strong promoter (ldhp) and the B. subtilis Shine-Dalgarno consensus sequence (AAGGAAG) of B. subtilis placed at an optimum distance from the ATG start codon were included (16). In addition, to avoid metabolic burden and to have a genetically stable strain, the heterologous pet operon was integrated by disrupting the native ldh gene. At the same time, the ldh promoter (ldhp) would drive pdc and adhB expression. The combination of all these factors allowed the generation of a B. subtilis strain with relatively high enzymatic activities for PDC and ADHII. The enzymatic activity levels were comparable to those of the patented ethanologenic E. coli KO11 (24, 28), indicating adequate transcription, translation, and protein folding of PDC and ADHII in B. subtilis under nonaerated conditions.
When alsS was inactivated, the resulting ethanologenic B. subtilis strain reached an ethanol yield of 88.8% of the maximum theoretical value from the glucose consumed in 48 h of batch cultures, although the ethanologenic B. subtilis consumed only 3 g/liter during this time; this is because the specific rate of glucose consumption was low. Interestingly, besides the partial restoration of the growth rate with udhA expression, the onset of ethanol production was shortened, similar to the onset of lactate production in the parental CH1 strain. With the aforementioned strategy, the ethanologenic B. subtilis BS37 produced 9 g/liter of ethanol with an ethanol yield on glucose close to 90% during 9 days of fermentation culture. These results clearly demonstrated that it is possible to engineer B. subtilis to produce ethanol as a single fermentation product. The specific rate of ethanol production was relatively low and was very closely related to the specific rate of glucose consumption. In conclusion, this is the first time that a recombinant strain of B. subtilis could produce ethanol in vivo as a sole fermentation product with a high ethanol yield from glucose. These results open new possibilities to develop a process that takes advantage of the B. subtilis secretion system by using complex substrates for ethanol production.
Acknowledgments
We are grateful to Georgina Hernández for high-pressure liquid chromatography analysis and to Mercedes Enzaldo for technical support, as well as to Eugenio López and Santiago Becerra for the synthesis of oligonucleotides. We thank S. L. Wong (Calgary University, Canada) for kindly providing the B. subtilis WB700 strain.
This research was supported by the Mexican Council of Science and Technology (CONACyT) grants CONACyT—SAGARPA 2004-C01-224 and CONACyT—Estado de Morelos MOR-2004-C02-048. Susana Romero was supported by CONACyT scholarship 184798.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Aarnikunnas, J., N. von Weymarn, K. Rönnholm, M. Leisola, and A. Palva. 2003. Metabolic engineering of Lactobacillus fermentum for production of mannitol and pure l-lactic acid or pyruvate. Biotechnol. Bioeng. 82:653-663. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa, M. F. S., and L. O. Ingram. 1994. Expression of the Zymomonas mobilis alcohol dehydrogenase II (adhB) and pyruvate decarboxylase (pdc) genes in Bacillus. Curr. Microbiol. 28:279-282. [Google Scholar]
- 3.Beall, D. S., K. Ohta, and L. O. Ingram. 1991. Parametric studies of ethanol production from xylose and other sugars by recombinant Escherichia coli. Biotechnol. Bioeng. 38:296-303. [DOI] [PubMed] [Google Scholar]
- 4.Boonstra, B., C. E. French, I. Wainwright, and N. C. Bruce. 1999. The udhA gene of Escherichia coli encodes a soluble pyridine nucleotide transhydrogenase. J. Bacteriol. 181:1030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway, T., Y. A. Osman, J. I. Konnan, E. M. Hoffmann, and L. O. Ingram. 1987. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J. Bacteriol. 169:949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conway, T., G. W. Sewell, Y. A. Osman, and L. O. Ingram. 1987. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J. Bacteriol. 169:2591-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz-Ramos, H., T. Hoffmann, M. Marino, H. Nedjari, E. Presecan-Siedel, O. Dreesen, P. Glaser, and D. Jahn. 2000. Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression. J. Bacteriol. 182:3072-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting, S. M., and P. B. Vander-Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 9.Dien, B. S., M. A. Cotta, and T. W. Jeffries. 2003. Bacteria engineered for fuel ethanol production: current status. Appl. Microbiol. Biotechnol. 63:258-266. [DOI] [PubMed] [Google Scholar]
- 10.Espinosa-de-los-Monteros, J., A. Martinez, and F. Valle. 2001. Metabolic profiles and aprE expression in anaerobic cultures of Bacillus subtilis using nitrate as terminal electron acceptor. Appl. Microbiol. Biotechnol. 57:379-384. [DOI] [PubMed] [Google Scholar]
- 11.Ferain, T., J. N. Hobbs, Jr., J. Richardson, N. Bernard, D. Garmyn, P. Hols, N. E. Allen, and J. Delcour. 1996. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J. Bacteriol. 178:5431-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferain, T., A. N. Schanck, and J. Delcour. 1996. 13C nuclear magnetic resonance analysis of glucose and citrate end products in an ldhL-ldhD double-knockout strain of Lactobacillus plantarum. J. Bacteriol. 178:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garay-Arroyo, A., A. A. Covarrubias, I. Clark, I. Niño, G. Gosset, and A. Martinez. 2004. Response to different environmental stress conditions of industrial and laboratory Saccharomyces cerevisiae strains. Appl. Microbiol. Biotechnol. 63:734-741. [DOI] [PubMed] [Google Scholar]
- 14.Garvie, E. I. 1980. Bacterial lactate dehydrogenases. Microbiol. Rev. 44:106-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gold, R. S., M. M. Meagher, S. Tong, R. W. Hutkins, and T. Conway. 1996. Cloning and expression of the Zymomonas mobilis “production of ethanol” genes in Lactobacillus casei. Curr. Microbiol. 33:256-260. [DOI] [PubMed] [Google Scholar]
- 16.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis σA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingram, L. O., H. C. Aldrich, A. C. Borges, T. B. Causey, A. Martinez, F. Morales, A. Saleh, S. A. Underwood, L. P. Yomano, S. W. York, J. Zaldivar, and S. Zhou. 1999. Enteric bacterial catalysts for fuel ethanol production. Biotechnol. Prog. 15:855-866. [DOI] [PubMed] [Google Scholar]
- 18.Ingram, L. O., P. F. Gomez, X. Lai, M. Moniruzzaman, B. E. Wood, L. P. Yomano, and S. W. York. 1998. Metabolic engineering of bacteria for ethanol production. Biotechnol. Bioeng. 58:204-214. [DOI] [PubMed] [Google Scholar]
- 19.Inui, M., H. Kawaguchi, S. Murakami, A. A. Vertes, and H. Yukawa. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 8:243-254. [DOI] [PubMed] [Google Scholar]
- 20.Jan, J., F. Valle, F. Bolivar, and E. Merino. 2001. Construction of protein overproducer strains in Bacillus subtilis by an integrative approach. Appl. Microbiol. Biotechnol. 55:69-75. [DOI] [PubMed] [Google Scholar]
- 21.Liu, S., N. N. Nichols, B. S. Dien, and M. A. Cotta. 2006. Metabolic engineering of a Lactobacillus plantarum double ldh knockout strain for enhanced ethanol production. J. Ind. Microbiol. Biotechnol. 33:1-7. [DOI] [PubMed] [Google Scholar]
- 22.Lynd, L. R., W. H. van Zyl, J. E. McBride, and M. Laser. 2005. Consolidated bioprocessing of cellulosic biomass: an update. Curr. Opin. Biotechnol. 16:577-583. [DOI] [PubMed] [Google Scholar]
- 23.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Martinez, A., S. W. York, L. P. Yomano, V. L. Pineda, F. C. Davis, J. C. Shelton, and L. O. Ingram. 1999. Biosynthetic burden and plasmid burden limit expression of chromosomally integrated heterologous genes (pdc, adhB) in Escherichia coli. Biotechnol. Prog. 15:891-897. [DOI] [PubMed] [Google Scholar]
- 25.Mielenz, J. R. 2001. Ethanol production from biomass: technology and commercialization status. Curr. Opin. Microbiol. 4:324-329. [DOI] [PubMed] [Google Scholar]
- 26.Nakano, M. M., Y. P. Dailly, P. Zuber, and D. P. Clark. 1997. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J. Bacteriol. 179:6749-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols, N. N., B. S. Dien, and R. J. Bothast. 2003. Engineering lactic acid bacteria with pyruvate decarboxylase and alcohol dehydrogenase genes for ethanol production from Zymomonas mobilis. J. Ind. Microbiol. Biotechnol. 30:315-321. [DOI] [PubMed] [Google Scholar]
- 28.Ohta, K., D. S. Beall, J. P. Mejia, K. T. Shanmugam, and L. O. Ingram. 1991. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl. Environ. Microbiol. 57:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonenshein, A. L., J. A. Hoch, and R. Losick. 1993. Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC.
- 30.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 31.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 32.Talarico, L. A., M. A. Gil, L. P. Yomano, L. O. Ingram, and J. A. Maupin-Furlow. 2005. Construction and expression of an ethanol production operon in gram-positive bacteria. Microbiology 151:4023-4031. [DOI] [PubMed] [Google Scholar]
- 33.Tao, H., R. Gonzalez, A. Martinez, M. Rodriguez, L. O. Ingram, J. F. Preston, and K. T. Shanmugam. 2001. Engineering a homo-ethanol pathway in Escherichia coli: increased glycolytic flux and levels of expression of glycolytic genes during xylose fermentation. J. Bacteriol. 183:2979-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong, H. C., and S. Chang. 1986. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc. Natl. Acad. Sci. USA 83:3233-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye, R., L. P. Yang, and S. L. Wong. 1996. Construction of protease deficient Bacillus subtilis strains for expression studies: inactivation of seven extracelluar proteases and the intracellular LonA protease, p. 160-169. In Proceedings of the International Symposium on Recent Advances in Bioindustry. The Korean Society for Applied Microbiology, Seoul, Korea.
- 36.Yoshida, A. 1965. Enzymatic properties of lactate dehydrogenase of Bacillus subtilis. Biochim. Biophys. Acta 99:66-77. [DOI] [PubMed] [Google Scholar]