Abstract
The family 8 glycoside hydrolase (RexA) from Bifidobacterium adolescentis was expressed in Escherichia coli. The recombinant enzyme was characterized as a reducing-end xylose-releasing exo-oligoxylanase. Apart from giving insights into this new class of enzymes, knowledge of the RexA enzyme helps to postulate a mechanism for the B. adolescentis breakdown of prebiotic xylooligosaccharides.
Xylooligosaccharides (XOS) are sugar oligomers that consist of β-1,4-linked xylose residues, which appear naturally in fruits, vegetables, bamboo shoots, milk, and honey. As nonstarch polysaccharides, they resist digestion in the upper part of the human gastrointestinal tract and are subsequently fermented by the gut microbiota in the colon. XOS fermentation especially stimulates the growth of bifidobacteria, as shown in vivo in rats and humans (2, 6, 7, 11). As a nondigestible food ingredient that beneficially affects the host by selectively stimulating the growth of health-promoting bacteria in the colon, XOS fulfill the requirements of prebiotics. In 1996, the Japanese government listed XOS as “foods for special health use” (3), and today several companies are marketing a wide range of XOS-supplemented foods (16).
Despite the increasing use of XOS as bifidogenic prebiotics, remarkably little is known about the XOS-degrading enzymes from bifidobacteria. Only one β-d-xylosidase (EC 3.2.1.37; stepwise removing xylose residues from the nonreducing end of a XOS) has been purified from Bifidobacterium breve K-110 (14). Known XOS-degrading enzymes from other organisms include endoxylanases (EC 3.2.1.8; cleaving randomly between β-1,4-linked xylose residues) and an only recently described reducing-end xylose-releasing exo-oligoxylanase (EC 3.2.1.156; releasing xylose residues from the reducing end of a XOS) (5). In silico, the presence of several XOS-degrading enzymes have been predicted to occur in the genomes of bifidobacteria (13, 15).
To gain insight into the XOS-metabolizing system of Bifidobacterium adolescentis, we recombinantly expressed and characterized one of these enzymes. The family 8 glycoside hydrolase (GH8) gene from B. adolescentis, previously annotated as an endoxylanase gene (15), in fact encodes a reducing-end xylose-releasing exo-oligoxylanase. This is the second report of a reducing-end xylose-releasing exo-oligoxylanase and the first one describing its occurrence in a potentially probiotic bacterium. Our results provide further insight into this new enzyme class and improve our understanding of how B. adolescentis metabolizes prebiotic XOS.
For experimental details, please see the supplemental material. The GH8 coding sequence was amplified from the genomic DNA of B. adolescentis LMG10502 by PCR and was ligated in a pEXP5-CT/TOPO expression vector (Invitrogen, Carlsbad, CA), introducing it in frame with the C-terminal His6 sequence. The coding sequence of the GH8 enzyme from B. adolescentis LMG10502 was found to be identical to the BAD_0300 gene from B. adolescentis ATCC 15703 (GenBank accession no. NC_008618; GeneID no. 4556993) and the xylA gene from B. adolescentis DSM20083 (GenBank accession no. AY233379). The amino acid sequence of this B. adolescentis enzyme showed the highest homology to the reducing-end xylose-releasing exo-oligoxylanase (Rex) from Bacillus halodurans C-125 (69% similarity and 56% identity) and was therefore named RexA. It shares the three catalytic residues with Rex (Glu66, Asp124, and Asp259; RexA numbering) (5) (Fig. 1) as well as the loop (Leu312, His313, and Pro314; RexA numbering) responsible for the exo-activity of Rex by blocking the +2 substrate binding subsite (4). Also, the absence of a signal peptide (SignalP analysis) (1) is in congruence with the Rex enzyme (5).
FIG. 1.
Amino acid sequence alignment of RexA of B. adolescentis and Rex of B. halodurans. The three catalytic residues (E66, D124, and D259; RexA numbering) are boxed, as is the loop (L312, H313, and P314; RexA numbering) responsible for blocking substrate binding subsite +2 in Rex.
Recombinant expression was induced in transformed Escherichia coli BL21(DE3)pLysS cells (Invitrogen) by 1 mM isopropyl-β-d-thiogalactopyranoside, followed by a 4-h incubation at 37°C. A single-step HisTrap (GE Healthcare, Uppsala, Sweden) purification, performed on an Äkta fast protein liquid chromatograph (GE Healthcare), yielded as much as 140 mg recombinant enzyme per liter cell culture. Based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (8), the protein was estimated to be ∼99% pure (Fig. 2). Apart from a major band at ∼42 kDa, two weaker bands were seen at approximately 84 and 126 kDa. These were confirmed to be di- and trimeric forms, as the bands disappeared after the addition of β-mercaptoethanol (Fig. 2). The pI of the enzyme was determined to be 4.7, slightly below the theoretical pI (5.2). The purified enzyme was stable for at least 4 months at 4°C in 25 mM sodium acetate buffer, pH 5.0.
FIG. 2.
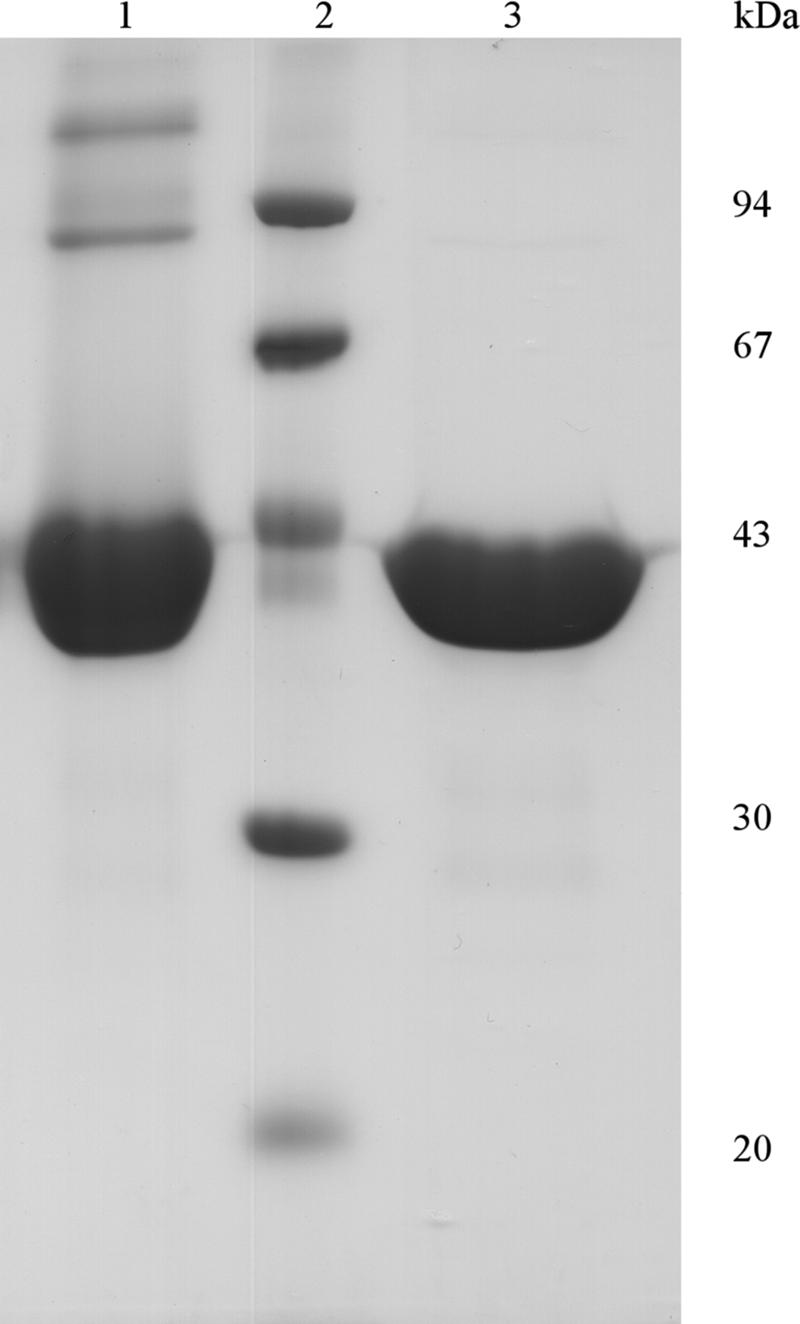
SDS-PAGE analysis of the recombinant B. adolescentis GH8 enzyme. SDS-PAGE of purified RexA shows a large protein band around 42 kDa (lane 1); the two weaker protein bands at ∼84 and ∼126 kDa disappear after 1% β-mercaptoethanol treatment (lane 3). Molecular masses of the low-molecular-mass markers (lane 2) are indicated on the right.
In contrast to Rex from B. halodurans, RexA was able to hydrolyze birchwood xylan and oat spelt xylan, as shown by the dinitrosalicylic acid method (10), but the enzymatic activity was relatively low (0.13 and 0.25 U/mg, respectively). It was also active on Xylazyme AX tablets (Megazyme, Bray, Ireland), with an optimal activity at 40°C and pH 5.0 (data not shown).
Enzyme activity was further assessed using XOS (xylobiose [X2], xylotriose [X3], xylotetraose [X4], xylopentaose [X5], and xylohexaose [X6]) as a substrate in 25 mM sodium acetate buffer, pH 5.0, at 40°C. At regular time intervals, aliquots were taken and heat inactivated at 100°C for 30 min. The different substrates and products were analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection. The recombinant enzyme constitutively hydrolyzed a xylose residue from Xn when n was >2, until the final products were (n − 2)X1 and X2 (Fig. 3). Just like Rex of B. halodurans, the B. adolescentis enzyme displayed no activity on X2 and p-nitrophenyl-β-d-xylopyranoside, even at an extended incubation time (24 h). These cleavage patterns establish the enzyme as a reducing-end xylose-releasing exo-oligoxylanase and not as an endoxylanase (as no endo mode of action is observed) or as a β-xylosidase (as there is no activity on xylobiose or p-nitrophenyl-β-d-xylopyranoside).
FIG. 3.
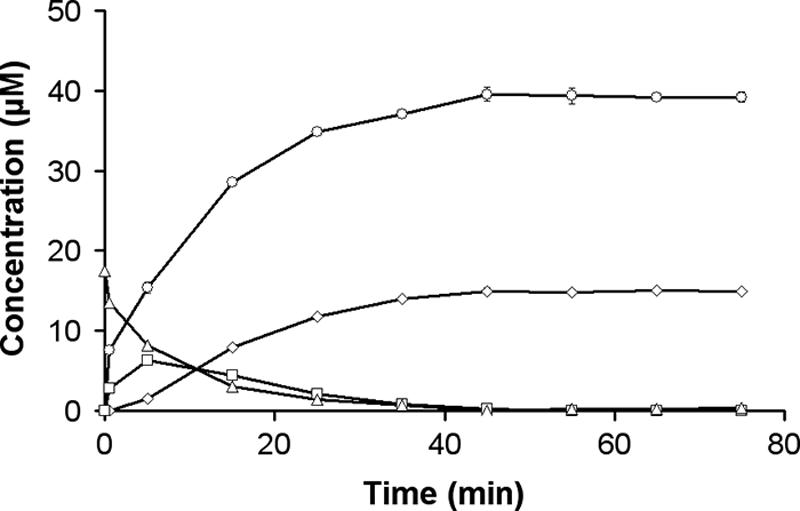
Constitutive hydrolysis of a xylose residue from XOS. Hydrolysis of X4 (triangles) generated X3 (squares) and xylose (X) (circles). X3 is further hydrolyzed to yield the final products X and X2 (diamonds). Error bars show standard deviations of the means of triplicate experiments.
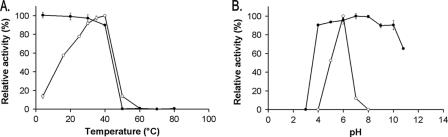
The pH and temperature stabilities were determined by incubating the enzyme for 120 min at a specific pH or for 40 min at a specific temperature, respectively. Next, activity was assayed by measuring the enzymatic hydrolysis of X3 upon addition of RexA in 25 mM sodium acetate buffer, pH 5.0, at 40°C for 30 min. The pH and temperature optima were determined by performing the activity test under different pH and temperature conditions, respectively. The enzyme exhibited an optimal activity on X3 at pH 6.0 and was stable between pH 4 and 10 for 120 min (Fig. 4). RexA was stable at temperatures up to 40°C, which was also found to be its optimal temperature.
FIG. 4.
Thermal and pH optima and stability of RexA. (A) Effect of temperature on enzymatic activity (open circles) and stability (solid circles). (B) Effect of pH on enzymatic activity (open circles) and stability (solid circles). Error bars show standard deviations of the means of triplicate experiments.
Progress curves of XOS cleavage were used to determine the kcat/Km of the reactions according to the equation of Matsui et al. (9). The specificity constant kcat/Km, calculated for X3 (65.7 s−1 mM−1), X4 (43.9 s−1 mM−1), X5 (29.5 s−1 mM−1), and X6 (25.5 s−1 mM−1), showed a decrease when the degree of polymerization of the XOS increased. These data show that X3 is the preferred substrate for the enzyme.
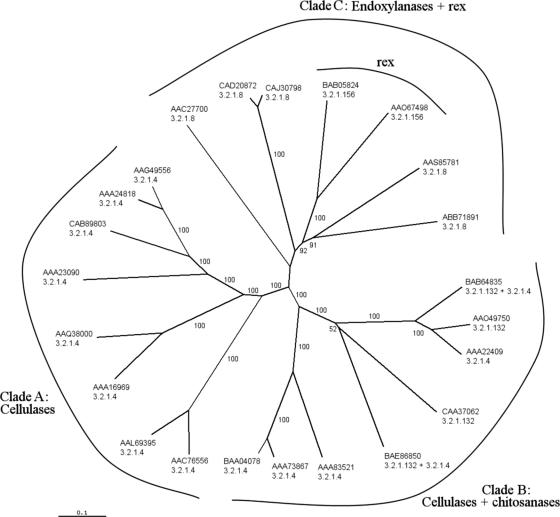
Amino acid sequences from all GH8 family members with experimentally determined enzyme activities were obtained from the GenPept database, using the GenPept identification numbers from the CAZy database (www.cazy.org [accessed February 2007]). Sequences were manually restricted to the catalytic domain, and phylogenetic trees were calculated by the neighbor-joining method (12) with bootstrap analysis. The resulting tree is composed of three clades (Fig. 5). The first clade consists solely of cellulases, whereas the second clade consists of cellulases and chitosanases. Xylanases are restricted to the third clade. The reducing-end xylose-releasing exo-oligoxylanases constitute a separate branch of enzymes within the xylanase clade. These data suggest that reducing-end xylose-releasing exo-oligoxylanases have evolved from endoxylanases rather than from a common ancestor enzyme. This evolution most likely arose from the insertion of a loop, present in both Rex and RexA, resulting in the blockage of substrate binding subsite +2 (4).
FIG. 5.
Phylogenetic tree of GH8 catalytic domain sequences. GenPept identification numbers are given together with EC numbers 3.2.1.4 (cellulase), 3.2.1.8 (xylanase), 3.2.1.132 (chitosanase), and 3.2.1.156 (reducing-end xylose-releasing exo-oligoxylanase or Rex). Apart from chitosanase activity, the CAA37062 enzyme also has lichenase activity (EC 3.2.1.73). Numbers at the branches are percentage bootstrap values for 1,000 resamplings. The three different clades are indicated. The enzymes mentioned in this article are accessible under GenPept identification no. BAB05824 (Rex) and AAO67498 (RexA) and are positioned in clade C.
In this report, we show that a putative GH8 endoxylanase gene of B. adolescentis in fact encodes a reducing-end xylose-releasing exo-oligoxylanase. To date, only one other reducing-end xylose-releasing exo-oligoxylanase is known, namely, Rex from B. halodurans C-125. RexA shows high homology to Rex and comparable activities on XOS. However, while Rex does not show any activity on birchwood xylan, we observed activity (albeit low) of RexA on both birchwood and oat spelt xylan. Crystallographic data from RexA should provide more insight into the structural differences between Rex and RexA and allow exploration of why the latter has some extra activity on polymeric xylan.
The presence of a reducing-end xylose-releasing exo- oligoxylanase may help to explain how B. adolescentis uses XOS as a carbon source. XOS are likely transported into the cell by a MalEFG-type oligosaccharide transporter, homologous to one of the numerous MalEFG-type oligosaccharide transporters found in Bifidobacterium longum (13). Once inside the cell, XOS are hydrolyzed by RexA to xylose and xylobiose. The xylobiose may be further hydrolyzed by a β-xylosidase, possibly homologous to the β-xylosidase from B. breve K-110.
Supplementary Material
Acknowledgments
We thank C. Grootaert (Laboratory for Microbial Ecology and Technology, Ghent University, Ghent, Belgium) for providing a genomic DNA sample of B. adolescentis LMG10502.
We gratefully acknowledge the financial support from the Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (I.W.T.) (SBO IMPAXOS project funding) and the Research Fund K.U. Leuven (IDO/03/005 project and postdoctoral fellowship of S. Van Campenhout).
Footnotes
Published ahead of print on 22 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, J. M., J. Fahey, and B. W. Wolf. 1997. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J. Nutr. 127:130-136. [DOI] [PubMed] [Google Scholar]
- 3.Crittenden, R. G., and M. J. Playne. 1996. Production, properties and applications of food-grade oligosaccharides. Trends Food Sci. Technol. 7:353-361. [Google Scholar]
- 4.Fushinobu, S., M. Hidaka, Y. Honda, T. Wakagi, H. Shoun, and M. Kitaoka. 2005. Structural basis for the specificity of the reducing end xylose-releasing exo-oligoxylanase from Bacillus halodurans C-125. J. Biol. Chem. 280:17180-17186. [DOI] [PubMed] [Google Scholar]
- 5.Honda, Y., and M. Kitaoka. 2004. A family 8 glycoside hydrolase from Bacillus halodurans C-125 (BH2105) is a reducing end xylose-releasing exo-oligoxylanase. J. Biol. Chem. 279:55097-55103. [DOI] [PubMed] [Google Scholar]
- 6.Hsu, C. K., J. W. Liao, Y. C. Chung, C. P. Hsieh, and Y. C. Chan. 2004. Xylooligosaccharides and fructooligosaccharides affect the intestinal microbiota and precancerous colonic lesion development in rats. J. Nutr. 134:1523-1528. [DOI] [PubMed] [Google Scholar]
- 7.Jaskari, J., P. Kontula, A. Siitonen, H. Jousimies-Somer, T. Mattila-Sandholm, and K. Poutanen. 1998. Oat β-glucan and xylan hydrolysates as selective substrates for Bifidobacterium and Lactobacillus strains. Appl. Microbiol. Biotechnol. 49:175-181. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 9.Matsui, I., K. Ishikawa, E. Matsui, S. Miyairi, S. Fukui, and K. Honda. 1991. Subsite structure of Saccharomycopsis α-amylase secreted from Saccharomyces cerevisiae. J. Biochem. (Tokyo) 109:566-569. [DOI] [PubMed] [Google Scholar]
- 10.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 11.Okazaki, M., S. Fujikawa, and N. Matsumoto. 1990. Effect of xylo-oligosaccharide on the growth of bifidobacteria. Bifidobact. Microflora 9:77-86. [Google Scholar]
- 12.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 13.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin, H. Y., J. H. Lee, J. Y. Lee, Y. O. Han, M. J. Han, and D. H. Kim. 2003. Purification and characterization of ginsenoside Ra-hydrolyzing β-d-xylosidase from Bifidobacterium breve K-110, a human intestinal anaerobic bacterium. Biol. Pharm. Bull. 26:1170-1173. [DOI] [PubMed] [Google Scholar]
- 15.van den Broek, L. A. M., R. M. Lloyd, G. Beldman, J. C. Verdoes, B. V. McCleary, and A. G. J. Voragen. 2005. Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 67:641-647. [DOI] [PubMed] [Google Scholar]
- 16.Vázquez, M. J., J. L. Alonso, H. Dominguez, and J. C. Parajo. 2000. Xylooligosaccharides: manufacture and applications. Trends Food Sci. Technol. 11:387-393. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.