Abstract
Aculeacin A acylase from Actinoplanes utahensis produced by Streptomyces lividans revealed acylase activities that are able to hydrolyze penicillin V and several natural aliphatic penicillins. Penicillin K was the best substrate, showing a catalytic efficiency of 34.79 mM−1 s−1. Furthermore, aculeacin A acylase was highly thermostable, with a midpoint transition temperature of 81.5°C.
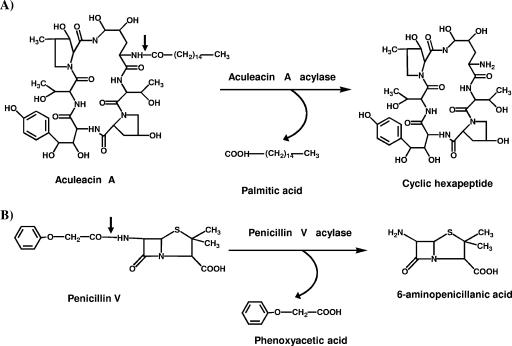
Aculeacin A acylase (EC 3.5.1.70) from Actinoplanes utahensis NRRL 12052 (AuAAC) catalyzes the hydrolyses of the acyl moieties of antifungal echinocandin antibiotics produced by Aspergillus species (13) (Fig. 1A). These cyclic hexapeptide antibiotics contain long fatty acid side chains (e.g., linoleic, myristic, or palmitic acid) and exhibit high antifungal activity. They have been used to produce several potential therapeutic agents after enzymatic hydrolysis of their acidic moieties, which releases a cyclic hexapeptide that can be further reacylated (3, 6).
FIG. 1.
(A) Hydrolysis of aculeacin A catalyzed by AAC from A. utahensis. (B) Hydrolysis of penicillin V by penicillin V acylase. The small arrows indicate the positions of hydrolysis.
AuAAC is largely extracellular and consists of two dissimilar subunits with molecular masses of 60 kDa and 19 kDa (8, 18). Remarkably, although this acylase shares sequence similarity with β-lactam acylases (8, 11), it has not been reported to show penicillin acylase activity. Preliminary analyses carried out in our laboratory showed that AuAAC was very similar to the β-lactam acylase from Streptomyces lavendulae (AY611030), which shows a preference for penicillins with long hydrophobic acyl moieties (21). On the other hand, although AuAAC has been purified (8, 9, 18), few data on its structure function relationships have been provided so far and many biochemical properties of these proteins remain to be analyzed.
Taking into account these observations, we have investigated whether AuAAC would behave as a new β-lactam acylase with novel properties. In this work, we describe newly discovered hydrolytic activities of AuAAC overproduced and purified from a recombinant Streptomyces lividans strain harboring the aac gene.
Overproduction of AuAAC.
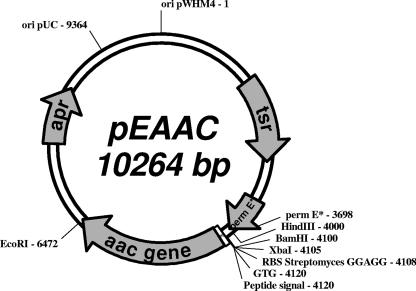
To overproduce AuAAC, an engineered aac gene was constructed and cloned into the bifunctional expression vector pEM4, which contains the ermE* promoter from Saccharopolyspora erythraea (15). The aac gene, including its signal peptide-encoding sequence, was amplified by PCR using chromosomal DNA from A. utahensis NRRL 12052 as a template (8, 18). The PCR primers were designed according to the DNA sequence of aac (8). The primers used were AAC53 (5′TGCTCTAGAGGAGGTGCCGCCGTGACGTCCTCGTACATGCGCC-3′) and AAC31 (5′CCGGAATTCCTCAGCGTCCCCGCTGTGCCAC-3′). The restriction sites XbaI and EcoRI are shown in italics, the ribosome binding site sequence is underlined, and the start codon is shown in bold. PCR amplification was carried out under standard conditions with a Mastercycler gradient thermocycler (Eppendorf, Germany). Purified PCR products (2.5 kb) were digested with XbaI and EcoRI endonucleases and cloned into the pEM4 vector. The resulting plasmid, pEAAC (Fig. 2), which includes the Streptomyces ribosome binding site consensus sequence and the putative GTG start codon (8), was introduced in S. lividans 1326 by standard procedures (10). The highest AuAAC production (1.264 U/ml) was reached after 96 h of incubation of the recombinant S. lividans (pEAAC) strain. Remarkably, the AuAAC yield was 21-fold higher in the recombinant strain than in A. utahensis (Table 1). More important, a heterodimeric form of homogeneous AuAAC was present in the fermentation broth of S. lividans (pEAAC), in contrast to the incomplete, processed forms of heterogeneous AuAAC described previously (8, 9).
FIG. 2.
Scheme of the recombinant plasmid pEAAC. Numbers indicate the positions of relevant sequences. The length of the plasmid is indicated in base pairs. Abbreviations: apr, ampicillin resistance gene; tsr, thiostrepton resistance gene; pemrE*, ermE* promoter; RBS, ribosome binding site; GTG, start codon; ori pUC, replication origin from pUC plasmid; ori pWHM4, replication origin from pWHM4 plasmid.
TABLE 1.
Production of AuAAC by different microorganisms
| Microorganism | AuAAC production (IU/ml)a at indicated time
|
||||
|---|---|---|---|---|---|
| 0 h | 72 h | 96 h | 120 h | 140 h | |
| A. utahensis NRRL 1205b | 0.060 | ||||
| S. lividans 1326b | 0 | 0 | 0 | 0 | 0 |
| S. lividans (pEM4)b | 0 | 0 | 0 | 0 | 0 |
| S. lividans (pEAAC)b | 0 | 0.695 | 1.264 | 0.849 | 0.861 |
IU, μmol of 6-APA produced per minute under the standard assay conditions. Acylase activity was routinely assayed using penicillin V as a substrate. The activities of A. utahensis NRRL 12502 and of the recombinant S. lividans culture supernatants were determined according to the method previously described (20).
For enzymatic assays, A. utahensis was cultured in liquid medium containing 3% sucrose, 0.5% peptone, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO4·7H2O, and 0.0002% FeSO4·7H2O (pH 6.5) (18) at 30°C and 250 rpm for 96 h, whereas S. lividans was cultured aerobically under submerged conditions in trypticase soy broth with thiostrepton (5 μg/ml) at 30°C and 250 rpm.
Purification and characterization of recombinant AuAAC.
The recombinant AuAAC produced by S. lividans (pEAAC) was purified by two chromatographic steps (Table 2), a procedure considerably faster than those previously described which used five purification steps (8, 9). Culture supernatants from recombinant S. lividans (pEAAC) cells, grown as described above, were adjusted to pH 6.0 and applied onto an S-Sepharose fast-flow column (Amersham Biosciences, United Kingdom) equilibrated with 10 mM sodium phosphate buffer, pH 6.0. The column was washed with the same buffer, and bound proteins were eluted with a linear gradient of 0 to 1.5 M NaCl in the same buffer. The fractions showing deacylase activity were pooled, concentrated with polyethylene glycol 35000 (reverse dialysis), and loaded onto a Superose 12 fast flow column (Amersham Biosciences, United Kingdom). The column was both equilibrated and eluted with 50 mM potassium phosphate buffer, pH 7.0 (isocratic elution). Routinely, protein concentration was determined according to Bradford (5) and the purity of AuAAC was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12).
TABLE 2.
Purification of recombinant AuAAC produced by S. lividans (pEAAC)
| Purification step | Total activity (IU) | Sp act (IU/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|
| Cell-free broth | 172.10 | 3.15 | 100 | |
| S-Sepharose | 66.73 | 33.60 | 39 | 10.7 |
| Superose 12 | 35.80 | 42.80 | 21 | 13.6 |
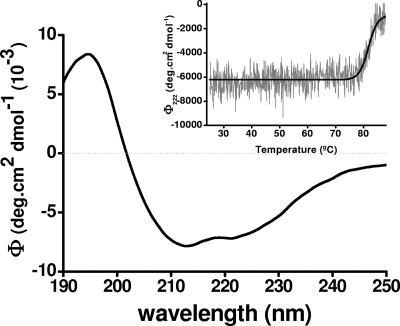
Several properties of recombinant AuAAC have been further investigated by using the purified enzyme. First, the molar extinction coefficient of AAC, 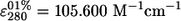 , was determined by the method of Edelhoch (7). Second, based on the far-UV circular dichroism (CD) spectrum of AuAAC (Fig. 3), we have determined that the protein contains 29% α-helix, 18% β-sheet, 17% β-turns, and 36% random coil. Remarkably, CD-thermal denaturation experiments revealed that the AuAAC global structure was highly thermostable, showing a midpoint transition temperature of 81.5°C (Fig. 3, inset).
, was determined by the method of Edelhoch (7). Second, based on the far-UV circular dichroism (CD) spectrum of AuAAC (Fig. 3), we have determined that the protein contains 29% α-helix, 18% β-sheet, 17% β-turns, and 36% random coil. Remarkably, CD-thermal denaturation experiments revealed that the AuAAC global structure was highly thermostable, showing a midpoint transition temperature of 81.5°C (Fig. 3, inset).
FIG. 3.
Far-UV CD spectra of pure AuAAC. Spectra were recorded at 0.28 mg/ml in 50 mM potassium buffer, pH 7.0, at 25°C between 175 and 240 nm under thermostated conditions by using a JASCO 715 (Japan) spectropolarimeter. The CD readings were expressed as mean residue molar ellipticities (deg.cm2 dmol−1), assuming a residue molecular weight of 110. (Inset) Thermal unfolding of AuAAC was studied by recording the molar ellipticity variations at 220 nm for temperatures between 25 and 85°C scanned at 20°C/hour. Secondary-structure information was obtained from CD spectra by the CCA (14) and CDNN V2.0.3.188 (4) programs.
Furthermore, the thermal stability of β-lactam acylase activity was studied by preincubating the recombinant AAC for 20 min at different temperatures (30 to 80°C). The activity was stable up to 50°C, slowly decreasing up to 75°C and becoming drastically reduced at higher temperatures. These results correlate with those for the CD-thermal denaturation studies, where unfolding was observed at high temperatures. In addition, the effect of temperature on AAC was examined by measuring its activity in the temperature range of 30 to 80°C, using penicillin V as a substrate. Surprisingly, the highest hydrolytic activity was achieved at 75°C.
Finally, AAC activity was assayed at different pH levels. Using penicillin V as a substrate, we observed that the optimal pH ranged between 8.0 and 8.5.
Substrate specificity of recombinant AuAAC.
As pointed out above, complementary analyses carried out in our laboratory showed that this enzyme was very similar to the penicillin V acylase from S. lavendulae, strongly suggesting that AuAAC might behave as a true β-lactam acylase. Remarkably, in agreement with this hypothesis, we have determined that purified recombinant AuAAC is able to hydrolyze penicillin V as penicillin V acylase does (Fig. 1B), in contrast to the echinochandin B acylase from A. utahensis (an isoenzyme of AuAAC), which is apparently unable to hydrolyze penicillin V (11). For such hydrolysis, the enzyme does not require any external cofactor, metal ion, or reducing agent for maximal activity, although a slight stimulation of activity was found in the presence of 0.6 M KCl (data not shown). This result demonstrated that AuAAC can certainly be reclassified as a β-lactam acylase, not only according to its primary structure but also based on its enzymatic activity, and therefore, we decided to study in detail its catalytic parameters, using β-lactam antibiotics as substrates.
To examine the hydrolytic specificity of AuAAC, we determined the kinetic parameters for the hydrolyses of different natural β-lactam antibiotics, i.e., penicillins V, K, F, dihydroF, and G (Table 3). The pure recombinant enzyme (0.5 μg) was incubated with increasing concentrations of the corresponding penicillins (V, K, F, dihydroF, and G) in 100 mM potassium phosphate buffer, pH 8.0, at 45°C for 15 min in a final volume of 100 μl. The reaction was stopped by addition of 400 μl of 0.5 M sodium acetate. After centrifugation of the samples, the released 6-APA (6-aminopenicillanic acid) was monitored with fluorescamine (16, 19). The reaction was linear under these assay conditions. Values of kinetic constants were determined by fitting initial velocity data to the Hanes-Woolf equation (17), using a hyperbolic regression program (Hyper.exe 1.01, by J. S. Easterby [http://homepage.ntlworld.com/john.easterby/software.html]). Substrate saturation kinetics were fitted to the equation v = (Vmax × S)/(Km + S).
TABLE 3.
Kinetic parameters of recombinant AuAAC assayed with different natural penicillins
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|
| Penicillin V | 15.4 | 70.3 | 4.55 |
| Penicillin K | 1.0 | 33.3 | 34.79 |
| Penicillin dihydroF | 5.6 | 4.2 | 0.75 |
| Penicillin F | 15.1 | 1.9 | 0.13 |
| Penicillin G | 155.8 | 2.2 | 0.01 |
Remarkably, AuAAC shows the same substrate specificity on these natural penicillins as penicillin V acylase from S. lavendulae (21). Thus, penicillin K was the best penicillin substrate for AuAAC, showing the highest bimolecular constant (specificity constant) value, kcat/Km = 34.79 mM−1 s−1. This value is within the range reported for its natural substrate aculeacin A (18). It is worth noticing that other β-lactam acylases of the penicillin G acylase family so far cloned and assayed do not display such hydrolytic capability (1, 2). Likewise, the enzyme hydrolyzes penicillin G at an extremely low rate (2 to 3% of the rate for hydrolysis of penicillin V). These structural preferences for aliphatic chains suggest the existence of a peculiar hydrophobic pocket in the active center, as described for penicillin acylase from S. lavendulae (21).
These results taken together suggest that AAC from A. utahensis and penicillin acylase from S. lavendulae can be considered the first members of a new subfamily of β-lactam acylases showing high specificity for penicillin K, and we propose to classify them as penicillin K acylases. Likewise, AuAAC should be considered an industrial biocatalyst with high potential in the production of semisynthetic penicillins.
Acknowledgments
We express our gratitude to J. A. Salas from the University of Oviedo for providing the pEM4 expression vector.
This work was supported by grant BIO 2003-04832 from the Spanish Ministry of Education and Science.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Arroyo, M., I. de la Mata, C. Acebal, and M. P. Castillón. 2003. Biotechnological applications of penicillin acylases: state-of-art. Appl. Microbiol. Biotechnol. 60:507-514. [DOI] [PubMed] [Google Scholar]
- 2.Arroyo, M., I. de la Mata, D. Hormigo, M. P. Castillón, and C. Acebal. 2005. Production and characterization of microbial β-lactam acylases, p 130-151. In J. L. Barredo and E. Mellado (ed.), Microorganisms for industrial enzymes and biocontrol. Research Signpost, Kerala, India.
- 3.Boeck, L. D., D. S. Fukuda, B. J. Abbott, and M. Debono. 1989. Deacylation of echinocandin B by Actinoplanes utahensis. J. Antibiot. 42:382-388. [DOI] [PubMed] [Google Scholar]
- 4.Böhm, G., R. Muhr, and R. Jaenicke. 1992. Quantitative analysis of protein far-UV circular dichroism spectra by neural networks. Protein Eng. 5:191-195. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-252. [DOI] [PubMed] [Google Scholar]
- 6.Debono, M., B. J. Abbott, D. S. Fukuda, M. Barnhart, K. E. Willard, R. M. Molloy, K. H. Michel, J. R. Turner, T. F. Butler, and A. H. Hunt. 1989. Synthesis of new analogues of echinocandin B by enzymatic deacylation and chemical reacylation of the echinocandin B peptide: synthesis of the antifungal agent cyclofungin (LY121019). J. Antibiot. 42:389-397. [DOI] [PubMed] [Google Scholar]
- 7.Edelhoch, H. 1967. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6:1948-1954. [DOI] [PubMed] [Google Scholar]
- 8.Inokoshi, J., H. Takeshima, H. Ikeda, and S. Omura. 1992. Cloning and sequencing of the aculeacin A acylase-encoding gene from Actinoplanes utahensis and expression in Streptomyces lividans. Gene 119:29-35. [DOI] [PubMed] [Google Scholar]
- 9.Inokoshi, J., H. Takeshima, H. Ikeda, and S. Omura. 1993. Efficient production of aculeacin A acylase in recombinant Streptomyces strains. Appl. Microbiol. Biotechnol. 39:532-536. [Google Scholar]
- 10.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. A laboratory manual. John Innes Foundation, Norwich, England.
- 11.Kreuzman, A. J., R. L. Hodges, J. R. Swartling, T. E. Pohl, S. K. Ghag, P. J. Baker, D. McGilvray, and W. K. Yeh. 2000. Membrane-associated echinocandin B deacylase of Actinoplanes utahensis: purification, characterization, heterologous cloning and enzymatic deacylation reaction. J. Ind. Microbiol. Biotechnol. 24:173-180. [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Mizuno, K., A. Yagi, S. Satoi, M. Takada, and M. Hayashi. 1977. Studies on aculeacin. I. Isolation and characterization of aculeacin A. J. Antibiot. 30:297-302. [DOI] [PubMed] [Google Scholar]
- 14.Perczel, A., M. Hollosi, G. Tusnady, and G. D. Fasman. 1991. Convex constrain analysis: a natural deconvolution of circular dichroism curves of proteins. Protein Eng. 4:669-679. [DOI] [PubMed] [Google Scholar]
- 15.Quiros, L. M., I. Aguirrezabalaga, C. Olano, C. Mendez, and J. A. Salas. 1998. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol. Microbiol. 28:1177-1185. [DOI] [PubMed] [Google Scholar]
- 16.Reyes, F., J. Martinez, and J. Salvery. 1989. Determination of cephalosporin C amidohydrolase activity with fluorescamine. J. Pharm. Pharmacol. 4:136-137. [DOI] [PubMed] [Google Scholar]
- 17.Segel, I. H. 1975. Enzyme kinetics. Behaviour and analysis of rapid equilibrium and steady-state enzyme systems. Wiley-Interscience Publications, New York, NY.
- 18.Takeshima, H., J. Inokoshi, Y. Takada, H. Tanaka, and S. Omura. 1989. A deacylation enzyme for aculeacin A, a neutral lipopeptide antibiotic, from Actinoplanes utahensis: purification and characterization. J. Biochem. 105:606-610. [DOI] [PubMed] [Google Scholar]
- 19.Torres, R., F. Ramón, I. de la Mata, C. Acebal, and M. P. Castillón. 1999. Enhanced production of penicillin V acylase from Streptomyces lavendulae. Appl. Microbiol. Biotechnol. 53:81-84. [DOI] [PubMed] [Google Scholar]
- 20.Torres, R., I. de la Mata, M. P. Castillón, M. Arroyo, J. Torres, and C. Acebal. 1998. Purification and characterization of penicillin V acylase from Streptomyces lavendulae, p. 719-724. In A. Ballesteros, F. J. Plou, J. L. Iborra, and P. J. Halling (ed.), Progress in biotechnology, vol. 15. Stability and stabilization of biocatalysts. Elsevier Science B.V., Amsterdam, The Netherlands. [Google Scholar]
- 21.Torres-Guzmán, R., I. de la Mata, J. Torres-Bacete, M. Arroyo, M. P. Castillón, and C. Acebal. 2002. Substrate specificity of penicillin acylase from Streptomyces lavendulae. Biochem. Biophys. Res. Commun. 291:593-597. [DOI] [PubMed] [Google Scholar]