Abstract
The goal of this study was to evaluate methanogen diversity in animal hosts to develop a swine-specific archaeal molecular marker for fecal source tracking in surface waters. Phylogenetic analysis of swine mcrA sequences compared to mcrA sequences from the feces of five animals (cow, deer, sheep, horse, and chicken) and sewage showed four distinct swine clusters, with three swine-specific clades. From this analysis, six sequences were chosen for molecular marker development and initial testing. Only one mcrA sequence (P23-2) showed specificity for swine and therefore was used for environmental testing. PCR primers for the P23-2 clone mcrA sequence were developed and evaluated for swine specificity. The P23-2 primers amplified products in P23-2 plasmid DNA (100%), pig feces (84%), and swine waste lagoon surface water samples (100%) but did not amplify a product in 47 bacterial and archaeal stock cultures and 477 environmental bacterial isolates and sewage and water samples from a bovine waste lagoon and a polluted creek. Amplification was observed in only one sheep sample out of 260 human and nonswine animal fecal samples. Sequencing of PCR products from pig feces demonstrated 100% similarity to pig mcrA sequence from clone P23-2. The minimal amount of DNA required for the detection was 1 pg for P23-2 plasmid, 1 ng for pig feces, 50 ng for swine waste lagoon surface water, 1 ng for sow waste influent, and 10 ng for lagoon sludge samples. Lower detection limits of 10−6 g of wet pig feces in 500 ml of phosphate-buffered saline and 10−4 g of lagoon waste in estuarine water were established for the P23-2 marker. This study was the first to utilize methanogens for the development of a swine-specific fecal contamination marker.
Swine waste is a significant source of fecal pollution (1) and can cause contamination of soil, groundwater, and surface waters from lagoon overflow and the use of lagoon surface water for irrigation (19, 27). Studies have shown that spills from swine waste lagoons have high pollution potential with increased levels of nitrogen, phosphorus, and Clostridium perfringens counts of 40,000 CFU·ml−1 (23). A similar study found that addition of pig manure or fecal slurries to agricultural soils led to persistence of pathogens (Salmonella, Listeria, and Campylobacter spp.) on the soil surface (13). Because swine waste can lead to watershed pollution due to runoff from rain events or leaching into groundwater systems, it is important to develop swine-specific fecal markers to identify the source of pollution for effective remediation efforts.
Only two potential methods currently exist for identifying swine waste (7, 17). The STII swine biomarker assay developed by Khatib et al. (17) shows specificity, sensitivity, and geographic stability. However, targeting toxin genes for host-specific source tracking may be problematic due to horizontal gene transfer events occurring in eubacterial populations (25), which may account for the presence of this gene in animals other than swine and humans with diarrhea (17). A swine-specific marker developed using Bacteroides spp. also shows potential specificity for swine (7), although no tests have been conducted to determine the efficacy of this primer for microbial source tracking.
Although many methanogens appear to be specific to the intestinal tract of animals and have the potential for use as host-specific markers of fecal pollution (36), there are no known archaeal markers of swine fecal pollution. Methanogens have been isolated in high numbers from the swine gastrointestinal system in counts of 106 to 108 methanogens per g wet feces (14). Only two methanogen species have been isolated and characterized from swine (5, 14), but molecular studies of swine fecal slurries and waste lagoons have indicated the presence of several unknown methanogens (32, 37) that, if host specific, may be useful for swine-specific marker development.
Because characterizing host distribution patterns of methanogens is essential to delineate potential host-specific archaeal indicators of fecal pollution, a large-scale examination of methanogen-specific mcrA genes in the feces of different host animals was conducted to identify sequences for swine-specific molecular marker development. The mcrA gene, encoding the α subunit of methyl coenzyme M reductase, was targeted due to the conserved nature of the gene, the specificity of the mcrA gene to methanogens, and the use of this gene as an environmental marker for methanogens (10). This study describes the development of an archaeal swine-specific marker of fecal pollution based on host distribution patterns of methanogens in the feces of six different animals (pig, deer, cow, sheep, horse, and chicken) and sewage.
MATERIALS AND METHODS
Prokaryotic species and strains tested for P23-2 assay specificity were as follows: Escherichia coli ATCC 25922; Pseudomonas aeruginosa ATCC 27853; Staphylococcus aureus ATCC 49476; Streptococcus pneumoniae ATCC 49619; Streptococcus bovis ATCC 9809; Serratia marcescens ATCC 13880; Proteus vulgaris ATCC 13315; Shigella sonnei ATCC 9290; Shigella flexneri ATCC 12022; Salmonella enterica subsp. enterica serovar choleraesuis ATCC 14028; Enterococcus faecalis ATCC 29212; Enterococcus faecium ATCC 35667; Enterococcus hirae ATCC 8043; Enterococcus casseliflavus ATCC 700327; Enterococcus durans ATCC 6056; Enterococcus gallinarum ATCC 49573; Enterococcus saccharolyticus ATCC 43076; Enterococcus avium ATCC 14025; Lactococcus lactis ATCC 11454; Methanococcus maripaludis DSMZ 2067; Methanocalculus pumilus DSMZ 12632; Methanocorpusculum aggregans DSMZ 3027; Methanosphaera stadtmaniae DSMZ 3091; Methanocaldococcus infernus DSMZ 11812; Methanofollis liminatans DSMZ 4140; Methanomicrobium mobile DSMZ 1539; Methanomicrococcus blatticola DSMZ 13328; Methanosarcina barkeri DSMZ 800; Methanolobus oregonensis DSMZ 5435; Methanosaeta concilii DSMZ 2139; Methanobacterium bryantii DSMZ 863; Methanobrevibacter oralis DSMZ 7256; Methanobrevibacter ruminantium OCM 146; Methanobrevibacter acididurans OCM 804; Methanobrevibacter wolinii OCM 814; Methanobrevibacter gottschalkii OCM 813; Methanobrevibacter arboriphilus OCM 147; Methanobrevibacter curvatus DSMZ 11111; Methanobrevibacter cuticularis DSMZ 11139; Methanobrevibacter filiformis DSMZ 11501; Methanobrevibacter thaueri DSMZ 11995; Methanobrevibacter woesei OCM 815; and Methanobrevibacter smithii OCM 144, DSMZ 861, DSMZ 2374, DSMZ 2375, and DSMZ 11975.
Bacterial cultures.
Live cultures of methanogens were obtained from the OCM (Oregon Collection of Methanogens; Portland State University, OR) and DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany) for use as positive and negative controls. Methanobrevibacter gottschalkii (strain GP1) was purchased from the Wadsworth Center of the New York State Department of Health (Albany, NY). The DNA was extracted from each of these cultures directly (see below). Cultures of gram-positive and gram-negative organisms were purchased from Chrisope Technologies (Lenexa, KS) and MicroBioLogics, Inc. (St. Cloud, MN), as negative controls for methanogen-specific primers. Aerobic organisms were grown in brain heart infusion (BHI) broth at 37°C for 24 h.
Cloning and sequencing of mcrA genes from animal fecal samples and sewage for swine primer development. (i) PCR.
DNA was extracted from three fecal samples of six different animals (sheep, deer, cow, pig, chicken, and horse) and sewage using the Power Soil DNA extraction kit (MO BIO, Carlsbad, CA). A 470-bp region of the mcrA gene was amplified using the ML primer pair (22) following a modified protocol from the work of Juottonen et al. (15). Each 50-μl PCR assay mixture contained 20 pmol of each ML primer (ML-f, 5′-GGTGGTGTMGGATTCACACARTAYGCWACAGC-3′, and ML-r, 5′-TTCATTGCRTAGTTWGGRTAGTT-3′; IDT, Coralville, IA), 200 μM deoxynucleoside triphosphate (New England Biolabs, Ipswich, MA), 1.5 U Taq polymerase (New England Biolabs), 1× ThermoPol buffer (New England Biolabs), 0.1% bovine serum albumin, and 5 μl fecal DNA template (ranging from 50 to 100 ng DNA). Thermal cycler conditions for the reactions were an initial denaturation for 5 min at 95°C; 40 cycles of 95°C for 40 s, 55°C for 1 min, and 72°C for 1.5 min; and a final elongation for 1 min at 72°C (15). PCR products were separated on a 1.5% agarose gel, excised, and purified using the Zymoclean gel DNA recovery kit (Zymo Research, Orange, CA), and the DNA was quantified.
(ii) Cloning of methanogen PCR Products, RFLP analysis, and sequencing.
Methanogen mcrA PCR products from animal samples were cloned into the pGEM-T vector using the pGEM-T Vector System II (Promega, Madison, WI). Clones were screened for positive transformants using the PCR conditions described above (ML primer pair and corresponding PCR conditions). Restriction fragment length polymorphism (RFLP) analysis was conducted on each of 384 clones to elucidate preliminary differences in animal and sewage clones. The mcrA insert in each clone was amplified using the ML primers and then digested with MspI and TaqI. The restriction fragments were separated on a 3% Synergel (Diversified Biotech, Boston, MA), and each image was digitized. Band matching comparisons were performed for each of the RFLP fingerprints using Bionumerics software, v.3.5 (Applied Maths, Austin, TX). Clones with unique RFLP patterns were identified, and at least three clones of each pattern were chosen for sequencing for a total of 206 clones. Plasmids were purified using the Zyppy Plasmid Miniprep II kit (Zymo Research) and sequenced commercially by Macrogen USA using the T7 promoter primer.
(iii) Sequence comparisons for mcrA gene diversity and primer development.
The mcrA sequences amplified from fecal DNA of different animals were aligned using ClustalW (DNAStar v.5.0) with manual inspection of the alignments. Phylogenetic trees were developed for each animal and for interanimal comparisons using the MEGA 3.1 program (20) with 1,000 bootstrap pseudoreplicates to confirm branching order (Fig. 1). BLAST searches were performed for sequences in each clade to determine the closest relatives. Swine-specific sequences were chosen from swine-specific clades for potential primer development and diagnostic testing.
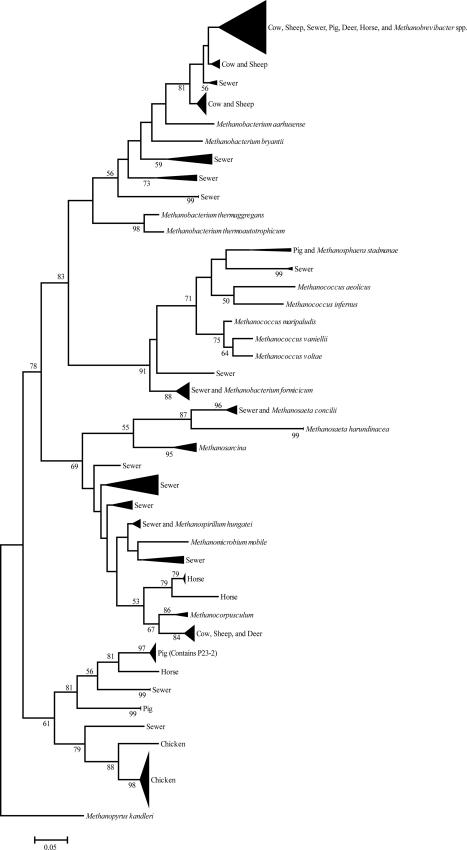
FIG. 1.
Phylogenetic relationships among partial mcrA DNA sequences (470 bp) of clones recovered from fecal and sewer samples (this study) and previously sequenced mcrA genes from methanogen species (italicized). The tree was inferred by the neighbor joining method with 1,000 bootstrap pseudoreplicates using the MEGA 3.1 tree building program and was rooted using the mcrA sequence from Methanopyrus kandleri. The scale bar represents 5% estimated sequence divergence.
Primers were designed for swine clone sequences using DNAStar PrimerSelect v. 5.0 and the Integrated DNA Technologies PrimerQuest program. Primers were subjected to BLAST searches and manual comparison with sequences from clones in each clade, and only those primers with little or no overlap with known sequences were synthesized by Integrated DNA Technologies. Six swine primers were chosen for synthesis initially (P23-1, P23-2, P23-6, P23-7, P23-14, and P1-13) (Table 1) based on negative BLAST search results and manual sequence comparison. Each pair was screened for specificity against pig fecal DNA samples (n = 25) following the amplification conditions listed below. Only the P23-2 primer pair showed swine specificity, and it was the only pair selected for further analysis.
TABLE 1.
Initial primers screened for use as swine molecular markers and the P23-2 IAC developed for diagnostic testing
| Primer clone | Primer sequence | No. of samples positive in pig feces (n = 25 samples) |
|---|---|---|
| P1-13 | f-5′-TTAGCACAAACCGAATCCA-3′ | 1 |
| r-5′-AAGACCTGCGTTTGAGTTACC-3′ | ||
| P23-1 | f-5′-TCTGCGACACCGGTAGCCATTGA-3′ | 16 |
| r-5′-AACATTCTTGAGGATTACACATACTA-3′ | ||
| P23-2 | f-5′-TCTGCGACACCGGTAGCCATTGA-3′ | 21 |
| r-5′-ATACACTGGCGACATTCTTGAGGATTAC-3′ | ||
| P23-6 | f-5′-CCGCAATCATGGAGGCACACTT-3′ | 24 |
| f-5′-CCTCAATTCCATGGGCAGACC-3′ | ||
| P23-7 | f-5′-AGCAGTTCAGTCCAACGTCTGCAA-3′ | 19 |
| f-5′-AGATCAAATCCAAGTACGGCGGCT-3′ | ||
| P23-14 | f-5′-AACTCGGTGACGACATCAACTCCT-3′ | 21 |
| r-5′-TACCAGCAGTTCAGTCCAACGTCT-3′ | ||
| P23-2IAC | 5′-GAGACCGCAGTCTGCGACACCGGTAGCCATTGAACCTGCGATACCGGTTGCTGCTGCTGCAACTGTTGCCTTGTCCTTGATGTAGTCGATTGCATAGTATGTGTAATCCTCAAGAATGTCGCCAGTGTATGCTGCTGTAG-3′ |
Primer specificity. (i) PCR methods.
Samples were tested using two different primer pairs, one for swine specificity (P23-2) and a universal bacterial primer pair used to determine viability of the DNA template prior to diagnostic testing (26). PCR analysis of the primers was carried out in 20-μl amplification reaction mixtures containing 1× PCR buffer, 0.1% bovine serum albumin, 200 μM deoxynucleoside triphosphate, 1 U Taq polymerase (New England BioLabs), 0.5 μM (each) primer, and various amounts of DNA template. The cycling conditions consisted of an initial 92°C step for 2 min and 30 cycles of amplification at 92°C for 30 s, 60°C for 15 s, and 72°C for 30 s. A final elongation was performed at 72°C for 6 min. Positive controls contained purified P23-2 plasmid DNA, and negative controls contained an internal amplification control (IAC) but no other DNA.
(ii) IAC.
An IAC was designed for the P23-2 primer pair as a PCR control (Table 1). The IAC (purchased from IDT, Coralville, IA) was designed by deleting all but 140 bp of the original P23-2 clone mcrA sequence to amplify a 120-bp product using the same forward and reverse primers as those in the P23-2 assay.
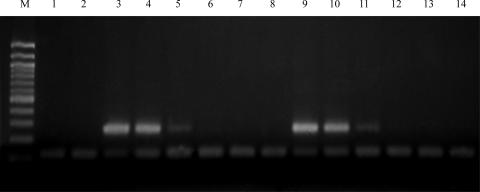
To determine the appropriate concentration of the P23-2 IAC, serial dilutions of the IAC (100 μM to 10−12 μM) were tested with various amounts of P23-2 plasmid control ranging from 100 ng to 0.1 pg plasmid DNA in a 20-μl reaction mixture. The P23-2 IAC was also tested with dilutions of pig fecal DNA to determine a suitable IAC concentration for the lowest level of detection in feces. Three swine fecal DNA samples were diluted to 1 pg·μl−1 and used in PCRs containing dilutions of IAC. The lowest amount of IAC detectable in the presence of swine fecal DNA (10−9 μM) was chosen for diagnostic testing (Fig. 2).
FIG. 2.
PCR amplification of swine fecal dilutions using the P23-2 primer pair developed for the mcrA gene of swine clone P23-2 and 10−9 mM IAC. The expected product is 257 bp; the IAC product is 120 bp. Both swine fecal samples were considered positive controls at high concentrations; no DNA template was added to the negative control; IAC was added to the negative control. Lanes: M, 100-bp ladder; 1, negative control; 2, P6, 0.001 ng; 3, P8, 25 ng; 4, P8, 10 ng; 5, P8, 1 ng; 6, P8, 0.1 ng; 7, P8, 0.01 ng; 8, P8, 0.001 ng; 9, P14, 25 ng; 10, P14, 10 ng; 11, P14, 1 ng; 12, P14, 0.1 ng; 13, P14, 0.01 ng; 14, P14, 0.001 ng.
(iii) Bacterial isolates.
The primers were tested against 15 species of Methanobrevibacter and 12 additional methanogen genera (see above) to determine specificity. A 100-μl sample of each methanogen culture was applied to a Whatman FTA Classic Card (Whatman, Florham Park, NJ) and allowed to dry for 1 hour at room temperature. A 2.0-mm punch from each methanogen sample was prepared as described previously and used directly in a 50-μl PCR mixture (36).
The specificity of each primer pair was further determined by testing a total of 477 gram-positive and gram-negative bacteria isolated from water samples from Hattiesburg Sewage Lagoons in Hattiesburg, MS; two sampling sites along Turkey Creek in Gulfport, MS; the Bouie and Leaf rivers in Hattiesburg, MS; and three Mississippi Department of Environmental Quality coastal sampling sites. Water samples were filtered through a 47-mm-diameter, 0.45-μm-pore-size cellulose acetate filter (Pall Corporation, East Hills, NY); placed onto 50-mm plates containing BHI agar plus 0.02% NaN3 and eosin methylene blue agar; and incubated for 24 to 48 h at 37°C. Isolated colonies (207 from eosin methylene blue agar and 270 from BHI plus NaN3) were picked and grown in BHI at 37°C for 24 h. Whole-cell PCR (36) was performed using 1 μl cells (approximately 1 × 109 cells·ml−1) in a 20-μl reaction mixture as described above.
(iv) Fecal samples.
Fecal samples were collected from individual animals in sterile 50-ml conical centrifuge tubes and transported on ice to the laboratory. Twenty-four goat and sheep fecal samples were collected from different farms. Bovine and swine fecal samples were collected from the South Mississippi Sale Barn in Hattiesburg, MS. Horse (n = 20), swine (n = 20), and bovine (n = 20) samples were collected from farms in northern Mississippi and sent overnight to the lab for DNA extraction. Twenty-four deer fecal samples from captive wild deer fed a pelleted diet for less than a year were collected from the Mississippi State University Wildlife and Fisheries Department. Chicken feces (n = 24) were collected from a chicken processing plant in Mississippi, and dog feces (n = 24) were collected from a local animal shelter. DNA was extracted from fecal samples using the UltraClean soil DNA extraction kit and the Power soil DNA kit (MO BIO) following the manufacturer's instructions. Fecal DNA samples were amplified in 20-μl reaction mixtures as described above using 50 ng DNA as template.
(v) Sewer and animal waste lagoon samples.
Animal waste lagoon and sewage samples (500 ml each) were tested to determine the presence or absence of the P23-2 marker in composite samples. Two samples each (500 ml each) from a bovine waste lagoon and an adjacent creek contaminated with lagoon water were collected, and each sample was centrifuged at 3,000 × g for 15 min. DNA was extracted from the resultant pellet using the MO BIO Power Soil DNA extraction kit (MO BIO).
Samples (50 ml each) from three different swine waste lagoons were collected from nursery (hogs up to 18 days old), sow (breeding sows), and finishing (hogs up to 250 lb) hog farms in northern Mississippi. Sow farm influent (flowing at the time of collection directly from the confinement building), lagoon surface water, the anaerobic layer at the bottom of the lagoon, and sludge samples from the edge of the lagoon were collected. The nursery lagoon samples were collected from the surface water and anaerobic layers of the lagoon. Sludge, surface layer, and anaerobic layer samples were collected from the finishing farm lagoon. Each sample was centrifuged in a 50-ml conical tube at 3,000 × g for 15 min; DNA was extracted from the pellets as described above.
Sewer samples (n = 22) were collected from seven different sewers in Gulfport, MS, each week for a period of 2 months. Sewer samples (500 ml) were prefiltered through a 47-mm-diameter, 3.0-μm-pore-size cellulose acetate filter (Pall Corporation) and then concentrated onto a 47-mm-diameter, 0.2-μm-pore-size Supor-200 membrane (Pall Corporation). Each filter was placed in a sterile 50-ml beaker containing 5 ml of sterile phosphate-buffered saline (PBS) and agitated with a magnetic stir bar for 5 minutes to dislodge bacteria. The samples were then centrifuged for 15 min at 13,000 × g, the cell pellet was resuspended in 100 μl sterile water, and DNA was extracted using the Power Soil DNA kit (MO BIO) or BIO 101 FastDNA spin kit for soil (MP Biomedicals, Solon, OH).
Environmental samples (sewage-contaminated water, swine waste lagoon, and bovine waste lagoon and contaminated creek) were amplified in 20-μl reaction mixtures following the protocol described above with various concentrations of environmental DNA. Bovine waste lagoon and adjacent creek DNA samples were added to a 20-μl reaction mixture using serial dilutions of total DNA to determine the presence or absence of the swine-specific methanogen.
(vi) Environmental water and sediment samples.
Environmental water samples (n = 111) were collected from coastal sampling stations in Harrison County, MS, for two 4-month periods in 2004 and 2005 and analyzed with the P23-2 primer pair. Ten fluvial water samples were also collected from the Bouie River and its tributaries in the Hattiesburg, MS, area for PCR analysis using the P23-2 marker. Water samples (500 ml) were processed in a manner similar to that of the sewage samples by prefiltering and concentrating the bacteria onto a 0.2-μm Supor-200 membrane (Pall Corporation). DNA was extracted from the processed water samples using either the MO BIO Power soil DNA kit (MO BIO) or the BIO 101 FastDNA spin kit for soil (MP Biomedicals). Environmental water samples were amplified in 20-μl and 50-μl reaction mixtures both containing 25 ng and 50 ng of DNA template.
Coastal sediment samples (n = 17) were also taken at four Mississippi Department of Environmental Quality sampling sites (36). DNA was extracted directly from 0.25 g of surface sediments at 15- and 30-cm increments in the sediment cores using the Ultraclean soil DNA extraction kit (MO BIO). Sediment DNA samples were amplified with 1 μl (50 ng) DNA in a 20-μl reaction mixture.
Sequencing of PCR products.
To verify the identity of the PCR products amplified by the P23-2 primers, amplified DNA from five swine fecal samples collected from two different locations was purified using the Zymoclean DNA recovery kit (Zymo Research) and sequenced by Macrogen USA. The sequences were subjected to a BLAST search and aligned using the NCBI bl2seq alignment program (35).
Limits of detection.
The limit of detection for the P23-2 plasmid was determined using serial dilutions of the purified plasmid DNA from 1 to 10−10 ng. One microliter of the plasmid DNA dilution was added to a 20-μl reaction mixture using the conditions described above. In addition, the sensitivity of the P23-2 assay was examined by testing serial dilutions of fresh swine fecal material in PBS. Fecal samples from four pigs (0.25 g [wet weight] each) were combined and added to 500 ml sterile PBS. The samples were blended in a Waring blender at top speed for 2 min to completely resuspend the feces. Diluted samples were processed, and DNA was extracted using the procedure described above for fecal samples. DNA was used in amounts of 10 ng, 25 ng, and 50 ng in 50-μl amplification reaction mixtures as described above.
Method validity.
The usefulness of the P23-2 PCR assay was determined by testing various amounts of DNA extracted from swine waste lagoon samples and dilutions of lagoon waste in filter-sterilized estuarine water. Dilutions of total DNA from each swine lagoon sample (nursery surface, nursery anaerobic, sow surface, sow influent, finishing surface, finishing anaerobic, and finishing surface sludge) were assayed in a 20-μl PCR mixture using the protocol above.
To determine the level of detection of swine lagoon waste in the environment, swine lagoon surface water (50 ml) was centrifuged at 3,000 × g, and the pellet (0.25 g) was added to 500 ml of filter-sterilized estuarine water. The sample was diluted in 10-fold increments and processed in a manner similar to that for the environmental water samples, and the DNA was extracted using the BIO 101 spin kit for soil. Each dilution was assayed in a 20-μl PCR mixture with various concentrations of DNA using the P23-2 protocol above.
Nucleotide sequence accession numbers.
Sequences have been submitted to GenBank under the accession numbers EF628013 to EF628203.
RESULTS
Preliminary diversity results and sequence comparison for primer development.
The mcrA clones formed many host-specific clades easily distinguishable among animal species (Fig. 1). Pig fecal clones showed four distinct clades, with one showing similarity to cow, sheep, deer, and sewer. Chicken clones formed two clades distinct from other animal and sewage clones. Sewage clones formed 16 distinct clades, with only one showing similarity to other animal fecal clones. Ruminant (deer, sheep, and cow) sequences generally clustered together, with three ruminant-specific clusters observed. Four horse clusters were observed, with three clusters being horse specific and one being phylogenetically similar to other animals and Methanobrevibacter species.
Specificity.
P23-2 was the only primer pair among the six developed that was useful for the detection of swine feces. The P23-2 primers produced an amplicon of 258 bp, and BLAST searches did not reveal any homology with bacterial or methanogen sequences in any of the available databases. The P23-2 primer pair produced the expected product in swine fecal DNA from two different farms (84%), swine lagoon surface water (100%), swine lagoon sludge samples (100%), and positive plasmid DNA but not in eubacterial cultures found in feces or environmental bacteria tested (Table 2). Of the 25 methanogens tested to determine cross-amplification with mcrA genes of related genera, no methanogen mcrA genes were amplified with the P23-2 primer pair. More importantly, the primers did not amplify products in human or animal feces (cow, dog, sheep, goat, deer, rat, horse, and chicken), cow waste lagoon water, creek water contaminated with cow lagoon waste, or sewage. Only one sheep sample out of a total of 260 human and nonswine animal fecal DNA samples showed amplification using the P23-2 primers. Environmental samples (nonpolluted estuarine water and sediments and fluvial water) showed no amplification of the P23-2 mcrA gene with 25 ng and 50 ng total DNA added to the reaction mixture (Table 2).
TABLE 2.
Environmental samples tested for P23-2 specificity
| Expectation status and sample source or type | No. of samples
|
|
|---|---|---|
| Total tested | P23-2 positive | |
| Expected to be positive: swine waste lagoon | ||
| Surface samples | 3 | 3 |
| Sludge samples | 1 | 1 |
| Sow influent | 1 | 1 |
| Anaerobic layer samples | 3 | 0 |
| Not expected to be positive | ||
| Bovine waste lagoon | 2 | 0 |
| Bovine waste-contaminated creek | 2 | 0 |
| Sewer | 22 | 0 |
| Coastal water and creek samples | 111 | 0 |
| Coastal sediment | 17 | 0 |
| Environmental bacteria | 477 | 0 |
| Fluvial water samples | 10 | 0 |
The P23-14 primer pair amplified 84% of swine fecal DNA samples and no sewer samples; however, the expected amplicon was also observed in 96% of sheep and 75% of goat samples (Table 3). Because P23-14 was not swine specific, it was removed from subsequent testing. All of the other four initial primer pairs chosen were removed after initial testing due to lack of amplification in pig feces (P1-13 and P23-1), production of multiple bands (P23-7), or amplification of sewer samples (P23-6).
TABLE 3.
Fecal samples tested for P23-2 and P23-14 primer specificity
| Source of tested fecal sample | No. of samples tested | No. of samples in which a product was amplified by primer/total no. of samples tested
|
|
|---|---|---|---|
| P23-2 | P23-14 | ||
| Swine | 25 | 21/25 | 21/25 |
| Human | 50 | 0/50 | 0/50 |
| Cow | 50 | 0/50 | 0/50 |
| Sheep | 24 | 1/24 | 23/24 |
| Rat | 20 | 0/20 | 0/20 |
| Horse | 20 | 0/20 | 0/20 |
| Deer | 24 | 0/24 | 0/20 |
| Goat | 24 | 0/24 | 18/24 |
| Chicken | 24 | 0/24 | 0/24 |
| Dog | 24 | 0/24 | 0/24 |
Detection limits and method validation.
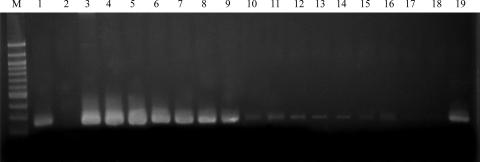
The sensitivity of the P23-2 PCR was evaluated by amplifying P23-2 mcrA genes from serial dilutions of plasmid with the addition of IAC. The minimal amount of DNA required for amplification of the P23-2 plasmid was 1 pg (Fig. 2 and 3). PCR amplification of serial dilutions of total DNA from pig feces with IAC showed a detection limit of 1 ng in pig feces (data not shown). Serial dilutions of pig feces in PBS showed a lower detection limit of the P23-2 mcrA gene to 10 μg of feces in 500 ml of PBS (Fig. 2). Swine lagoon waste diluted in estuarine water showed a lower detection limit to 10−4 g in 500 ml estuarine water using 10 ng DNA template (data not shown).
FIG. 3.
PCR amplification of P23-2 plasmid dilutions and pig fecal dilutions in PBS for detection limits for diagnostic testing using the P23-2 primer pair. The expected product is 257 bp. The positive control is swine fecal sample P23; the negative control contains no DNA template. Lane M, 100-bp ladder. Lanes 1 to 10 correspond to P23-2 plasmid dilutions: 1, positive control; 2, negative control; 3, P23-2 plasmid, 100 ng; 4, P23-2 plasmid, 10 ng; 5, P23-2 plasmid, 1 ng; 6, P23-2 plasmid, 0.1 g; 7, P23-2 plasmid, 0.01 ng; 8, P23-2 plasmid, 0.001 ng; 9, P23-2 plasmid, 1 pg; 10, P23-2 plasmid, 0.1 pg. Lanes 11 to 19 correspond to pig fecal dilutions in PBS: 11, 100 mg feces; 12, 10 mg feces; 13, 1 mg feces; 14, 100 ng feces; 15, 10 ng feces; 16, 1 ng feces; 17, 0.1 ng feces; 18, 0.01 ng feces; 19, positive control.
When the sequenced PCR products were aligned using bl2seq, the products amplified with the P23-2 primer pair from pig fecal DNA were 100% identical to the P23-2 mcrA clone sequence. Amplification was observed in all swine waste surface water samples from hog farms tested (nursery, sow, and finishing farms) with a detection limit of 50 ng. Detection was also observed to a limit of 1 ng and 10 ng in sow waste influent and sludge samples from the finishing farm, respectively. No amplification was seen in samples from the anaerobic layers of the lagoon.
DISCUSSION
Problems with identifying the sources of fecal pollution in surface waters hamper efforts to adequately remediate areas of high contamination. Although many methods exist to quickly and accurately identify human and ruminant sources of contamination (3, 6, 16, 30, 36), there are only two methods, both based on the detection of eubacteria, designed to identify possible swine sources of fecal pollution (7, 17). The effectiveness of these two methods in microbial source tracking is not known at the present time, although one disadvantage of the swine STII biomarker (17) is that the method requires cultivation to remove possible false-negative results. The present study documents the development of a new swine-specific fecal pollution marker that is culture independent and is the first report targeting archaea.
The usefulness of a host-specific indicator of fecal contamination is dependent on the specificity of the marker to the host animal. The P23-2 marker developed in this study showed high specificity to swine fecal samples (84%) collected from different farms. More importantly, the P23-2 assay amplified all swine waste lagoon surface samples from three different farms but did not amplify samples from a bovine waste lagoon, a creek contaminated with bovine waste, or sewage samples. In addition, sequencing confirmed that the expected products from swine fecal samples showed 100% identity to the P23-2 clone mcrA sequence. Inhibitory factors present in DNA extracts from environmental samples including fecal, sewage, animal waste lagoon, and environmental water and sediment samples routinely cause false-negative results using PCR (12). Therefore, an IAC was developed and used in this assay as a positive control to ensure that negative results from environmental samples were not due to PCR inhibition. These data show the usefulness of the P23-2 assay as a method for detecting swine-specific fecal contamination.
The P23-2 assay amplified one sheep sample out of a total of 260 human and animal fecal samples. Results from the present study indicate that exclusivity in a particular animal is not a requirement for a fecal indicator to be useful in microbial source tracking. An associated study proposed Bacteroides thetaiotamicron as a human-specific marker for sewage pollution, even though 16% of dog fecal samples were positive using the assay (6). This suggests that host-specific markers of fecal pollution are useful despite their occasional occurrence in nontarget animal sources. Therefore, the weak amplification of only one sheep fecal sample would not compromise the usefulness of the P23-2 swine assay.
Because methanogens are found in a multitude of environmental niches including fresh and marine water and sediments (10), it was important to test nonpolluted surface waters for the presence of the uncultured swine-specific methanogen to determine if the organism is found outside the swine gastrointestinal tract. The lack of P23-2 amplification in 138 nonpolluted environmental samples with up to 50 ng of total DNA indicates that the methanogen targeted by the P23-2 marker is not a normal inhabitant of the marine or fluvial environments and that the assay developed is not detecting other organisms in these environments.
Methanogenic archaeal communities can change from the storage tanks to the storage pond in a swine waste system (27); therefore, samples taken at several locations in three lagoon systems at two different hog farms were tested with the P23-2 markers developed in this study. Swine fecal samples were also collected at two different farms in a different location than that of the farms sampled for lagoon waste. Amplification using the P23-2 marker in swine feces, surface lagoon water from all lagoons, waste lagoon influent, and lagoon sludge samples showed that the marker is not replaced by another methanogen and is stable in different swine populations. The absence of signal in the anaerobic layers of the swine lagoons may be due to the amount of product used for DNA extraction. Only 0.25 g of each anaerobic layer pellet was used for extraction, thereby limiting the amount of product for detection. Also, the surface layer of the swine lagoon system is the important target for microbial source tracking because it is used for field irrigation and fertilization. This study showed amplification with the P23-2 marker in all surface lagoon water samples, and the marker is therefore a potential indicator for swine lagoon waste runoff.
Samples collected in swine waste lagoon surface waters showed sensitivity of the P23-2 assay with detection to 50 ng total DNA. PCR sensitivity for the P23-2 plasmid was measured to 10−12 g of DNA, which is comparable to the plasmid detection limit for a previously identified ruminant primer (3). The P23-2 marker successfully amplified DNA extracted from pig fecal dilutions to 10−6 g fecal material in 500 ml PBS. This level of detection was comparable to the detection of cow feces using the Bacteroides ruminant primers described previously (3). Further, swine lagoon waste was detected to 10−4 g in estuarine water, which suggests that the swine-specific P23-2 marker developed in this study is sensitive and specific and has potential as an indicator of swine lagoon waste in the environment.
An interesting feature of the phylogenetic analysis of mcrA genes in this study was the methanogen diversity present in each of the host animals. Previous studies have hypothesized greater methanogen diversity in ruminants than in monogastric animals (29). In contrast, this study showed higher levels of methanogen diversity in monogastric animals than in ruminant animals. Of 24 mcrA sequences obtained from horse feces, four different clades were observed, showing 17% diversity. This level of diversity was the highest observed in this study for individual animals and may correlate with high levels of methane production seen previously in horses (14), indicating the presence of unknown or uncharacterized methanogens in the horse cecum. To date only Methanobrevibacter gottschalkii strain HO has been isolated and characterized (24).
In this study, swine fecal mcrA sequences were most similar to Methanobrevibacter and Methanosphaera spp. and uncultured methanogens. Presently, only Methanobrevibacter gottschalkii strain PG and Methanosarcina spp. have been isolated and characterized from the swine gastrointestinal tract (4, 24), indicating the presence of several uncultured and uncharacterized methanogen species. Of 45 swine fecal clones, five different mcrA clades were observed, corresponding to 11% diversity. This is lower than that observed among clones originating from a pig slurry in a previous study using the 16S rRNA gene (18.8% [32]). This difference in sequence diversity is likely due to either a difference in the number of pigs studied or a possible difference in the rate of divergence between the genes for mcrA and 16S rRNA. The mcrA sequences in the present study originated from three individual pigs whereas the 16S rRNA sequences originated from waste slurries containing the feces of a large number of pigs (32). This relationship is also seen in the high level of diversity observed in sewage in this study (19%) compared to a study showing only one phylotype (Methanobrevibacter smithii) observed out of 1524 archaeal 16S rRNA sequences from the feces of three individual humans (9).
Only two different clades were observed in 48 chicken clones (4% diversity). This is similar to a previous study in which only 2.6% diversity was observed in chicken fecal samples using the 16S rRNA gene (29). Most of the methanogen species previously identified and characterized from the animal gastrointestinal tract were Methanobrevibacter spp. (8), and it has been hypothesized that chickens harbor Methanobrevibacter woesei as the predominant species (29). This study, however, showed no sequences similar to Methanobrevibacter spp. in 46 chicken fecal mcrA sequences. Chicken methanogen clones in this study showed no similarity to previously characterized methanogens and formed clades specific only to chicken. This discrepancy in chicken methanogen diversity may be due to use of the mcrA gene (this study) versus the 16S rRNA gene (29), although the ML primers for the mcrA gene have been shown to be comparable to the 16S rRNA gene for phylogenetic studies (22). Alternatively, the differences between the two studies may be due to the breed of chicken studied or factors such as the age of the bird, the diet, or antibiotic use (18).
Ruminant fecal mcrA sequences from this study showed closest similarity to Methanocorpusculum, Methanobacterium, and Methanobrevibacter species. Methanobrevibacter is a dominant genus found in ruminants (31), but to date no Methanocorpusculum species have been cultured from the rumen. Only four different clades were observed among 190 cow, sheep, and deer clones, corresponding to only a 2% overall level of diversity. This is lower than levels observed in previous studies of cow and sheep methanogen diversity. Tatsuoka et al. (34) observed four different groups of mcrA clones in the cattle rumen representing 11% diversity, and Whitford et al. (38) showed 41 clones grouped into four clusters representing 9.8% diversity. Wright et al. (39) observed 65 methanogen phylotypes of 733 total clones in sheep (based on the 16S rRNA gene) corresponding to 8.9% clonal diversity. The lower level of diversity may be due to use of cattle feces rather than rumen fluid or the number of individual fecal samples studied or may be due to age, health, and diet differences in the individual animals (33, 38).
The distribution of mcrA sequences among the different animal groups suggests both endemic and cosmopolitan methanogen populations with implications for microbial source tracking. This study showed that most methanogen sequences clustered as endemic populations specific to host animals. The few cosmopolitan populations from each animal clustered together and were phylogenetically similar to Methanobrevibacter species. This finding is not surprising since Methanobrevibacter is the predominant methanogen genus in the animal gastrointestinal tract (11, 21). Methanobrevibacter species include M. ruminantium, M. millerae, and M. olleyae (ruminant specific); M. arboriphilus (plant specific); M. cuticularis, M. curvatus, and M. filiformis (termite specific); M. oralis (human mouth); M. gottschalkii and M. thaueri (horse and pig); M. woesei (rat and goose); and M. acididurans and M. wolinii (sheep) (8, 28). Although many species of Methanobrevibacter are found in more than one animal (i.e., M. gottschalkii in horses and pigs and M. woesei in different gallinaceous birds [8, 29]), many are believed to inhabit host-specific niches. M. ruminantium occupies a ruminant-specific niche due to a strict growth requirement found only in ruminal fluid, which precludes growth of this organism outside the rumen (2). M. smithii occupies a niche specific to the human gastrointestinal system and is considered the predominant methanogen in this system (9, 36). Sequence similarity among fecal clones in this study suggests that cosmopolitan sequences were related to Methanobrevibacter and are likely separate species of Methanobrevibacter inhabiting specific niches in different animal systems.
It has been suggested that differences in host animal gastrointestinal systems create unique environments allowing for host-specific bacterial niches that may be useful for microbial source tracking (7). This study showed distinct endemic populations of methanogen sequences, suggesting the presence of uncultured methanogens inhabiting host-specific niches in chickens, horses, pigs, sewage, and ruminants. Endemic distributions were observed in pig fecal mcrA sequences with several swine-specific clades allowing for microbial source tracking marker development. Several horse-specific and ruminant-specific clusters were also observed, representing endemic distribution.
Endemic distribution was also represented in the chicken and the sewage environments, with no chicken mcrA sequences similar to other animals and only one sewage mcrA sequence shared with other animal sequences (similar to Methanobrevibacter species). The presence of endemic populations in these different systems suggests evidence for host animal-specific methanogen niches in animals with different gastrointestinal systems.
Conclusions.
This study showed the potential for using the methanogen-specific mcrA gene to identify host-specific methanogens for microbial source tracking, as well as using methanogens as swine-specific markers of fecal pollution. The P23-2 assay developed in this study shows promise as a sensitive, rapid, reliable, and specific method for identifying swine contamination in the environment, although further testing is required to determine the applicability of the assay in different geographical settings.
This study is the first to identify phylogenetic relationships between mcrA genes in sewage and the feces of different animals. Understanding the nature and host distribution patterns of methanogens in intestinal systems of different animals will allow for a greater appreciation of host-methanogen interactions, knowledge of uncultured methanogens in different environments, and design of microbial source tracking host-specific markers. Future studies will concentrate on the ecological implications and host distribution patterns of the methanogens in different intestinal environments.
Acknowledgments
This work was supported by the National Oceanic and Atmospheric Administration Oceans and Human Health Program under the grant NA04OAR4600216 and by the U.S. Environmental Protection Agency, Gulf of Mexico Program Office, through grants MX964295 and MX964012.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Ackerman, E. O., and A. G. Taylor. 1995. Stream impact due to feedlot runoff, p. 119-125. In K. Steele (ed.), Animal waste and the land-water interface. Lewis Publishers, Boca Raton, FL.
- 2.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard, A. E., and K. G. Field. 2000. PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boopathy, R. 1996. Isolation and characterization of a methanogenic bacterium from swine manure. Bioresour. Technol. 55:231-235. [Google Scholar]
- 5.Butine, T. J., and J. A. Leedle. 1989. Enumeration of selected anaerobic bacterial groups in cecal and colonic contents of growing-finishing pigs. Appl. Environ. Microbiol. 55:1112-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson, C. A., J. M. Christiansen, H. Yampara-Iquise, V. W. Benson, C. Baffaut, J. V. Davis, R. R. Broz, W. B. Kurtz, W. M. Rogers, and W. H. Fales. 2005. Specificity of a Bacteroides thetaiotamicron marker for human feces. Appl. Environ. Microbiol. 71:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dighe, A. S., K. Jangid, J. M. Gonzalez, V. J. Pidiyar, M. S. Patole, D. R. Ranade, and Y. S. Shouche. 2004. Comparison of 16S rRNA gene sequences of genus Methanobrevibacter. BMC Microbiol. 4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedrich, M. W. 2005. Methyl-coenzyme M reductase gene: unique functional markers for methanogenic and anaerobic methane-oxidizing Archaea. Methods Enzymol. 397:428-442. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, J. L., B. K. C. Patel, and B. Ollivier. 2000. Taxonomic, phylogenetic, and ecological diversity of methanogenic archaea. Anaerobe 6:205-226. [DOI] [PubMed] [Google Scholar]
- 12.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchison, M. L., L. D. Walters, T. Moore, D. J. Thomas, and S. M. Avery. 2005. Fate of pathogens present in livestock wastes spread onto fescue plots. Appl. Environ. Microbiol. 71:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, B. B. 1996. Methanogenesis in monogastric animals. Environ. Monit. Assess. 42:99-112. [DOI] [PubMed] [Google Scholar]
- 15.Juottonen, H., P. E. Galand, E. S. Tuittila, J. Laine, H. Fritze, and K. Yrjala. 2005. Methanogen communities and bacteria along an ecohydrological gradient in a northern raised bog complex. Environ. Microbiol. 7:1547-1557. [DOI] [PubMed] [Google Scholar]
- 16.Khatib, L. A., Y. L. Tsai, and B. H. Olson. 2002. A biomarker for the identification of cattle fecal pollution in water using the LTIIa toxin gene from enterotoxigenic Escherichia coli. Appl. Microbiol. Biotechnol. 59:97-104. [DOI] [PubMed] [Google Scholar]
- 17.Khatib, L. A., Y. L. Tsai, and B. H. Olson. 2003. A biomarker for the identification of swine fecal pollution in water, using the STII toxin gene from enterotoxigenic Escherichia coli. Appl. Microbiol. Biotechnol. 63:231-238. [DOI] [PubMed] [Google Scholar]
- 18.Knarreborg, A., M. A. Simon, R. M. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 68:5918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krapac, I. G., W. S. Dey, W. R. Roy, C. A. Smyth, E. Storment, S. L. Sargent, and D. Steele. 2002. Impacts of swine manure pits on groundwater quality. Environ. Pollut. 120:475-492. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 21.Lin, C., and T. L. Miller. 1998. Phylogenetic analysis of Methanobrevibacter isolated from feces of humans and other animals. Arch. Microbiol. 169:397-403. [DOI] [PubMed] [Google Scholar]
- 22.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 23.Mallin, M. A., J. M. Burkholder, M. R. McIver, G. C. Shank, H. B. Glasgow, Jr., B. W. Touchette, and J. Springer. 1997. Comparative effects of poultry and swine waste lagoon spills on the quality of receiving streamwaters. J. Environ. Qual. 26:1622-1631. [Google Scholar]
- 24.Miller, T. L., and C. Lin. 2002. Description of Methanobrevibacter gottschalkii sp. nov., Methanobrevibacter thaueri sp. nov., Methanobrevibacter woesei sp. nov. and Methanobrevibacter wolinii sp. nov. Int. J. Syst. Evol. Microbiol. 52:819-822. [DOI] [PubMed] [Google Scholar]
- 25.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 26.Ovreas, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peu, P., H. Brugere, A. M. Pourcher, M. Kerouredan, J. J. Godon, J. P. Delgenes, and P. Dabert. 2006. Dynamics of a pig slurry microbial community during anaerobic storage and management. Appl. Environ. Microbiol. 72:3578-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rea, S., J. P. Bowman, S. Popovski, C. Pimm, and A. G. Wright. 2007. Methanobrevibacter millerae sp. nov. and Methanobrevibacter olleyae sp. nov., methanogens from the ovine and bovine rumen that can utilize formate for growth. Int. J. Syst. Evol. Microbiol. 57:450-456. [DOI] [PubMed] [Google Scholar]
- 29.Saengkerdsub, S., R. C. Anderson, H. H. Wilkinson, W. K. Kim, D. J. Nisbet, and S. C. Ricke. 2007. Identification and quantification of methanogenic archaea in adult chicken ceca. Appl. Environ. Microbiol. 73:353-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott, T. M., T. M. Jenkins, J. Lukasik, and J. B. Rose. 2004. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol. 39:283-287. [PubMed] [Google Scholar]
- 31.Smith, P. H., and R. E. Hungate. 1958. Isolation and characterization of Methanobacterium ruminantium n. sp. J. Bacteriol. 75:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snell-Castro, R., J. J. Godon, J. P. Delgenès, and P. Dabert. 2005. Characterization of the microbial diversity in a pig manure storage pit using small subunit rDNA sequence analysis. FEMS Microbiol. Ecol. 52:229-242. [DOI] [PubMed] [Google Scholar]
- 33.Tajima, K., T. Nagamine, H. Matsui, M. Nakamura, and R. I. Aminov. 2001. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol. Lett. 200:67-72. [DOI] [PubMed] [Google Scholar]
- 34.Tatsuoka, N., N. Mohammed, M. Mitsumori, M. Kurihara, and H. Itabashi. 2004. Phylogenetic analysis of methyl coenzyme-M reductase detected from the bovine rumen. Lett. Appl. Microbiol. 39:257-260. [DOI] [PubMed] [Google Scholar]
- 35.Tatusova, T. A., and T. L. Madden. 1999. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 36.Ufnar, J. A., S. Wang, J. M. Christiansen, H. Yampara-Iquise, C. A. Carson, and R. D. Ellender. 2006. Detection of the nifH gene of Methanobrevibacter smithii: a potential tool to identify sewage pollution in recreational waters. J. Appl. Microbiol. 101:44-52. [DOI] [PubMed] [Google Scholar]
- 37.Whitehead, T. R., and M. A. Cotta. 1999. Phylogenetic diversity of methanogenic archaea in swine waste storage pits. FEMS Microbiol. Lett. 179:223-226. [DOI] [PubMed] [Google Scholar]
- 38.Whitford, M. F., R. M. Teather, and R. J. Forster. 2001. Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright, A. D. G., A. J. Williams, B. Winder, C. T. Christophersen, S. L. Rodgers, and K. D. Smith. 2004. Molecular diversity of rumen methanogens from sheep in Western Australia. Appl. Environ. Microbiol. 70:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]