Abstract
Prior research revealed that Polaromonas naphthalenivorans CJ2 carries and expresses genes encoding the gentisate metabolic pathway for naphthalene. These metabolic genes are split into two clusters, comprising nagRAaGHAbAcAdBFCQEDJI′-orf1-tnpA and nagR2-orf2I″KL (C. O. Jeon, M. Park, H. Ro, W. Park, and E. L. Madsen, Appl. Environ. Microbiol. 72:1086-1095, 2006). BLAST homology searches of sequences in GenBank indicated that the orf2 gene from the small cluster likely encoded a salicylate 5-hydroxylase, presumed to catalyze the conversion of salicylate into gentisate. Here, we report physiological and genetic evidence that orf2 does not encode salicylate 5-hydroxylase. Instead, we have found that orf2 encodes 3-hydroxybenzoate 6-hydroxylase, the enzyme which catalyzes the NADH-dependent conversion of 3-hydroxybenzoate into gentisate. Accordingly, we have renamed orf2 nagX. After expression in Escherichia coli, the NagX enzyme had an approximate molecular mass of 43 kDa, as estimated by gel filtration, and was probably a monomeric protein. The enzyme was able to convert 3-hydroxybenzoate into gentisate without salicylate 5-hydroxylase activity. Like other 3-hydroxybenzoate 6-hydroxylases, NagX utilized both NADH and NADPH as electron donors and exhibited a yellowish color, indicative of a bound flavin adenine dinucleotide. An engineered mutant of P. naphthalenivorans CJ2 defective in nagX failed to grow on 3-hydroxybenzoate but grew normally on naphthalene. These results indicate that the previously described small catabolic cluster in strain CJ2 may be multifunctional and is essential for the degradation of 3-hydroxybenzoate. Because nagX and an adjacent MarR-type regulatory gene are both closely related to homologues in Azoarcus species, this study raises questions about horizontal gene transfer events that contribute to operon evolution.
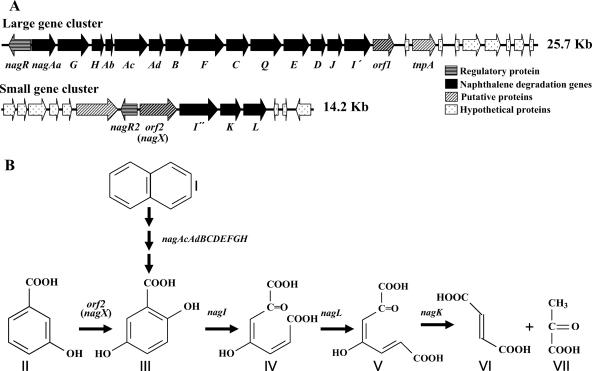
Polaromonas naphthalenivorans CJ2, previously found to be responsible for the degradation of naphthalene in situ at a coal tar waste-contaminated site (10), is able to utilize naphthalene as the sole carbon source. Recent sequencing and analysis of the entire naphthalene degradation pathway of P. naphthalenivorans CJ2 (12) revealed that the naphthalene-catabolic (nag) genes in P. naphthalenivorans CJ2 are homologous to those in Ralstonia sp. strain U2, which metabolizes naphthalene via the gentisate pathway, converting naphthalene into fumarate and pyruvate via salicylate (2-hydroxybenzoate) and gentisate (2,5-dihydroxybenzoate) (5). This finding has recently been confirmed biochemically (G. M. Pumphrey and E. L. Madsen, submitted for publication). In contrast to the nag genes of strain U2, which are organized in a single operon, the naphthalene-catabolic genes of strain CJ2 are organized into two gene clusters (Fig. 1A), comprising nagRAaGHAbAcAdBFCQEDJI′- orf1-tnpA (large cluster) and nagR2-orf2I″KL (small cluster). LysR-type (nagR) and MarR-type (nagR2) transcriptional regulators act, respectively, to up-regulate and down-regulate the growth of strain CJ2 on naphthalene in mineral salts basal (MSB) medium (12).
FIG. 1.
(A) Physical maps of naphthalene degradation genes from P. naphthalenivorans CJ2. (B) Gentisate pathway for the degradation of naphthalene and 3-hydroxybenzoate. I, naphthalene; II, 3-hydroxybenzoate; III, gentisate; IV, maleylpyruvate; V, fumarylpyruvate; VI, fumarate; VII, pyruvate.
The nag genes in strain U2 have been sequenced and cloned and their functions have been characterized previously (5, 14, 36, 37). However, the role of the second gene in the small gene cluster, orf2, in naphthalene catabolism by strain CJ2 is still uncertain. BLAST homology searches of sequences in GenBank indicated that the orf2-encoded protein (Orf2) was most closely related to a putative salicylate 5-hydroxylase (88.3% amino acid sequence identity) from Azoarcus sp. strain EbN1 that catalyzes the conversion of salicylate into gentisate. Recently, two related proteins (MhbM from Klebsiella pneumoniae M5a1 [18] and XlnD from Pseudomonas alcaligenes NCIMB 9867 [6]) with low levels of amino acid sequence identity to Orf2 (55.6 and 72.2%, respectively) have been reported to act as 3-hydroxybenzoate 6-hydroxylases. Additional information available on the genetics and biochemistry of 3-hydroxybenzoate 6-hydroxylase is limited largely to that from studies using five model organisms, K. pneumoniae (13, 18, 30), P. alcaligenes (6, 21, 22), Burkholderia (formerly Pseudomonas) cepacia (23), Micrococcus sp. (33), and Pseudomonas aeruginosa (8).
Here, we show that orf2 from strain CJ2 encodes 3-hydroxybenzoate 6-hydroxylase, not salicylate 5-hydroxylase. The orf2 gene product was expressed in Escherichia coli and characterized. Furthermore, when an orf2 knockout mutant of P. naphthalenivorans CJ2 was constructed, we found that orf2 is involved in the degradation of 3-hydroxybenzoate, not naphthalene (Fig. 1B). For this reason, we renamed orf2 nagX. These findings mean that orf2 (nagX) is essential for the degradation of 3-hydroxybenzoate and suggest that in strain CJ2, the small cluster may be multifunctional.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains, vectors, and plasmids used in the present study are listed in Table 1. P. naphthalenivorans CJ2 was grown at 20°C and maintained on MSB medium (28) with either naphthalene vapor or a 0.5% (wt/vol) suspension of naphthalene crystals (MSB-N), 0.3% (wt/vol) pyruvate (MSB-P), or 0.3% (wt/vol) 3-hydroxybenzoate as the sole carbon source. Spontaneously rifampin-resistant P. naphthalenivorans CJ2 colonies were selected on R2A plates (Difco) containing a rifampin gradient (0 to 200 μg/ml) (29). All E. coli strains were grown at 37°C in Luria-Bertani (LB) medium on a rotary incubator (180 rpm). When required, the appropriate antibiotics and reagents were added to the medium: X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 30 μg/ml), IPTG (isopropyl-β-d-thiogalactopyranoside; 40 μg/ml), kanamycin (20 μg/ml), rifampin (200 μg/ml), salicylate (200 μM), and 3-hydroxybenzoate. Growth was monitored by measuring the optical densities of the cultures at a wavelength of 600 nm (OD600s) in a UV-visible spectrophotometer (DU series 530; Beckman).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| P. naphthalenivorans | ||
| CJ2 | Naphthalene degrader | 11 |
| CJM110 | P. naphthalenivorans CJ2 ΔnagR2::Km lacZ+ | 12 |
| CJM112 | P. naphthalenivorans CJ2 nagR2-nagX::lacZ+ | 12 |
| CJ2ΔnagXKm | P. naphthalenivorans CJ2 ΔnagX::Km lacZ+sacB | This study |
| CJ2ΔnagX | P. naphthalenivorans CJ2 ΔnagX | This study |
| E. coli | ||
| S17-1/λpir | Carries RK2 tra regulon and pir; host for pir-dependent plasmids | 15 |
| BL21(DE3) | F−dcm ompT hsdS(rB− mB−) gal λ (DE3) | Novagen |
| Plasmids | ||
| pET21b | Expression vector; Apr; carries T7 tag, multiple cloning site, His tag | Novagen |
| pBluescript II SK(−) | Cloning vector; carries multiple cloning site in lacZα; Apr | Stratagene |
| pK19mobsacB | Integration vector; Kmr oriVEc oriT sacB | 25 |
| pET21bnagX | NdeI-BamHI-cut PCR fragment containing nagX inserted into pET21b | This study |
| pBSIInagX | 1,072-bp XbaI-SalI-cut PCR fragment containing nagX inserted into pBluescript II SK(−) | This study |
| pBSIIΔnagX | Carries deletion of the 14-bp fragment of nagX from pBSIInagX | This study |
| pK19mobsacBΔnagX | Carries insertion of the 1,058-bp XbaI-SalI insert from pBSIIΔnagX into pK19mobsacB | This study |
DNA manipulation, sequencing, and sequence data analysis.
DNA manipulations and other molecular biology techniques were carried out by using established standard protocols (24). PCR primers were designed with the aid of the Lasergene software package (DNAStar, Inc., Madison, WI). DNA sequencing was performed by using an ABI model 3700 instrument at Genotech (Korea). DNA sequences were compiled and analyzed with the Lasergene software package (DNAStar). BLASTX and blastn were used, respectively, for the deduced-amino-acid-sequence and nucleotide sequence identity determinations (1).
Overexpression and identification of nagX in E. coli.
To overexpress nagX in E. coli, the nagX gene was cloned into the expression vector pET21b (Novagen). The nagX gene was PCR amplified from wild-type CJ2 genomic DNA by using Pfu DNA polymerase (Solgent, Korea) and primer pair nagX-forward (5′-AAACATATGAGCGACAACCCTGCAGATTT-3′) and nagX-reverse (5′-CTGGGATCCCGGGTGCCGGTCAGTCTTTG-3′), which contained NdeI (underlined) and BamHI (underlined) restriction sites, respectively. The amplified PCR product obtained was then excised with NdeI and BamHI and ligated into pET21b, resulting in the recombinant plasmid designated pET21bnagX. E. coli BL21(DE3) was subsequently transformed with the pET21bnagX construct to produce the protein NagX.
Biochemical analysis of the NagX protein.
E. coli BL21(DE3) cells carrying pET21bnagX were grown overnight in 5 ml of LB broth containing kanamycin. A 2-ml aliquot of the overnight culture was inoculated into 200 ml of fresh LB broth, and the cells were grown with shaking at 37°C (to an OD600 of ca. 0.6). The culture was induced with IPTG (1 mM) and incubated 4 h, and the cells were harvested (9,300 rpm for 10 min at 4°C). Crude cell extracts were prepared by resuspending the bacterial pellets into ice-cold 100 mM potassium phosphate buffer (pH 7.4) and disrupting them by sonication in an ice-water bath for three periods of 30 s with 30-s intervals, after which cell debris was removed by centrifugation at 24,650 rpm for 30 min at 4°C.
Crude cell extracts containing the NagX protein were applied to a DEAE column (Amersham Biosciences). The column containing the crude cell extract was washed with binding buffer (30 mM sodium phosphate [pH 7.5]), and the protein was released with elution buffer (30 mM sodium phosphate, 1 M NaCl [pH 7.5]) at a flow rate of 5.0 ml/min. Fractions (0.5 ml) were collected, and the desired fractions were concentrated by using Centriprep (Millipore). Gel filtration of the partially purified recombinant NagX for additional purification was carried out by using Superdex-200 10/300 GL column (Amersham Biosciences) chromatography with blue dextran (Mr, 2,000,000), thyroglobulin (Mr, 669,000), ferritin (Mr, 440,000), catalase (Mr, 232,000), aldolase (Mr, 158,000), albumin (Mr, 67,000), ovalbumin (Mr, 43,000), chymotryptonase A (Mr, 25,000), and RNase A (Mr, 13,700) as molecular weight standards. The column was equilibrated and eluted with 50 mM HEPES (pH 8.0) containing 150 mM NaCl at a flow rate of 0.5 ml/min.
The in vitro activity of the NagX enzyme was assessed spectrophotometrically via a decrease in the absorbance at 340 nm due to the substrate-dependent oxidation of NADH or NADPH. The molar extinction coefficient was taken as 6,200 M−1 cm−1 (18, 33). Assays were carried out with 100 mM potassium phosphate buffer (pH 8.0) at 30°C by using a UV-visible spectrophotometer (DU series 800; Beckman). The sample cuvette contained 100 mM potassium phosphate buffer, 0.2 μmol of the substrate, and 0.2 μmol of NADH or NADPH. The reference cuvette contained the same components except for the substrate. The assay was initiated by adding the NagX enzyme to both cuvettes. One unit of enzyme activity was defined as the conversion of 1 μmol of NADH into NAD+ per min at 30°C. Protein concentrations were determined by the method of Bradford (3) with bovine serum albumin as the standard.
The reaction converting 3-hydroxybenzoate into gentisate was initiated by adding purified recombinant NagX protein to a final volume of 4 ml containing 100 mM potassium phosphate buffer with 1 mM 3-hydroxybenzoate and 200 μM NADH (pH 8.0; the final protein concentration was approximately 3.209 μg/ml), and the absorbance of NADH at 340 nm was monitored with a UV spectrophotometer. When the oxidation of NADH (indicated by a decrease in the absorbance at 340 nm) stopped, the mixture was respiked with 80 μl of 10 mM NADH in three consecutive doses. During the course of conversion, concentrations of 3-hydroxybenzoate and gentisate were monitored by a high-performance liquid chromatography (HPLC) system (Shimadzu). For HPLC analysis, 20 μl of each sample was injected onto a reversed-phase C18 column (250 by 4.6 mm; Kromasil; Akzo Nobel) and run at room temperature with 20% methanol in 50 mM potassium phosphate buffer (pH 4.0) as the eluant at a flow rate of 1.0 ml/min. Detection was by UV spectroscopy at 235 nm. 3-Hydroxybenzoate and gentisate concentrations were assessed using standards of known concentrations.
A series of 3-hydroxybenzoate solutions, ranging from 5 to 200 μM, were prepared for the determination of Km values of the purified 3-hydroxybenzoate 6-hydroxylase. Spectrophotometric assays (absorbance at 340 nm) were completed while maintaining a constant enzyme concentration in 100 mM potassium phosphate buffer (pH 8.0). Initial reaction velocity determinations were used in Lineweaver-Burk plots to determine Km values.
Construction of a nagX knockout mutant of strain CJ2 and tests of its growth.
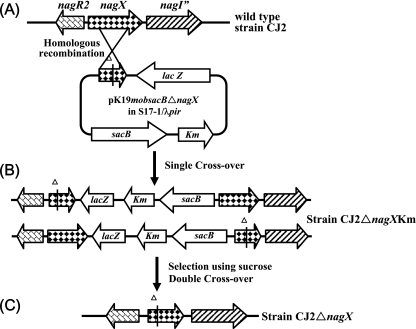
Plasmid pK19mobsacB (25), containing oriT of plasmid RP4 for conjugative mobilization and sacB (encoding the Bacillus subtilis levansucrase) as a counterselectable marker, was used for the construction of a mutagenic plasmid, pK19mobsacBΔnagX, for the introduction of a nagX deletion into the P. naphthalenivorans CJ2 chromosome. Primers nagX-FD1 (5′-CCCTCTAGAGGCTTTTCCGTCAAGGTG-3′) and nagX-RD2 (5′-CCCGTCGACCATTCCATGGCGTCATAG-3′) were designed to target internal regions of the nagX gene. The amplified 1,072-bp fragment of the nagX gene was cloned into XbaI- and SalI-digested pBluescript II SK(−), creating pBSIInagX. The resulting plasmid was digested with EcoRI, polished with a Klenow fragment (New England Biolabs), treated with SmaI, and then religated to produce pBSIIΔnagX. This plasmid was subsequently used for the construction of pK19mobsacBΔnagX by cloning nagX with a 14-bp deletion into the XbaI-SalI site of pK19mobsacB, after which pK19mobsacBΔnagX was introduced by electroporation into E. coli S17-1/λpir cells carrying the tra region of RP4. Then, for the construction of the mutant, pK19mobsacBΔnagX was mobilized from E. coli S17-1/λpir cells (≈5.0 × 108 to 1.0 × 109 grown overnight in LB broth with 20 μg of kanamycin/ml) and introduced into rifampin-resistant P. naphthalenivorans CJ2 cells (≈1.0 × 109 to 2.0 × 109 grown overnight in MSB-P broth with 200 μg of rifampin/ml) by mating on R2A agar medium at 25°C for 8 to 16 h as previously described (9, 20). The transconjugant (strain CJ2ΔnagXKm [CJ2 ΔnagX::Km]) was selected on R2A plates containing kanamycin (20 μg/ml) and rifampin (200 μg/ml) at 20°C. Confirmation of the transconjugant was conducted by using PCR and sequencing. The deletion of nagX was achieved by growing P. naphthalenivorans strain CJ2ΔnagX, a transconjugant, in R2A broth containing 10% sucrose and subsequently plating the culture onto R2A agar medium. The Sucr colonies obtained were replica plated onto R2A medium supplemented with kanamycin. The Sucr colony, which lost its kanamycin resistance (Sucr Kans), was confirmed by colony PCR (outer primer sc-f [5′-TCGGTGTTCGGGTGGTTGT-3′] and nagX-RD2), EcoRI digestion, and sequencing. Figure 2 schematically shows the theoretically expected molecular organization resulting from the genomic integration of pK19mobsacBΔnagX via homologous recombination into the P. naphthalenivorans CJ2 chromosome.
FIG. 2.
Schematic overview of the molecular organization of the region encoding NagX in wild-type P. naphthalenivorans strain CJ2 (A), in strain CJ2 after the integration of pK19mobsacBΔnagX by a single homologous recombination event at the targeted locus downstream (upper structure) and upstream (lower structure) from the nagX gene deletion (B), and in strain CJ2 after sacB counterselection for the second crossover event and plasmid excision resulting in the nagX deletion mutant strain CJ2ΔnagX (lower structure in panel B) (C). Km, kanamycin resistance gene.
For the growth tests with naphthalene and 3-hydroxybenzoate as the sole carbon sources, cells of strains CJ2, mutant CJ2ΔnagX, and mutant CJM110 (12) grown in MSB-P medium for 24 h at 20°C were inoculated (5%, vol/vol) into MSB-N and MSB-3-hydroxybenzoate media, respectively, and cultivated at 20°C in a shaking incubator at 180 rpm. Growth was monitored by measuring the OD600s of the cultures.
RESULTS
Overexpression and purification of the NagX protein.
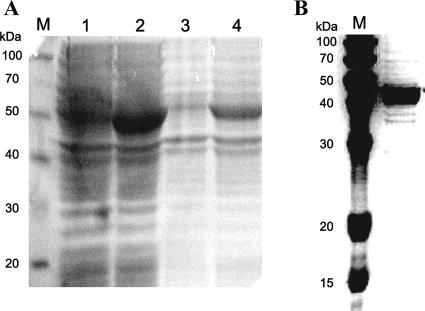
The nagX gene was cloned under the transcriptional regulation of the IPTG-inducible T7 promoter in the expression vector pET21b. The pET21bnagX clone was expressed in its native state. Cell extracts of IPTG-induced E. coli BL21(DE3) harboring the construct were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 3A), and a band corresponding to an overexpressed protein with an apparent molecular mass of 43 kDa, the expected size of NagX, was observed.
FIG. 3.
Analysis of the expressed NagX enzyme by PAGE. (A) Crude protein extracts of E. coli BL21(DE3) overexpressing NagX were separated by SDS-10% PAGE. Lane M, protein molecular mass markers; lane 1, crude extract of BL21(DE3) carrying no plasmid; lane 2, crude extract of BL21(DE3) carrying pET21bnagX; lane 3, suspended cell debris from BL21(DE3) carrying no plasmid; lane 4, suspended cell debris from BL21(DE3) carrying pET21bnagX. (B) Purified recombinant NagX separated by SDS-10% PAGE. For SDS-PAGE, the recombinant blue-prestained standard (ELPIS, Korea) was used as a molecular mass standard (lane M).
Recombinant NagX was purified from cell extracts of IPTG-induced E. coli BL21(DE3) harboring pET21bnagX by using a DEAE column and gel filtration chromatography. The eluted fractions which showed 3-hydroxybenzoate 6-hydroxylase activity were separated by SDS-PAGE, and a single protein band corresponding to an approximate molecular mass of 43 kDa was found and verified by gel filtration (Fig. 3B). From these results, we inferred that the active recombinant NagX was likely to be a monomeric protein.
Substrate specificity of the NagX protein in an in vitro assay.
The activity of the purified protein product encoded by nagX toward several substrates was investigated on the basis of the substrate-linked oxidation of NADH as measured at 340 nm (18, 33). The potential problem of NADH oxidase contamination from the extracts of the E. coli host was overcome by using a blank cuvette containing all assay components except for the substrate. The NagX recombinant exhibited 3-hydroxybenzoate 6-hydroxylase-specific activity of 14.0 U/mg at 30°C. However, no enzyme activity was detected when 4-hydroxybenzoate, benzoate, or salicylate was used as the substrate.
In vitro conversion of 3-hydroxybenzoate by the NagX enzyme.
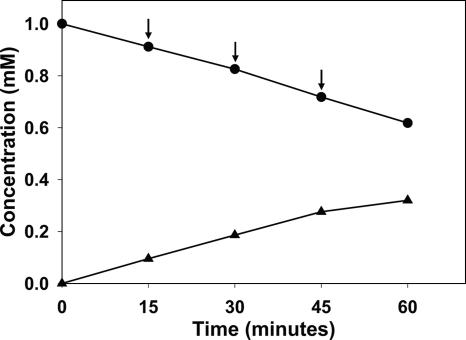
To confirm that NagX is indeed a 3-hydroxybenzoate 6-hydroxylase converting 3-hydroxybenzoate into gentisate, we carried out an in vitro conversion assay using the purified recombinant NagX enzyme. The reaction was completed within 5 min, and a series of consecutive NADH doses were added to extend activity in the reaction mixture. Concentrations of 3-hydroxybenzoate and gentisate in the reaction mixture were measured by HPLC. Data in Fig. 4 clearly show that in response to the addition of NADH, the concentration of 3-hydroxybenzoate decreased and that of gentisate increased. Thus, the NagX enzyme is a 3-hydroxybenzoate 6-hydroxylase (Fig. 4).
FIG. 4.
Enzymatic activity of the nagX gene product in E. coli. Data show the conversion of 3-hydroxybenzoate (top line, •) into gentisate (bottom line, ▴) as determined by HPLC. Arrows indicate the addition of NADH.
Effects of temperature, pH, and flavin adenine dinucleotide (FAD) on enzymatic activity.
The pH dependence of the purified recombinant 3-hydroxybenzoate 6-hydroxylase was investigated by assaying for enzyme activity in 0.1 M potassium phosphate buffers at pHs ranging from 5.0 to 9.4. The enzyme exhibited optimum activity at a slightly alkaline pH of 8.4. The optimal temperature of the recombinant 3-hydroxybenzoate 6-hydroxylase was observed to be 30°C, with no activity detected at temperatures higher than 50°C. However, 3-hydroxybenzoate 6-hydroxylase was found to retain 57% of maximum activity at 10°C, which is consistent with strain CJ2's relatively low optimum growth temperature of 20°C (11).
The effect of FAD on the activity of the purified NagX protein was investigated because the recombinant NagX protein showed a yellowish color, indicative of a bound flavin, and FAD has been shown to be a prosthetic group in other 3-hydroxybenzoate 6-hydroxylases (8, 23, 30, 33, 35). The addition of 10 μM FAD increased the activity of the purified recombinant 6-hydroxylase approximately fourfold. Such an increase in activity implies that a part of the flavin cofactor may have been lost during enzyme purification and that the loss was corrected by the added exogenous FAD.
Kinetic properties.
The purified recombinant NagX enzyme displayed typical Michaelis-Menten kinetics, and Lineweaver-Burk plots of enzyme activity yielded an apparent Km of 64.9 μM for 3-hydroxybenzoate when the concentration of NADH was fixed at 200 μM and the concentrations of 3-hydroxybenzoate were varied. At a constant level of 3-hydroxybenzoate (0.2 mM) and various amounts of NADH or NADPH, the NagX protein exhibited an apparent Km of 493 μM for NADH, compared to 860 μM for NADPH, indicating a preference for NADH over NADPH.
Effects of a nagX knockout in P. naphthalenivorans strain CJ2.
To investigate the role of nagX in its original host, P. naphthalenivorans CJ2, a nagX knockout mutant designated CJ2ΔnagX was constructed by unmarked gene deletion mutagenesis (Fig. 2). Integration by homologous recombination may occur at either side of the nagX deletion present on pK19mobsacBΔnagX, resulting in two types of transconjugants, as shown in Fig. 2B. We verified (by PCR and EcoRI digestion) that integration to the right of the nagX deletion on the mutagenic plasmid (second gene structure in Fig. 2B) occurred by a single homologous recombination event. The conjugation frequency of pK19mobsacBΔnagX in P. naphthalenivorans CJ2 after overnight mating at 25°C on the filter was often below the detection limit (10−9), which may be caused by an inappropriate growth temperature for the donor strain (E. coli) or the extracellular polysaccharide production of the recipient strain (P. naphthalenivorans CJ2). The deletion of nagX was obtained by growing strain CJ2ΔnagX in R2A broth containing 10% sucrose and subsequently plating the culture onto R2A agar medium. Four Sucr Kans colonies were subjected to colony PCR with nagX-FD1 and nagX-RD2 and subsequent digestion with EcoRI. The results revealed that only one of the colonies, or 25% of the total Sucr population, contained the truncated nagX gene. Gene deletion was confirmed by sequencing. The elevated probability (3:1 ratio) of recovering the wild-type nagX after the second crossover and excision events was expected because the DNA fragment on the right side of the nagX deletion was 2.5 times larger than that on the left side.
The cell growth of wild-type CJ2, mutant CJ2ΔnagX, and mutant CJM110 (the latter mutant features derepressed expression of nagX [12]) in MSB medium with 0.5% naphthalene crystals or 0.3% 3-hydroxybenzoate as the sole carbon source was monitored by measuring the OD600s.
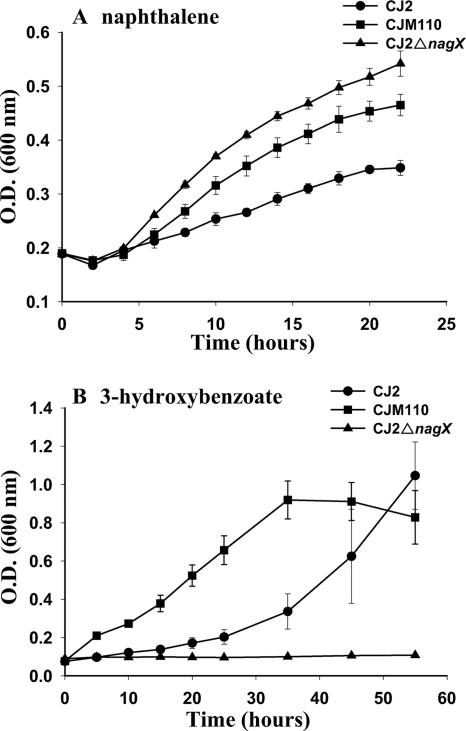
The mutant CJ2ΔnagX retained its ability to grow on naphthalene; in fact, the nagX mutant grew faster in MSB-N medium than both the wild-type and the mutant CJM110 strains (Fig. 5A); discovering the basis for this enhancement was beyond the scope of this investigation. The data clearly indicated that nagX is not involved in the degradation of naphthalene. The nagX knockout strain CJ2ΔnagX was not able to grow in the MSB-0.3% (wt/vol) 3-hydroxybenzoate medium (Fig. 5B), meaning that the nagX gene is required for the degradation of 3-hydroxybenzoate. To investigate if 3-hydroxybenzoate can serve as an inducing chemical, we performed an additional induction experiment using strain CJM112 (CJ2 nagR2-nagX::lacZ+) as described previously (12). However, surprisingly, 3-hydroxybenzoate did not cause any detectable induction of β-galactosidase activity (data not shown), indicating that 3-hydroxybenzoate is not an inducer of the small cluster, although the cluster is involved in 3-hydroxybenzoate biodegradation.
FIG. 5.
Growth of the P. naphthalenivorans CJ2 wild type and its mutant derivatives, CJM110 and CJ2ΔnagX, in liquid minimal media containing naphthalene (A) and 3-hydroxybenzoate (B) as the sole carbon sources. Growth was determined by measuring the OD600. Strain CJΔnagX is the nagX deletion mutant. Strain CJM110 has a truncated nagR2 gene, which encodes a MarR-type repressor. This mutation results in the elevated expression of nagX.
DISCUSSION
In our prior study of the operon structure of P. naphthalenivorans CJ2 (12), we pointed out that the peculiar arrangement of naphthalene-catabolic genes suggested that horizontal gene transfer from Azoarcus to strain CJ2 may have occurred. Three observations served as clues for this hypothesis.
(i) Gene insertion has occurred.
In Ralstonia sp. strain U2 (which carries a contiguous complete nag operon), the order of genes is nagAaGHAbAcAdBFCQEDJIKLMN. Relative to strain U2, strain CJ2's small operon features a two-gene insertion (comprising nagR2, a MarR-type regulator, and nagX) at the 5′ end of a duplicated nagI′ gene (Fig. 1A).
(ii) Gene sequence similarities suggest that transferred genes originated in Azoarcus.
BLAST searching and nucleotide analyses of three loci showed genes in Azoarcus sp. strain EbN1 to be the closest matches to genes in strain CJ2's catabolic operons. The product of the nagX gene (putative salicylate 5-hydroxylase) in the second gene cluster exhibited 89% amino acid identity to its homologue in Azoarcus sp. strain EbN1. Also, in the region just beyond the putative terminator in the large gene cluster, the products of the putative transposase-related open reading frames tnpA′ and istB exhibited 56 and 65% amino acid identity, respectively, to the products of the corresponding genes in Azoarcus sp. strain EbN1.
(iii) MarR in Azoarcus provides an example of MarR control of catabolism.
Schuehle et al. (26) have shown that the genes encoding the aerobic metabolism of 2-aminobenzoate by Azoarcus evansii include a MarR-type regulator. Thus, there is a precedent for MarR control of catabolism in Azoarcus.
nagX occurs in a small cluster containing three genes considered essential for naphthalene metabolism: genes for gentisate 1,2-dioxygenase (nagI″), fumarylpyruvate hydrolase (nagK), and maleylpyruvate isomerase (nagL) for naphthalene degradation. nagK and nagL are not present in the large nag catabolic cluster in strain CJ2. Due to its association with these well-characterized naphthalene-catabolic genes, we presumed that nagX also played a functional role in naphthalene catabolism. However, data presented in the present study clearly demonstrated that a nagX knockout mutant of strain CJ2 retained its ability to grow on naphthalene. This means that nagX has no direct role in naphthalene degradation. But we cannot rule out indirect roles. For instance, Fig. 1B illustrates an indirect role of nagX in the broad physiological process of aromatic hydrocarbon metabolism: gentisate produced by the NagX protein feeds into the naphthalene-catabolic pathway.
Surprisingly, by knocking out nagX, we eliminated the ability of strain CJ2 to grow on 3-hydroxybenzoate. Thus, here we provided clear physiological and genetic evidence that, despite the high level of amino acid identity of the nagX product to the salicylate-5-hydroxylase of Azoarcus, nagX encodes 3-hydroxybenzoate 6-hydroxylase, the enzyme which introduces an additional hydroxyl group at the para position (relative to the existing hydroxyl group) and yields gentisate. For this reason, a new name for orf2, nagX, was proposed.
Although 3-hydroxybenzoate 6-hydroxylase is a key enzyme in the gentisate pathway of 3-hydroxybenzoate degradation, very few studies of this para-hydroxylation reaction have been carried out (36). Presently, the sequences of only two genes known to encode 3-hydroxybenzoate 6-hydroxylase are available: those of mhbM from K. pneumoniae M5a1 (18) and xlnD from P. alcaligenes NCIMB 9867 (6). The NagX enzyme from strain CJ2 was able to convert 3-hydroxybenzoate into gentisate without salicylate 5-hydroxylase activity. Active recombinant NagX had an approximate molecular mass of 43 kDa, as estimated by gel filtration, and was probably a monomeric protein, similar to the 3-hydroxybenzoate 6-hydroxylases from P. cepacia (44 kDa) (33, 35) and K. pneumoniae M5a1 (42 kDa) (30). Like other characterized 3-hydroxybenzoate 6-hydroxylase enzymes, NagX utilizes NADH as well as NADPH as the electron donor for the reduction of the flavin cofactor, FAD. With 3-hydroxybenzoate as the substrate, NagX had a higher affinity for NADH than NADPH. This trait is shared with 3-hydroxybenzoate 6-hydroxylase activity in Micrococcus sp. (23), P. alcaligenes NCIMB 9867 (6), and P. aeruginosa (8).
Pumphrey and Madsen (submitted) have shown, via gas chromatography-mass spectrometry detection of gentisate and a cell-free gentisate-dioxygenase assay, that strain CJ2 grows on naphthalene by using the gentisate pathway. Thus, salicylate-5-hydroxylase activity is expressed. Prior to the results presented here, there were two potential sources for salicylate-5-hydroxylase activity in strain CJ2: nagX in the small gene cluster and nagGH (encoding, respectively, the large and small salicylate-5-hydroxylase subunits and exhibiting ≥86% identity to the corresponding genes in Ralstonia sp. strain U2) in the large gene cluster (12). By revealing the true metabolic function of nagX, we have eliminated questions about redundant salicylate-5-hydroxylase genes in the operon pair. But simultaneously, we have raised new questions about operon evolution and the regulation of growth on 3-hydroxybenzoate in strain CJ2. Was 3-hydroxybenzoate a growth substrate for ancestral strain CJ2 prior to the putative horizontal transfer of two genes (nagR2 and nagX) from Azoarcus? If so, why is the prior 3-hydroxybenzoate 6-hydroxylase now inactive? Are the remaining structural genes for 3-hydroxybenzoate metabolism in an operon under MarR-type regulatory control?
The modular or mosaic nature of catabolic operons is well recognized (2, 17, 19, 31, 32, 36). It is clear that variations in catabolic capabilities in prokaryotes have evolved and continue to evolve via a series of acquisitions and rearrangements at the DNA level ranging from a few nucleotides (e.g., transition and inversion) to many kilobases (e.g., gene transfer, duplication, and deletion) (31, 34). Documenting the mechanism of operon development is not facile. We must rely upon retrospective inspection of nucleotide sequences for traits that include G+C content, codon usage, levels of identity of noncoding homologous DNA regions, and remnants of transposons or insertion sequence elements (see, for example, reference 2). These properties sometimes allow historical steps in the development of operons and entire plasmids to be inferred (4, 7, 16, 27). As demonstrated by Fuenmayor et al. (5), cloning and expression assays can also be insightful and have led to the suggestion that the functionality of nagGH (encoding salicylate monooxygenase) in strain U2 developed by the insertion of nagGH within the otherwise continuous functional cluster nagAaAbAcAd. Future studies may reveal additional details about the origin, regulation, and multifunctionality (degradation of both naphthalene and 3-hydroxybenzoate) of the small naphthalene-catabolic cluster in strain CJ2.
Acknowledgments
These efforts were supported by grants from the MOST/KOSEF to the Environmental Biotechnology National Core Research Center (grant no. R15-2003-012-02002-0) and to the 21C Frontier Microbial Genomics and Application Center Program (grant no. MG05-0104-4-0), Ministry of Science and Technology, Republic of Korea. M.P. and Y.J. were supported by scholarships from the BK21 program of the Ministry of Education and Human Resources Development in Korea.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, R., E. Garcia-Valdes, and E. R. B. Moore. 1999. Genetic characterization and evolutionary implication of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 236:149-157. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Dennis, J. J., and G. J. Zylstra. 2004. Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J. Mol. Biol. 341:753-768. [DOI] [PubMed] [Google Scholar]
- 5.Fuenmayor, S. L., M. Wild, A. L. Boyers, and P. A. Williams. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao, X., C. L. Tan, C. C. Yeo, and C. L. Poh. 2005. Molecular and biochemical characterization of the xlnD-encoded 3-hydroxybenzoate 6-hydroxylase involved in the degradation of 2,5-xylenol via the gentisate pathway in Pseudomonas alcaligenes NCIMB 9867. J. Bacteriol. 187:7696-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greated, A., L. Lambertsen, P. A. Williams, and C. M. Thomas. 2002. Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4:856-871. [DOI] [PubMed] [Google Scholar]
- 8.Groseclose, E. E., D. W. Ribbons, and H. Hughes. 1973. 3-Hydroxybenzoate 6-monooxygenase from Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 55:897-903. [DOI] [PubMed] [Google Scholar]
- 9.Hohnstock, A. M., K. G. Stuart-Keil, E. E. Kull, and E. L. Madsen. 2000. Naphthalene and donor cell density influence field conjugation of naphthalene catabolism plasmids. Appl. Environ. Microbiol. 66:3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon, C. O., W. Park, P. Padmanabhan, C. DeRito, J. R. Snape, and E. L. Madsen. 2003. Discovery of a previously undescribed bacterium with distinctive dioxygenase that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. USA 100:13591-13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeon, C. O., W. Park, W. C. Ghiorse, and E. L. Madsen. 2004. Polaromonas naphthalenivorans sp. nov., a naphthalene-degrading bacterium from naphthalene-contaminated sediment. Int. J. Syst. Evol. Microbiol. 54:93-97. [DOI] [PubMed] [Google Scholar]
- 12.Jeon, C. O., M. Park, H. Ro, W. Park, and E. L. Madsen. 2006. The naphthalene catabolic (nag) genes of Polaromonas naphthalenivorans CJ2: evolutionary implications for two gene clusters and novel regulatory control. Appl. Environ. Microbiol. 72:1086-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, D. C., and R. A. Cooper. 1990. Catabolism of 3-hydroxybenzoate by the gentisate pathway in Klebsiella pneumoniae M5a1. Arch. Microbiol. 154:489-495. [DOI] [PubMed] [Google Scholar]
- 14.Jones, R. M., B. Britt-Compton, and P. A. Williams. 2003. The naphthalene catabolic (nag) genes of Ralstonia sp. strain U2 are an operon that is regulated by NagR, a LysR-type transcriptional regulator. J. Bacteriol. 185:5847-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalogeraki, V. S., and S. C. Winans. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188:69-75. [DOI] [PubMed] [Google Scholar]
- 16.Kulakov, L. A., S. Chen, C. C. R. Allen, and M. J. Larkin. 2005. Web-type evolution of Rhodococcus gene clusters associated with utilization of naphthalene. Appl. Environ. Microbiol. 71:1754-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: horizontal transfer may drive evolution of gene clusters. Genetics 143:1843-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, D. Q., H. Liu, X. L. Gao, D. J. Leak, and N. Y. Zhou. 2005. Arg169 is essential for catalytic activity of 3-hydroxybenzoate 6-hydroxylase from Klebsiella pneumoniae M5a1. Microbiol. Res. 160:53-59. [DOI] [PubMed] [Google Scholar]
- 19.Osborn, A. M., and D. Böltner. 2002. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48:202-212. [DOI] [PubMed] [Google Scholar]
- 20.Park, W., C. O. Jeon, A. M. Hohnstock-Ashe, S. C. Winans, G. J. Zylstra, and E. L. Madsen. 2003. Identification and characterization of the conjugal transfer region of the pCg1 plasmid from naphthalene-degrading Pseudomonas putida Cg1. Appl. Environ. Microbiol. 69:3263-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poh, C. L., and R. C. Bayly. 1980. Evidence for isofunctional enzymes used in m-cresol and 2,5-xylenol degradation via the gentisate pathway in Pseudomonas alcaligenes. J. Bacteriol. 143:59-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poh, C. L., and R. C. Bayly. 1988. Regulation of isofunctional enzymes in Pseudomonas alcaligenes mutants defective in the gentisate pathway. J. Appl. Bacteriol. 64:451-458. [DOI] [PubMed] [Google Scholar]
- 23.Rajasekharan, S., R. Rajasekharan, and C. S. Vaidyanathan. 1990. Substrate-mediated purification and characterization of a 3-hydroxybenzoic acid-6-monooxygenase from Micrococcus. Arch. Biochem. Biophys. 278:21-25. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and K. J. Janssen. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pűhler. 1994. Small mobilizable multi-purpose cloning vectors derived from Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 26.Schuehle, K., M. Jahn, S. Ghisla, and G. Fuchs. 2001. Two similar gene clusters coding for enzymes of a new type of aerobic 2-aminobenzoate (anthranilate) metabolism in the bacterium Azoarcus evansii. J. Bacteriol. 183:5268-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sota, M., H. Yano, A. Ono, R. Miyazaki, H. Ishii, H. Genka, E. M. Top, and M. Tsuda. 2006. Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J. Bacteriol. 188:4057-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanier, R. Y., N. J. Palleroni, and M. Doudorhoff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 29.Stuart-Keil, K. G., A. M. Hohnstock, K. P. Drees, J. B. Herrick, and E. L. Madsen. 1998. Plasmid responsible for horizontal transfer of naphthalene catabolism genes between bacteria at a coal tar-contaminated site are homologous to pDTG1 from Pseudomonas putida NCIB9816-4. Appl. Environ. Microbiol. 64:3633-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suárez, M., E. Ferrer, A. Garrido-Pertierra, and M. Martin. 1995. Purification and characterization of the 3-hydroxybenzoate 6-monooxygenase from Klebsiella pneumoniae M5a1. FEMS Microbiol. Lett. 126:283-290. [DOI] [PubMed] [Google Scholar]
- 31.van der Meer, J. 1997. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie Leeuwenhoek 71:159-178. [DOI] [PubMed] [Google Scholar]
- 32.van der Meer, J. R., W. M. deVos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, L.-H., R. Y. Hamzah, Y. Yu, and S.-C. Tu. 1987. Pseudomonas cepacia 3-hydroxybenzoate 6-monooxylase: induction, purification, and characterization. Biochemistry 26:1099-1104. [DOI] [PubMed] [Google Scholar]
- 34.Williams, P. A., and J. R. Sayers. 1994. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation 5:195-217. [DOI] [PubMed] [Google Scholar]
- 35.Yu, Y., L.-H. Wang, and S.-C. Tu. 1987. Pseudomonas cepacia 3-hydroxybenzoate 6-monooxylase: stereochemistry, isotope effects, and kinetic mechanism. Biochemistry 26:105-110. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, N.-Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, N.-Y., J. Al-Dulayymi, M. S. Baird, and P. A. Williams. 2002. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J. Bacteriol. 184:1547-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]