Abstract
When filter-feeding shellfish are consumed raw, because of their ability to concentrate and store waterborne pathogens, they are being increasingly associated with human gastroenteritis and have become recognized as important pathogen vectors. In the shellfish industry, UV depuration procedures are mandatory to reduce pathogen levels prior to human consumption. However, these guidelines are based around more susceptible fecal coliforms and Salmonella spp. and do not consider Cryptosporidium spp., which have significant resistance to environmental stresses. Thus, there is an urgent need to evaluate the efficiency of standard UV depuration against the survival of Cryptosporidium recovered from shellfish. Our study found that in industrial-scale shellfish depuration treatment tanks, standard UV treatment resulted in a 13-fold inactivation of recovered, viable C. parvum oocysts from spiked (1 × 106 oocysts liter −1) Pacific oysters. Depuration at half power also significantly reduced (P < 0.05; ninefold) the number of viable oocysts recovered from oysters. While UV treatment resulted in significant reductions of recovered viable oocysts, low numbers of viable oocysts were still recovered from oysters after depuration, making their consumption when raw a public health risk. Our study highlights the need for increased periodic monitoring programs for shellfish harvesting sites, improved depuration procedures, and revised microbial quality control parameters, including Cryptosporidium assessment, to minimize the risk of cryptosporidiosis.
Cryptosporidium spp. are obligate intracellular Apicomplexa protozoan parasites which infect a wide range of vertebrates and undergo endogenous development, culminating in the production of an encysted stage (oocyst), which is discharged in the feces of their host (38). There are currently 16 recognized species of Cryptosporidium (43, 44), and cryptosporidiosis in humans and livestock is caused mainly by C. parvum and C. hominis (39). Cryptosporidiosis is usually associated with drinking contaminated water, visits to swimming pools, or animal exposure (34). Globally, Cryptosporidium is responsible for the majority of gastrointestinal parasitic infections, representing a significant cause of morbidity and mortality in its hosts (32, 42). In immunocompetent humans, cryptosporidiosis normally causes a self-limited diarrhea, but it can cause life-threatening conditions in immunocompromised patients due to dehydration by chronic diarrhea. Due to its widespread occurrence in drinking water supplies and its significant resistance to environmental stresses, Cryptosporidium is regarded as one of the most important waterborne microbial parasites (41). Cryptosporidium oocysts can retain infectivity for months outside of hosts, in both freshwater and salt water (40). Since the year 2000, the global production of several shellfish types, including the bivalve Pacific oyster (Crassostrea gigas), which is generally consumed raw, has increased dramatically by several million metric tons (3). This increased consumption directly correlates to the significant rise of reported seafood-borne infections (2, 27, 28).
Raw shellfish, e.g., molluscs, oysters, clams, scallops, and other marine life, in costal waters are increasingly being implicated as potential reservoirs for the transmission of the viable and infective stages of Cryptosporidium (2, 8, 13, 16, 19). Contamination is apparent mainly in near-shore waters and estuaries where human sewage is discharged and runoff from land exposed to animal manure occurs (18, 25). While no development of the pathogen in tissue has been observed, shellfish concentrate and retain viable oocysts within their hemocytes and gills (16, 19), and viable oocysts recovered from shellfish have been shown to be infectious (10). Thus, raw shellfish have potentially important roles in spreading Cryptosporidium infection in depuration plants and areas where aquatic organisms are cultivated (15, 41). Chemical disinfection with chlorine is not effective against oocysts of Cryptosporidium and Giardia spp., and ozone applied at low cycle threshold values to limit the formation of bromate has relatively little effect on oocyst viability (1, 20, 21). UV disinfection technology is of growing interest to water industries because the treatment can be readily applied, is low maintenance, results in significant viability reductions of all waterborne pathogens, and produces no hazardous by-products (20, 30, 33). Previous studies have found that the effects of UV irradiation on C. parvum oocysts, as determined by animal infectivity and DNA repair, can conclusively be considered irreversible (24, 30).
The European Community (EC) directive 91/492 demands periodic checks on the microbial quality of live bivalve mollusc relaying and in production areas (7). However, currently these microbial parameters do not include monitoring Cryptosporidium (7). Depending on the water quality (measured by the amount of fecal coliforms), bivalve molluscs are either directly placed on the market, undergo purification, or are relayed for a minimum of 2 months in clean water, which can be combined with a final depuration purification step to remove contaminants (7). EC legislation demands the depuration of oysters for 48 h to remove potentially harmful contaminants, which is based upon the removal of fecal coliforms to within safe levels for human consumption; <300 coliform bacteria per 100 g of oyster tissue (7, 12, 14). Novel methods such as E-beam irradiation, microwaves, and exposure to high pressure have been evaluated as alternative means for the removal of contaminants from shellfish, but UV depuration is still the only method which is commercially practiced (4, 5).
To our knowledge, no research has been previously performed to assess whether current industrial protocols for UV depuration are sufficient to inactivate C. parvum recovered from oysters to within safe limits for human consumption. Thus, we replicated Pacific oyster commercial depuration conditions, artificially spiked the oysters with C. parvum, and assessed the depuration efficiency of standard UV treatment at oocyst inactivation.
MATERIALS AND METHODS
Harvesting and enumerating Cryptosporidium.
A modification of a previously described method was used to purify and collect oocysts from fecal material (31). Fecal material from calves with a positive diagnosis of cryptosporidiosis was obtained from the Department of Parasitology, Department of Agriculture and Food, Central Veterinary Laboratory, Dublin, Ireland. To enumerate oocysts, aliquots (100 μl) of Cryptosporidium-positive stool samples were mixed with 250 μl of double-distilled water (ddH2O) by vortexing for 30 s. Approximately 150 μl of ether was added, and samples were shaken vigorously by hand for 30 s. Tubes were then centrifuged at 14,000 × g for 1 min, and the fluid both above and below the oocysts was aspirated and discarded. The sediment (oocysts) was then washed three times with 1 ml of ddH2O. Harvested oocysts were counted using a hemocytometer and genotyped as described below.
DNA extractions.
An additional two washes in 1 ml of lysis buffer (LB medium, 50 mM Tris-HCl [pH 8.5; Bio-Rad, Hercules, CA], 1 mM EDTA (Sigma, Dorset, United Kingdom), 0.5% sodium dodecyl sulfate [Sigma]), followed by centrifuging and careful aspiration with a pipette, were required to prepare collected oocysts for DNA extraction (31). Sediment with oocysts was resuspended in 100 μl LB medium and subjected to five freeze-thaw cycles, as follows: 5 min in liquid nitrogen, then 5 min at 65°C, followed by 10 s of vortexing and centrifugation at 2,000 × g for 1 min. Samples were then treated with proteinase K (200 μg ml−1; Sigma) at 55°C for 3 h, followed by 20 min at 90°C for inactivation (31). Samples were then placed on ice and centrifuged at 10,000 × g for 5 min, and supernatants were removed and replaced with equal amounts of 70% ethanol. Samples were then vortexed and centrifuged at 10,000 × g for 2 min (31). This step was repeated, supernatants were removed, samples were air dried, and the DNA pellets were then resuspended in 30 μl of ddH2O, and stored at −20°C (31).
Oocyst identification.
Oocysts isolated from fecal material were identified using a previously described nested PCR-restriction fragment length polymorphism (RFLP) technique targeting a polymorphic region of the 18S rRNA gene of Cryptosporidium species (23, 43). Briefly, PCR conditions for primary amplification reaction primer pair 5′-TTCTAGAGCTAATACATGCG-3′ and 5′-CCCTAATCCTTCGAAACAGGA-3′ were used to obtain amplicons of approximately 1,325 bp (43). The internal forward primer 5′-GGAAGGGTTGTATTTATTAGATAAAG-3′ and reverse primer 5′-AAGGAGTAAGGAACAACCTCCA-3′ were then used to amplify an 820-bp region of the primary PCR product (23). Cryptosporidium genotyping was then carried out using RFLP analysis following digestion of secondary products using endonucleases VspI and SspI (Promega Ltd., Southampton, United Kingdom) as per the manufacturer's instructions (23, 43). Digested products were fractionated on a 1.5% agarose gel and visualized using ethidium bromide staining. Only C. parvum oocysts were isolated and genotyped as bovine genotype 2.
Oyster sample preparation and oocyst enumeration and viability.
For oocyst recovery, whole-oyster tissue was homogenized using a stomacher (Lab Blender 400, Seward, London, United Kingdom). Using immunomagnetic separation (IMS) (26), oocysts were recovered from 1-ml aliquots of homogenized tissue by using Cryptosporidium-specific Dynal beads (Dynal Ltd., Bromborough, United Kingdom) according to the manufacturer's instructions. IMS-recovered oocysts were dissociated from magnetic beads and then diluted in ddH2O, and oocyst counts were determined using a hemocytometer.
To assess oocyst viability, sporozoite nuclei were stained with 4′,6′-diamidino-2-phenyl indole (DAPI; Roche Diagnostics Ltd., Sussex, United Kingdom) fluorogenic dye and inclusion/exclusion propidium iodide (PI) vital dye (Sigma) (1). Aliquots (30 μl) of phosphate-buffered saline (PBS; Sigma) containing DAPI (1 mg ml−1) and PI (1 mg ml−1) were added to samples and incubated for 2 min at 37°C in darkness. Samples were washed twice with PBS (pH 7.4). Samples were air dried and fixed with methanol. Samples were then mounted with glycerol-PBS-based antibleaching agent AF1 (Citifluor Ltd., London, United Kingdom). Oocyst numbers and viability were determined (at a magnification of ×400) using a Nikon Eclipse E400 microscope. A blue filter block (480-nm excitation, 520-nm emission) was used for viewing fluorescein isothiocyanate emissions, a UV filter block (350-nm excitation, 450-nm emission) for DAPI emissions, and a UV filter block (560- to 580-nm excitation, 600- to 650-nm emission) for PI emissions. Low numbers of doubtful “oocyst-like” entities were examined at a magnification of ×1,000 to confirm their identity by the identification of sporozoite nuclei (DAPI), suture lines, and if possible, internal structures (37).
Oyster collection, contamination checks, and spiking.
Bivalve Pacific oysters (Crassostrea gigas) were grown for 28 months (median weight including shell, 100 g; meat weight, 25 g) and harvested from commercial sites in Northern Ireland. Five oysters (a representative sample) were assessed for the presence of contaminating Cryptosporidium organisms and their levels of coliform. Both standard immunofluorescence fluorescein (1, 11) and nested 18S rRNA PCR (23, 43) were used to confirm the absence of Cryptosporidium. A 24-h direct plating method for fecal coliform enumeration with a resuscitation step (preincubation for 2 h at 37 ± 1°C and transfer to 44 ± 1°C for 22 h) using fecal coliform agar (Difco, Detroit, MI) (9, 22) was also used to quantify oyster coliform contamination levels, both before and after depuration.
Artificial seawater was produced using Seamix (Peacock Salt, Ayr, United Kingdom) to approximately 32 ppt salinity for all experiments. Seawater (400 liters) containing oysters was spiked with Cryptosporidium bacteria-positive bovine stool samples to final concentrations of 1 × 106 oocysts liter−1 in 400-liter-capacity commercial-size (1 m by 1 m by 0.6 m) shellfish depuration tanks (Depur Systems Ltd., Dundrum, Co. Down, United Kingdom). Oysters were allowed to filter circulation water and uptake Cryptosporidium oocysts for 4 h under the continuous circulation of spiked seawater. The health of oysters was frequently assessed (every 4 h) by checking their respiration (36). In order to remove oocysts which were not taken up by the oysters, oysters were rinsed for 15 min with fresh, uncontaminated artificial seawater prior to depuration.
Industrial-scale standard UV depuration.
All of the following experiments were carried out in triplicate, and UV depuration was carried according to current United Kingdom legislation requirements (36). Batches of 100 spiked and nonspiked (negative control) oysters were positioned in the middle of commercial-size depuration tanks containing 200 liters of freshly made, circulated, artificial seawater (Fig. 1A to C). Previous studies have shown that oocysts remain viable for up to 2 weeks during storage in artificial seawater (30 ppt) at 20°C and for even longer periods when stored at lower temperatures (Fayer et al. [8]). Thus, to avoid oyster sporulation, all depuration was carried out at <15°C (36). Briefly, in commercial-size tanks, water was circulated at 1,200 liter min−1 and was exposed to continuous UV treatments, with full-power tanks at 50 W per pass and half-power tanks at 25 W per pass. Circulation water samples were analyzed both during (24 h) and after depuration (48 h), and oocyst viability was assessed.
FIG. 1.
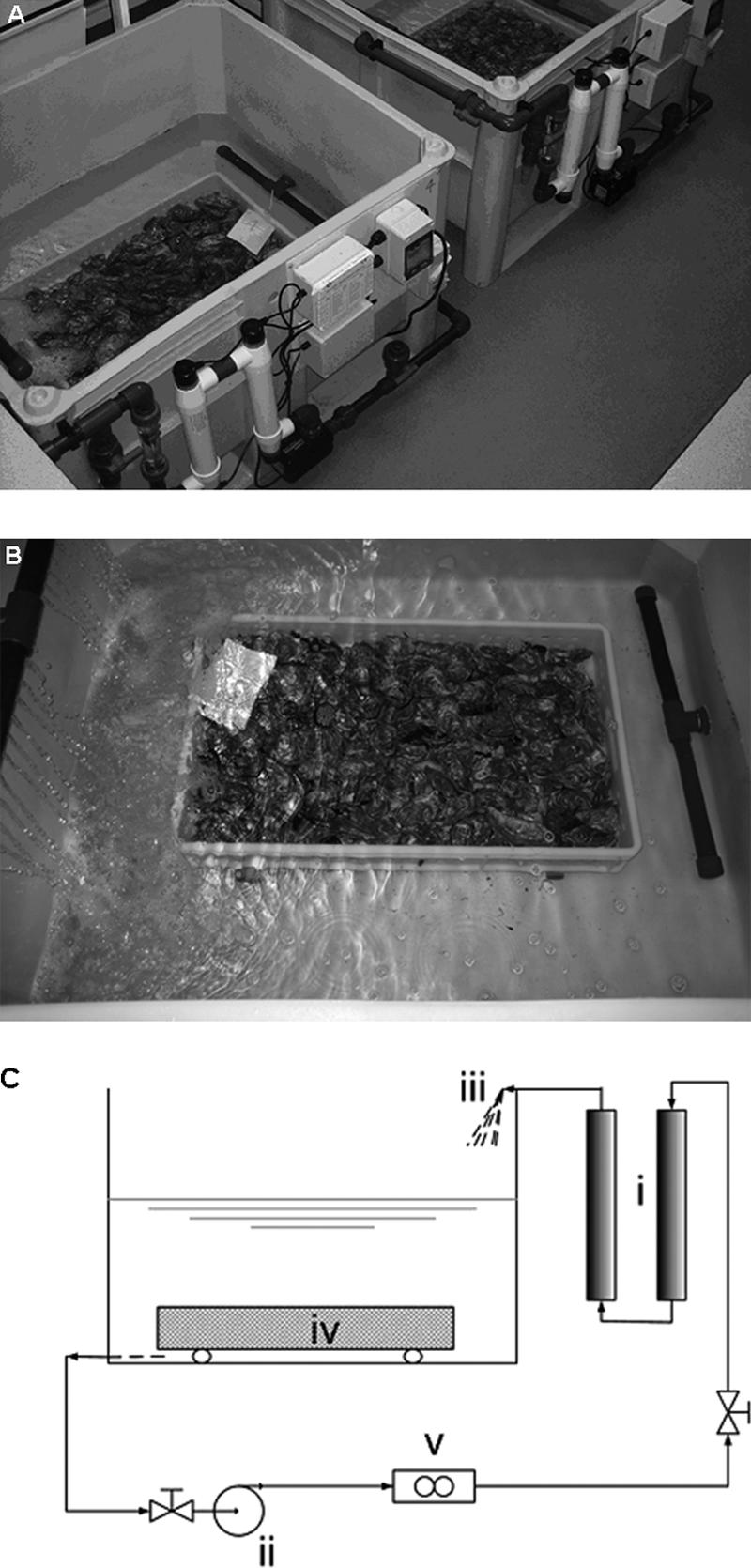
(A) Commercial-size depuration unit. (B) Oysters in a tray undergoing depuration. (C) Diagram of a depuration unit: (i) UV filter for treatment of circulation water; (ii) circulation water pump; (iii) spray for aeration of water; (iv) oysters in a tray; (v) flow meter.
During depuration, salinity, water temperature, and oxygen levels were all measured using TetraCon 325 and OxyCal SL 197 probes (WWT, Limonest, France) and maintained within industrially acceptable levels (36). Samples of five oysters were taken before and after depuration to measure the efficacy of depuration. From each sample, determination of oocyst numbers and viability was carried out in triplicate. Additionally, nested PCR targeting the 18S rRNA gene (23, 43), followed by sequencing, was carried out to check for cross-contamination and verification of C. parvum (formerly genotype 2).
Circulation water sampling.
Oocysts were concentrated using immunomagnetic separation according to the manufacturer's instructions. Briefly, anti-Cryptosporidium magnetic beads were added to 10 ml of water sample, and oocysts were captured for 4 h on a Dynal rotary mixer (26). Bound oocysts were collected and recovered from beads in 20 μl of 0.1 M HCl (Sigma). Samples were neutralized, and oocysts were stained with DAPI and PI as described previously. Oocyst counts and viability assessments were performed in triplicate. After spiking, oysters were washed to remove fecal debris and excess oocysts not taken up by oysters. Oyster oocyst uptake rates were based on the differences between counts from spiked water and the numbers of oocysts retained in oyster tissue.
Statistical analysis.
Data from individual experiments were collected and examined for significant differences (P < 0.05) using Excel-based statistical Analyze-It software (Analyze-It Software Ltd., Leeds, United Kingdom). The effect of UV dosage on oocyst viability in experimentally spiked Pacific oysters was analyzed by one-way analysis of variance (ANOVA). When the effect of treatment was significant (P < 0.05), the Bonferroni t test with 95% confidence interval for multiple measurements was carried out. Two-way ANOVA tests were used to analyze the effect of standard depuration conditions on oocyst viability in circulation water over the 48-h period.
RESULTS
Oysters of satisfactory microbial quality prior to spiking and coliform inactivation.
Before spiking, Cryptosporidium was not detected in oysters, and coliform levels were within category B levels for shellfish, i.e., <6,000 coliform bacteria per 100 g of oyster tissue (14). Control experiments confirmed the efficiency of standard UV depuration at totally inactivating Escherichia coli in both circulation water and oyster flesh. Genotype analysis of recovered oocysts prior to and postdepuration confirmed that only C. parvum was present in fecal material, oysters, and circulation water, ruling out the possibility of cross-contamination during experimentation (data not shown).
Significant inactivation of Cryptosporidium from oysters using UV depuration.
In this study, we assessed the ability of commercial depuration units to inactivate C. parvum oocysts recovered from oysters. As expected, standard UV treatment (at full power) for 48 h resulted in the largest and most significant reduction (P < 0.05; 13-fold) of viable oocysts recovered from oysters (Table 1). Depuration at half-power for 48 h also significantly reduced (P < 0.05; ninefold) amounts of viable oocysts recovered from oysters (Table 1). While significant amounts (P < 0.05) of viable oocysts were inactivated with no UV treatment (Table 1), as expected, significantly more (P < 0.05) oocysts were inactivated when both full- and half-power UV depuration treatments were administered. During UV treatment, oysters remained in a healthy condition during spiking, washing, and depuration, as they showed normal respiration rates as determined by monitoring soluble oxygen levels, water temperature, salinity, and visual checks (data not shown).
TABLE 1.
Survival of C. parvum oocysts recovered from experimentally contaminated oysters during UV depuration treatmentsa
| Time (h) | No UV treatment
|
Half UV dosage
|
Full UV dosage
|
|||
|---|---|---|---|---|---|---|
| Avg count g−1 | Avg viability (%) | Avg count g−1 | Avg viability (%) | Avg count g−1 | Avg viability (%) | |
| 0 | 46.0 ± 1.7 | 73.0 ± 5.2 | 46.5 ± 2.1 | 71.5 ± 6.4 | 47.7 ± 2.5 | 71.7 ± 4.5 |
| 48 | 14.0 ± 2.6 | 53.7 ± 2.5 | 11.0 ± 0.0 | 32.5 ± 0.7 | 14.7 ± 1.2 | 17.6 ± 3.7 |
Oocyst counts and viability were measured from oysters prior to and after 48 h of depuration at both full (standard) and half-power UV treatments. Each assay was performed in triplicate, and oocyst numbers and viability were assessed using standard immunofluorescent antibody and DAPI/PI staining with microscopic examination at magnifications of ×40 and ×100. Standard deviations were calculated using three separate oocyst counts from each trial, which were each performed in triplicate. UV dosage was administered according to United Kingdom guidelines for standard depuration.
Inactivation of Cryptosporidium from circulation water.
During depuration (24 h) and postdepuration (48 h), circulation water samples were taken from tanks to evaluate the effect UV treatment had towards oocyst viability. While the average viability of oocysts was significantly lower (P < 0.05) after 48 h of depuration, because oysters had more time to depurate, significantly more oocysts per ml were present in the circulation water (Table 2) where they were exposed to UV, significantly lowering their viability (P < 0.05).
TABLE 2.
Survival of C. parvum oocysts recovered from the circulation water of experimentally contaminated oystersa
| Time (h) | No UV treatment
|
Full UV dosage
|
||
|---|---|---|---|---|
| Avg viability (%) | Avg total oocyst count liter−1 | Avg viability (%) | Avg total oocyst count liter−1 | |
| 24 | 71.8 ± 2.3 | 51 ± 5.6 | 15.7 ± 2.5 | 17.3 ± 11.8 |
| 48 | 63.9 ± 8.0 | 922 ± 64.0 | 3.2 ± 4.0 | 766.7 ± 520.4 |
Total oocyst count and viability in circulation water with experimentally contaminated oysters during (24 h) and after (48 h) standard UV depuration were determined using standard immunofluorescent antibody and DAPI/PI staining, with microscopic examination at magnifications of ×40 and ×100. Each assay was performed in triplicate, and standard deviations were calculated on the basis of three separate oocyst counts. Values represent the average number of oocysts recovered per liter of circulation water. UV dosage was administered according to United Kingdom guidelines for standard depuration.
DISCUSSION
Our study is the first to report that both standard and half-power UV depuration significantly lowered quantities of recoverable, viable C. parvum oocysts from oysters in commercial-size depuration tanks. Importantly, concentrations of oocysts used for the current study (106 liter−1) were equivalent to quantities of C. parvum commonly found in surface waters due to urban runoff, wastewater discharges, and agricultural runoff from ruminant farms (14, 19). Therefore, by using fecal calf material which contained levels of oocysts commonly found in contaminated surface water, our experimental trials replicated environmental fecal contamination. While standard UV depuration treatment resulted in the largest degree of oocyst inactivation (P < 0.05; 13-fold), it still did not totally remove all viable oocysts recovered from oysters. Previous studies found that various depuration strategies, including UV, are unable to totally remove oocyst contamination in shellfish (10) and that no correlation exists between depuration times (14). Water type, temperature, and the concentration of oocysts in the suspension have all been found not to have significant impacts on oocyst inactivation during UV depuration (6).
We found that oocysts recovered from oysters were significantly inactivated during depuration without UV treatment. Depuration studies with C. parvum bacterium-spiked shellfish (the oyster Ostrea edulis and marine clam Tapes decussatus) have also found that sharp decreases in levels of recovered viable oocysts occurred during depuration in the first 4 days, with 15 to 25% viable oocysts remaining thereafter (10). Thus, longer depuration periods, both with and without UV, for shellfish are still not sufficient to totally remove all viable oocysts from shellfish. This persistence may be due to the immobilization and protection of oocysts within the shellfish gills and hemocytes (10, 14, 17).
As expected, during depuration with the application of a higher UV dosage (standard depuration), more oocysts were inactivated, both in circulation water and within oysters. When applying half the UV dosage during depuration, mimicking a less effective administration of UV intensity, the number of viable oocysts within oysters remained significantly higher. Previous studies have also shown that the amount of UV dosage during treatment is directly related to treatment effectiveness (30).
Shellfish (mussels and cockles) harboring infective C. parvum oocysts can transport >103 oocysts, and this contamination is strongly associated with shellfish recovered from areas located near the mouths of rivers with a high density of grazing ruminants on their banks (13). However, there is no relationship between the presence of Cryptosporidium oocysts in shellfish and fecal coliform levels, a common indicator of microbiological contamination (14). United Kingdom and EC guidelines for shellfish depuration have been based around the removal of fecal coliforms and Salmonella organisms (36). However, shellfish, including oysters, show different behavior when purging Cryptosporidium during depuration, and viable oocysts can survive within them (10, 14-16, 19). After depuration, the survival of Cryptosporidium in shellfish destined to be consumed raw by humans is a serious concern to public health, especially considering the low infective dose of Cryptosporidium (35). Our study showed that current United Kingdom and European Union guidelines for removing Cryptosporidium may not guarantee that shellfish are safe for raw consumption. Furthermore, the existence of viable oocysts in samples with microbiological contamination levels <300 fecal coliforms/100 g, which in accordance with current European Union legislation are considered suitable for human consumption, suggests the need to include parasitological analyses in the quality control parameters for these molluscs (14, 25, 29, 36).
Our findings support the need for establishing continuous Cryptosporidium monitoring programs at shellfish harvesting sites and for the implementation of periodic quality checks on oysters prior to human consumption. Thorough cooking should be sufficient to inactivate Cryptosporidium, although oysters are often eaten raw and are therefore a potential source of food-borne cryptosporidiosis.
Acknowledgments
O.S. was funded by a University of Ulster Vice-Chancellor's Scholarship. W.J.S. was supported by The Northern Ireland Centre of Excellence in Functional Genomics, with funding from the European Union Program for Peace and Reconciliation, under the Technology Support for the Knowledge-Based Economy.
We thank Martin Pyke for providing SEAFISH funding and advice during technical discussions. We acknowledge Valerie DeWeale for kindly providing Cryptosporidium-positive calf fecal samples.
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Bukhari, Z., M. M. Marshall, D. G. Korich, C. R. Fricker, H. V. Smith, J. Rosen, and J. L. Clancy. 2000. Comparison of Cryptosporidium parvum viability and infectivity assays following ozone treatment of oocysts. Appl. Environ. Microbiol. 66:2972-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butt, A. A., K. E. Aldridge, and C. V. Sanders. 2004. Infections related to the ingestion of seafood. Part II. Parasitic infections and food safety. Lancet Infect. Dis. 4:294-300. [DOI] [PubMed] [Google Scholar]
- 3.CEFAS (Centre for Environmental, Fisheries and Aquaculture Science). November 2005, posting date. Shellfish news, 20, DEFRA (Department for Environment food and Rural Affairs), page 34-35. http://www.cefas.co.uk/publications/shellfishnews/shellnews20.pdf.
- 4.Collins, M. V., G. J. Flick, S. A. Smith, R. Fayer, E. Rubendall, and D. S. Lindsay. 2005. The effects of E-beam irradiation and microwave energy on Eastern Oysters (Crassostrea virginica) experimentally infected with Cryptosporidium parvum. J. Eukaryot. Microbiol. 52:484-488. [DOI] [PubMed] [Google Scholar]
- 5.Collins, M. V., G. J. Flick, S. A. Smith, R. Fayer, R. Croonenberghs, S. O'Keefe, and D. S. Lindsay. 2005. The effect of high-pressure processing on infectivity of Cryptosporidium parvum oocysts recovered from experimentally exposed eastern oysters (Crassostrea virginica). J. Eukaryot. Microbiol. 52:500-504. [DOI] [PubMed] [Google Scholar]
- 6.Craik, S. A., D. Weldon, G. R. Finch, J. R. Bolton, and M. Belosevic. 2001. Inactivation of Cryptosporidium parvum oocysts using medium- and low-pressure ultraviolet radiation. Water Res. 35:1387-1398. [DOI] [PubMed] [Google Scholar]
- 7.European Communities. 1991. Council Directive 91/492 of 15 July 1991 laying down the health conditions for the production and the placing on the market of live bivalve molluscs. Off. J. Eur. Commun. L268:1-14. [Google Scholar]
- 8.Fayer, R., T. K. Graczyk, E. J. Lewis, J. M. Trout, and C. A. Farley. 1998. Survival of infectious Cryptosporidium parvum oocysts in seawater and eastern oysters (Crassostrea virginica) in the Chesapeake Bay. Appl. Environ. Microbiol. 64:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.FDA Form 2400a (3/01), Food and Drug Administration, Department of Health and Human Services. 2004. Public Health Service, Food and Drug Administration, milk laboratory evaluation form. http://www.fda.gov/opacom/morechoices/fdaforms/FDA-2400a.pdf.
- 10.Freire-Santos, F., H. Gomez-Couso, M. R. Ortega-Inarrea, J. A. Castro-Hermida, A. M. Oteiza-Lopez, O. Garcia-Martin, and M. E. Ares-Mazas. 2002. Survival of Cryptosporidium parvum oocysts recovered from experimentally contaminated oysters (Ostrea edulis) and clams (Tapes decussatus). Parasitol. Res. 88:130-133. [DOI] [PubMed] [Google Scholar]
- 11.Giangaspero, A., U. Molini, R. Iorio, D. Traversa, B. Paoletti, and C. Giansante. 2005. Cryptosporidium parvum oocysts in seawater clams (Chameleagallina) in Italy. Vet. Med. 69:203-212. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, R. J., J. de Louvois, T. Donovan, C. Little, K. Nye, C. D. Ribeiro, J. Richards, D. Roberts, and F. J. Bolton. 2000. Guidelines for the microbiological quality of some ready-to-eat foods sampled at the point of sale. PHLS Advisory Committee for Food and Dairy Products. Commun. Dis. Public Health 3:163-167. [PubMed] [Google Scholar]
- 13.Gomez-Bautista, M., L. M. Ortega-Mora, E. Tabares, V. Lopez-Rodas, and E. Costas. 2000. Detection of infectious Cryptosporidium parvum oocysts in mussels (Mytilus galloprovincialis) and cockles (Cerastoderma edule). Appl. Environ. Microbiol. 66:1866-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Couso, H., F. Freire-Santos, J. Martinez-Urtaza, O. Garcia-Martin, and M. E. Ares-Mazas. 2003. Contamination of bivalve molluscs by Cryptosporidium oocysts: the need for new quality control standards. Int. J. Food. Microbiol. 87:97-105. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Couso, H., F. Freire-Santos, M. R. Ortega-Inarrea, J. A. Castro-Hermida, and M. E. Ares-Mazas. 2003. Environmental dispersal of Cryptosporidium parvum oocysts and cross transmission in cultured bivalve molluscs. Parasitol. Res. 90:140-142. [DOI] [PubMed] [Google Scholar]
- 16.Gomez-Couso, H., F. Freire-Santos, G. A. Hernandez-Cordova, and M. E. Ares-Mazas. 2005. A histological study of the transit of Cryptosporidium parvum oocysts through clams (Tapes decussatus). Int. J. Food. Microbiol. 102:57-62. [DOI] [PubMed] [Google Scholar]
- 17.Graczyk, T. K., R. Fayer, E. J. Lewis, C. A. Farley, and J. M. Trout. 1997. In vitro interactions between hemocytes of the eastern oyster, Crassostrea virginica Gmelin, 1791 and Cryptosporidium parvum oocysts. J. Parasitol. 83:949-952. [PubMed] [Google Scholar]
- 18.Graczyk, T. K., and K. J. Schwab. 2000. Foodborne infections vectored by molluscan shellfish. Curr. Gastroenterol. Rep. 2:305-309. [DOI] [PubMed] [Google Scholar]
- 19.Graczyk, T. K., A. S. Girouard, L. Tamang, S. P. Nappier, and K. J. Schwab. 2006. Recovery, bioaccumulation, and inactivation of human waterborne pathogens by the Chesapeake Bay nonnative oyster, Crassostrea ariakensis. Appl. Environ. Microbiol. 72:3390-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hijnen, W. A., E. F. Beerendonk, and G. J. Medema. 2006. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: a review. Water Res. 40:3-22. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. H., M. S. Elovitz, U. von Gunten, H. M. Shukairy, and B. J. Marinas. 2007. Modeling Cryptosporidium parvum oocyst inactivation and bromate in a flow-through ozone contactor treating natural water. Water Res. 41:467-475. [DOI] [PubMed] [Google Scholar]
- 22.Leclercq, A., C. Wanegue, and P. P. Baylac. 2002. Comparison of fecal coliform agar and violet red bile lactose agar for fecal coliform enumeration in foods. Appl. Environ. Microbiol. 68:1631-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limor, J. R., A. A. Lal, and L. Xiao. 2002. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J. Clin. Microbiol. 40:2335-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linden, K. G., G. Shin, G. Faubert, W. Cairns, and M. D. Sobsey. 2002. UV disinfection of Giardia lamblia cysts in water. Environ. Sci. Technol. 36:2519-2522. [DOI] [PubMed] [Google Scholar]
- 25.Lipp, E. K., S. A. Farrah, and J. B. Rose. 2001. Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Mar. Pollut. Bull. 42:286-293. [DOI] [PubMed] [Google Scholar]
- 26.Lowery, C. J., P. Nugent, J. E. Moore, B. C. Millar, X. Xiru, and J. S. Dooley. 2001. PCR-IMS detection and molecular typing of Cryptosporidium parvum recovered from a recreational river source and an associated mussel (Mytilus edulis) bed in Northern Ireland. Epidemiol. Infect. 127:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacRae, M., C. Hamilton, N. J. Strachan, S. Wright, and I. D. Ogden. 2005. The detection of Cryptosporidium parvum and Escherichia coli O157 in UK bivalve shellfish. J. Microbiol. Methods 60:395-401. [DOI] [PubMed] [Google Scholar]
- 28.Millar, B. C., M. Finn, L. H. Xiao, C. J. Lowery, J. S. G. Dooley, and J. E. Moore. 2002. Cryptosporidium in foodstuffs: an emerging aetiological route of human foodborne illness. Trends Food Sci. Technol. 13:168-187. [Google Scholar]
- 29.Miller, W. A., E. R. Atwill, I. A. Gardner, M. A. Miller, H. M. Fritz, R. P. Hedrick, A. C. Melli, N. M. Barnes, and P. A. Conrad. 2005. Clams (Corbicula fluminea) as bioindicators of fecal contamination with Cryptosporidium and Giardia spp. in freshwater ecosystems in California. Int. J. Parasitol. 35:673-684. [DOI] [PubMed] [Google Scholar]
- 30.Morita, S., A. Namikoshi, T. Hirata, K. Oguma, H. Katayama, S. Ohgaki, N. Motoyama, and M. Fujiwara. 2002. Efficacy of UV irradiation in inactivating Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 68:5387-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nichols, R. A., J. E. Moore, and H. V. Smith. 2006. A rapid method for extracting oocyst DNA from Cryptosporidium-positive human faeces for outbreak investigations. J. Microbiol. Methods 65:512-524. [DOI] [PubMed] [Google Scholar]
- 32.Nydam, D. V., and H. O. Mohammed. 2005. Quantitative risk assessment of Cryptosporidium species infection in dairy calves. J. Dairy Sci. 88:3932-3943. [DOI] [PubMed] [Google Scholar]
- 33.Oppenheimer, J. A., J. G. Jacangelo, J. M. Laine, and J. E. Hoagland. 1997. Testing the equivalency of ultraviolet light and chlorine for disinfection of wastewater to reclamation standards. Water Environ. Res. 69:14-24. [Google Scholar]
- 34.Pandak, N., K. Zeljka, and A. Cvikovic. 2006. A family outbreak of cryptosporidiosis: probable nosocomial infection and person-to-person transmission. Wien. Klin. Wochenschr. 118:485-487. [DOI] [PubMed] [Google Scholar]
- 35.Pouillot, R., P. Beaudeau, J. B. Denis, and F. Derouin. 2004. A quantitative risk assessment of waterborne cryptosporidiosis in France using second-order Monte Carlo simulation. Risk Anal. 24:1-17. [DOI] [PubMed] [Google Scholar]
- 36.Seafish Industry Authority. 1997. Guidelines for the harvesting, handling and distribution of live bivalve molluscs. Seafish Industry Authority. Hull, United Kingdom. http://www.seafish.org/pdf.pl?file=seafish/Documents/guideline_bivalve_molluscs_facilities.pdf.
- 37.Smith, H. V., B. M. Campbell, C. A. Paton, and R. A. Nichols. 2002. Significance of enhanced morphological detection of Cryptosporidium sp. oocysts in water concentrates determined by using 4′,6′-diamidino-2-phenylindole and immunofluorescence microscopy. Appl. Environ. Microbiol. 68:5198-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sunnotel, O., C. J. Lowery, J. E. Moore, J. S. Dooley, L. Xiao, B. C. Millar, P. J. Rooney, and W. J. Snelling. 2006. Cryptosporidium. Lett. Appl. Microbiol. 43:7-16. [DOI] [PubMed] [Google Scholar]
- 39.Sunnotel, O., W. J. Snelling, L. Xiao, K. Moule, J. E. Moore, B. Cherie Millar, J. S. G. Dooley, and C. J. Lowery. 2006. Rapid and sensitive detection of single Cryptosporidium oocysts from archived glass slides. J. Clin. Microbiol. 44:3285-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamburrini, A., and E. Pozio. 1999. Long-term survival of Cryptosporidium parvum oocysts in seawater and in experimentally infected mussels (Mytilus galloprovincialis). Int. J. Parasitol. 29:711-715. [DOI] [PubMed] [Google Scholar]
- 41.Tufenkji, N., D. R. Dixon, R. Considine, and C. J. Drummond. 2006. Multi-scale Cryptosporidium/sand interactions in water treatment. Water Res. 40:3315-3331. [DOI] [PubMed] [Google Scholar]
- 42.Wetzel, D. M., J. Schmidt, M. S. Kuhlenschmidt, J. P. Dubey, and L. D. Sibley. 2005. Gliding motility leads to active cellular invasion by Cryptosporidium parvum sporozoites. Infect. Immun. 73:5379-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]


