Abstract
Our objective was to evaluate methods for identifying cattle with high concentrations of Escherichia coli O157 in their feces. In two experiments, feces were collected from cattle orally inoculated with nalidixic acid (Nal)-resistant E. coli O157, and direct plating of diluted feces on sorbitol MacConkey agar with cefixime and potassium tellurite (CT-SMAC) containing Nal was considered the gold standard (GS) method. In experiment 1, methods evaluated were preenrichment direct streak, immunomagnetic separation with most probable number (MPN), and postenrichment direct streak with MPN, all using CT-SMAC. The mean concentration of Nal-resistant E. coli O157 in samples (n = 59) by use of the GS was 3.6 log10 CFU/g. The preenrichment streak detected >3.0 log10 CFU/g samples with a 74.4% sensitivity and 68.8% specificity. Postenrichment direct streak-MPN and immunomagnetic separation-MPN concentrations were correlated significantly with GS concentrations (r = 0.53 and r = 0.39, respectively). In experiment 2 (480 samples), pre- and postenrichment direct streaking performed in triplicate and spiral plating on CT-SMAC were evaluated. For preenrichment streaks, sensitivity was 79.7% and specificity was 96.7% for detecting >3.0 log10 CFU/g when the criterion was positive cultures on at least two plates. For spiral plating at that concentration, sensitivity and specificity were 83.9% and 56.3%, respectively. Postenrichment streaking performed relatively poorly. Triplicate preenrichment streaks of 1:10-diluted feces on CT-SMAC may be useful for identifying cattle shedding high concentrations of E. coli O157. Estimates of sensitivity and specificity enable appropriate application of methods and interpretation of results and may enhance applied research, surveillance, and risk assessments.
Escherichia coli O157 is a food-borne pathogen that continues to cause severe outbreaks of human infection despite many years of research. Cattle are a known carrier of E. coli O157, and previous evidence suggests that prevalence of the organism on carcasses at harvest is correlated with prevalence in feces or on hides (1). More recent literature suggests that not only prevalence but also concentration of E. coli O157 in the feces is indicative of a risk of carcass contamination (3). There is evidence that within a given population of cattle most animals shed E. coli O157 intermittently and at relatively low concentrations, but some animals may shed at high concentrations (i.e., greater than 3.0 log10 CFU/g of feces) (14). Additionally, animals shedding E. coli O157 at higher concentrations are thought to transmit infection to a greater number of animals than animals shedding the organism at low levels (8, 9), thus effectively increasing prevalence within the population. These facts suggest the importance of identifying high-shedding animals prior to harvest as well as identifying these animals when evaluating intervention strategies. Furthermore, quantifying the prevalence of these high-shedding animals in a given population may improve the accuracy of microbial risk assessments (4).
Many methods for determining the concentration of E. coli O157 have been described previously. Direct plating of serially diluted fecal samples is a popular method of enumerating E. coli O157 organisms (6, 7, 11). The most-probable-number (MPN) technique also is used to enumerate E. coli O157 organisms, and immunomagnetic separation (IMS) often is combined with this method (2, 16, 17). Spiral plating of diluted fecal samples (13, 15) and real-time PCR-based quantification (5) are additional methods that have been evaluated for enumerating E. coli O157 organisms in feces.
These methods may lead to quantitative or semiquantitative estimates of E. coli O157 organisms in feces, but equipment, materials, and labor costs for some methods may become substantial when large numbers of samples are considered. In addition, many previous evaluations of these methods have failed to report sensitivity and specificity estimates, which can severely limit the accurate application of diagnostic tests and interpretation of subsequent results for research and risk assessments (4). Our objectives were to evaluate different methods for detecting cattle shedding high concentrations of E. coli O157 in their feces and compare these methods to a commonly used and accepted method (i.e., the gold standard method).
MATERIALS AND METHODS
Overall approach and methods.
Objectives were accomplished with two separate experiments, both utilizing fecal samples from cattle orally inoculated with nalidixic acid (Nal)-resistant E. coli O157. The first experiment evaluated direct streaking of samples prior to enrichment, IMS with MPN enumeration, postenrichment direct streaking with MPN enumeration, and standard IMS. Results from the first experiment led to the reevaluation of the preenrichment direct streak technique with modifications and standard IMS. A spiral plating technique also was included in experiment 2. Gold standard methods used to assess the precision and accuracy of other methods in both experiments were based on direct plating of diluted feces on selective medium containing nalidixic acid. Culture media and methods to confirm E. coli O157 were similar in both experiments.
Gram-negative broth (Becton Dickinson Co., Franklin Lakes, NJ) containing cefixime (50 ng/ml), cefsulodin (10 μg/ml), and vancomycin (8 μg/ml) (GNccv) was used for enrichment. Sorbitol MacConkey agar (Becton Dickinson Co., Franklin Lakes, NJ) supplemented with cefixime (50 ng/ml) and potassium tellurite (2.5 μg/ml) (CT-SMAC) was used for plating samples. For determination of gold standard concentrations of nalidixic acid-resistant E. coli O157, CT-SMAC was supplemented with nalidixic acid (Sigma-Aldrich, St. Louis, MO), at either 20 (CT-SMACNal20; experiment 1) or 50 (CT-SMACNal50; experiment 2) μg/ml. For confirmation of E. coli O157 on CT-SMAC, isolates were grown on blood agar (Remel, Lenexa, KS) and tested for indole production and latex agglutination of the O157 antigen (Oxoid Limited, Basingstoke, United Kingdom).
Experiment 1. (i) Animals and bacterial inoculation.
Cattle (n = 10) were orally inoculated via a stomach tube with a mixture of three bovine fecal strains (FRIK920, FRIK1123, and FRIK2000) of E. coli O157 made resistant to 20 μg/ml of nalidixic acid (Sigma). Cattle were commercial beef calves of mixed breeds, of either sex, between 200 and 300 kg in body weight. Fecal samples were collected via rectal grab sampling from these animals two times per week for 3 weeks, giving a total of 60 fecal samples for evaluation of the procedures. Samples were transported in sterile plastic bags to the laboratory, and further procedures were performed within 1 h of collection. Each sample was considered as an independent experimental unit for assessment of the enumeration procedures.
(ii) Gold standard.
For each sample, approximately 2 g of feces was placed into 18 ml of GNccv in large, preweighed test tubes. Tubes were then weighed again to determine the amount of feces and vortexed for 1 min. Two-hundred-microliter aliquots from the fecal slurry tubes (preenrichment) were placed into 1.8 ml GNccv in a 96-well assay block (Corning Inc., Corning, NY) in triplicate. Each of these subsamples underwent four serial 10-fold dilutions of 200 μl into 1.8 ml GNccv to yield dilutions from 10−2 to 10−6 in the assay block. The gold standard concentration of E. coli O157 in each fecal sample was determined by spread plating 100 μl of the original fecal slurry in triplicate and each dilution in the assay block onto CT-SMACNal20. Following overnight incubation (37°C) of plates, sorbitol-negative colonies on CT-SMACNal20 plates were counted to determine the concentration of E. coli O157 (CFU/g) in each sample. Two sorbitol-negative colonies from each plate were tested for confirmation of E. coli O157, as described above.
(iii) Direct streaking of preenriched sample.
Prior to enrichment, loopfuls (broad tip [10 μl] of a sterile bacterial loop) (catalog no. 13-075-4a; Fisher Scientific, Palantine, IL) of sample slurry in GNccv were streaked onto CT-SMAC plates. Following overnight incubation of plates at 37°C, up to two sorbitol-negative colonies were tested for confirmation of E. coli O157.
(iv) Standard IMS.
Tubes containing feces in GNccv were incubated for 6 h at 37°C. Following enrichment, tubes were vortexed for 1 min and 1 ml of the mixture was pipetted into a 1.5-ml microcentrifuge tube (Fisher Scientific) containing 20 μg of Dynabeads (anti-E. coli O157; Dynal Biotech ASA, Oslo, Norway). Following IMS, 50 μl was spread plated onto CT-SMAC and incubated (37°C) overnight. Up to six sorbitol-negative colonies were tested to confirm E. coli O157.
(v) IMS-MPN.
Following sample dilution in assay blocks and transfer of inocula onto plates for gold standard enumeration, blocks were incubated for 6 h at 37°C. After enrichment, 1 ml from 10−2, 10−4, and 10−6 dilutions was subjected to IMS as described above and plated on CT-SMAC. Assay blocks were refrigerated (4°C) during the overnight incubation of plates. After overnight incubation, if sorbitol-negative colonies were evident on CT-SMAC from the 10−2 or 10−4 dilution, then 1 ml of the next-higher dilution (10−3 or 10−5, respectively) was subjected to IMS on the following day (2). Isolation and identification of E. coli O157 were performed as described above. Based upon the number of wells that were positive for E. coli O157 in each dilution, MPN values were determined with MPN Build 23 (Mike Curiale, http://i2workout.com/mcuriale/mpn/index.html) and concentrations were expressed as MPN/g.
(vi) Postenrichment direct streak-MPN.
Following enrichment (6 h at 37°C), loopfuls of GNccv-fecal slurry and of each dilution and replication in the assay block were then streaked onto CT-SMAC plates. Confirmation of E. coli O157 was performed as described above. Again, based upon the number of wells that tested positive per dilution, MPN was determined as described above.
Experiment 2. (i) Animals and bacterial inoculation.
Commercial, mixed-breed beef calves (n = 30) weighing approximately 180 kg (5 to 6 months of age) were orally inoculated via a stomach tube with a mixture of five bovine fecal strains (FRIK920, FRIK1123, FRIK2000, 01-2-10561, and 01-2-08970) of E. coli O157 made resistant in the laboratory to 50 μg/ml of nalidixic acid. Fecal samples were obtained from the animals three times a week for approximately 5 weeks for a total of 480 samples, and as in experiment 1, samples were transported in plastic bags and further procedures were performed within 1 h of collection. Each sample was again identified as an independent experimental unit for assessment of the enumeration procedures.
(ii) Gold standard.
In contrast to the gold standard determination in experiment 1, serial dilution of feces in GNccv was not performed in triplicate, yet spread plating of each dilution onto CT-SMACNal50 was done in triplicate. Procedures for determining the concentration of E. coli O157 in each sample were similar to procedures used for experiment 1. In order to detect E. coli O157 at levels below the threshold of detection (102) by this method, fecal slurry tubes were incubated for 6 h at 37°C and then tubes were vortexed and 1 ml of the mixture was transferred to another test tube containing 9.0 ml GNccv. This tube was incubated (37°C) for an additional 18 to 24 h and then plated onto CT-SMACNal50. Similar procedures were used to confirm E. coli O157 on these plates.
(iii) Direct streaking of pre- and postenrichment samples.
Prior to enrichment, a loopful of GNccv-fecal slurry was streaked onto CT-SMAC in triplicate. This procedure was then repeated after tubes were incubated for 6 h at 37°C (postenrichment direct streak). Up to two sorbitol-negative colonies were tested for the confirmation of E. coli O157 for each method.
(iv) Standard IMS.
Standard IMS was performed as described for experiment 1.
(v) Spiral plating.
Spiral plating was performed on 150 samples collected during the first 2 weeks following inoculation. Prior to enrichment, a 100-μl aliquot from the GNccv-fecal slurry was spiral plated onto one CT-SMAC plate and one CT-SMACNal50 plate. The plating was done using a WASP 2 Spiral plater (Microbiology International, Frederick, MD). After overnight incubation at 37°C, sorbitol-negative colonies (if present) were counted using an aCOLyte SuperCount colony counter (Microbiology International). Two colonies from each plate were tested for indole production and latex agglutination for the O157 antigen. E. coli O157 counts from spiral plates were expressed as CFU per g of feces.
Data analysis.
The Shapiro-Wilk test was used to determine if concentration data were normally distributed using Proc UNIVARIATE of SAS (SAS version 9.1, Cary, NC). Pearson correlation coefficients were generated between the log-transformed concentration of E. coli O157, as determined by the gold standard method, and the log-transformed MPN values for the IMS and postenrichment direct streak methods. Correlations were determined using Proc CORR of SAS. Linear trend lines, equations, and R2 values were determined with Microsoft Excel (Microsoft Corporation, Redmond, WA). For determining sensitivity and specificity estimates of methods to identify samples with high concentrations of E. coli O157, we used 3.0 and 4.0 log10 CFU per g of feces as threshold values to separate low (below-threshold) and high (above-threshold) concentrations. Sensitivity estimates were the proportions of truly positive samples (based on the gold standard) that tested positive (either overall, in a specified category, or above specified threshold values). Specificity estimates were calculated similarly based on truly negative samples that tested negative. Exact 95% binomial confidence intervals (CI) were calculated for sensitivity and specificity estimates by use of the BETAINV function of Microsoft Excel. Exact CI are given in parentheses for all presented sensitivity and specificity estimates. Positive predictive values (PPV) were calculated as the proportion of test-positive samples that were true positives.
RESULTS
Experiment 1. (i) Sample distribution.
In total, 60 fecal samples were collected in the experiment, but 1 was excluded because there was less than 0.5 g of fecal material. Descriptive statistics for samples are shown in Table 1. E. coli O157 was detected and CFU were quantified in all samples. The minimum and maximum concentrations of E. coli O157 were 2.5 × 101 CFU/g and 2.3 × 105 CFU/g, respectively. Concentrations of E. coli O157 were not normally distributed; however, log-transformed concentrations were not significantly different from normal (P = 0.42).
TABLE 1.
Distribution of E. coli O157 concentrations in fecal samples in experiment 1
| E. coli O157 concn range (log10 CFU/g) (no. of samples) | Mean original concn (CFU/g) (SD) | Mean transformed concn (log10 CFU/g) (SD) |
|---|---|---|
| 1 to 2 (3) | 46 (18) | 1.63 (0.20) |
| 2 to 3 (13) | 497 (281) | 2.62 (0.28) |
| 3 to 4 (23) | 4,730 (2,871) | 3.59 (0.29) |
| 4 to 5 (18) | 28,561 (16,337) | 4.40 (0.22) |
| 5 to 6 (2) | 175,996 (75,189) | 5.22 (0.19) |
| Overall (59) | 16,635 (35,036) | 3.58 (0.88) |
(ii) Direct streaking of preenriched sample.
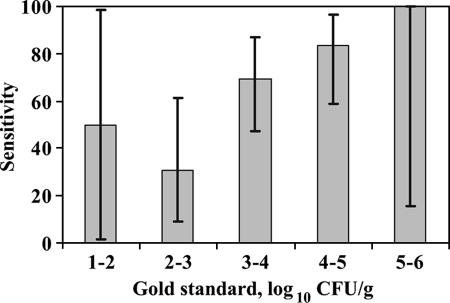
E. coli O157 was isolated from 37 of the 59 (62.7%) direct streaked, preenriched samples when a maximum of two colonies present on CT-SMAC were evaluated per plate. Sensitivity estimates categorized by the concentrations of E. coli O157 in samples are shown in Fig. 1. By use of this method to identify samples with E. coli O157 concentrations above 3.0 log10 CFU/g, sensitivity and specificity estimates were 74.4% (58.8 to 86.5%) and 68.8% (41.3 to 89.0%), respectively. Sensitivity and specificity estimates for this method when used to detect samples above 4.0 log10 CFU/g were 85.0% (62.1 to 96.8%) and 48.7% (32.4 to 65.2%), respectively.
FIG. 1.
Sensitivity estimates and corresponding 95% CI (error bars) of the preenrichment streak method, categorized by the gold standard concentration (experiment 1). Estimates are presented as the percentages of samples with a confirmed E. coli O157 colony on the preenrichment direct streak plate when only two colonies were tested.
(iii) Standard IMS.
By use of IMS, E. coli O157 was detected in 52 of 59 known positive fecal samples (sensitivity estimate, 88.1% [77.1 to 95.1%]). The means and standard errors of the gold standard concentration of E. coli O157 in samples testing positive and negative by this method were 3.62 ± 0.12 and 3.27 ± 0.29 CFU/g, respectively. Sensitivity and specificity estimates for identifying samples with >4.0 log10 CFU per g were 95.0% (75.1 to 99.9%) and 15.4% (5.9 to 30.5%), respectively.
(iv) MPN.
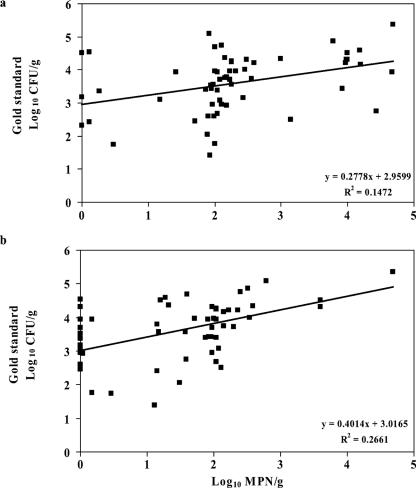
Mean MPN values for the IMS-MPN and postenrichment direct streak-MPN methods were 3.7 × 103 and 1.0 × 103 MPN/g, respectively. Log10 transformations were made on the MPN values, but normality was not present in either original or transformed concentrations of E. coli O157. However, linear relationships were evaluated in an attempt to examine the association of these methods with the gold standard (Fig. 2). The gold standard was correlated (P < 0.01) with both IMS-MPN and postenrichment direct streak-MPN methods. Additionally, the IMS-MPN method was correlated with the direct streak method (r = 0.54; P < 0.01).
FIG. 2.
Relationships between the log-transformed gold standard concentration of E. coli O157 in fecal samples and the log-transformed MPN concentration of E. coli O157 in the same sample, as determined by the IMS-MPN method (a) and the direct streak-MPN method (b). Equations and R2 values for linear trend lines are presented.
The sensitivity and specificity of MPN methods were estimated by creating categories of log10 groups of E. coli O157 concentrations derived from linear equations, which were generated by plotting gold standard concentrations versus nonzero MPN values (Fig. 2). If MPN values were zero, then zero was used. The IMS-MPN method identified samples with >3.0 log10 CFU/g with sensitivity and specificity estimates of 90.7% (77.9 to 97.4%) and 25.0% (7.3 to 52.4%), respectively. At this concentration threshold, the postenrichment direct streak-MPN method had sensitivity and specificity estimates of 76.7% (61.4 to 88.2%) and 56.2% (29.9 to 80.3%), respectively. Using 4.0 log10 CFU/g as the threshold, sensitivity and specificity estimates for the IMS-MPN and postenrichment direct streak-MPN methods were 40.0% (19.1 to 64.0%) and 92.3% (79.1 to 98.4%) and 35.0% (15.4 to 59.2%) and 100% (91.0 to 100%), respectively.
Experiment 2. (i) Sample distribution.
In experiment 2, concentrations of E. coli O157 were handled as categorical data (Table 2). The gold standard detected E. coli O157 in 397 of 480 (82.7%) fecal samples collected. Based on the gold standard concentration or detection by enrichment, samples were grouped into one of eight categories: zero, when samples were negative on all tests; <1, when samples were positive only by secondary enrichment; and 1 to 2, 2 to 3, 3 to 4, 4 to 5, 5 to 6, and 6 to 7, corresponding to the log10 E. coli O157 CFU/g.
TABLE 2.
Distribution of samples in experiment 2 categorized by the gold standard E. coli O157 concentration (log10 CFU/g)
| Categorya | No. of samples (% of total) |
|---|---|
| 0 (not detected) | 83 (17.3) |
| <1 | 167 (34.8) |
| 1 to 2 | 36 (7.5) |
| 2 to 3 | 46 (9.6) |
| 3 to 4 | 66 (13.8) |
| 4 to 5 | 46 (9.6) |
| 5 to 6 | 28 (5.8) |
| 6 to 7 | 8 (1.7) |
Categories are described in Results.
(ii) Direct streaking of pre- and postenriched samples.
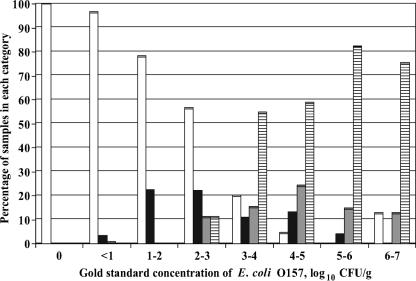
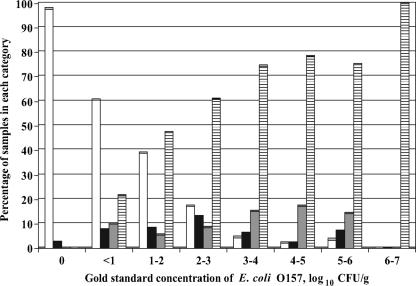
For direct streaking of pre- and postenriched samples, the results were categorized as zero, one, two, or three E. coli O157-positive CT-SMAC plates. Distributions of samples with these responses for both pre- and postenrichment direct streak methods, categorized by the gold standard concentration of E. coli O157, are shown in Fig. 3 and 4. Sensitivities, specificities, and PPV for pre- and postenrichment direct streak methods identifying samples with high levels of E. coli O157 (threshold concentration values of 3.0 or 4.0 log10 CFU/g) are presented in Table 3.
FIG. 3.
Percentages of samples with zero (white bars), one (black bars), two (gray bars), or three (hatched bars) of three preenrichment plates testing positive for E. coli O157 in experiment 2. Percentages are categorized by the gold standard concentration. A positive plate had at least one confirmed E. coli O157 colony when only two suspect colonies were tested.
FIG. 4.
Percentages of samples with zero (white bars), one (black bars), two (gray bars), or three (hatched bars) of three postenrichment plates testing positive for E. coli O157 in experiment 2. Percentages are categorized by the gold standard concentration. A positive plate had at least one confirmed E. coli O157 colony when only two suspect colonies were tested.
TABLE 3.
Sensitivity and specificity estimates with corresponding PPV for direct streak methodsa
| Direct streak method and no. of positive plates | E. coli O157 concn (log10 CFU/g) | % Sensitivity (95% CI) | % Specificity (95% CI) | PPV
|
||
|---|---|---|---|---|---|---|
| At study prevalence | At 10% prevalence | At 20% prevalence | ||||
| Preenrichment | ||||||
| ≥1 | 3 | 89.2 (83.0-93.7) | 89.8 (86.0-92.8) | 0.80 | 0.49 | 0.69 |
| 4 | 96.3 (89.7-99.2) | 78.1 (73.8-82.1) | 0.48 | 0.33 | 0.52 | |
| ≥2 | 3 | 79.7 (72.3-86.0) | 96.7 (94.2-98.3) | 0.92 | 0.73 | 0.86 |
| 4 | 87.8 (78.7-94.0) | 85.7 (81.9-89.0) | 0.56 | 0.41 | 0.61 | |
| 3 | 3 | 62.2 (53.8-70.0) | 98.5 (96.5-99.5) | 0.95 | 0.82 | 0.91 |
| 4 | 68.3 (57.1-78.1) | 89.7 (86.3-92.5) | 0.58 | 0.42 | 0.62 | |
| Postenrichment | ||||||
| ≥1 | 3 | 96.6 (92.3-98.9) | 61.4 (56.0-66.7) | 0.53 | 0.22 | 0.39 |
| 4 | 97.6 (91.5-99.7) | 52.0 (47.0-57.0) | 0.30 | 0.18 | 0.34 | |
| ≥2 | 3 | 91.9 (86.3-95.7) | 68.7 (63.4-73.6) | 0.57 | 0.25 | 0.42 |
| 4 | 93.9 (86.3-98.0) | 59.1 (54.0-63.9) | 0.32 | 0.20 | 0.36 | |
| 3 | 3 | 77.0 (69.4-83.5) | 75.6 (70.6-80.1) | 0.59 | 0.26 | 0.44 |
| 4 | 79.3 (68.9-87.4) | 67.3 (62.5-71.9) | 0.33 | 0.21 | 0.38 | |
Direct streak methods used one or more, two or more, or three positive plates to classify samples as positive at concentrations of greater than 3 or 4 log10 E. coli O157 CFU per g of feces. Samples were considered positive if there was at least one confirmed E. coli O157 colony when two colonies per plate were evaluated. Corresponding PPV are the proportions of direct-streak-positive samples that were positive at the given concentration by the gold standard. These values were calculated at the observed prevalence and 10 and 20% prevalences by use of sensitivity and specificity estimates from the study.
(iii) Standard IMS.
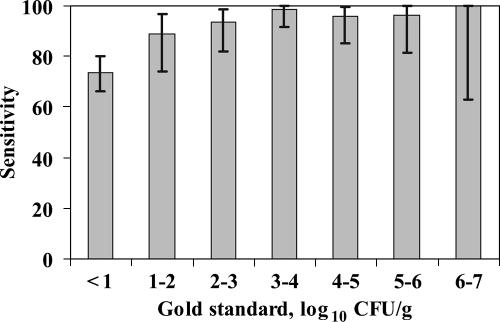
By use of the standard IMS procedure, E. coli O157 was detected in 353 of 480 total samples. This technique detected the organism in 342 of the 397 samples (sensitivity estimate, 86.1% [82.4 to 89.4%]) found positive by the gold standard and in 11 of 83 samples that were negative by the gold standard. Sensitivity was greater than 90% when the concentration of E. coli O157 was greater than 2.0 log10 CFU/g (Fig. 5). The method identified samples with high levels of E. coli O157 (3.0 log10 CFU/g) with a 97.3% (93.2 to 99.3%) sensitivity and a 37.0% (31.8 to 42.5%) specificity. At a threshold of 4.0 log10 CFU/g, sensitivity and specificity estimates were 96.3% (89.7 to 99.2%) and 31.2% (26.6 to 36.0%), respectively.
FIG. 5.
Sensitivity estimates and corresponding 95% CI (error bars) for the standard IMS procedure, categorized by the gold standard concentrations of E. coli O157 in experiment 2. Estimates are presented as the percentages of samples with a confirmed E. coli O157 colony when up to six colonies were tested.
(iv) Spiral plating.
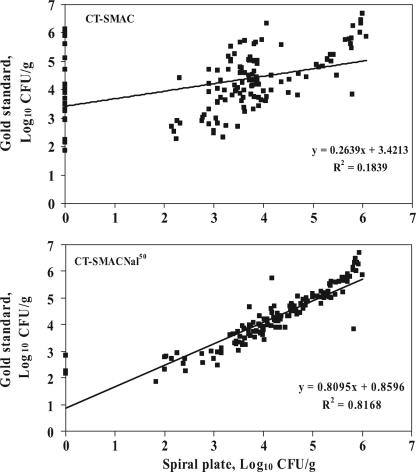
The spiral plating technique was performed on 150 samples. However, only samples with gold standard counts (n = 138) were evaluated, and samples with sorbitol-negative isolates picked from the CT-SMAC spiral plate but not confirmed as E. coli O157 (n = 10) were considered to have a spiral plate concentration of zero. E. coli O157 CFU were detected and counted for 115 of 138 samples plated on CT-SMAC and 135 of 138 samples plated on CT-SMACNal50. The linear relationships between log-transformed gold standard counts of E. coli O157 and log-transformed spiral plate counts of E. coli O157 are shown in Fig. 6. Pearson correlation coefficients indicated that log10 spiral plate counts on both CT-SMAC (r = 0.43) and CT-SMACNal50 (r = 0.90) were significantly correlated with the log10 gold standard concentration. If samples that did not have counts on the CT-SMAC spiral plate (n = 23) are removed, the correlation coefficient for that comparison improves (r = 0.66). The spiral plate CT-SMAC method identified samples with high concentrations of E. coli O157, defined as >3.0 log10 CFU/g, with sensitivity and specificity estimates of 79.0% (70.6 to 85.9%) and 63.2% (38.4 to 83.7%), respectively. Utilizing 4.0 log10 CFU/g as the threshold value, sensitivity and specificity estimates were 34.2% (23.9 to 45.7%) and 88.1% (77.1 to 95.1%), respectively.
FIG. 6.
Relationships between the log-transformed gold standard concentration of E. coli O157 in fecal samples (n = 138) and the log-transformed spiral plate concentration in the same sample when plated on CT-SMAC and CT-SMACNal50. Equations and R2 values are presented for linear trend lines.
DISCUSSION
Cattle shedding E. coli O157 at high concentrations may play an important role in transmission of the organism to people and other cattle. Methods targeted to detect these animals are more useful if estimates of test performance are available. For our experiments, we used feces from cattle that were inoculated experimentally with E. coli O157. This ensured the presence of the organism in most samples, while retaining the complex nature of the microbial ecology that can present a challenge when detecting E. coli O157 from bovine feces. We felt that this was a more appropriate model for evaluating the detection methods than was using fecal samples inoculated with E. coli O157. However, the fecal shedding patterns of inoculated cattle in our studies may not be entirely reflective of patterns in populations of naturally shedding cattle.
In our study, we used approximately 2 g of feces instead of a larger amount, such as 10 g, which has been used previously (1, 6). The smaller amount was selected for two reasons. First, using 10 g of feces for detection in very large numbers of animals may not always be economically or logistically feasible due to the increased cost of media and the incubator space required. Additionally, acquiring 10 g of feces via rectal grab at any set time during the day is not always possible. Because our intention was to compare these methods as they would be used in large-scale research or surveillance, the smaller sample seemed more applicable. Second, we wanted to evaluate all methods by using identical specimens to ensure that variation in concentration within the feces collected from the animal did not represent an additional source of variation when comparing experimental methods to the gold standard. This variation has been shown previously by Naylor et al. (10) and Pearce et al. (12).
The sensitivity of detecting E. coli O157 by the standard IMS method in our study was similar to results from previous studies where sensitivity was enhanced with increasing concentrations of E. coli O157 and a plateau effect was seen as concentrations approached 2.0 log10 CFU/g (6, 11). In experiment 2 of the current study, the standard IMS method detected E. coli O157 in 11 samples that were not detected by the gold standard. Three of these samples were from the same animal on three consecutive sampling dates, two were nonconsecutive samples from the same animal, and the remaining six samples were from six different animals. All 11 samples were collected within a 2-week period. These results may be due to the presence of noninoculated strains of E. coli O157 that were susceptible to nalidixic acid and therefore not detected by the gold standard method or simply stem from the fact that the gold standard method did not detect inoculated strains with 100% sensitivity. This is a potential source of error when comparing enumeration procedures with a gold standard that may be imperfect, but in this study the effect was negligible.
Direct plating (prior to enrichment) is a common methodology used to quantify naturally occurring E. coli O157 organisms in bovine feces (6, 7, 11). LeJeune et al. (6) employed a similar animal challenge model and found a strong correlation (r = 0.88) between their gold standard E. coli O157 concentrations and concentrations identified by direct spread plating of diluted samples (300 μl) onto CT-SMAC. This correlation was determined using samples testing positive on CT-SMAC, representing 130 of 224 (58.0%) samples positive by the gold standard (6). In the current study, we used direct streaking instead of direct spread plating. In experiment 1, with only one direct streak plate, we observed sensitivity estimates above 70% and specificity at or above 50%, depending on the definition of high concentration (threshold). We pursued this method further in experiment 2 and added replications (three plates) in an attempt to improve sensitivity and specificity. With the additional plates, we observed relatively high sensitivity, with comparable specificity. An added benefit to this method is that it can be used for many applications by changing only the interpretation of the results. For instance, if the goal is to maximize specificity with less concern about sensitivity, then a threshold value of 3.0 log10 CFU/g and three positive plates could be used as the criteria for high concentration. However, if the goal was to optimize both sensitivity and specificity, then using a threshold of 4.0 log10 CFU/g and two or more positive plates as the criteria would be better. Other advantages to this method are that it does not require special equipment (IMS, spiral plating, and PCR), the required culture materials are relatively inexpensive (bacterial loops, culture medium, and petri dishes), and it is a feasible method for handling large numbers of samples.
IMS with MPN has been used in previous studies to enumerate E. coli O157 organisms in bovine feces (2, 16, 17). Stephens (16) compared the technique to a gold standard by utilizing fecal samples spiked with streptomycin-resistant E. coli O157 strains and observed that IMS-MPN values were consistently lower than gold standard values. In our study, the IMS-MPN method was positively correlated with the gold standard, but relatively low estimates of sensitivity or specificity may prevent this method from being applied successfully for distinguishing samples with high concentrations of E. coli O157. The correlation between gold standard and IMS-MPN concentrations was lower in our study than in the previous study (16). One key difference between this previous study and our study was the amount of sample used for each test. Stephens (16) performed serial dilutions of 1 ml into 9 ml, whereas we performed serial dilutions of 0.2 ml into 1.8 ml. Even though both are 1:10 dilutions, this difference may impact the results given that samples were enriched after dilution. Refrigerating samples overnight before performing IMS on 10−3 and 10−5 dilutions may also have impacted our results in a negative fashion, but previous experiments have shown that storing fecal slurry samples at 2°C for up to 3 days did not affect inoculated E. coli O157 counts (2). This procedure was implemented to reduce the number of IMS tests, because costs can become quite high with multiple tests per sample. The postenrichment direct streak-MPN method also was correlated with the gold standard, and the cost of materials needed for this method is low, but either sensitivity or specificity estimates were relatively poor depending on the threshold used to define a high concentration of E. coli O157.
Spiral plating is another popular method of enumerating E. coli O157 organisms in feces (13, 15). While this method yielded accurate counts at higher levels (>4.0 log10 CFU/g), the precision was reduced as concentrations declined below this threshold (15). It is important to note that these previous studies utilize a selective agar for spiral plating different than that used in the current study. Categorizing samples into low and high E. coli O157 concentrations with the spiral plating method leads to compromises in either sensitivity or specificity depending on the threshold value utilized. Utilizing 3.0 log10 CFU as the threshold value for a high concentration of E. coli O157 compromises specificity, whereas using 4.0 log10 CFU compromises sensitivity. The initial equipment costs of the spiral plating technique may be substantial and represent an additional disadvantage of the technique unless a laboratory already has the equipment available. The strong linear relationship between gold standard E. coli O157 concentrations and spiral plate concentrations on CT-SMACNal50 (Fig. 6) shows that spiral plating can be highly effective for enumeration. However, the use of resistant strains and selective agar is very advantageous given the microbial ecology of bovine feces, and the results from the CT-SMAC plates illustrate that the effectiveness of the method can be diminished substantially without the ability to select isolates based on resistance.
We evaluated multiple culture procedures that may be used for identifying high concentrations of E. coli O157 in bovine feces, and we conclude that different methods could be useful depending on the objectives of testing and the resources available. When the intent is to identify animals shedding high levels of E. coli O157 in a field situation, then predictive values, which indicate the probability of a high concentration given the test result, would be of primary interest. Thus, it is important to consider the population prevalence in addition to the diagnostic sensitivity and specificity of the testing scheme. For example, we have illustrated that decreasing test specificity, prevalence of high shedders, or both will lead to lower PPV, which equates with an increase in the proportion of positive tests that are false positive (Table 3). In Table 3, we included predictive value calculations at 10 and 20% prevalences since the observed study prevalence depended on the threshold concentration used to define a high concentration of E. coli O157 and the shedding patterns of inoculated cattle in our study may not be entirely reflective of patterns in populations of naturally shedding cattle. Of the methods that we evaluated, preenrichment direct streaking of 1:10-diluted feces in triplicate appeared to be the most useful, with multiple potential applications for identifying fecal samples with high levels of E. coli O157. Regardless of the method utilized, the data on test sensitivity and specificity generated are essential for appropriate application of detection methods and for interpretation of testing results of E. coli O157 shedding patterns in bovine populations.
Acknowledgments
Financial support for this research was provided in part by a U.S. Department of Agriculture grant (2006-34359-06177).
We extend special thanks to the staff at the KSU College of Veterinary Medicine Preharvest Food Safety Lab.
This is contribution number 07-183-J from the Kansas Agricultural Experiment Station.
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fegan, N., G. Higgs, P. Vanderlinde, and P. Desmarchelier. 2004. Enumeration of Escherichia coli O157 in cattle faeces using most probable number technique and automated immunomagnetic separation. Lett. Appl. Microbiol. 38:56-59. [DOI] [PubMed] [Google Scholar]
- 3.Fegan, N., G. Higgs, P. Vanderlinde, and P. Desmarchelier. 2005. An investigation of Escherichia coli O157 contamination of cattle during slaughter at an abattoir. J. Food Prot. 68:451-457. [DOI] [PubMed] [Google Scholar]
- 4.Gardner, I. A. 2004. An epidemiologic critique of current microbial risk assessment practices: the importance of prevalence and test accuracy data. J. Food Prot. 67:2000-2007. [DOI] [PubMed] [Google Scholar]
- 5.Ibekwe, A. M., and C. M. Grieve. 2003. Detection and quantification of Escherichia coli O157:H7 in environmental samples by real-time PCR. J. Appl. Microbiol. 94:421-431. [DOI] [PubMed] [Google Scholar]
- 6.LeJeune, J. T., D. D. Hancock, and T. E. Besser. 2006. Sensitivity of Escherichia coli O157 detection in bovine feces assessed by broth enrichment followed by immunomagnetic separation and direct plating. J. Clin. Microbiol. 44:872-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Low, J. C., I. J. McKendrick, C. McKechnie, D. Fenlon, S. W. Naylor, C. Currie, D. G. E. Smith, L. Allison, and D. L. Gally. 2005. Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. Appl. Environ. Microbiol. 71:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthews, L., J. C. Low, D. L. Gally, M. C. Pearce, D. J. Mellor, J. A. P. Heesterbeek, M. Chase-Topping, S. W. Naylor, D. J. Shaw, S. W. J. Reid, G. J. Gunn, and M. E. J. Woolhouse. 2006. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl. Acad. Sci. USA 103:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews, L., I. J. McKendrick, H. Ternent, G. J. Gunn, B. Synge, and M. E. J. Woolhouse. 2006. Super-shedding cattle and the transmission dynamics of Escherichia coli O157. Epidemiol. Infect. 134:131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, D. G. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omisakin, F., M. MacRae, I. D. Ogden, and N. J. C. Strachan. 2003. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Appl. Environ. Microbiol. 69:2444-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearce, M. C., D. Fenlon, J. C. Low, A. W. Smith, H. I. Knight, J. Evans, G. Foster, B. A. Synge, and G. J. Gunn. 2004. Distribution of Escherichia coli O157 in bovine fecal pats and its impact on estimates of the prevalence of fecal shedding. Appl. Environ. Microbiol. 70:5737-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson, S. E., P. E. Brown, E. J. Wright, M. Bennett, C. A. Hart, and N. P. French. 2005. Heterogeneous distributions of Escherichia coli O157 within naturally infected bovine faecal pats. FEMS Microbiol. Lett. 244:291-296. [DOI] [PubMed] [Google Scholar]
- 14.Robinson, S. E., E. J. Wright, C. A. Hart, M. Bennett, and N. P. French. 2004. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J. Appl. Microbiol. 97:1045-1053. [DOI] [PubMed] [Google Scholar]
- 15.Robinson, S. E., E. J. Wright, N. J. Williams, C. A. Hart, and N. P. French. 2004. Development and application of a spiral plating method for the enumeration of Escherichia coli O157 in bovine faeces. J. Appl. Microbiol. 97:581-589. [DOI] [PubMed] [Google Scholar]
- 16.Stephens, T. P. 2006. Improvements, challenges, and validation of enumeration of Escherichia coli O157 and sampling methods for Escherichia coli O157 and Salmonella in feedlot cattle. Ph.D. dissertation. Texas Tech University, Lubbock, TX.
- 17.Widiasih, D. A., N. Ido, K. Omoe, S. Sugii, and K. Shinagawa. 2004. Duration and magnitude of faecal shedding of Shiga toxin-producing Escherichia coli from naturally infected cattle. Epidemiol. Infect. 132:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]