Abstract
Black band disease (BBD) is a pathogenic, sulfide-rich microbial mat dominated by filamentous cyanobacteria that infect corals worldwide. We isolated cyanobacteria from BBD into culture, confirmed their presence in the BBD community by using denaturing gradient gel electrophoresis (DGGE), and demonstrated their ecological significance in terms of physiological sulfide tolerance and photosynthesis-versus-irradiance values. Twenty-nine BBD samples were collected from nine host coral species, four of which have not previously been investigated, from reefs of the Florida Keys, the Bahamas, St. Croix, and the Philippines. From these samples, seven cyanobacteria were isolated into culture. Cloning and sequencing of the 16S rRNA gene using universal primers indicated that four isolates were related to the genus Geitlerinema and three to the genus Leptolyngbya. DGGE results, obtained using Cyanobacteria-specific 16S rRNA primers, revealed that the most common BBD cyanobacterial sequence, detected in 26 BBD field samples, was related to that of an Oscillatoria sp. The next most common sequence, 99% similar to that of the Geitlerinema BBD isolate, was present in three samples. One Leptolyngbya- and one Phormidium-related sequence were also found. Laboratory experiments using isolates of BBD Geitlerinema and Leptolyngbya revealed that they could carry out sulfide-resistant oxygenic photosynthesis, a relatively rare characteristic among cyanobacteria, and that they are adapted to the sulfide-rich, low-light BBD environment. The presence of the cyanotoxin microcystin in these cultures and in BBD suggests a role in BBD pathogenicity. Our results confirm the presence of Geitlerinema in the BBD microbial community and its ecological significance, which have been challenged, and provide evidence of a second ecologically significant BBD cyanobacterium, Leptolyngbya.
Coral disease has been implicated as one of the major factors contributing to the recent global decline of coral cover and diversity (22, 34, 44). Approximately 30 different diseases have been suggested to affect corals worldwide (21, 49), of which 18 have been characterized (49). One of the most important of these is black band disease (BBD), which was first identified in 1973 (2) and is now distributed globally, affecting at least 25 Caribbean and 45 Indo-Pacific coral species (49, 53). BBD is characterized as a dark-colored band that is between 1 mm and several cm wide and up to 1 mm thick (3, 37, 46), separating apparently healthy tissue from freshly exposed coral skeleton. The band migrates across coral colonies at rates averaging 3 mm per day and can kill entire colonies in a matter of months (3, 4, 46).
BBD comprises a complex microbial consortium dominated by phycoerythrin-rich, gliding, filamentous cyanobacteria (2, 39, 41, 46, 50) whose identity has been the subject of recent controversy (12, 14, 15, 47, 48). It also contains populations of sulfide-oxidizing bacteria proposed to belong to the genus Beggiatoa (20, 30), a group of sulfate-reducing bacteria that includes members of the genus Desulfovibrio (20, 52), more than 60 species of heterotrophic bacteria (12, 14, 16, 47), and marine fungi (36). Many of these microorganisms have been suggested as potential primary pathogens that cause BBD (12, 13, 20, 30, 45, 46, 50). However, Koch's postulates have not been fulfilled for any of the BBD consortium members (38). A more recent hypothesis is that BBD is not caused by one particular pathogen but is a polymicrobial infection (5, 38).
In 1973, Antonius identified the dominant black band disease cyanobacterium as Oscillatoria submembranacea, based on analysis by light microscopy (2). Using light and transmission electron microscopy, Rützler and Santavy (45) determined that the morphology of the black band cyanobacterium was different from that of O. submembranacea. The authors reported the distinguishing characteristics of the BBD cyanobacterium as including cylindrical trichomes with one end cell tapered and the other usually hemispherical, a cell width of 4 to 4.5 μm, a cell length of 2.5 to 5 μm, gliding motility, and the presence of a thin sheath (45). Based on their morphological description and the association of this cyanobacterium with a microbial community that lysed coral tissue, they proposed that the dominant BBD cyanobacterium be reclassified as Phormidium corallyticum (45).
In 1991, a cyanobacterium was isolated from BBD on the coral Montastraea annularis in the Florida Keys and was identified as Phormidium corallyticum, based on microscopic analysis (41). However, more recent molecular results revealed that this isolate is most closely related to the genus Geitlerinema (12, 35). The 16S rRNA gene of this cultured BBD cyanobacterium has been sequenced and appears in the GenBank database as Geitlerinema sp. BBD (accession no. DQ151461) (35). A second laboratory (12) also sequenced the 16S rRNA gene for this isolate (culture provided by our group) and incorrectly deposited the sequence in GenBank as “uncultured cyanobacterium LR-3L” (accession no. AF474001). In this paper we refer to this isolate as Geitlerinema sp. BBD.
In addition to the above-described studies targeting individual BBD cyanobacteria, the presence of multiple cyanobacterial species within the BBD community has been documented. Rützler et al. (46) studied gorgonians with BBD by using microscopy techniques (light microscopy and scanning electron microscopy) and determined that BBD samples often contained the cyanobacteria Schizothrix mexicana, Schizothrix calcicola, and Spirulina subsalsa. Ducklow and Mitchell (13) used scanning electron microscopy to examine BBD on Diploria strigosa and reported cyanobacteria of the genus Oscillatoria along with several filaments that resembled those of Spirulina sp.
Several investigators have used molecular approaches to study cyanobacteria associated with BBD. Cooney and colleagues (12) used denaturing gradient gel electrophoresis (DGGE) with universal 16S rRNA gene primers and amplified 16S rRNA gene restriction analysis of BBD clone libraries to characterize BBD samples from 11 Caribbean coral colonies of the species D. strigosa, M. annularis, and Colpophyllia natans. Their results indicated the presence of a single cyanobacterial sequence common to all of the BBD samples. The sequence of the uncultured BBD Oscillatoria (accession no. AF473936) was 92% similar to that of Oscillatoria cf. corallinae (accession no. X84812) (12). Frias-Lopez et al. (14) used cloning and sequencing to compare the bacterial communities associated with seawater, apparently healthy coral, BBD, and dead coral skeleton on reefs of Curaçao. Cyanobacterial sequences that were 93% similar to that of Trichodesmium tenue were found in the clone libraries of BBD from M. annularis and D. strigosa coral hosts (14). However, this is a common planktonic species and probably a contaminant from the water column rather than a member of the BBD community. In a second study, Frias-Lopez et al. (15) used cyanobacterium-specific primers along with terminal restriction fragment length polymorphism (T-RFLP) and cloning and sequencing to identify cyanobacteria present in 11 BBD samples from Curaçao and New Britain, Papua New Guinea. Their results revealed the presence of one dominant cyanobacterial sequence in nine of the BBD samples from Curaçao (15). The 16S rRNA gene sequence for this cyanobacterium, which was designated CD1C11 (accession no. AY038527) (15), was 98% similar to that of the uncultured BBD Oscillatoria identified by Cooney et al. (12). A different cyanobacterial sequence, for PNG-50, was detected in the two Indo-Pacific samples, and the partial sequence for this cyanobacterium was most closely related to that of the planktonic genus Trichodesmium (15). The T-RFLP results in that study suggested the presence of at least four additional cyanobacterial ribotypes in the Caribbean BBD samples and at least six additional cyanobacterial ribotypes in the Indo-Pacific BBD samples; however, the identities of these cyanobacteria were not determined (15). Sekar et al. (47) analyzed the microbial community associated with BBD from Siderastrea siderea in the Bahamas, using cloning and sequencing of the 16S rRNA gene with universal primers. Sequencing results revealed the presence of three cyanobacterial sequences. Two of these sequences were 91% similar to that of Lyngbya hieronymusii var. hieronymusii (accession no. AB045906) (47). The third cyanobacterial sequence was 93% similar to that of the uncultured BBD Oscillatoria (accession no. AF473936). Sussman et al. (48) reported only one cyanobacterial ribotype associated with black (and red) bands on reefs of Palau. This ribotype was 99% similar to that of the Caribbean uncultured BBD Oscillatoria. The authors report (48) that they isolated the Palau BBD cyanobacterium into culture, but no physiological studies have been published. All of the above studies failed to detect sequences related to members of the genus Phormidium or Geitlerinema (12, 14, 15, 47, 48).
Due to the variability among the results of these studies, the identity of the dominant black band cyanobacterium has become controversial. In particular, the 1991 Geitlerinema sp. BBD isolate has been challenged as a member of the BBD community (12, 14, 15, 16, 25) since molecular studies by these groups did not detect a Geitlerinema species-related sequence in BBD samples.
In this study we examined the diversity of cyanobacteria present in BBD samples from widely separated regions and nine different coral hosts, using complementary cultivative and noncultivative techniques. We included BBD samples from three regions of the Caribbean and one Indo-Pacific location which had not previously been investigated in studies of BBD cyanobacterial diversity and four new coral host species.
In addition to investigating BBD cyanobacterial diversity using molecular techniques, we used culture-based methods to investigate BBD cyanobacterial physiology in the context of the BBD environment. BBD microorganisms, including cyanobacteria, are subjected to an environment with a migrating oxygen/sulfide interface that frequently results in simultaneous light- and sulfide-rich conditions (5, 40). Sulfide is toxic to eukaryotes and inhibits oxygenic photosynthesis of all phototrophs, including that of most cyanobacteria (10, 17). Therefore, BBD cyanobacteria must have a relatively rare ability to tolerate sulfide to exist within the sulfide-rich BBD environment. Only one study has assessed the physiological capabilities of a BBD cyanobacterium in terms of sulfide exposure (41). Our laboratory demonstrated that the 1991 Geitlerinema sp. BBD isolate could conduct oxygenic photosynthesis in the presence of sulfide (41). We report here further results indicating that in addition to the Geitlerinema sp. BBD isolate, a second BBD cyanobacterium, Leptolyngbya BBD, can tolerate sulfide. The ecological significance of these findings is discussed.
MATERIALS AND METHODS
Sample collection.
Samples with active BBD were collected from reefs located offshore of Key Largo, Florida (n = 10), Lee Stocking Island, the Bahamas (n = 9), St. Croix, the U.S. Virgin Islands (n = 7), and Negros Island, Philippines (n = 3). Sample sites, host species, date of sampling, and sample designations referred to in this study are presented in Table 1.
TABLE 1.
Locations of BBD sample collection sites and dates, host species, and sample designations
| Location | Reef | Host coral species (no. of samples) | Sample designation | Sample date (mo/yr) |
|---|---|---|---|---|
| Florida Keys, FL | Conch Shallow | Meandrina meandrites (2) | FLK7, FLK20 | 5/05 |
| Davis | Solenastrea sp. (1) | D-Sol | 7/06 | |
| French | Diploria strigosa (1) | FR | 10/06 | |
| Molasses | Dichocoenia stokesi (1) | FLK37 | 6/04 | |
| Dendrogyra cylindrus (1) | MR | 10/06 | ||
| Watson's | Siderastrea siderea (1) | W-1 | 5/05 | |
| White Banks | Montastraea annularis (1) | FLK1 | 6/04 | |
| Diploria strigosa (1) | WB-Ds | 10/06 | ||
| Siderastrea siderea (1) | WB-Ss | 10/06 | ||
| St. Croix, U.S. Virgin Islands | Frederiksted | Diploria strigosa (3) | SC-D1, SC-D2, SC-D3 | 8/05 |
| Diploria strigosa (1) | 105 | 8/06 | ||
| Montastraea cavernosa (1) | 106 | 8/06 | ||
| Siderastrea siderea (2) | SC-1 | 6/05 | ||
| 107 | 8/06 | |||
| Lee Stocking Island, Bahamas | Horseshoe | Siderastrea siderea (5) | HS22, HS214, HS216, HS217 | 7/04 |
| HS223 | 8/04 | |||
| Rainbow | Siderastrea siderea (2) | R1, R2 | 7/05 | |
| South Perry | Siderastrea siderea (2) | S1, S2 | 7/05 | |
| Negros Island, Philippines | Agan-an | Porites lutea (3) | P1, P2a, P2b | 8/05 |
Samples were collected via SCUBA, using sterile 10-ml syringes, and kept under low-light conditions at ambient temperatures in a cooler during transport to the laboratory. Once in the laboratory, samples were subsampled for culture and molecular analysis. For molecular analysis, a portion of each sample was aseptically transferred to a 2-ml cryovial. DNA was extracted either immediately or after storage at −20°C. The remaining portion of each live BBD sample was aseptically transferred to either sterile seawater or ASN III medium (43). Cyanobacteria were isolated from BBD samples by using the gliding method described by Castenholz (7). This method involves inoculating an agar plate with a cyanobacterial sample and using a dissecting microscope to locate filaments that have glided out from the initial point of inoculation (7). Single filaments from each sample were cut out of the agar using a sterile watchmaker's forceps and introduced into ASN III medium in test tubes and monitored for growth. All BBD cyanobacterial cultures were grown and maintained in a temperature-controlled incubator at 30°C with 30 μE m−2 s−1 of cool-white fluorescent light on a 12-h-light/12-h-dark cycle (41). Newly inoculated test tubes were wrapped in paper towel to reduce light exposure in order to prevent photoinhibition and assessed visually for 2 to 3 weeks until enough biomass accumulated to allow for self-shading (clump formation). The wrapped tubes were exposed to light levels of 18.3 μE m−2 s−1.
DNA extraction and 16S rRNA gene amplification.
Genomic DNA was extracted from BBD samples and from cyanobacterial isolates using a FastDNA SPIN kit for soil (Q-Biogene, Vista, CA), with slight modification of the protocol as previously described (29). DNA extracts were verified by electrophoresis in a 1% agarose gel, followed by staining with ethidium bromide. The DNA was quantified with a Bio-Rad VersaFluor fluorometer (Hercules, CA) and subsequently diluted to 10 ng/μl solution.
Bacterial 16S rRNA genes were amplified with universal bacterial primers 27F (5′-AGA GTT TGA TCM TGG GTC AG-3′) and 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) (32). Final PCR concentrations were as follows: 1× PCR buffer, 2.5 mM MgCl2, 0.25 mM of each deoxynucleoside triphosphate, 0.5 μM of each primer, 0.5 U AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), 0.1% bovine serum albumin, 10 ng of genomic DNA, and nuclease-free water added to a final volume of 20 μl. PCRs were carried out in a PTC-200 model Peltier thermal cycler (MJ Research, Waltham, MA), with an initial denaturing step at 95°C for 5 min, followed by 25 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 10 min. PCR products were run on an agarose gel, along with a negative control (DNA-grade water), for verification prior to purification with a QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA).
Cloning and sequencing.
Purified PCR products were cloned with a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) per the manufacturer's protocol and transformed into One Shot TOP10 chemically competent Escherichia coli. Clones were screened for inserts by PCR amplification with primers M13F and M13R (28). Plasmids were extracted from positive clones with a QIAprep spin miniprep kit (QIAGEN, Valencia, CA), and inserts were sequenced with the M13F primer (28). Sequencing was performed at the DNA Core Facility at Florida International University, Miami, FL, on an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Two additional primers, M13R (28) and 518F (31), were used to sequence the full 16S rRNA gene for cyanobacterial sequences. Sequences were edited and assembled using ContigExpress (Invitrogen, Carlsbad, CA). Each sequence was checked for vector contamination using VecScreen (http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html) and for chimeras using Chimera Detection (11). Sequences were analyzed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) in order to identify their closest relatives.
Phylogenetic analysis.
Phylogenetic analysis was performed for the cyanobacterial 16S rRNA gene sequences, using ARB software (http://www.arb-home.de) (27). Sequences were manually aligned using the ARB Edit_Tool by including closest relatives. Maximum likelihood and maximum parsimony analyses were performed, and a consensus tree was produced. Bootstrap analysis (100 resamplings) was performed with PHYLIP DNA Parsimony (DNAPARS) of ARB software to estimate the confidence of the 16S rRNA gene tree topology.
Denaturing gradient gel electrophoresis.
DNA was extracted from cyanobacterial cultures and BBD field samples as described above and amplified via PCR with the cyanobacterium-specific primers GC-CYA359F and CYA781R(b) (33), which have been shown to specifically target cyanobacterial 16S rRNA gene fragments and retrieve them from complex microbial communities (1, 19, 33). The final concentrations for PCRs were as described above, except nuclease-free water was added to a final volume of 25 μl. PCR amplifications were performed in an Eppendorf Master Cycler (Eppendorf, Westbury, NY) with an initial denaturing step at 94°C for 3.5 min, followed by 40 cycles of 94°C for 30 s, 55°C for 45 s (which was decreased by 0.5°C every cycle until a touchdown at 52°C, at which 34 cycles were carried out), and 72°C for 30 s, with a final extension step at 72°C for 10 min. The touchdown amplification protocol was used to reduce nonspecific binding during the amplification process. DNA extracted from one isolate (Table 2, isolate HS223-C) was used as a positive control for all PCRs. The PCR products were analyzed by agarose gel electrophoresis prior to separation by DGGE on a vertical polyacrylamide gel with a denaturant gradient of 45 to 90%. Electrophoresis was conducted at 110 V for 16 h in 1× Tris-acetate-EDTA (TAE), after which the gel was stained for 20 min with 10 μl of 1× TAE and 2 μl of SybrGreen (Invitrogen, Carlsbad, CA). Each gel was visualized on a FOTO/Prep UV transilluminator (Fotodyne, Hartland, WI), and individual bands were excised from the gel and eluted in 0.5 ml of nuclease-free water for 48 h. Eluted DNA was reamplified with 0.5 μM of each of the primers CYA359F and CYA781R(b). PCR products were verified and subsequently purified as described above. The purified products were sequenced using the CYA359F primer. Sequences were edited and analyzed using BLAST.
TABLE 2.
Cyanobacteria isolated into culture from BBDa
| Isolate ID | Source | Sequence length (bp) | GenBank accession no. | Closest relatives in the GenBank database (accession no.) | Similarity (%) |
|---|---|---|---|---|---|
| FLK1-C | Montastraea annularis, White Banks, | 1,458 | EF110975 | Leptolyngbya sp. strain PCC7375 (AF132786) | 98 |
| Florida Keys | Leptolyngbya sp. strain PCC7375 (AB039011) | 98 | |||
| HS217-C | Siderastrea siderea, Horseshoe Reef, | 1,419 | EF110974 | Geitlerinema sp. BBD (DQ151461) | 99 |
| Bahamas | Geitlerinema sp. strain PCC7105 (AF132780) | 97 | |||
| HS223-C | Siderastrea siderea, Horseshoe Reef, | 1,439 | DQ680351 | Geitlerinema sp. BBD (DQ151461) | 99 |
| Bahamas | Geitlerinema sp. strain PCC7105 (AF132780) | 97 | |||
| P2b-1-C | Porites lutea, Negros Island, Philippines | 1,461 | EF372580 | Geitlerinema sp. BBD (DQ151461) | 99 |
| Geitlerinema sp. strain PCC7105 (AF132780) | 97 | ||||
| P2b-2-C | Porites lutea, Negros Island, Philippines | 1,454 | EF372581 | Leptolyngbya sp. strain PCC7375 (AF132786) | 98 |
| Leptolyngbya sp. strain PCC7375 (AB039011) | 98 | ||||
| SC-1-C | Siderastrea siderea, St. Croix, U.S. | 1,427 | EF372582 | Leptolyngbya sp. strain PCC7375 (AF132786) | 94 |
| Virgin Islands | Leptolyngbya sp. strain PCC7375 (AB039011) | 94 | |||
| W-1-C | Siderastrea siderea, Watson's Reef, | 1,396 | EF154084 | Geitlerinema sp. BBD (DQ151461) | 99 |
| Florida Keys | Geitlerinema sp. strain PCC7105 (AF132780) | 97 |
The host coral species, the reef location, the length of 16S rRNA gene sequence obtained, and the closest relatives are presented. Isolate identification (ID) consists of the field sample designation (see Table 1) with a “C” to designate a culture.
Photosynthetic capabilities of cyanobacterial isolates.
Photosynthetic growth rates of cyanobacterial cultures were determined experimentally by measuring the photo incorporation of [14C]NaHCO3 over a 2-h period, using a photosynthetron, which generates a photosynthesis-versus-irradiance curve in a single experiment by using replicate samples over a range of light intensities (26). Light was supplied by a reflective fixture with four cool-white fluorescent bulbs (F40/CW) and two very-high-output fluorescent bulbs (FR40T12/VHO). In each experiment, three replicate scintillation vials were incubated for each of 15 simultaneous light intensities (45 vials total), which were controlled by using neutral-density filters to provide light in 7.1% increments from darkness to 100% light (150 μE m−2 s−1). Light was measured using a Biospherical Instruments Inc. (San Diego, CA) model QSL-100 averaging quantum meter. Experiments were conducted at a constant temperature of 30°C.
To measure oxygenic photosynthesis, an equal volume of cell suspension (1 ml) from a culture that was inoculated and incubated overnight was added to each experimental vial by using a sterile self-repeating syringe. This was accomplished by first dispersing the filaments by pumping with the syringe. Each of the 45 scintillation vials contained an equal volume (5 ml) of sterile ASN III medium. After inoculation, [14C]NaHCO3 was added to a specific activity level of 0.05 μCi ml−1. After the 2-h incubation, each sample was killed by the addition of formalin to a final concentration of 1.5%. The samples were filtered onto 934-AH glass fiber filters, washed with sterile ASN III medium, and rinsed with a solution of 2.0% HCl. Each filter was placed in 7 ml of Ecolume scintillation cocktail (MP Biomedicals, Solon, OH) to measure the photo incorporation of 14C, using a Beckman LS-6500 liquid scintillation counter (Beckman Coulter, Fullerton, CA). The amount of 14C incorporated was expressed as CO2 fixation per biomass by use of dry weight, which was obtained from three vials containing only ASN III medium (without [14C]NaHCO3) that were inoculated at the same time as the experimental vials, immediately filtered onto preweighted filters, dried, weighed, and averaged. Each experiment was performed in triplicate and included a dark aerobic control.
Photosynthesis in the presence of sulfide was measured using the same experimental procedures as described above, with the following modifications. Anaerobic media were prepared by bubbling ASN III medium with a steady stream of reagent-grade N2 gas for 30 min to remove the oxygen. In order to maintain anaerobic conditions, 7-ml scintillation vials with Hungate caps and septa were used. Sulfide was added from a stock solution of Na2S·9H2O through the septum of each vial to a final concentration of 0.5 mM. Each sulfide experiment included the following two controls: anaerobic without the addition of sulfide and anaerobic with sulfide and the addition of 3-(3′,4′-dichlorophenyl)-1,1-dimethylurea (DCMU; final concentration of 10 μM), which blocks electron transport to photosystem II. These controls were performed in triplicate at the highest experimental light intensity.
Photomicroscopy.
Photomicrographs were taken using a Leitz DMR microscope (Rockleigh, NJ) equipped with a Leica DC500 digital camera interfaced with a computer equipped with digital imaging software.
Nucleotide sequence accession numbers.
Full-length nucleotide sequences have been deposited in the GenBank database under the following accession numbers: DQ680351, EF110974, EF110975, EF154084, EF372580, EF372581, and EF372582.
RESULTS
Cultured BBD cyanobacteria.
Five cyanobacteria were isolated into culture from freshly collected Caribbean BBD samples and two from BBD samples from the Philippines (Table 2). Cloning and sequencing of the 16S rRNA gene of the three Caribbean isolates (HS217-C, HS223-C, and W-1-C), followed by BLAST analysis, revealed that they were 99% similar to that of the Geitlerinema sp. BBD isolate (accession no. DQ151461) and 97% similar to that of the Geitlerinema sp. strain PCC7105 (accession no. AF132780). Isolates HS217-C and HS223-C were from BBD samples from the Bahamas, and isolate W-1-C was cultured from a BBD sample from the Florida Keys, all from the host coral species S. siderea. Isolate FLK1-C was cultured from BBD on M. annularis in the Florida Keys. The full-length sequence for this isolate was 98% similar to that of Leptolyngbya sp. strain PCC7375 (accession numbers AF132786 and AB039011). One cyanobacterial isolate, SC-1-C, was obtained from BBD on S. siderea in St. Croix. The nearly complete 16S rRNA gene sequence for this isolate showed 94% similarity that of Leptolyngbya sp. strain PCC7375 (accession numbers AF132786 and AB039011).
Two cyanobacteria (P2b-1-C and P2b-2-C) were isolated from one Philippine BBD sample (P2b). BLAST analysis revealed that the sequence for isolate P2b-1-C was 99% similar to that of the Geitlerinema sp. BBD isolate and 97% similar to that of Geitlerinema sp. strain PCC7105 (Table 2). The full-length sequence for isolate P2b-2-C was 98% similar to that of Leptolyngbya sp. strain PCC7375 (accession numbers AF132786 and AB039011).
Phylogenetic analysis of cyanobacterial sequences.
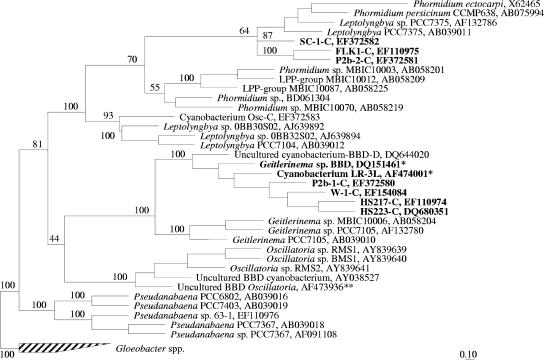
Phylogenetic analysis (Fig. 1) confirmed that the sequences for isolates W-1-C, HS217-C, HS223-C, and P2b-1-C were closely related to each other and to that of the Geitlerinema sp. BBD isolate. Bootstrap analysis supported (100%) a clade that included the sequences for these isolates, the two sequences for the Geitlerinema sp. BBD isolate (accession no. DQ151461) and the incorrectly deposited “uncultured cyanobacterium LR-3L” (12) discussed previously (AF474001), and several Geitlerinema sequences (accession numbers AB058204, AF132780, and AB039010). BLAST analysis of the sequences for isolates SC-1-C, FLK1-C, and P2b-2-C showed 94 to 98% similarity to that of Leptolyngbya sp. strain PCC7375 (accession numbers AF132786 and AB039011). Detailed phylogenetic analysis showed that the sequences for these isolates clustered with several Leptolyngbya and Phormidium sequences (Fig. 1). Phylogenetic analysis also revealed that none of the BBD cyanobacterial cultures isolated in this study was closely related to the uncultured BBD Oscillatoria (accession no. AF473936) (12) reported in Caribbean BBD by other research groups (12, 14, 15, 16).
FIG. 1.
Phylogenetic tree of 16S rRNA gene sequences of BBD cyanobacterial isolates and their most closely related neighbors. Tree topology is based on maximum-parsimony analysis. The values shown at the nodes indicate the bootstrap values (percentages) based on 100 resamplings. The scale bar corresponds to 10% estimated sequence divergence. Sequences from the current study are indicated by boldface type. Single asterisks indicate sequences for the 1991 culture of Geitlerinema sp. BBD. The double asterisk indicates the sequence for the uncultured BBD Oscillatoria. The 16S rRNA gene sequence of Gloeobacter spp. (indicated by the hatched bar) was used as an outgroup to root the tree.
DGGE analysis of BBD field samples.
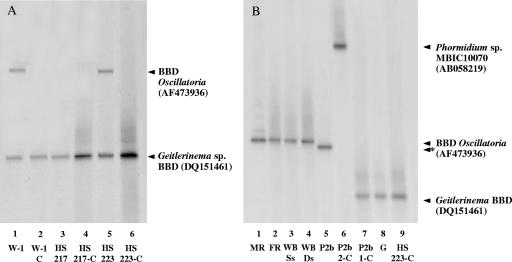
BBD field samples (community DNA) and the corresponding cyanobacteria isolated from the sampled bands were analyzed using DGGE with 16S rRNA gene primers specific for cyanobacteria (33). An example of this data set is shown in Fig. 2. The DGGE profiles of three cyanobacteria isolated from Caribbean BBD and their corresponding field samples produced bands that migrated to identical positions (Fig. 2A, lanes 1 to 6). Excision and sequencing of these bands confirmed that they were identical, and BLAST analysis revealed that they were all 99% similar to that of the Geitlerinema sp. BBD isolate (accession no. DQ151461). Two of these field samples (isolates W-1 and HS223) also produced a second band that migrated to the same position (Fig. 2A, lanes 1 and 5). BLAST analysis of the sequences from these bands revealed a 99% similarity to that of the uncultured BBD Oscillatoria (accession no. AF473936) found in other studies (12, 14, 15, 16).
FIG. 2.
DGGE analysis of cyanobacterial 16S rRNA gene fragments from BBD field samples and cyanobacterial cultures isolated from the BBD samples. Samples were as follows: (A) lane 1, BBD on Siderastrea siderea, Watson's Reef, Florida Keys (W-1); lane 2, isolate W-1-C; lane 3, BBD on S. siderea, Horseshoe Reef, Bahamas (HS217); lane 4, isolate HS217-C; lane 5, BBD on S. siderea, Horseshoe Reef, Bahamas (HS223); lane 6, isolate HS223-C; (B) lane 1, BBD on Dendrogyra cylindrus, Molasses Reef, Florida Keys (MR); lane 2, BBD on Diploria strigosa, French Reef, Forida Keys (FR); lane 3, BBD on Siderastrea siderea, WhiteBanks, Florida Keys (WB-SS); lane 4, BBD on D. strigosa, WhiteBanks, Florida Keys (WB-Ds); lane 5, BBD on Porites lutea, Agan-an Reef, Philippines (P2b); lane 6, isolate P2b-2-C; lane 7, isolate P2b-1-C; lane 8, Geitlerinema sp. BBD (G); lane 9, isolate HS223-C. Closest relatives identified by BLAST analysis are shown. The asterisk indicates a band that was most closely related to that of Caribbean BBD Oscillatoria based on sequencing but migrated to a position that was different from the band in lanes 1 to 4 (panel B) due to a difference in two bases (see text).
DGGE analysis of 23 Caribbean BBD field samples revealed the presence of a single frequently occurring band (for example, samples MR, FR, WB-Ss, and WB-Ds shown in Fig. 2B, lanes 1 to 4). The sequence for this band was 99% similar to that of the uncultured BBD Oscillatoria (accession no. AF473936) previously found in other BBD samples from the Caribbean (12, 14, 15, 16). Six of the Caribbean BBD samples had additional bands corresponding to other cyanobacterial genera, specifically Geitlerinema (n = 2), Leptolyngbya (n = 1), and Phormidium (n = 1), and two unidentified cyanobacterial ribotypes (see Table S1 in the supplemental material). A cyanobacterial ribotype that was 99% similar to that of Leptolyngbya sp. strain PCC7375 (accession no. AF132786) was detected in the DGGE profile of a sample (R1) from BBD on S. siderea in the Bahamas. The DGGE profile of a sample (S1) from BBD on an S. siderea colony at a different reef in the Bahamas produced an additional band that, when sequenced, was most closely related to that of Phormidium sp. strain MBIC10070 (accession no. AB058219), with a similarity of 93%. The DGGE profile of two BBD samples (isolates HS22 and HS216) from separate colonies of S. siderea from the Bahamas produced an additional weak band, which we were unable to identify.
The DGGE profile of sample P2b collected from BBD on the coral Porites lutea in the Philippines (Fig. 2B, lane 5) revealed the presence of a distinctive band which, when excised and sequenced, was 99% similar to that of the uncultured BBD Oscillatoria (accession no. AF473936) (12). This sequence was also 99% similar to the cyanobacterial ribotype analyzed in the DGGE profiles of our Caribbean BBD samples. However, Fig. 2B shows that the band for the cyanobacterial ribotype found in the Philippines sample (Fig. 2B, lane 5) migrated to a slightly different position than that of the Caribbean samples (Fig. 2B, lanes 1 to 4). Sequence analysis revealed that the ribotype found in the Philippines sample was two bases different from the ribotype found in the Caribbean samples. Thus, the cyanobacterium found in the Philippines BBD sample may be a different, but closely related, strain of the cyanobacterium found in the Florida Keys, St. Croix, and the Bahamas. Differences between these sequences may reflect biogeographic variation.
Two cyanobacterial isolates (P2b-1-C and P2b-2-C) were obtained from sample P2b. The DGGE profile of isolate P2b-2-C (Fig. 2B, lane 6) showed the presence of a band that was 94% similar to that of Phormidium sp. strain MBIC10070, based on the partial 16S rRNA gene sequence obtained with the cyanobacterium-specific primers used for DGGE. However, as previously mentioned, the full-length 16S rRNA gene sequence for this isolate was more closely related to the genus Leptolyngbya. DGGE analysis of isolate P2b-1-C produced a band that migrated to the same position as that of the Geitlerinema BBD and HS223-C isolates (Fig. 2B, lanes 7 to 9). Sequencing of this band showed that it was 99% similar to that of the Geitlerinema sp. BBD isolate (accession no. DQ151461). Isolates P2b-1-C and P2b-2-C were not detected in the DGGE profile of the sample from which they were isolated, which may be due to low abundance of these cyanobacteria in sample P2b.
Photosynthetic capabilities of BBD Geitlerinema and Leptolyngbya isolates.
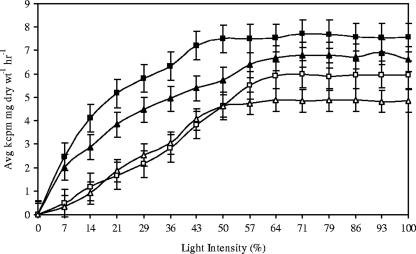
Photosynthetic incorporation of [14C]NaHCO3 by the Geitlerinema BBD (HS217-C) and Leptolyngbya BBD isolates (FLK1-C) was measured under aerobic and anaerobic conditions, the latter with sulfide added, to determine rates of (oxygenic) photosynthesis. Figure 3 presents photosynthesis-versus-irradiance curves for these experiments. Both of the cyanobacterial isolates were able to photosynthesize under anaerobic conditions with 0.5 mM sulfide but with slightly lower photosynthetic maximum (Pmax) values than without sulfide. Pmax for the Geitlerinema BBD isolate occurred at 81 μE m−2 s−1 (50% of the maximum light provided) under aerobic conditions and at 108 μE m−2 s−1 (64%) in the presence of sulfide. For the Leptolyngbya BBD isolate, the Pmax was attained at a light intensity of 108 μE m−2 s−1 (64% of the maximum light provided) under both aerobic and anaerobic/sulfidic conditions (Fig. 3). Photosynthetic activity was not detected in the presence of DCMU or in the presence of DCMU plus 0.5 mM sulfide (data not shown). The latter result indicates that these isolates cannot carry out (DMCU-forced) anoxygenic photosynthesis using sulfide as the electron donor.
FIG. 3.
Photosynthesis-versus-irradiance curves for the cyanobacterial isolates Geitlerinema (HS217-C) and Leptolyngbya (FLK-1-C) under aerobic conditions and anaerobic conditions with 0.5 mM sulfide. Curves are labeled as follows: ▪, Geitlerinema, aerobic; □, Geitlerinema, anaerobic with 0.5 mM sulfide; ▴, Leptolyngbya, aerobic; ▵, Leptolyngbya, anaerobic with 0.5 mM sulfide. Each data point represents the average of nine measurements (three experiments with three replicates for each experiment). Dark controls were subtracted. Maximum light intensity (100%) provided was 150 μE m−2 s−1. Error bars indicate standard deviations.
DISCUSSION
Molecular results for BBD cyanobacterial diversity.
Four different BBD cyanobacterial ribotypes were identified in our molecular analysis of BBD communities. These were most closely related to the genera Oscillatoria (n = 26), Geitlerinema (n = 3), Leptolyngbya (n = 1), and Phormidium (n = 1). The most abundant cyanobacterial ribotype, detected in 26 out of 29 BBD samples, was closely related (99% sequence similarity) to that of the uncultured BBD Oscillatoria (accession no. AF473936) (12) discussed previously. We detected this ribotype in Caribbean BBD samples from St. Croix, the Bahamas, and the Florida Keys. Previous studies in this region have detected this ribotype in BBD samples from Barbados, Curaçao, and St. Croix (12, 14, 15). Additionally, a sequence closely related to this ribotype was detected in our Philippines BBD samples. To date, this uncultured BBD Oscillatoria sequence has been detected in the BBD community of nine Caribbean coral species (12, 14, 15) and two Indo-Pacific species (48) and is the most commonly reported BBD-associated cyanobacterium. Thus, this species may be an integral member of the BBD consortium. Although, to our knowledge, no one has obtained the Caribbean BBD Oscillatoria in culture, as noted above, a closely (99%) related Oscillatoria isolate was reportedly cultured from reefs of Palau (48). While no physiological studies have been published for this BBD cyanobacterial isolate, it would be interesting to determine if this culture is able to tolerate sulfide by using the same strategy (sulfide-resistant oxygenic photosynthesis) as that used by the Geitlerinema BBD and Leptolyngbya BBD isolates reported here.
The second most common BBD cyanobacterium we detected, Geitlerinema, exhibited 99% sequence homology to that of the Geitlerinema sp. BBD isolated by our laboratory in 1991 from BBD in the Florida Keys (41). As discussed above, molecular analyses by other groups failed to identify the presence of a Geitlerinema species-related sequence in BBD communities (12, 14, 15, 16, 25, 48), and our earlier results (41) have been challenged in the literature (12, 14, 15, 16, 25, 48).
The results we present here show that we have successfully identified cyanobacterial sequences with 99% similarity to that of the cultured Geitlerinema sp. BBD isolate in BBD field samples, using molecular techniques. Furthermore, we have isolated four new BBD Geitlerinema strains into culture. To confirm our results, the DGGE profile of the original (1991) cultured Geitlerinema BBD isolate was compared to the DGGE profiles of 27 BBD field samples and to those of our new isolates. A band at the same position as the Geitlerinema sp. BBD isolate was detected in one Florida Keys sample (W-1) and two Bahamian samples (HS217 and HS223). Sequencing of the DGGE bands from these BBD samples confirmed that they were 99% similar to that of the Geitlerinema sp. BBD isolate. Thus, we provide evidence that this cyanobacterium is a member of the BBD community on reefs in Florida and in the Bahamas, and we propose that the presence of Geitlerinema in the BBD community should no longer be considered controversial.
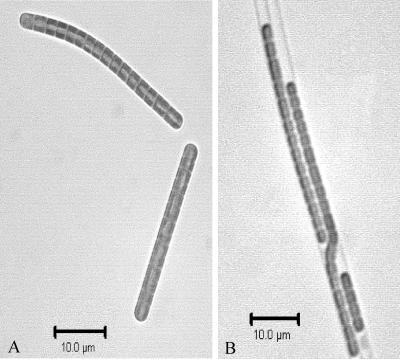
The four new Geitlerinema BBD isolates obtained from BBD samples collected during 2004 and 2005 are in culture in our laboratory. Detailed phylogenetic analysis of the 16S rRNA gene obtained from these isolates (Fig. 1) confirmed that they are closely related to each other and to that of the Geitlerinema sp. BBD isolate. DGGE analysis also showed that three of these isolates (W-1-C, HS217-C, and HS223-C) were present in the BBD samples from which they were isolated. Morphologically, all four isolates appear similar. They are all filamentous and nonheterocystous, contain phycoerythrin as the dominant light-harvesting pigment, and exhibit trichomes that are 4 to 5 μm wide, with cells 4 μm long (Fig. 4A).
FIG. 4.
Photomicrographs of cyanobacterial (A) Geitlerinema (HS217-C) and (B) Leptolyngbya (FLK1-C) isolates. Images were taken at a magnification of ×1,000.
In addition to Geitlerinema, we isolated two cyanobacteria, designated FLK1-C and SC-1-C, from Caribbean BBD samples (Florida Keys and the U.S. Virgin Islands) that are most closely related to Leptolyngbya sp. strain PCC7375. We were unable to show that these cyanobacteria were present in the BBD samples from which they were isolated, as we were unable to successfully amplify cyanobacterial DNA from the corresponding field samples. However, we did identify a cyanobacterial ribotype in a BBD sample (Table 1, sample R1) from the Bahamas that was also 99% similar to that of Leptolyngbya sp. strain PCC7375 (accession no. AF132786). Thus, our molecular and culture-based findings are in agreement and indicate that cyanobacteria related to Leptolyngbya are present in the Caribbean BBD microbial consortium.
A Philippines BBD cyanobacterial isolate, P2b-2-C, was most closely related to Leptolyngbya sp. strain PCC7375, with 98% similarity (Table 2). Detailed phylogenetic analysis showed that the sequence for isolate P2b-2-C clustered together with sequences of the Caribbean Leptolyngbya isolates FLK1-C and SC-1-C and was grouped in the Leptolyngbya and Phormidium clade (Fig. 1). The morphologies of all three isolates are similar. Each is filamentous, nonheterocystous, and phycoerythrin rich and has trichomes that are 2 μm wide and cells that are 2 μm long (Fig. 4B). None of the seven new BBD cyanobacterial isolates (nor the 1991 isolate) has trichomes with a tapered cell at one end, reported as characteristic of Phormidium corallyticum (45). However, it is very common for cyanobacteria to change morphology in culture.
Ecological significance of BBD cyanobacteria.
Results of the photosynthesis experiments indicate that both Geitlerinema and Leptolyngbya isolated from BBD can tolerate sulfide by continuing to conduct oxygenic photosynthesis in the presence of sulfide, but they cannot perform anoxygenic photosynthesis with sulfide as the electron donor. This physiological capability of sulfide tolerance is important for BBD cyanobacteria, because they live in an environment that contains a migrating oxygen and sulfide interface which routinely exposes them to sulfide (5, 40). The laboratory experiments we present here were conducted using a sulfide concentration (0.5 mM) that we have measured directly in the band (5, 40) and which is therefore ecologically relevant. In general, the ability to conduct sulfide-resistant oxygenic photosynthesis is rare among cyanobacteria (10, 17). Cohen et al. (10) described four metabolic (photosynthetic) strategies that cyanobacteria exhibit in the presence of sulfide and light. One strategy is sulfide-sensitive oxygenic photosynthesis, which involves the inhibition of photosynthesis in the presence of even low amounts of sulfide. When sulfide is removed, oxygenic photosynthesis resumes. Most cyanobacteria fall into this category and are considered to be obligate oxygenic phototrophs. The second strategy is sulfide-resistant oxygenic photosynthesis, which allows cyanobacteria to continue conducting oxygenic photosynthesis in the presence of sulfide. Cyanobacteria that utilize the second strategy are not able to use sulfide as an electron donor for anoxygenic photosynthesis (10). The third strategy consists of the cooccurrence of sulfide-insensitive oxygenic photosynthesis and sulfide-dependent anoxygenic photosynthesis. This strategy allows cyanobacteria to simultaneously carry out both oxygenic and anoxygenic photosynthesis with electrons donated to both photosystems (10). The final strategy involves shutting down electron flow in oxygenic photosynthesis and performing only sulfide-dependent anoxygenic photosynthesis, which is photosystem I dependent. Cyanobacteria that use this method can tolerate high concentrations of sulfide for long periods of time while retaining the ability to conduct oxygenic photosynthesis (10). Our eight BBD cyanobacterial isolates (one isolated in 1991, seven newly reported here) all carry out the second strategy of sulfide-resistant oxygenic photosynthesis, which is relatively rare among cyanobacteria (10). This strategy would allow these cyanobacteria to continue photosynthesizing when they are present below the oxygen/sulfide interface in BBD. Such an occurrence is common within the 1-mm-thick band and has been documented using sulfide-sensitive microelectrodes (5, 40).
The ability of BBD cyanobacteria to tolerate sulfide may have an additional ecological importance. The presence of sulfide may prevent other marine cyanobacteria from colonizing the nutrient-rich BBD environment of lysing coral tissue and thus confer a competitive advantage. Additional experiments have shown that cyanobacteria isolated from mats on the coral reef, but from sources other than BBD, do not exhibit sulfide tolerance (J. L. Myers, unpublished data).
Results of the photosynthesis-versus-irradiance experiments indicated that both of the BBD cyanobacterial isolates (HS217-C and FLK-1-C) investigated in this study attained Pmax values at relatively low light levels of 81 and 108 μE m−2 s−1, respectively. This is ecologically relevant in that it is well known that many gliding, filamentous cyanobacteria do not tolerate high light levels and have the ability to self-shade and/or migrate to lower light levels that support Pmax values (6, 9, 18). In terms of BBD, we have previously documented the in situ vertical migration patterns of coexisting populations of BBD cyanobacteria and BBD Beggiatoa organisms (37, 51). These studies revealed a highly unusual pattern in which Beggiatoa was present on the band surface during the periods of the highest light level (238 to 308 μE m−2 s−1), the opposite of that usually seen with other sulfide-rich microbial mats (23, 24). Furthermore, we determined that the migration pattern was due to the cyanobacteria moving down into the mat when light levels increased to 238 μE m−2 s−1 and above (51). These field results agree with the light levels at which Pmax is attained, which we determined in the photosynthesis-versus-irradiance experiments. Thus, our laboratory and field results are in agreement with the results of others who have examined low-light preferences and photosynthetic abilities of mat-forming cyanobacteria (8, 9, 18, 24).
The light responses of BBD cyanobacteria may also be important in terms of the physicochemical structure of the band. Many filamentous cyanobacteria, including our BBD isolates, prefer low light and form clumps based on self-shading, negative phototaxis, and step-up photophobic responses that allow them to generate a low-light environment (6). In terms of the ecology of BBD, such clumping behavior by the BBD cyanobacteria on the surface of the coral host would result in the formation of a dense community in which diffusion is limited (23, 24). This would allow development of zones of anoxia in the band, which would select for populations of BBD sulfate-reducing bacteria, another group of BBD microorganisms that are important in BBD ecology (39).
There is yet another aspect of the ecological significance of BBD Geitlerinema and Leptolyngbya isolates, specifically in terms of BBD pathogenicity. We have recently detected the cyanobacterial toxin microcystin (42) in our laboratory cultures of both the 1991 Geitlerinema BBD isolate and a new Leptolyngbya isolate (FLK1-C) as well as in field samples of BBD. Experiments using the BBD cyanobacterial cultures indicated that they were positive for protein phosphatase inhibition (42), a measure of microcystin toxic activity. Thus, Geitlerinema BBD and Leptolyngbya BBD may play a key role in the pathogenicity of BBD by producing cyanotoxins that may target corals and/or their associated zooxanthellae.
In summary, the results of this study confirm the presence of Geitlerinema in the BBD community by using molecular techniques. With molecular methods, we also demonstrate for the first time the presence of BBD cyanobacterial sequences that are most closely related to those of Phormidium and Leptolyngbya. We present physiological data showing the ecological significance of the BBD cyanobacteria Geitlerinema and Leptolyngbya in terms of sulfide tolerance and light preference that advance our understanding of their ability to survive in the extreme environment of the BBD mat and to contribute to the band formation. Together with our recently reported results revealing the presence of the cyanotoxin microcystin in both the BBD field samples and the cultures of BBD Geitlerinema and Leptolyngbya isolates, our findings further our understanding of the ecological physiology of BBD cyanobacteria as well as the ecology and etiology of black band disease.
Supplementary Material
Acknowledgments
We thank L. Kaczmarsky, J. Pinzón, E. Remily, and J. Voss for sample collection and D. Mills for helpful discussions. We also thank T. LaJeunesse for assistance with DGGE and the Florida Keys National Marine Sanctuary for boat support.
Sample collection in the Florida Keys National Marine Sanctuary was conducted under permit numbers FKNMS-2003-011 and FKNMS-2005-010.
This research was supported by the NIH (NIH/NIGMS SO6GM8205), the NOAA's National Undersea Research Center (FKRP-2004-11A), and the NOAA's Caribbean Marine Research Center (CMRC-04-PRJV-01-04C).
This report is contribution 127 of the Tropical Biology Program at Florida International University.
Footnotes
Published ahead of print on 29 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abed, R. M., F. Garcia-Pichel, and M. Hernandez-Marine. 2002. Polyphasic characterization of benthic, moderately halophilic, moderately thermophilic cyanobacteria with very thin trichomes and the proposal of Halomicronema excentricum gen. nov., sp. nov. Arch. Microb. 177:361-370. [DOI] [PubMed] [Google Scholar]
- 2.Antonius, A. 1973. New observations on coral destruction in reefs, p. 3. Abstr. 10th Meet. Assoc. Isl. Mar. Lab. Caribb. University of Puerto Rico, Mayaguez, Puerto Rico.
- 3.Antonius, A. 1981. Coral reef pathology: a review, p. 3-6. In E. D. Gomez, C. E. Birkeland, R. W. Buddemeier, R. E. Johannes, J. A. Marsh, Jr., and R. T. Tsuda (ed.), The reef and man. Proceedings of the Fourth International Coral Reef Symposium, vol. 2. Marine Sciences Center, University of Philippines, Quezon City, Philippines. [Google Scholar]
- 4.Antonius, A. 1981. The “band” diseases in coral reefs, p. 7-14. In E. D. Gomez, C. E. Birkeland, R. W. Buddemeier, R. E. Johannes, J. A. Marsh, Jr., and R. T. Tsuda (ed.), The reef and man. Proceedings of the Fourth International Coral Reef Symposium, vol. 2. Marine Sciences Center, University of Philippines, Quezon City, Philippines. [Google Scholar]
- 5.Carlton, R. G., and L. L. Richardson. 1995. Oxygen and sulfide dynamics in a horizontally migrating cyanobacterial mat: black band disease of corals. FEMS Microbiol. Ecol. 18:155-162. [Google Scholar]
- 6.Castenholz, R. W. 1982. Motility and taxes. p. 414-439. In N. G. Carr and B. A. Whitton (ed.), Biology of cyanobacteria. Blackwell Scientific Publications, Ltd., Oxford, United Kingdom.
- 7.Castenholz, R. W. 1988. Culturing methods for cyanobacteria. Methods Enzymol. 167:68-93. [Google Scholar]
- 8.Castenholz, R. W., and H. Utkilen. 1984. Physiology of sulfide tolerance in a thermophilic Oscillatoria. Arch. Microbiol. 138:299-305. [Google Scholar]
- 9.Castenholz, R. W., B. B. Jørgensen, E. D’ Amelio, and J. Bauld. 1991. Photosynthetic and behavioral versatility of the cyanobacterium Oscillatoria boryana in a sulfide-rich microbial mat. FEMS Microb. Ecol. 86:43-58. [Google Scholar]
- 10.Cohen, Y., B. B. Jørgensen, N. P. Revsbech, and R. Poplawski. 1986. Adaptation to hydrogen sulfide of oxygenic and anoxygenic photosynthesis among cyanobacteria. Appl. Environ. Microbiol. 51:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The ribosomal database project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooney, R. P., O. Pantos, M. D. A. Le Tissier, M. R. Barer, A. G. O'Donnell, and J. C. Bythell. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401-413. [DOI] [PubMed] [Google Scholar]
- 13.Ducklow, H. W., and R. Mitchell. 1979. Observations on naturally and artificially diseased tropical corals: a scanning electron microscopy study. Microb. Ecol. 5:215-223. [DOI] [PubMed] [Google Scholar]
- 14.Frias-Lopez, J., A. L. Zerkle, G. T. Bonheyo, and B. W. Fouke. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68:2214-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frias-Lopez, J., G. T. Bonheyo, Q. S. Jin, and B. W. Fouke. 2003. Cyanobacteria associated with coral black band disease in Caribbean and Indo-Pacific Reefs. Appl. Environ. Microbiol. 69:2409-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frias-Lopez, J., J. S. Klaus, G. T. Bonheyo, and B. W. Fouke. 2004. Bacterial community associated with black band disease in corals. Appl. Environ. Microbiol. 70:5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Pichel, F., and R. W. Castenholz. 1990. Comparative anoxygenic photosynthetic capacity in 7 strains of a thermophilic cyanobacterium. Arch. Microbiol. 153:344-351. [Google Scholar]
- 18.Garcia-Pichel, F., M. Mechling, and R. W. Castenholz. 1994. Diel migrations of microorganisms within a benthic, hypersaline mat community. Appl. Environ. Microbiol. 60:1500-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Pichel, F., A. Lopez-Cortes, and U. Nübel. 2001. Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado plateau. Appl. Environ. Microbiol. 67:1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrett, P., and H. Ducklow. 1975. Coral diseases in Bermuda. Nature 253:349-350. [Google Scholar]
- 21.Green, E. P., and A. W. Bruckner. 2000. The significance of coral disease epizootiology for coral reef conservation. Biol. Conserv. 96:347-361. [Google Scholar]
- 22.Hughes, T. P., A. H. Baird, D. R. Bellwood, M. Card, S. R. Connolly, C. Folke, R. Grosberg, O. Hoegh-Guldberg, J. B. C. Jackson, J. Kleypas, J. M. Lough, P. Marshall, M. Nyström, S. R. Palumbi, J. M. Pandolfi, B. Rosen, and J. Roughgarden. 2003. Climate change, human impacts, and the resilience of coral reefs. Science 301:929-933. [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen, B. B. 1982. Ecology of the bacteria of the sulphur cycle with special reference to anoxic-oxic interface environments. Philos. Trans. R. Soc. Lond. B 298:543-561. [DOI] [PubMed] [Google Scholar]
- 24.Jørgensen, B. B., N. P. Revsbech, T. H. Blackburn, and Y. Cohen. 1979. Diurnal cycle of oxygen and sulfide microgradients and microbial photosynthesis in a cyanobacterial mat sediment. Appl. Environ. Microbiol. 38:46-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klaus, J. S., I. Janse, J. M. Heikoop, R. A. Sanford, and B. W. Fouke. 19 February 2007, posting date. Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ. Microbiol. doi: 10.1111/j. 1462-2920.2007.01249.x. [DOI] [PubMed]
- 26.Lewis, M. R., and J. C. Smith. 1983. A small volume, short-incubation time method for measurement of photosynthesis as a function of incident irradiance. Mar. Ecol. Prog. Ser. 13:99-102. [Google Scholar]
- 27.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101: 20-78. [DOI] [PubMed] [Google Scholar]
- 29.Mills, D. K., K. Fitzgerald, C. D. Litchfield, and P. M. Gillevet. 2003. A comparison of DNA profiling techniques for monitoring nutrient impact on microbial community composition during bioremediation of petroleum-contaminated soils. J. Microbiol. Methods 54:57-74. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell, R., and I. Chet. 1975. Bacterial attack of coral in polluted seawater. Microb. Ecol. 2:227-233. [DOI] [PubMed] [Google Scholar]
- 31.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationship of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 33.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandolfi, J. M., R. H. Bradbury, E. Sala, T. P. Hughes, K. A. Bjorndal, R. G. Cooke, D. McArdle, L. McClenachan, M. J. H. Newman, G. Paredes, R. R. Warner, and J. B. C. Jackson. 2003. Global trajectories of the long-term decline of coral reef ecosystems. Science 301:955-958. [DOI] [PubMed] [Google Scholar]
- 35.Ragoonath, D. N. 2005. Heterotrophic capabilities and the molecular identification of a cyanobacterium found in black band disease of coral reefs. M.S. thesis. Florida International University, Miami.
- 36.Ramos-Flores, T. 1983. Lower marine fungus associated with black line disease in star corals (Montastrea annularis). Biol. Bull. 165:429-435. [DOI] [PubMed] [Google Scholar]
- 37.Richardson, L. L. 1996. Horizontal and vertical migration patterns of Phormidium corallyticum and Beggiatoa spp. associated with black-band disease of corals. Microb. Ecol. 32:323-335. [DOI] [PubMed] [Google Scholar]
- 38.Richardson, L. L. 2004. Black band disease, p. 325-349. In E. Rosenberg and Y. Loya (ed.), Coral health and disease. Springer-Verlag, Berlin, Germany.
- 39.Richardson, L. L., K. G. Kuta, S. Schnell, and R. G. Carlton. 1997. Ecology of the black band disease microbial consortium, p. 597-600. In H. A. Lessios and I. G. Macintyre (ed.), Proceedings of the 8th International Coral Reef Symposium, Smithsonian Tropical Research Institute, Balboa, Panama.
- 40.Richardson, L. L., G. W. Smith, K. B. Ritchie, and R. G. Carlton. 2001. Integrating microbiological, microsensor, molecular, and physiologic techniques in the study of coral disease pathogenesis. Hydrobiologia 460:71-89. [Google Scholar]
- 41.Richardson, L. L., and K. G. Kuta. 2003. Ecological physiology of the black band disease cyanobacterium Phormidium corallyticum. FEMS Microbiol. Ecol. 43:287-298. [DOI] [PubMed] [Google Scholar]
- 42.Richardson, L. L., R. Sekar, J. L. Myers, M. Gantar, J. D. Voss, L. Kaczmarsky, E. R. Remily, G. L. Boyer, and P. V. Zimba. 2007. The presence of the cyanobacterial toxin microcystin in black band disease of corals. FEMS Microbiol. Lett. 272:182-187. [DOI] [PubMed] [Google Scholar]
- 43.Rippka, R., J. Dervelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 44.Rosenberg, E., and Y. Loya. 2004. Coral health and disease. Springer-Verlag, Berlin, Germany.
- 45.Rützler, K., and D. Santavy. 1983. The black band disease of Atlantic reef corals. I. Description of a cyanophyte pathogen. Mar. Ecol. 4:301-319. [Google Scholar]
- 46.Rützler, K., D. L. Santavy, and A. Antonius. 1983. The black band disease of Atlantic reef corals. III. Distribution, ecology and development. Mar. Ecol. 4:329-358. [Google Scholar]
- 47.Sekar, R., D. K. Mills, E. R. Remily, J. D. Voss, and L. L. Richardson. 2006. Microbial communities in the surface mucopolysaccharide layer and the black band microbial mat of black band-diseased Siderastrea siderea. Appl. Environ. Microbiol. 72:5963-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sussman, M., D. G. Bourne, and B. L. Willis. 2006. A single cyanobacterial ribotype is associated with both red and black bands on diseased corals from Palau. Dis. Aquat. Organ. 69:111-118. [DOI] [PubMed] [Google Scholar]
- 49.Sutherland, K. P., J. W. Porter, and C. Torres. 2004. Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar. Ecol. Prog. Ser. 266:273-302. [Google Scholar]
- 50.Taylor, D. L. 1983. The black band disease of Atlantic reef corals. II. Isolation, cultivation, and growth of Phormidium corallyticum. Mar. Ecol. 4:321-328. [Google Scholar]
- 51.Viehman, T. S., and L. L. Richardson. 2002. Motility patterns of Beggiatoa and Phormidium corallyticum in black band disease, p. 1251-1255. In M. K. Moosa, S. Soemodihardjo, A. Soegiarto, K. Romimohtarto, A. Nontji, Soekarno, and Suharsono (ed.), Proceedings of the Ninth International Coral Reef Symposium, vol. 2. Bali, Indonesia. [Google Scholar]
- 52.Viehman, S., D. K. Mills, G. W. Meichel, and L. L. Richardson. 2006. Culture and identification of Desulfovibrio spp. from black band disease of corals on reefs of Florida Keys and Dominica. Dis. Aquat. Organ. 69:119-127. [DOI] [PubMed] [Google Scholar]
- 53.Weil, E. 2004. Coral reef diseases in the wider Caribbean, p. 35-67. In E. Rosenberg and Y. Loya (ed.), Coral health and disease. Springer-Verlag, Berlin, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.