Abstract
Pseudomonas pseudoalcaligenes CECT5344 grows in minimal medium containing cyanide as the sole nitrogen source. Under these conditions, an O2-dependent respiration highly resistant to cyanide was detected in cell extracts. The structural genes for the cyanide-resistant terminal oxidase, cioA and cioB, are clustered and encode the integral membrane proteins that correspond to subunits I and II of classical cytochrome bd, although the presence of heme d in the membrane could not be detected by difference spectra. The cio operon from P. pseudoalcaligenes presents a singular organization, starting upstream of cioAB by the coding sequence of a putative ferredoxin-dependent sulfite or nitrite reductase and spanning downstream two additional open reading frames that encode uncharacterized gene products. PCR amplifications of RNA (reverse transcription-PCR) indicated the cyanide-dependent up-regulation and cotranscription along the operon. The targeted disruption of cioA eliminates both the expression of the cyanide-stimulated respiratory activity and the growth with cyanide as the nitrogen source, which suggests a critical role of this cytochrome bd-related oxidase in the metabolism of cyanide by P. pseudoalcaligenes CECT5344.
Cyanide has been present in the Earth since the origin of life, where it probably played an essential role as an N-group donor for nonbiotic synthesis of elemental biomolecules (22). Since then, bacteria and plants have developed biochemical processes for synthesis of cyanide to poison competitors, phagocytes, pathogens, or herbivores (9). Today, the vast majority of cyanide present in the biosphere has an anthropogenic origin, the consequence of combustion processes and mainly due to its use in chemical synthesis. One million tonnes of cyanide (about 80% of the annual production) is used in the chemical industry for the production of organic chemicals such as nylon and acrylics, in the electroplating industry, in the metal processing industry, and in some photographic applications (31). Therefore, the risk of contamination by volatile or soluble cyanide is a major ecological concern that is dramatically confirmed by the frequency and health impact of gas emissions and effluent escape events (20). Cyanide is one of the most rapidly acting lethal poisons known to humankind; its main molecular target is the cytochrome c oxidase, blocking irreversibly the aerobic respiration of mitochondria. However, at least two different biochemical systems can bypass cyanide inhibition, driving cell respiration. Widely distributed among plants and fungi, and now also identified in animals, protists, and bacteria, the cyanide-resistant alternative oxidase is an ubiquinol-O2 oxide-reductase that is not coupled to proton translocation and therefore to ATP synthesis (19, 29). It belongs to the diiron carboxylate protein family that also includes ribonucleotide reductase, among other enzymes (2). Although still controversial, the role of alternative oxidase, and that of the closely related plastoquinol terminal oxidase, seems to be the regulation of the redox balance for adaptation to changing environmental conditions (2). In contrast, cytochrome bd is an unrelated terminal oxidase also resistant to cyanide and widely distributed among prokaryotes (14). The cytochrome bd is a heterodimeric complex, with large (I) and small (II) subunits that hold together the three heme components of the oxidase, which are b558, b595, and d. Different approaches including topological studies, mutagenesis, and spectroscopic and electrochemical analyses are giving clues about the molecular architecture of this complex. By analogy to the unrelated heme-copper oxidases, the O2 reduction site in the cytochrome bd would be the binuclear center heme d-heme b595 (14). A model has been proposed for the location and orientation of the transmembrane and globular domains of the subunit I, in which the heme ligands and ubiquinol binding determinants are placed (32). The cyanide resistance of the cytochrome bd oxidase activity is variable among species, although their Ki values for cyanide (mM range) usually remain 1 order of magnitude higher than those for the cytochrome oxidase (μM range) (14). Beside cyanide sensitivity, the main difference among both oxidases is the higher affinity for O2 shown by the cytochrome bd. This characteristic might provide a biological role to cytochrome bd, enabling aerobic respiration under microaerophilic conditions (14). In addition, an active cytochrome bd increases the tolerance to low O2 concentrations in strict anaerobes of the Bacteroides group (1) and could alleviate the O2 sensitivity of certain anaerobic processes such as N2 fixation (15).
Two genes named cioA and cioB (cyanide-insensitive oxidase in Pseudomonas species) or cydA and cydB (cytochrome bd in enterobacteria) are cotranscribed and encode the subunits I and II of the cytochrome bd complexes found in bacteria (14). An ABC-type glutathione transporter can be also found in the vicinity of or within the cio operon of some bacterial strains. It has been proposed that in Escherichia coli the glutathione must be translocated to the periplasm, where it is required for the assembly and maturation of the cytochrome bd and other periplasmic cytochromes (23). Other genes of unknown function have been detected within the cio operon of some organisms (21), and even an unknown protein has been found in association with the cytochrome bd complex extracted from membranes of E. coli (27).
The bacterium Pseudomonas pseudoalcaligenes CECT5344, which was isolated from a cyanide-contaminated sludge, is highly resistant to cyanide, utilizing it as the sole nitrogen source in minimal medium at alkaline pH (18). In this work, we describe the genetic structure of the operon that encodes a cytochrome bd-related oxidase in P. pseudoalcaligenes CECT5344. Targeted mutagenesis and disruption of cioA block the expression of cyanide-resistant terminal oxidase activity and, accordingly, eliminate both the cyanide tolerance and its potential use as a nitrogen source. Moreover, the finding of three additional genes coexpressed within the cio operon, including a putative sulfite or nitrite reductase gene, might provide new targets for genetic engineering focused on the characterization of cytochrome bd function and the improvement of the cyanide-detoxifying process.
MATERIALS AND METHODS
Bacterial isolation and culture conditions.
Pseudomonas pseudoalcaligenes CECT5344 cells were grown either in standard minimal medium adjusted to pH 8.5 (18) or in LB medium (25) adjusted to pH 8.5, and incubations were performed on a shaker at 190 rpm and 30°C. For minimal medium, sodium acetate at a concentration of 50 mM was used as the C source, and the appropriate nitrogen sources were included at the indicated concentrations. All studies in this work were performed with a single spontaneous mutant selected on LB medium containing rifampin (40 μg/ml), and the growth of the cells for inoculation of experimental cultures was always performed in rifampin-selective media.
Electroporation of P. pseudoalcaligenes CECT5344 cells was performed by using a protocol slightly different from that described elsewhere (25). Electrocompetent cells were prepared by growth of cultures up to an optical density at 600 nm (OD600) of 0.35 and centrifugation for 20 min at 1,000 × g, followed by three successive washes (4°C) in 1:1, 1:2, 1:50, and 1:500 volumes of 10% glycerol. The last solution, which yields the stock of electrocompetent cells, contained yeast extract (0.125%) and tryptone (0.25%). A mixture of 50 μl of cells (2 × 1010 to 3 × 1010 CFU/ml) and 1 to 10 ng of DNA was electroporated in 2-mm cuvettes with a Bio-Rad Gene Pulser II apparatus, operated at 2.5 kV, 25 μF, and 200 Ω (4- to 5-ms time constants).
Nucleic acid purification and manipulation.
Genomic DNA was purified from P. pseudoalcaligenes CECT5344 cells grown in liquid LB medium at pH 8.5 by using the Gnome DNA isolation kit (Qbiogene). Plasmidic DNA was purified from E. coli cells grown on liquid cultures of LB media supplemented with the selective antibiotic and using the High Pure plasmid isolation kit (Roche). The E. coli XL1 Blue MRF′ strain (Stratagene) was used for cloning of recombinant DNA. Hybridization analysis was performed with the digoxigenin-High Prime labeling and detection kit and positively charged nylon membranes (Roche). Restriction and modification enzymes (Invitrogen) were utilized under standard conditions (25) by following the manufacturer's instructions. RNA extraction and purification were carried out with the Aurum total RNA mini kit (Bio-Rad). DNA synthesis and sequencing reactions for plasmids and PCR products were performed using services provided by Stab Vida (Oeiras, Portugal).
PCR. (i) Degenerate primers.
Two degenerate oligonucleotides were devised from the multiple alignments generated with CIOA sequences from Pseudomonas aeruginosa PA01 (O07440) (5), P. putida KT2440 (NP_746760), P. fluorescens pf5 (AAY94574 and AAY92997), P. syringae DC3000 (NP_794399), and Escherichia coli K-12 (P11026 and P26459) (6, 11). The upper primer, ciA [TACCAGTTCGGNACNAA(T/C)TGG], and the lower primer, ciB [CCGTA(G/C)AC(G/C)ACCCANGG(T/C)TG], correspond to identity blocks “YQFGTNW” and “QPWVVYG,” respectively. Final degeneracy was reduced to 32 sequences by using the codon usage table for P. pseudoalcaligenes provided by the Kazusa DNA Research Institute (http://www.kazusa.or.jp/codon/). PCR samples were prepared with the following components: 0.5 ng/μl genomic DNA, 0.5 μM each primer, 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), 0.05 U/μl EcoTaq DNA polymerase, and 1× buffer as recommended by the manufacturer (Ecogen). The PCR conditions were 3 min at 94°C, followed by 35 cycles of 1 min at 94°C, 30 s at 54°C, and 1.5 min at 72°C, with a final extension of 10 min at 72°C.
(ii) iPCR.
Genomic DNA (4 μg) was digested with 10 U of the selected restriction enzyme in a volume of 20 μl. After 12 h of digestion, the enzyme and digestion buffer were eliminated by phenol extraction and ethanol precipitation. DNA fragments were self-ligated for 3 h at 20°C in a sample containing 0.2 μg/μl of digested DNA and 0.05 U/μl of T4-DNA ligase. Digested and self-ligated genomic DNA was used at 2 ng/μl for inverse PCR (iPCR) in a mixture that contained each primer at 0.5 μM, 0.2 mM (each) dNTPs, and 0.01 U/μl of EcoTaq-Plus polymerase (Ecogen). The primers used in this work are numbered according to their 5′ positioning in the full fragment of the 7,046-bp sequence (see Fig. 4). Their polarity is indicated as U (upper) or L (lower), and their length is indicated at the end. The PCR conditions were 20 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 6 min, followed by 15 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 6 min, with an increase in 20 s per cycle in the extension time. The first round of iPCR was performed on XhoI digestion and self-ligation and with amplification of a 3.0-kb DNA fragment driven by primers 4524U21 and 3793L21. The second round of iPCR was performed on SalI or KpnI digestions and self-ligations, for left-end or right-end chromosome walks, respectively. The primers used for chromosome walking in the second round of iPCR are 2731U21 and 2250L21, which amplified a fragment of 2.5 kb on SalI-iPCR (left-end walk), and 5994U21 and 3953L21, which amplified a fragment of 4 kb on KpnI-iPCR (right-end walk).
FIG. 4.
Map of the cio genomic region from P. pseudoalcaligenes CECT534. Genes identified in the region are represented by arrows, and the line below corresponds to the restriction map. Sl, SalI; N, NcoI; E, EcoRI; Sc, SacI; X, XhoI; P, PstI; and K, KpnI. The lower scheme shows the positions of the primers used in this work. Boxed agarose gel images contain the RT-PCR results obtained with RNA purified from ammonium-grown cells (A) or cyanide-grown cells (CN−).
(iii) RT-PCR experiments.
RNA (1 μg) preparations were treated with 1 U of RNase-free DNase (Promega) in a 10-μl final volume containing 40 mM Tris-HCl (pH 8.0), 10 mM MgSO4, and 1 mM CaCl2. The reaction was carried out for 30 min at 37°C before denaturing the enzyme at 70°C for 15 min. Reverse transcription (RT) was performed using the Moloney murine leukemia virus (MMLV) reverse transcriptase enzyme (Promega) under the following conditions: 0.5 μg of each DNase-treated RNA, 70 pmol of each downstream primer (lower primer in RT-PCR), and diethyl pyrocarbonate (DEPC)-treated water up to a final volume of 15 μl. RNAs were then denaturized for 5 min at 70°C and kept on ice. A master mix was then prepared containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 1.25 μl of a dNTP mix (10 mM each), 30 U of RNase OUT RNase inhibitor (Promega), 1 U of the MMLV reverse transcriptase and diethyl pyrocarbonate-H2O up to a final volume of 10 μl. After addition of the master mix to the RNA, the mixture was ramp heated to 42°C for 5 min, and the reaction mixture was maintained at this temperature for 1 h, after which the enzyme was denatured at 90°C for 5 min before placing the resulting cDNA on ice. For spanning nir/sir and cioA coding sequences, primers 2731U21 and 3793L21 were used in RT-PCR under the following conditions: 96°C for 2 min, followed by 40 cycles of 94°C 30 for s, 60°C for 30 s, and 72°C for 1 min. Transcripts spanning cioA and cioB were detected using primers 4524U21 and 5730L30 and RT-PCR as follows: 96°C for 2 min followed by 30 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 1.5 min. To span the cioA-serC coding sequences, primers 4524U21 and 6263L25 were used in RT-PCR under the following conditions: 96°C for 2 min, followed by 30 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C 2 min. In all cases, 2 μl of the cDNA product was used as the template in 20 μl of final volume, and a parallel reaction mixture lacking the MMLV reaction mixture, as a negative control, was included. All reactions were carried out using 0.1 U/μl of EcoTaq-Plus polymerase (Ecogen) with the recommended buffer (Optibuffer), 0.2 mM dNTP, 1 pmol/μl of each primer, and 2 mM of Cl2Mg. The correct sizes of RT-PCR fragments were verified by amplifying them from genomic DNA.
Bioinformatic tools.
Sequence analyses were performed using the programs available on line at Centre de Ressources INFOBIOGEN, now implemented under the BIOSUPPORT project (http://bioinfo.hku.hk/services/menuserv.html), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), Expert Protein Analysis System (www.expasy.ch), and Comprehensive Microbial Resource of The Institute for Genomic Research (www.cmr.tigr.org). Oligonucleotides were designed by using Oligo Primer Analysis software v6.57.
Biochemical analysis.
Cell extracts were prepared by treatment of bacterial suspensions in 100 mM Tris-HCl, pH 8.5, with a French press at 20,000 lb/in2 and centrifugation at 8,000 × g for 20 min. When indicated, the membrane-containing fraction was pelleted by ultracentrifugation at 68,000 × g for 1 h. The pelleted material was recovered in 100 mM Tris-HCl, pH 8.5, and used for further analysis.
Measurements of NADH oxidation activities were performed in a Helios-β spectrophotometer by recording the variation in A340 at 30°C in a mixture containing 0.2 mM NADH-50 mM Tris-HCl, pH 8.0, and aliquots of cell extracts. The extinction coefficient (mM−1 cm−1) used was 6.22. When indicated, cyanide was used at a final concentration of 2 mM. In every case, the 2:1 expected stoichiometry of NADH to O2 consumption was confirmed by respirometric analysis of O2 reduction carried out in a Clark-type O2 electrode.
Room temperature difference spectra were recorded in a double-beam Cary-300Bio Varian spectrophotometer by subtracting the reduced minus oxidized absorbance of membrane samples. A few grains (<5 mg) of sodium dithionite or ammonium persulfate were added to the sample and reference cuvette, respectively, before measurements.
The protein contents of cell extracts were quantified by using the Bio-Rad reactive protein assay, normalized with bovine serum albumin (Sigma). The protein concentrations in the membrane preparations were determined by using the Lowry protocol (17) with bovine serum albumin as a standard.
Nucleotide sequence accession number.
The sequence described in this work has been deposited in the EMBL database under the provisional accession no. AM115695.
RESULTS
Cyanide-resistant respiration of P. pseudoalcaligenes CECT5344.
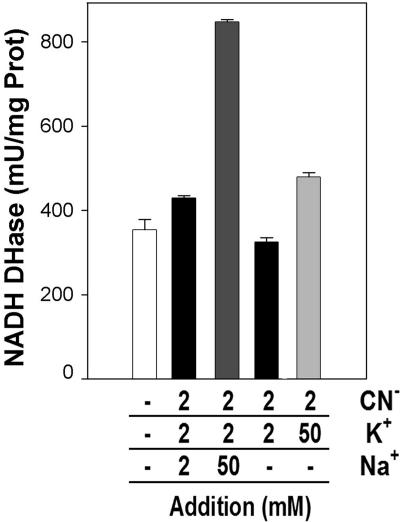
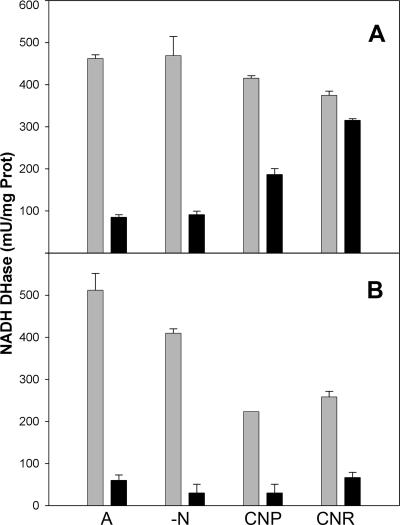
P. pseudoalcaligenes CECT5344 grows in minimal medium supplemented with an industrial residue from the electroplating industry (hereafter called “residue”) that contains cyanide as the main nitrogen source and the composition of which has been described elsewhere (18). When commercial sodium cyanide was utilized, the cell growth rate was lower than that with the residue or with ammonium but significantly higher than that under nitrogen starvation (Fig. 1A). Moreover, the higher cell yield was obtained with the residue, probably due to the presence of metallic cyanide complexes (18), although a higher cell growth rate was observed with ammonium as the nitrogen source. The respiratory activity of membranes extracted from ammonium-grown cells was severely inhibited by the addition of sodium or potassium cyanide. In contrast, the respiratory activity from cells grown with cyanide (either residue or sodium cyanide) was resistant. In order to avoid its volatilization from the stock, the cyanide used as inhibitor in all cases was KCN dissolved in a solution containing an equimolar concentration of KOH or NaOH. Interestingly, the effect was different depending on the alkali used. While the NADH oxidase activity from cyanide-grown cells was stimulated by the addition of NaOH-dissolved cyanide, it was almost imperceptibly inhibited by cyanide dissolved in its potassium salt (Fig. 2). Mainly sodium but also potassium salts produced a concentration-dependent stimulation of the respiratory activity. That is the reason why the effect of cyanide in the respiratory activity was determined by adjusting the same concentration of potassium between measurements, including the corresponding controls (Fig. 2 and 3). Taking into account these considerations, it was found that cyanide weakly inhibits the respiratory activity of cyanide-grown cells from P. pseudoalcaligenes CECT5344, while it strongly blocks the NADH oxidation of membranes extracted from ammonia-grown or nitrogen-starved cells (Fig. 3A). The cyanide-resistant respiratory chain represents up to ∼50% or ∼90% of total respiration in cultures supplied with pure cyanide or cyanide from the residue, respectively. These results indicate that the respiration in cells cultured in minimal medium without cyanide is mainly driven by a cyanide-sensitive terminal oxidase, whereas a cyanide-insensitive oxidase is induced in cells grown with cyanide as a nitrogen source.
FIG. 1.
Growth of P. pseudoalcaligenes CECT5344 (A) and the CIO1 mutant (B) in minimal media. Cells were precultured in 2 mM ammonium media until nitrogen was exhausted (OD600 ≈ 0.3). Arrows indicate the addition of 1 mM of nitrogen source in the form of ammonium (•), cyanide from jewelry residue (○), or sodium cyanide (▾). One flask was kept without N addition as a control of nitrogen-free medium (▵).
FIG. 2.
Effect of cyanide, potassium, and sodium in the respiratory activity of P. pseudoalcaligenes CECT5344 membrane cell extracts. Cells were precultured in 2 mM ammonium medium until nitrogen was exhausted, and after 4 h of growth with 2 mM of cyanide from jewelry residue, the membrane fractions were obtained by ultracentrifugation. NADH oxidation activities were determined as described in Materials and Methods with the additions indicated. The bars represent the average of three independent experiments.
FIG. 3.
Induction of the cyanide-insensitive NADH oxidase activity in P. pseudoalcaligenes CECT5344 (A) and the CIO1 mutant (B). Cells were precultured in 2 mM ammonium medium until nitrogen was exhausted. The cultures were separated in four flasks, and after treatment for 4 h with 2 mM of ammonium (NH4Cl), nitrogen free medium (−N), 2 mM of potassium cyanide (KCN), or 2 mM of cyanide from jewelry residue (RCN), the membrane fractions were obtained as described in Materials and Methods. The NADH oxidase activities were determined after addition of 2 mM KCN alkalinized with KOH (black bars) or 4 mM KCl as controls (gray bars). Prot, protein.
Cloning the cio genes from P. pseudoalcaligenes CECT5344.
Two terminal oxidases that share cyanide-resistant activity have been found in prokaryotes, the alternative oxidase and the cytochrome bd (14, 19), although only the latter seems to be commonly present in bacteria. Thus, a PCR strategy was devised to identify the putative presence of the cytochrome bd-encoding genes in P. pseudoalcaligenes CECT5344.
Two clustered genes, cioA and cioB, encode the two subunits of cytochrome bd, the cyanide-resistant terminal oxidase found ubiquitously in most bacteria (14). The cioA sequences were identified in the genome databases from Pseudomonas aeruginosa PA01, Pseudomonas putida KT2440, Pseudomonas syringae DC3000, Pseudomonas fluorescens pf5, and Escherichia coli K-12 genomes, and they were aligned to detect residue conservation within the group (see the figure in the supplemental material). Two identity blocks were selected to design the degenerate primers ciA and ciB (see Materials and Methods and Fig. 4), which were used in PCR on genomic DNA from P. pseudoalcaligenes CECT5344. This experiment resulted in the amplification of a 945-bp DNA fragment corresponding to the central region of the cioA gene. Finally, two rounds of chromosome walking by iPCR (see Materials and Methods) allowed the coverage and sequencing of a 7.0-kb DNA genomic region. Seven clustered genes were detected in the amplified region by searching homologous sequences in nonredundant protein databases. Five coding sequences, including cioA and cioB, are tightly linked or even shortly overlapped, indicating that they are probably cotranscribed. The first gene from the 5′ end of the operon is sir/nir, which encodes a putative ferredoxin-dependent sulfite or nitrite reductase, and two additional genes with unknown function are located at the 3′ end of sir/nir (orf1) and cioB (orf2). At 125 nucleotides downstream from the 3′ end of the operon, the DNA fragment spans a 1.0-kb sequence of the gene serC, encoding the N-terminus fragment of a phosphoserine aminotransferase (8). In the opposite strand, and divergently transcribed from the operon, the left end of the sequenced fragment encodes the N terminus of MocR, a member of the GntR family (24) of transcriptional regulators (Fig. 4).
Cotranscription of the cio operon was evidenced by RT-PCR, indicating the presence of mRNA molecules spanning from sir/nir to cioA and from cioA to cioB (Fig. 4). The operon structure might also include orf2, the start codon of which is immediately adjacent to the cioB stop codon. However, the RT-PCR designed to detect potential transcripts that would span cioA-serC genes was unsuccessful, which suggests that a serC-specific promoter should be located in the orf2-serC intergenic sequence.
The cio operon seems to be regulated at the transcriptional level since the corresponding messenger was not detected by RT-PCR with RNA obtained from ammonium-grown cells (Fig. 4). Moreover, it is worth remarking that the cyanide-insensitive respiration activity was only detected in cyanide-grown cells (Fig. 3).
Up-regulation by cyanide of the cyanide-resistant respiration provides the conditions to investigate the heme d content of cytochrome bd in this bacterium. The spectroscopic analysis of membranes from cells of P. pseudoalcaligenes CECT5344 grown on ammonium or cyanide (Fig. 5A and B, respectively) produced spectra similar to those previously described for P. aeruginosa (4), which shares peaks in the alpha and beta regions at 551 and 558 nm that might correspond to hemes c and b. The lack of the characteristic peaks of hemes b595 and d628 described in E. coli seems to be typical rather than exceptional for certain prokaryotes (4, 5, 13) and could indicate a different redox center composition for the product of cio genes from those organisms.
FIG. 5.

Reduced minus oxidized difference spectra of membranes from P. pseudoalcaligenes CECT5344 (A and B) and CIO1 mutant (C and D). Cells were precultured in 2 mM ammonium medium until nitrogen was exhausted. The cultures were separated in two flasks, and after treatment for 4 h with 2 mM of ammonium (A and C) or 2 mM potassium cyanide (B and D), the membrane fractions were obtained as described in Materials and Methods. The spectra were recorded in 100 mM Tris (pH 8.0), and the protein concentration was 3 mg/ml in all cases.
Mutagenesis of cioA.
The role of cytochrome bd-related oxidase from P. pseudoalcaligenes CECT5344 has been investigated by directed mutagenesis of the cioA coding sequence found in the bacterial genome. A cioA internal sequence was cloned into pKmob18 (see Materials and Methods), a suicide plasmid for Pseudomonas since it confers kanamycin resistance but lacks a functional replication origin for this bacterium (26). The cioA fragment amplified by the ciA and ciB primers (see the previous section) was cloned into the SmaI site of pBluescriptII (Stratagene) and then was excised by KpnI-PstI double digestion and cloned into pK18mob (26). The strain named CIO1, which was isolated after electroporation and antibiotic selection, was shown to have the cioA gene disrupted by a single exchange and homologous recombination that integrates the plasmid structure into the targeted locus, as determined by Southern hybridization against a cioA probe (not shown). The phenotype of the mutant strain evidenced that CIO1 undergoes complete loss of growth with cyanide as the N source, although N assimilation from ammonium was as efficient as that of the wild-type strain (Fig. 1B). The CIO1 mutant was also unable to grow in LB medium supplemented with 1 mM cyanide. Moreover, the NADH oxidation activity was dramatically inhibited by cyanide in the CIO1 mutant, and no cyanide-insensitive oxidase was induced after cyanide addition (Fig. 3B).
The difference spectra of membranes from CIO mutant were identical to those obtained for the wild type (Fig. 5). The peak attributable to heme b558 was not affected by mutation of cioA, encoding the subunit of cytochrome bd where this group might be attached (14), suggesting that some additional redox center(s) could contribute to the absorbance in the beta region of the spectrum in both P. aeruginosa (4) and P. pseudoalcaligenes (this work).
DISCUSSION
The aim of this work was to investigate the possible role of cytochrome bd-related oxidase in cyanide metabolism by the cyanotrophic bacterium P. pseudoalcaligenes CECT5344. Cloning of the cio operon and targeted disruption of cioA have provided evidence for the involvement of the cyanide-resistant terminal oxidase in cyanide tolerance and, therefore, in cyanide assimilation.
The biochemical pathway for cyanide assimilation remains unknown in this bacterium (18). The cyanide hydratase activity that has been described in a few prokaryotes (16) is not detected in P. pseudoalcaligenes CECT5344 cell extracts, as well as other enzymatic activities related to cyanide assimilation (18). However, Pseudomonas fluorescens NCIMB 11764 presents a cytosolic NADH-specific cyanide monooxygenase that is induced by cyanide and plays a critical role in cyanide assimilation (10). In preliminary experiments, we observed that sodium cyanide produced an increment in the O2-dependent NADH-oxidase activity catalyzed by cell extracts from cells grown with cyanide but not with ammonium as a nitrogen source. These results could be interpreted in the context of a cyanide monooxygenase activity, as recently reported (10). Nevertheless, we conclude that this is not the case based on two set of experiments: (i) neither cyanide was consumed nor ammonium was produced during the experiments, and (ii) the activity was located in the membrane fraction (Fig. 2). The stimulatory effect of sodium cyanide on the NADH-oxidase activity can be attributed to the sodium, not to the cyanide (Fig. 2). These results are consistent with the presence of a sodium-dependent NADH dehydrogenase, most probably corresponding to the redox-driven Na+ pump that has been described in prokaryotes (28).
Cloning of cioA has allowed the generation of a directed mutant unable to use cyanide as the sole nitrogen source (Fig. 1B) or even to grow in LB medium supplemented with 1 mM cyanide. In addition, the CIO1 mutant didn't induce any cyanide-insensitive respiration when exposed to the poison (Fig. 3B). These results provide evidence for the involvement of the cio operon in the cyanotrophic growth of Pseudomonas pseudoalcaligenes CECT5344 and suggest that this is the unique terminal oxidase conferring a cyanide-resistant respiration to the bacterium.
The genes encoding cytochrome bd-related oxidase from P. pseudoalcaligenes CECT5344 have been cloned and sequenced. CIOA, the subunit I of the complex, is closely related to its orthologs from the Pseudomonas group (see the figure in the supplemental material) and presents all of the structural determinants previously found in other members of the protein family (14, 32). Those include protein topology, the nine predicted transmembrane α-helices, and the three heme ligands that are positioned at H-159, H-234, and H-339 in the primary sequence of the P. pseudoalcaligenes CECT5344 cioA gene. However, it should be noted that conservation of these motifs does not constitute a signature for the binding of the protein to all heme groups b558, b595, and d, since the difference spectra of membranes from P. pseudoalcaligenes CECT5344 do not show all of the corresponding peaks (Fig. 5). Similar spectroscopic properties have been argued to support the absence of heme d in the CIO complex from some other bacteria (4, 5, 13). In addition, the periplasmic loop between helices 3 and 4 is longer in proteins from other bacteria than in those from the Pseudomonas group, where resistance to the inhibitor is variable among strains, indicating that this structural element is not related to cyanide tolerance. The sequence of cioB, the gene encoding subunit II of cytochrome bd-related oxidase, is also well conserved, although in this case the only known structural determinants are the eight transmembrane α-helices predicted from the hydrophobic profile (14; data not shown). Phylogenetic analysis reveals an asymmetrical evolution of cioB, which shows an increased sequence divergence in comparison to CIOA, suggesting the existence of a higher selective pressure for the evolution of CIOB (12). Although CIOB's contribution to cyanide resistance evolution is not known at the moment, a recent study has shown that the introduction of cioB from a naturally occurring cyanide-resistant bacterium (Staphylococcus carnosus) into a cyanide-sensitive strain (Staphylococcus aureus) increases the cyanide tolerance of the latter (30). Expression of CIOA and/or CIOB from P. pseudoalcaligenes CECT5344 into cyanide-sensitive strains is in progress. This could serve to evaluate a possible role of CIOB as a key determinant for cyanide resistance in this bacterium, which tolerates unusually high cyanide concentrations (18).
The gene that initiates the sequence of the cio operon in P. pseudoalcaligenes CECT5344 encodes a chimeric protein with an intriguing structure. The 550 residues of the C-terminal domain correspond to SIR/NIR, a putative assimilatory-type and ferredoxin-dependent sulfite or nitrite reductase (3, 7), but the 119 residues of the N domain share homology with other open reading frames found within operons that also includes sir/nir and siroheme biosynthesis genes from a few bacterial genomes (not shown). Although both domains are fused, the coding sequence of the C domain starts by a potential initiation gtg codon, and a potential Shine-Dalgarno sequence at −12 that could drive the translation of a “classical form” SIR/NIR from Val-120. In addition, a putative promoter for the cio operon might control the coregulation of cytochrome bd-related oxidase and SIR/NIR, since there is evidence of cotranscription and coexpression for sir/nir and cio genes (Fig. 4) and cyanide-resistant respiration (Fig. 3). This specific coregulation or some unexpected and still unknown functional connections between SIN/NIR and the oxidase might explain the evolutive origin for the unique organization of the cio operon found in this bacterium.
The analysis of the cio operon from P. pseudoalcaligenes CECT5344 presented in this work provides evidences for the role of cytochrome bd-related oxidase in cyanide assimilation, since disruption of cioA transforms the bacterium in a cyanide-sensitive strain. In conjunction with the biotechnological potential of P. pseudoalcaligenes CECT5344 (18), the singular organization of cio genes found in this organism might provide new tools to investigate and improve cyanide biodegradation.
Supplementary Material
Acknowledgments
This work has been funded by projects BMC-2002-04126-C03-01 and BIO2005-07741-CO2-02 from the Spanish Ministry of Science and Education and 2PR04A022 from the Junta de Extremadura, Spain.
We gratefully acknowledge the support of the BIOSUPPORT project (http://bioinfo.hku.hk) for providing bioinformatics resources and computational services from the HKU Computer Centre.
Footnotes
Published ahead of print on 15 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baughn, A. D., and M. H. Malamy. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441-444. [DOI] [PubMed] [Google Scholar]
- 2.Berthold, D. A., and P. Stenmark. 2003. Membrane-bound diiron carboxylate proteins. Annu. Rev. Plant Biol. 54:497-517. [DOI] [PubMed] [Google Scholar]
- 3.Crane, B. R., and E. D. Getzoff. 1996. The relationship between structure and function for the sulfite reductases. Curr. Opin. Struct. Biol. 6:744-756. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham, L., and H. D. Williams. 1995. Isolation and characterization of mutants defective in the cyanide-insensitive respiratory pathway of Pseudomonas aeruginosa. J. Bacteriol. 177:432-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham, L., M. Pitt, and H. D. Williams. 1997. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24:579-591. [DOI] [PubMed] [Google Scholar]
- 6.Dassa, J., H. Fsihi, C. Marck, M. Dion, M. Kieffer-Bontemps, and P. L. Boquet. 1991. A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA). Mol. Gen. Genet. 229:341-352. [DOI] [PubMed] [Google Scholar]
- 7.Dhillon, A., S. Goswami, M. Riley, A. Teske, and M. Sogin. 2005. Domain evolution and functional diversification of sulfite reductases. Astrobiology 5:18-28. [DOI] [PubMed] [Google Scholar]
- 8.Dubnovitsky, A. P., E. G. Kapetaniou, and A. C. Papageorgiou. 2005. Enzyme adaptation to alkaline pH: atomic resolution (1.08 A) structure of phosphoserine aminotransferase from Bacillus alcalophilus. Protein Sci. 14:97-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evered, D., and S. Harnett (ed.). 1988. Cyanide compounds in biology. Ciba Foundation Symposium 140. John Wiley, Chichester. England. [PubMed]
- 10.Fernández, R. F., and D. A. Kunz. 2005. Bacterial cyanide oxygenase is a suite of enzymes catalyzing the scavenging and adventitious utilization of cyanide as a nitrogenous growth substrate. J. Bacteriol. 187:6396-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green, G. N., H. Fang, R. J. Lin, G. Newton, M. Mather, C. D. Georgiou, and R. B. Gennis. 1988. The nucleotide sequence of the cyd locus encoding the two subunits of the cytochrome d terminal oxidase complex of Escherichia coli. J. Biol. Chem. 263:13138-13143. [PubMed] [Google Scholar]
- 12.Hao, W., and G. B. Golding. 2006. Asymmetrical evolution of cytochrome bd subunits. J. Mol. Evol. 62:132-142. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, R. J., K. T. Elvers, L. J. Lee, M. D. Gidley, L. M. Wainwright, J. Lightfoot, S. F. Park, and R. K. Poole. 2007. Oxygen reactivity of both respiratory oxidases in Campylobacter jejuni: the cydAB genes encode a cyanide-resistant low-affinity oxidase that is not of the cytochrome bd type. J. Bacteriol. 189:1604-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jünemann, S. 1997. Cytochrome bd terminal oxidase. Biochim. Biophys. Acta 1321:107-127. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, M. J., R. K. Poole, M. G. Yates, and C. Kennedy. 1990. Cloning and mutagenesis of genes encoding the cytochrome bd terminal oxidase complex in Azotobacter vinelandii: mutants deficient in the cytochrome d complex are unable to fix nitrogen in air. J. Bacteriol. 172:6010-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunz, D. A., C. S. Wang, and J. L. Chen. 1994. Alternative routes of enzymic cyanide metabolism in Pseudomonas fluorescens NCIMB 11764. Microbiology 140:1705-1712. [DOI] [PubMed] [Google Scholar]
- 17.Lowry, O. H., N. J. Rosenbrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 18.Luque-Almagro, V. M., M. J. Huertas, M. Martinez-Luque, C. Moreno-Vivian, M. D. Roldan, L. J. Garcia-Gil, F. Castillo, and R. Blasco. 2005. Bacterial degradation of cyanide and its metal complexes under alkaline conditions. Appl. Environ. Microbiol. 71:940-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald, A. E., and C. G. Vanlerberghe. 2005. Alternative oxidase and plastoquinol terminal oxidase in marine prokaryotes of the Sargasso Sea. Gene 349:15-24. [DOI] [PubMed] [Google Scholar]
- 20.Mudder, T. I. (ed.). 1998. The cyanide monograph. Mining Journal Books Limited, London, United Kingdom.
- 21.Muller, M., and R. E. Webster. 1997. Characterization of the tol-pal and cyd region of Escherichia coli K-12: transcript analysis and identification of two new proteins encoded by the cyd operon. J. Bacteriol. 179:2077-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oró, J. 1961. Mechanism of synthesis of adenine from hydrogen cyanide under possible primitive Earth conditions. Nature 191:1193-1194. [DOI] [PubMed] [Google Scholar]
- 23.Pittman, M. S., H. C. Robinson, and R. K. Poole. 2005. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 280:32254-32361. [DOI] [PubMed] [Google Scholar]
- 24.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 27.Stenberg, F., P. Chovanec, S. L. Maslen, C. V. Robinson, L. L. Ilag, G. von Heijne, and D. O. Daley. 2005. Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280:34409-34419. [DOI] [PubMed] [Google Scholar]
- 28.Steuber, J. 2001. Na+ translocation by bacterial NADH:quinone oxidoreductases: an extension to the complex-I family of primary redox pumps. Biochim. Biophys. Acta 1505:45-56. [DOI] [PubMed] [Google Scholar]
- 29.Vanlerberghe, G. C., and L. McIntosh. 1997. Alternative oxidase: from gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:703-734. [DOI] [PubMed] [Google Scholar]
- 30.Voggu, L., S. Schlag, R. Biswas, R. Rosenstein, C. Rausch, and F. Götz. 2006. Microevolution of cytochrome bd oxidase in staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J. Bacteriol. 188:8079-8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization. 2004. Hydrogen cyanide and cyanides: human health aspects. Concise international chemical assessment document 61. World Health Organization, Geneva, Switzerland.
- 32.Zhang, J., B. Barquera, and R. B. Gennis. 2004. Gene fusions with beta-lactamase show that subunit I of the cytochrome bd quinol oxidase from E. coli has nine transmembrane helices with the O2 reactive site near the periplasmic surface. FEBS Lett. 561:58-62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.