Abstract
A spontaneous rpsL mutant of Thermus thermophilus was isolated in a search for new selection markers for this organism. This new allele, named rpsL1, encodes a K47R/K57E double mutant S12 ribosomal protein that confers a streptomycin-dependent (SD) phenotype to T. thermophilus. Models built on the available three-dimensional structures of the 30S ribosomal subunit revealed that the K47R mutation directly affects the streptomycin binding site on S12, whereas the K57E does not apparently affect this binding site. Either of the two mutations conferred the SD phenotype individually. The presence of the rpsL1 allele, either as a single copy inserted into the chromosome as part of suicide plasmids or in multicopy as replicative plasmids, produced a dominant SD phenotype despite the presence of a wild-type rpsL gene in a host strain. This dominant character allowed us to use the rpsL1 allele not only for positive selection of plasmids to complement a kanamycin-resistant mutant strain, but also more specifically for the isolation of deletion mutants through a single step of negative selection on streptomycin-free growth medium.
Extreme thermophiles constitute models of great biological interest because of their ancestral origin, the applicability of their enzymes, and the ability of thermostable enzymes and macromolecular complexes to crystallize better than their mesophilic counterparts (18, 30, 34, 35). However, very few extreme thermophiles are presently suitable as laboratory study models due to intrinsic growth difficulties and a general absence of genetic tools required for genetic manipulations. Thermus thermophilus represents an exception to this rule because of (i) its ability to grow under laboratory conditions with good yields, (ii) its aerobic or facultative mode of growth, and (iii) its constitutive expression of an efficient natural competence system (7, 14). Such properties have recently allowed the development of numerous tools and methods to manipulate this bacterium at the levels comparable to those available for most mesophilic bacteria (5, 16, 17, 19, 22) yet far from those currently available for Escherichia coli.
One of the breakthroughs in the field of genetic analysis of Thermus thermophilus was provided by a method of isolation of knockout mutants based on insertion of a gene cassette (kat) encoding a thermostable kanamycin (Kan) nucleotidyltransferase (16). However, in spite of being a rapid method for the isolation of directed knockout mutants, the insertion of the kat cassette blocks further selection procedures based on this marker. Two alternative methods for the selection of marker-free deletion mutants have been published. The multistep method of Tamakoshi et al. (32) requires the isolation of a pyrE (encoding orotate phosphoribosyltransferase) uracil auxotroph as the parental strain and uses complementation to select for a Δtarget gene::pyrE insertion mutant after transformation with the appropriate construct. Then, counterselection of this pyrE with 5-fluoroorotic acid as antimetabolite permits the subsequent isolation of Δtarget gene ΔpyrE double mutants that can be subjected again to further selection. More recently, a “pop-in/pop-out” method based on a suicide plasmid conferring Kan resistance (pK18) was used to isolate ΔgreA Δgfh1 double mutants of T. thermophilus (15). In this method, the insertion of the plasmid on the target by recombination is selected by Kan, and a further manual screening among thousands of colonies for spontaneous back recombinants allows the selection of the desired deletion mutant. Thus, although the first step of this pK18-based method is straightforward and can be applied to wild-type strains, the subsequent screening for antibiotic-sensitive clones requires a great deal of time and manual work. With this in mind, we hypothesized that a gene that could confer simultaneous resistance to and dependence on an antibiotic (e.g., streptomycin [Str]) could be used in a similar protocol to select the insertion and the excision of a target gene in the presence or the absence of the antibiotic, respectively, without the requirement of any tedious manual screening.
Str inhibits bacterial protein synthesis through binding to multiple structural elements of the 30S ribosomal subunit, including the S12 protein and the 16S rRNA helices 1, 18, 27, and 44 (1). Although Str-resistant (SR) and Str-dependent (SD) mutants of E. coli have been known for a long time, the molecular details of the interaction of the antibiotic with its binding site in the bacterial ribosome have been described only recently after the resolution of Str-30S complexes of T. thermophilus (1). Most SR and SD mutants of different bacterial groups, including T. thermophilus (8), present amino acid substitutions in their S12 ribosomal protein, which in E. coli is encoded by the rpsL gene. Therefore, we hypothesized that it could be possible to isolate SD alleles of the rpsL gene of T. thermophilus that could fulfill our requirements for a selectable and counterselectable gene marker. In fact, SR and SD alleles of the rpsL gene of T. thermophilus have been described previously (8), and DNA from SR strains of T. thermophilus are routinely used for the functional analysis of the natural competence system of this organism in Kan-resistant genetic backgrounds (7), thus supporting the feasibility of this hypothesis.
Here we describe the isolation of an rpsL allele (rpsL1) encoding a double mutant S12 protein that confers an unexpectedly dominant SD phenotype in T. thermophilus. We show that this allele can be used as a positive selection marker for suicide and replicative plasmids alternative to and compatible with the widely used Kan resistance gene (kat) and also as a negative selection marker for the isolation of marker-free deletion mutants of T. thermophilus in a single-step procedure. The likely molecular details of this dominant SD phenotype are also discussed based on the models of the described structures of the T. thermophilus ribosome (1, 23).
MATERIALS AND METHODS
Strains, growth conditions, and transformation.
T. thermophilus HB27 was a generous gift of Y. Koyama. The strain T. thermophilus HB27c is a facultative anaerobe obtained by transfer of the nitrate conjugative element (3) to the HB27 strain (27). Insertional narC::kat (39), phoA::kat (21), and greA::kat (15) mutants are derivatives of the HB27c strain in which the corresponding genes were knocked out by insertion of a gene (kat) coding for a thermostable resistance to Kan (16). The E. coli strain DH5α (supE44 ΔlacU169 φ80 lacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1) (10) was used for genetic constructions. E. coli and T. thermophilus were grown in LB at 37°C and in Thermus broth (TB) (25) at 70°C, respectively.
Transformation of T. thermophilus was achieved by natural competence on growing cells (5, 13), whereas standard protocols were used to transform E. coli (10). Selection was carried out on 1.5% (wt/vol) agar plates with Str (different concentrations), Kan (30 mg/liter), and/or ampicillin (Amp; 100 mg/liter).
Isolation and cloning of the rpsL1 gene.
Identical amounts of total DNA preparations from individual colonies of spontaneous mutants of T. thermophilus that grew on TB plates with Str (100 mg/liter) were used to transform competent cultures of the wild-type strain. Total DNA preparations of individual colonies that rendered the highest number of transformants on plates with Str (100 mg/liter) were used as templates for the amplification by PCR of a 759-bp DNA fragment containing sequences from positions −294 to +445 with respect to the GTG start codon of the rpsL gene (T. thermophilus HB27 genome code TTC1333) (11). Primers O-str1 (5′-AAAACATATGTCCAGGCCCTCCA-3′) and O-str2 (5′-AAAACATATGACGGACCTCTGCT-3′), each containing an NdeI site (underlined), were used for this amplification. Identical amounts of the PCR products were used to transform T. thermophilus HB27, and transformants that produced the highest number of colonies on plates with Str were selected for further studies. The comparison of the sequence of this allele (rpsL1 thereafter) with that of the wild type revealed two A-to-G transitions (positions +140 and +169) that produced the amino acid replacements K47R and K57E. To analyze the individual role of each amino acid substitution, a 470-bp 5′-end fragment (positions −294 to +166) and a 312-bp 3′-end fragment (+143 to +445) of rpsL1 were amplified with primer pairs O-str1/O-str6 (5′-CCACCTTACGGAGCGCCGAGTTGG-3′) and O-str5 (5′-CCAACTCGGCGCTCCGTAAGGTGG-3′) and O-str2, respectively, and the corresponding PCR products were used to transform the wild-type strain. The colonies that grew on TB plates with Str (100 mg/liter) were analyzed for the presence of each mutation by sequencing the whole rpsL gene once amplified by PCR with primers O-str1 and O-str2.
Plasmid construction.
DNA isolation, plasmid construction, and restriction analysis were performed as described elsewhere (28). Amplification of the rpsL gene was carried out by PCR with Tth or Pfu DNA polymerases (BIOTOOLS B & M, Madrid, Spain) and primers O-str1 and O-str2. The insertion of this PCR fragment at the NdeI site of plasmid pUC18 rendered the pS18a and pS18b constructs that differ only in the orientation of the rpsL1 gene (see map in Fig. 2A). The presence of the suicide plasmids in the chromosome of transformed cells of T. thermophilus was detected by PCR with primer pairs Ostr1/M13reverse (5′-GAAACAGCTATGACCATG-3′), Ostr1/Ostr8 (5′-GATGAAGGCCGTGACCAG-3′), and O-132pUC (5′-GGGGCTGGCTTAACTATG-3′)/Ostr8.
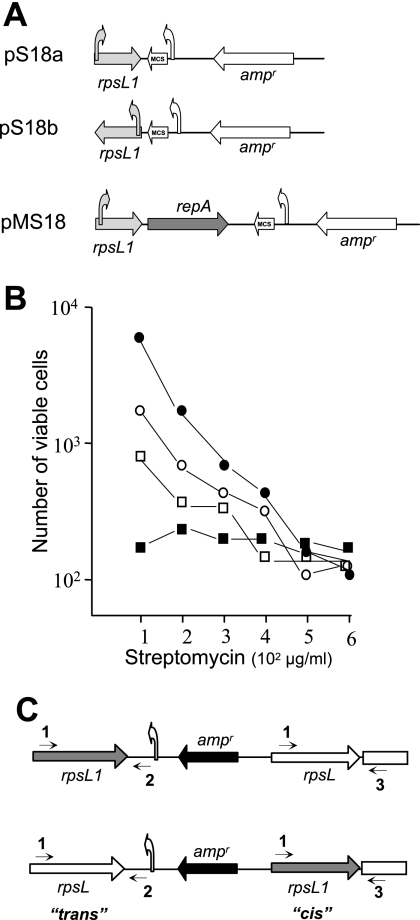
FIG. 2.
Suicide and replicative plasmids based on the rpsL1 allele. (A) Structure of suicide (pS18a and pS18b) and replicative (pMS18) plasmids based on the rpsL1 allele. (B) The number of colonies that grew on TB plates containing the indicated concentrations of Str before (filled squares) or after transformation with identical amounts of pS18a (open circles), pS18b (open squares), or pMS18 (filled circles). (C) Two putative results of the integration of pS18a into the rpsL target (white arrows) through recombination with the rpsL1 allele (gray arrows). Approximate positions of the Amp resistance gene (black) and the lacZp (vertical arrows) are indicated. Small arrows represent positions of primers O-str1 (1), M13-rev (2), and O-str8 (3).
Plasmid pMS18 is a derivative of pS18a that carries a replicative origin functional in Thermus spp. (6). For its construction, the pMK18 replicon was amplified by PCR with primers O-repNde2d (5′-CTGTTCATATGCCTCAGTTGA-3′) and O-repNder (5′-GGGACCATATGCCCCTGGA-3′), both containing NdeI sites (underlined), and ligated into pS18a (see map in Fig. 2A). The bgaA and phoA genes encode a thermostable beta-galactosidase (βgal) (14) and a hyperalkaline phosphatase (2, 21), respectively. The narC and gfh1 genes encode a periplasmic cytochrome c that constitutes the fourth subunit of the respiratory nitrate reductase (40) and the transcripition elongation factor Gfh1 (15) of T. thermophilus, respectively. Derivatives of pS18a and/or pMS18 carrying the narC, bgaA (22), and phoA (22) genes under the control of the nitrate reductase promoter (narp) (22) were obtained by cloning the respective genes between the XbaI and HindIII restriction sites of the plasmids. Derivatives of pS18a and pMS18 containing the gfh1 gene under the control of its own promoter (15) were obtained by cloning the gene between the restriction sites EcoRI and SalI of both plasmids.
Induction of narp and complementation assays.
Cells were grown at 70°C in TB under aerobic conditions in a rotational shaker bath (150 rpm), and transcription from the nitrate reductase operon promoter (narp) was activated by addition of KNO3 (40 mM) and the simultaneous arrest of the shaker. Under such static growth conditions, the low solubility of O2 at 70°C and its rapid consumption by the cells make the culture anoxic in a very short time (4, 22). Cells were incubated for 4 h at 70°C in such static conditions before being processed. The presence of the kat gene inserted into its target in the complementation experiments was confirmed by PCR with primer pairs FPPnarDIR (5′-CTGGACCAGGTGGGCGCA-3′) and KAT4 (5′-AGAAATTCTCTAGCGAT-3′) for the narC::kat mutant and O-AP1 (5′-CTACGTCACCGAGTCCA-3′) and KAT4 for the phoA::kat mutant.
The BgaA activity of soluble cell extracts of the T. thermophilus cultures was assayed at 70°C with o-nitrophenyl-β-d-galactopyranoside as chromogenic substrate by following previously described methods (22). The periplasmic hyperalkaline phosphatase (PhoA) of similar soluble cell extracts was measured with p-nitrophenyl-phosphate as described previously (22). The activity of the nitrate reductase was detected by the production of nitrite per min and mg of protein (31). Detection of Gfh1 on complete cell extracts from exponential cultures of T. thermophilus was carried out by Western blotting with specific rabbit antiserum as described previously (15).
Isolation of deletion mutants.
Plasmid pS18ΔnarC is a derivative of pS18a carrying upstream and downstream DNA regions of the narC gene (38) that was used to transform the wild-type strain. A group (around 50) of small-sized transformant colonies that grew on TB plates with Str (100 mg/liter) were coinoculated in 10 ml of TB medium and incubated for 2 h at 70°C before 50-μl aliquots of the concentrated and the 10−1 dilution of the cell suspension were plated onto TB plates without Str. As the rpsL1 allele confers a dominant SD phenotype, only those colonies in which this allele was deleted by back recombination could grow on these plates. The Str-sensitive clones were then assayed for the loss of nitrate reductase activity and further analyzed for the absence of the narC target gene by PCR with primers FPPnarDIR and O27-32 (5′-CCCAGCTCGCCCGGCGG-3′).
Bioinformatic analysis.
Crystal structures of the S12 wild-type protein and their 16S rRNA chain-contacting residues were obtained from the crystallographic structures of the ribosomal 30S subunit for comparative geometrical analysis. Calculation of the variation of the dihedral angles of the 16S rRNA phosphates trace and the S12 protein Cα atoms backbone were performed with the open (Protein Data Bank entry 1J5E) (37), closed tRNA/mRNA-bound (Protein Data Bank entry 1N32) (23), and Str-bound (Protein Data Bank entry 1FJG) (1) forms as described previously (20). Dihedral angles and root mean square deviation values between structures were calculated using the Insight II package. Plots and statistics of the three pairwise comparisons were made with SigmaPlot.
Building of three-dimensional structures for K47R and K57E mutants of S12 protein was performed using standard homology-based modeling procedures. Briefly, models were made using the SWISS-MODEL server (9, 24, 29). The structural quality was verified using WHAT-CHECK (12) of the WHAT IF package program (36). In order to perform a first geometry optimization and correct atomic clashes, the energy of the obtained structure was minimized with the implementation of the GROMOS 43B1 force field of the DeepView program (9) using 500 steps of steepest descent minimization and 500 steps of conjugate-gradient minimization.
RESULTS
Isolation and characterization of an rpsL gene allele conferring an SD phenotype.
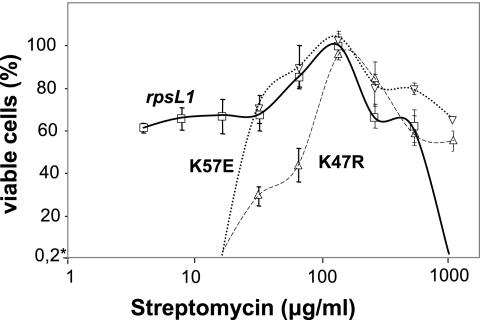
We designed a protocol to isolate a spontaneous rpsL mutant allele (rpsL1 thereafter) able to produce a high number of T. thermophilus colonies on plates with Str (Materials and Methods). When dilutions of these transformant colonies containing around 500 colonies were plated off, no growth was detected in the absence of Str, whereas colonies were detected on plates containing between 4 and 512 mg/liter of Str, with the maximum at 100 mg/liter and no growth at 1,024 mg/liter (Fig. 1). The parental strain was unable to produce colonies even at the lowest concentration of Str used. We concluded that the rpsL1 allele actually confers an SD phenotype to T. thermophilus.
FIG. 1.
Str dependence of the rpsL1 mutant and its single mutant derivatives. Overnight cultures of the wild-type strain grown on TB and of the rpsL1, K47R, and K57E mutants grown with 100 mg/liter of Str in the same medium were diluted (10−5 and 10−6) and plated (100 μl) on TB plates with increasing Str concentrations. After 48 h at 70°C, the number of colonies that grew in each plate was counted and is represented as the percentage of viable cells with respect to their respective maximum values. The data represent the mean values and the standard deviations of two independent assays. The y-axis minimum value (0.2%) is limited by the sensitivity of the assays.
The rpsL1 allele encodes a mutant S12 ribosomal protein with two amino acid substitutions, K47R and K57E (numbers correspond to the T. thermophilus S12 amino acid sequence). Sequence alignments of S12 proteins from other bacteria, including E. coli S12, revealed that K47 (corresponds to K43 in E. coli S12) is a conserved amino acid in all the bacterial S12 sequences analyzed. By contrast, the K57 residue corresponds to an arginine in most of the S12 proteins, including E. coli R53.
Single K47R and K57E mutants present SD phenotypes.
To examine the individual role of each mutation of the rpsL1 allele in the SD phenotype of T. thermophilus, we took advantage of the highly efficient natural competence system of this organism and transformed the wild-type T. thermophilus strain with identical amounts of two PCR-amplified fragments of the rpsL1 allele, coding for either the K47R or K57E mutation (Materials and Methods). Selection on plates with Str (100 mg/liter) revealed a similar number of transformants with both fragments (not shown), suggesting that each individual mutation confers an SD phenotype. This was further confirmed by sequence analysis of the rpsL gene, which was PCR amplified using genomic DNA derived from a number of colonies from each transformation group.
In order to compare the phenotype conferred by each mutation with that of the double mutant, we inoculated identical numbers of cells with each mutation on plates containing increasing concentrations of Str. As shown in Fig. 1, the K47R and the K57E single mutants showed a clear SD phenotype, although they required higher antibiotic concentrations to grow than did the double K47R K57E mutant. In fact, none of the single mutants grew at concentrations of Str lower than 32 mg/liter. The experiments also revealed that the K47R mutant required higher antibiotic concentrations than the K57E mutant to show an efficient growth; at 32 mg/liter, this strain showed only 30% of its maximum number of viable cells, reached at 100 mg/liter, compared to the 70% viability shown by the K57E strain. On the other hand, the individual mutants grew efficiently even at 1,024 mg/liter, a concentration on which the double mutant did not show a detectable growth.
We also compared the growth rates of each mutant with that of the wild-type strain in liquid medium. In exponential phase, all mutants grew in TB with Str (100 mg/liter) with longer doubling times (55 to 60 min), whereas the wild-type strain grew with doubling times around 40 min in the absence of the antibiotic.
Construction of suicide vectors based on the rpsL1 allele.
Following PCR amplification, the rpsL1 allele (including its promoter) was cloned into pUC18, rendering plasmids pS18a and pS18b, each carrying this gene in opposite orientations with respect to the lacZp promoter (Fig. 2A). Both plasmids ensured Amp resistance and had the multicloning site and the α-LacZ complementation system of their parental plasmid to facilitate the cloning procedures in E. coli. However, neither the pS18a nor the pS18b plasmid conferred the SR or SD phenotype to E. coli (not shown) as we expected based on the differences in amino acid sequences between the S12 proteins of E. coli and that of T. thermophilus (8). On the other hand, these plasmids could not replicate in Thermus spp. because of the absence of an appropriate replication origin. Thus, the only way to provide any detectable Str-related phenotype in T. thermophilus was through its integration into the chromosome by homologous recombination.
In order to analyze if this was the case, identical amounts (200 ng) of each plasmid were used to transform the wild-type strain, and selection was carried out on plates with different concentrations of Str. As shown in Fig. 2B, transformation with pS18a and pS18b allowed the growth on plates with Str (100 mg/liter) of a much higher number of colonies than the number of spontaneous mutants that grew on the untransformed control plates. In all cases, plasmid pS18a rendered more transformed colonies than pS18b. These data suggest that the plasmids were most likely inserted into the chromosome of the receptor strain after transformation and provided the host with the capability of growing on Str. At higher Str concentrations, the number of colonies decreased, while the number of spontaneous mutants remained more or less constant (frequency of around 10−6 to 10−7), even at the highest concentration assayed in these experiments (600 mg/liter). Moreover, whereas most of the untransformed colonies that grew on the control plates were large (as with the wild-type strain in the absence of the antibiotic), those that grew after transformation with pS18a presented a number of “large” colonies similar in size and number to that grown on the control plates and a much higher number of “small” colonies (not shown). This observation suggests that integration of pS18a into the chromosome produces a slow-growth phenotype similar to that shown by the rpsL1 mutant. In fact, when these two colony types from the pS18a transformation were assayed for their ability to grow in the absence of Str, most (95%) of the large colonies were SR, a percentage similar to that shown by the large, spontaneous mutant colonies grown on untransformed control plates. By contrast, all the small-sized colonies checked were SD.
The results described above supported the idea that the rpsL1 allele was inserted into the chromosome after transformation with pS18a, but they did not allow us to discriminate whether the whole plasmid remained linked to the chromosome after recombination or was further deleted through a back recombination. To distinguish between these two possibilities, PCR amplification assays were carried out on total DNA from 10 small-sized colonies transformed with pS18a. As positive amplification with primers Ostr-1 and M13-reverse (labeled 1 and 2 in Fig. 2C) was detected in all of them, we concluded that the plasmid was still present in the chromosome of all the transformants assayed. Consequently, these colonies had two copies of the rpsL gene, the wild type and its rpsL1 allele (Fig. 2C), leading to the conclusion that in these pS18a-transformed cells, the rpsL1 allele was dominant over its parental wild-type rpsL gene.
We subsequently analyzed whether the mutant S12 protein was expressed in these colonies from the rps operon (in “cis”) or from the copy generated after plasmid integration (in “trans”) (Fig. 2C) by sequencing the “cis” copy after PCR amplification with primers labeled 1 and 3 in Fig. 2C and the “trans” copy after PCR amplification with primers labeled 1 and 2. The two mutations corresponding to rpsL1 were identified in the sequence of the “cis” copy of all the colonies analyzed. Interestingly, the cells transformed with pS18a grew with doubling times indistinguishable from that of the rpsL1 mutant.
Construction of replicative vectors.
To analyze if the rpsL1 allele also was an appropriate selection marker when being expressed in a multicopy plasmid, we inserted the replicative origin of pMK18 (5, 6) into pS18a. As its parental pS18a plasmid, the new construct (pMS18) (Fig. 2A) could be selected in the presence of Amp in E. coli and of Str in T. thermophilus. As expected from its higher copy number per cell, selection of pMS18 on Str rendered 5- to 10-fold more colonies than those obtained by transformation with similar concentrations of its parental pS18a suicide vector (Fig. 2B). Moreover, the transformant colonies were unable to grow on plates without Str. Therefore, the cells transformed with pMS18 are SD, despite the presence of a chromosomal wild-type rpsL copy as revealed by PCR (primers 1 and 3 in Fig. 2C). Thus, the rpsL1 allele was also dominant over its parental wild type, even when expressed from the multicopy plasmid. Accordingly, the growth rate of cells transformed with pMS18 was similar to that of the rpsL1 mutant.
We compared the features of two plasmids, pMK18 (5) and pMS18, side by side. The copy numbers of both plasmids were similar, as could be evaluated by the total quantity of each plasmid purified from the same amount of cells transformed with each of them. The stability of pMS18 was also assayed through the analysis of the restriction map from colonies obtained after 30 generations of growth in liquid TB with Str (100 mg/liter): our data show that around 90% of the plasmids preserved the same restriction pattern as the original pMS18, supporting that the plasmid was stable enough to be used as a cloning vector.
pS18a and pMS18 as cloning vectors.
The results described above suggested that plasmids pS18a and pMS18 could be used as integrative (single-copy) and replicative (multicopy) cloning vectors for T. thermophilus, respectively. To check this, the bgaA gene, encoding a thermostable beta-galactosidase, was cloned into both plasmids to render pS18βgal and pMS18βgal. As the expression of the cloned bgaA gene was under the control of the promoter from the respiratory nitrate reductase (narp) (22), we used as a host the facultative strain T. thermophilus HB27c that contains the transcription factors required for the expression of this promoter under appropriate conditions (26).
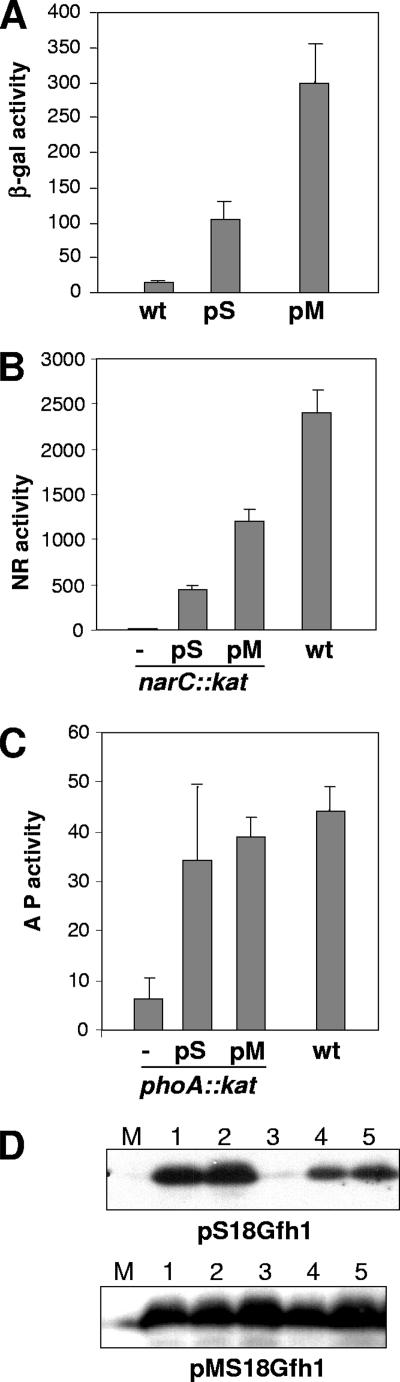
In a typical experiment, 18 out of 30 SD colonies transformed with pS18βgal analyzed showed a beta-galactosidase activity five- to sevenfold higher than that of the basal level of untransformed cells (Fig. 3A). By contrast, all the SD colonies transformed with pMS18βgal that were assayed expressed the thermostable beta-galactosidase up to 15- to 20-fold more than did untransformed cells (Fig. 3A). These findings allowed us to conclude that pS18a and pMS18 indeed can be used in T. thermophilus as single- and multicopy cloning vectors, respectively.
FIG. 3.
Applications of pS18a and pMS18 on Kan-sensitive and -resistant strains. (A) Expression of BgaA. Shown is the beta-galactosidase activity in Miller units of T. thermophilus HB27c before (wild type [wt]) or after transformation with pS18-bgaA (pS) and pMS18-bgaA (pM). (B) Nitrate reductase (NR) activity of the T. thermophilus HB27c narC::kat mutant before (-) or after transformation with plasmid pS18narC (pS) or pMS18narC (pM). The activity is expressed as nmol of nitrite produced per min and mg of protein. (C) Hyperalkaline phosphatase activity in Miller units of a T. thermophilus HB27c phoA::kat mutant before (-) or after transformation with pS18phoA (pS) or pMS18phoA (pM). The activity of the wild-type strain (wt) is also shown in panels B and C. (D) Western blot against Gfh1 of colonies (lanes 2 to 5) of a gfh1::kat mutant transformed with the indicated plasmids. Lanes M and 1 correspond to the mutant and the wild-type strains, respectively. The data presented in panels A to C correspond to mean values and standard deviations of triplicate samples from four to six independent assays.
Complementation of Kan-resistant mutants of T. thermophilus by pS18a and pMS18 derivatives.
As mentioned in the introduction, the use of the kat gene for the isolation of insertion mutants represents the easiest method for the functional analysis of genes in T. thermophilus. However, a complementation experiment with such Kan-resistant mutants indeed is a difficult task. Due to this, we assayed the capability of pS18a and pMS18 to complement three different Kan-resistant insertion mutants of T. thermophilus.
For this purpose, the genes encoding a cytochrome c (NarC) that constitutes the fourth subunit of the respiratory nitrate reductase (narC), the hyperalkaline phosphatase (phoA), and the Ghf1 transcription elongation factor (gfh1) of T. thermophilus were cloned into pS18a and pMS18, and the constructs were subsequently used to transform the respective narC::kat, phoA::kat, and ghf1::kat mutants. Selection was done on TB plates with Kan and Str to avoid the replacement of the kat gene from the target. The colonies were further analyzed by PCR for the presence of the kat insertion (Materials and Methods) and for the recovery of the respective wild-type phenotypes (βgal or PhoA activities or Gfh1 presence).
Figure 3B and C show the results of such complementation experiments. As can be observed, complementation of the nitrate reductase with the narC gene was only partial for both plasmids compared to the activity of the wild-type strain, although cells transformed with the pMS18 derivative (pM) reached higher activity than those transformed with the pS18a derivative (pS). As the growth rate of cells transformed with either of these plasmids was much lower than that of the wild type, we assumed that part of the difference in activity with respect to the wild type was due to the growth-dependent lower induction rates in the mutants. By contrast, complementation of the phoA::kat mutant with phoA derivatives of both plasmids resulted in activities similar to that of the wild type (Fig. 3C), most likely because of the early saturation of the secretion machinery that translocates PhoA to periplasm (2). Finally, complementation of the gfh1::kat mutant with the corresponding pS18a derivative allowed the expression of Gfh1 in three out of four colonies checked by Western blotting, although the expression levels were not identical in all of them (Fig. 3D). By contrast, expression from the pMS18gfh derivative produced wild-type levels of proteins in all the colonies assayed (Fig. 3D).
In addition, it is worthy to note that complementation of the three mutations with the pS18a derivatives occurs in about 70 to 80% of the SD transformant colonies, whereas 100% of the colonies showed the expected complementation when transformed with the pMS18 derivatives.
One-step selection of deletion mutants with pS18a.
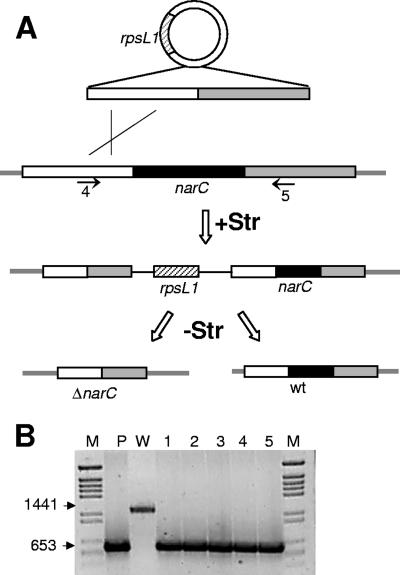
Taking advantage of the dominant SD phenotype conferred by the rpsL1 allele in plasmid pS18a, we hypothesized that this property could be useful for development of a rapid method for the positive selection of deletion mutants of a target gene. In the method proposed, the insertion into the chromosome of a pS18a derivative containing upstream and downstream homologous regions around a target gene, narC in this case, is selected on plates containing Str, and its deletion by back recombination is further selected by plating on a medium without the antibiotic. This is illustrated in Fig. 4 with the isolation of a ΔnarC mutant, in which the insertion of the pS18ΔnarC construct was selected on plates with Str. Then we coinoculated around 50 transformant colonies in a single tube with 10 ml of TB, and after 2 h at 70°C, the cell mix was plated on TB plates without the antibiotic. Under these conditions, less than 1% of the cells grew, compared to the percentage that grew on TB with Str. Moreover, these colonies were sensitive to Str, supporting that they have lost the rpsL1 allele. The analysis of a number of these Str-sensitive colonies revealed that around half of them lacked any detectable nitrate reductase activity, as was expected for mutants defective in narC (40). Further PCR assays on five of these nitrate reductase-negative colonies revealed that all of them were ΔnarC mutants, as demonstrated by the amplification of a 653-bp fragment instead of the 1,441 bp expected for a wild-type strain (Fig. 4B). In conclusion, around half of the Str-sensitive colonies isolated by this protocol carried the expected gene deletion. This one-step procedure represents the fastest and the most straightforward method described to date to obtain deletion mutants in this thermophilic bacterial model.
FIG. 4.
Isolation of deletion mutants by counterselection of the rpsL1 allele. (A) Scheme showing the protocol followed to isolate deletion mutants of the narC gene. Insertion of pS18ΔnarC derivative by homologous recombination is selected on plates with Str (+Str), and the plasmid presence is subsequently counterselected on plates without the antibiotic (−Str). Arrows labeled 4 and 5 correspond to primers FPPnarDIR and O27-32 used in panel B. (B) PCR with primers 4 and 5 of panel A on total DNA from colonies (lanes 1 to 5) selected from panel A that did not show nitrate reductase activity. Lanes P and W correspond to the PCR amplification of the pS18ΔnarC plasmid and to total DNA of the wild-type strain, respectively. Lane M corresponds to DNA size markers. Lateral numbers indicate the sizes in base pairs of the PCR products.
DISCUSSION
Thermus spp. constitute a major bacterial model in structural biology and in biotechnology, and high-resolution structures of ribosomes (30) or RNA polymerase (34) are clear examples of this. However, there are still few genetic tools and, specifically, a great limitation in selectable markers to elucidate structure-function relationships in these organisms. Here we describe a dominant gene marker encoding a double mutant S12 protein from T. thermophilus HB27 that confers not just resistance to, but dependence on, Str, making its use feasible for either positive or negative selection.
As the isolation of the rpsL1 allele was carried out in a single step, the presence of two mutations in the same gene would be unexpected because of its low probability (10−12 to 10−14) unless it occurred sequentially through some kind of effective selection. The most likely selective force could be a requirement for a secondary compensatory mutation that could alleviate a strong growth defect produced by a primary mutation. In this sense, it is interesting to compare the rpsL1 allele of T. thermophilus with the SD mutants of E. coli described by Timms and Bridges (33). These authors found that double rpsL mutants appeared with an unexpectedly high frequency among spontaneous or UV-induced SD mutants and suggested the existence of a transient hypermutability state of the organism that allowed the appearance of compensatory mutations during the first generations of a single-mutant strain. Interestingly, the authors distinguished between “primary” and “ancillary” mutations, in reference to the appearance of the former in some single SD alleles, whereas the latter were essentially found as accompanying mutations of the former. However, the putative individual effects of such ancillary mutations on the Str phenotype were not analyzed in detail. By contrast, in this work we have been able both to separate mutations in T. thermophilus by transformation with rpsL1 fragments and to observe that each individual mutation confers an independent SD phenotype, although both required higher Str concentrations than the rpsL1 double mutant to grow (Fig. 1). Therefore, we could not deduce which of these two SD mutations would be considered “primary” by this criterion. However, the comparison with the E. coli S12 protein favors the K47R replacement as the primary mutation: the position K43 in E. coli, equivalent to K47 in T. thermophilus, is a frequent target for a primary (K43E in E. coli) SD mutation, whereas the R53 residue in E. coli, corresponding to K57 in T. thermophilus, has been described as the target for different ancillary mutations (R53S, R53C, and R53L in E. coli) bound to the P90L primary SD mutation in E. coli (33). Therefore, it is likely that the rpsL1 allele was selected by the incorporation of a compensatory mutation (K57E) during the first division cycles on a K47R mutant.
The structural reasons by which these mutations, and especially K57E, confer a SD phenotype are difficult to define due to the huge number of interactions implicated. As shown elsewhere (see Fig. S1A and S1B in the supplemental material), evidence for a global displacement of residues 1490 to 1493 of the 16S RNA close to the Str binding site were detected from backbone dihedral trace comparisons between the “open,” the “closed,” and the “Str-bound” structures of the 30S ribosomal subunit of T. thermophilus, supporting that Str fixes the structure in a closed-like conformation. The K47R and K57E mutations independently affect the global electrostatic and steric configuration of this region, pushing out the 16S RNA in such a way that it seems difficult to form a “closed” functional conformation (see Fig. S1C and S1D in the supplemental material). In this scenario, the presence of Str could partially compensate for this, allowing the structure to recover its capability to accommodate the conformational changes required to be functional.
Whatever the reason underlying the SD phenotype, the most surprising result from this work was the dominant character of the rpsL1 allele over its wild-type rpsL parental gene when both are present in the cell. This dominant character was shown when the rpsL1 allele was expressed as a single copy in the chromosome because of the integration of either the suicide pS18a plasmid or its derivative pS18ΔnarC. In the first case, the rpsL1 allele was expressed as part of its natural operon (“cis” in Fig. 2C) in all the colonies checked. On the other hand, the pS18ΔnarC and pMS18 plasmids also confer an SD phenotype, despite the presence of a wild-type rpsL gene expressed from its operon. In consequence, we deduced that the rpsL1 allele produces a dominant SD phenotype, independently of being expressed as part of its operon (pS18a), from another locus in the chromosome (pS18ΔnarC) or from a plasmid (pMS18).
The reason for this dominance cannot be presently understood. In fact, the presence of both wild-type and mutant ribosomes could be expected in a strain carrying the rpsL and the rpsL1 alleles, and consequently, such cells could be expected to grow either with or without Str. As this was not the case, the dominant SD phenotype is necessarily related to some kind of interference during the synthesis of the wild-type ribosomes in the presence of the mutant S12 protein.
On a practical level, the rpsL1 allele has been successfully proven as a positive selection marker alternative to the kat gene for plasmid construction, allowing the expression of genes either on Kan-sensitive or Kan-resistant genetic backgrounds. In both cases, expression of the selected genes as a single copy was possible only in 70 to 80% of the SD transformant colonies. Such a percentage was actually expected because of putative integration into the chromosome through partially homologous regions, which could result in nonfunctional genes, or because of double recombination events that could result in the replacement of the rpsL gene by its rpsL1 allele with the consequent plasmid excision. By contrast, complementation success reached nearly 100% of the SD transformant colonies with the replicative plasmid that does not have to recombine to be maintained in the cell. In conclusion, the rpsL1-based plasmids constitute a new genetic tool for T. thermophilus useful for the complementation of Kan-resistant strains.
However, the most promising application of the rpsL1 allele as a genetic tool is its counterselectable character in Str-free growth medium. This allowed us to develop a fast and straightforward method for the isolation of deletion mutants of a target gene in a single step. The procedure has been applied here to the isolation of ΔnarC mutants in order to compare it with the procedures of Tamakoshi et al. (32) that we used previously to isolate a ΔnarC mutant (38). This method requires the production of a ΔpyrE mutant of the parental strain prior to the use of uracil complementation on mineral medium to isolate Δtarget gene::pyrE mutants by double recombination, which finally must be treated with fluoroorotic acid to counterselect the presence of the pyrE gene. The method that we propose here requires only two TB plates with and without Str and a single-plasmid construct to obtain the required deletion mutant. The new method will greatly facilitate the structure-function studies in this important bacterial model.
Supplementary Material
Acknowledgments
This work has been supported by grants of codes BIO2004-02671, cofunded by the Ministerio de Educación y Ciencia and the European Union, and S0505/PPQ/0344, from Comunidad Autónoma de Madrid. Financial support of Fundación Ramón Areces to CBMSO is acknowledged.
The bioinformatics support of Biomol Informatics S.L. (http://bioinfo.es) and the critical review of this article by Oleg Laptenko and Tony Barsotti are also acknowledged.
Footnotes
Published ahead of print on 29 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 2.Castan, P., O. Zafra, R. Moreno, M. A. de Pedro, C. Valles, F. Cava, E. Caro, H. Schwarz, and J. Berenguer. 2002. The periplasmic space in Thermus thermophilus: evidence from a regulation-defective S-layer mutant overexpressing an alkaline phosphatase. Extremophiles 6:225-232. [DOI] [PubMed] [Google Scholar]
- 3.Cava, F., and J. Berenguer. 2006. Biochemical and regulatory properties of a respiratory island encoded by a conjugative plasmid in the extreme thermophile Thermus thermophilus. Biochem. Soc. Trans. 34:97-100. [DOI] [PubMed] [Google Scholar]
- 4.Cava, F., M. A. de Pedro, H. Schwarz, A. Henne, and J. Berenguer. 2004. Binding to pyruvylated compounds as an ancestral mechanism to anchor the outer envelope in primitive bacteria. Mol. Microbiol. 52:677-690. [DOI] [PubMed] [Google Scholar]
- 5.de Grado, M., P. Castán, and J. Berenguer. 1999. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42:241-245. [DOI] [PubMed] [Google Scholar]
- 6.de Grado, M., I. Lasa, and J. Berenguer. 1998. Characterization of a plasmid replicative origin from an extreme thermophile. FEMS Microbiol. Lett. 165:51-57. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich, A., C. Prust, T. Hartsch, A. Henne, and B. Averhoff. 2002. Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68:745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory, S. T., J. H. Cate, and A. E. Dahlberg. 2001. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus. J. Mol. Biol. 309:333-338. [DOI] [PubMed] [Google Scholar]
- 9.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss- PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Henne, A., H. Bruggemann, C. Raasch, A. Wiezer, T. Hartsch, H. Liesegang, A. Johann, T. Lienard, O. Gohl, R. Martinez-Arias, C. Jacobi, V. Starkuviene, S. Schlenczeck, S. Dencker, R. Huber, H. P. Klenk, W. Kramer, R. Merkl, G. Gottschalk, and H. J. Fritz. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22:547-553. [DOI] [PubMed] [Google Scholar]
- 12.Hooft, R. W., G. Vriend, C. Sander, and E. E. Abola. 1996. Errors in protein structures. Nature 381:272. [DOI] [PubMed] [Google Scholar]
- 13.Koyama, Y., T. Hoshino, N. Tomizuka, and K. Furukawa. 1986. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koyama, Y., S. Okamoto, and K. Furukawa. 1990. Cloning of α- and β-galactosidase genes from an extreme thermophile, Thermus strain T2, and their expression in Thermus thermophilus HB27. Appl. Environ. Microbiol. 56:2251-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laptenko, O., S. S. Kim, J. Lee, M. Starodubtseva, F. Cava, J. Berenguer, X. P. Kong, and S. Borukhov. 2006. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 25:2131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasa, I., J. R. Caston, L. A. Fernandez-Herrero, M. A. de Pedro, and J. Berenguer. 1992. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus HB8. Mol. Microbiol. 6:1555-1564. [DOI] [PubMed] [Google Scholar]
- 17.Lasa, I., M. de Grado, M. A. de Pedro, and J. Berenguer. 1992. Development of Thermus-Escherichia shuttle vectors and their use for expression of the Clostridium thermocellum celA gene in Thermus thermophilus. J. Bacteriol. 174:6424-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malawski, G. A., R. C. Hillig, F. Monteclaro, U. Eberspaecher, A. A. Schmitz, K. Crusius, M. Huber, U. Egner, P. Donner, and B. Muller-Tiemann. 2006. Identifying protein construct variants with increased crystallization propensity-a case study. Protein Sci. 15:2718-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mather, M. W., and J. A. Fee. 1992. Development of plasmid cloning vectors for Thermus thermophilus HB8: expression of a heterologous, plasmid-borne kanamycin nucleotidyltransferase gene. Appl. Environ. Microbiol. 58:421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendieta, J., and F. Gago. 2004. In silico activation of Src tyrosine kinase reveals the molecular basis for intramolecular autophosphorylation. J. Mol. Graph. Model. 23:189-198. [DOI] [PubMed] [Google Scholar]
- 21.Moreno, R. 2004. Clonaje de enzimas termoestables de interés biotecnológico y desarrollo de herramientas para su expresión en Thermus thermophilus. Ph.D. dissertation. Universidad Autónoma de Madrid, Madrid, Spain.
- 22.Moreno, R., O. Zafra, F. Cava, and J. Berenguer. 2003. Development of a gene expression vector for Thermus thermophilus based on the promoter of the respiratory nitrate reductase. Plasmid 49:2-8. [DOI] [PubMed] [Google Scholar]
- 23.Ogle, J. M., F. V. Murphy, M. J. Tarry, and V. Ramakrishnan. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111:721-732. [DOI] [PubMed] [Google Scholar]
- 24.Peitsch, M. C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez-Arcos, S., L. A. Fernandez-Herrero, and J. Berenguer. 1998. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim. Biophys. Acta 1396:215-227. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez-Arcos, S., L. A. Fernandez-Herrero, I. Marin, and J. Berenguer. 1998. Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J. Bacteriol. 180:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramírez-Arcos, S., R. Moreno, O. Zafra, P. Castan, C. Valles, and J. Berenguer. 2000. Two nitrate/nitrite transporters are encoded within the mobilizable plasmid for nitrate respiration of Thermus thermophilus HB8. J. Bacteriol. 182:2179-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selmer, M., C. M. Dunham, F. V. Murphy, A. Weixlbaumer, S. Petry, A. C. Kelley, J. R. Weir, and V. Ramakrishnan. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313:1935-1942. [DOI] [PubMed] [Google Scholar]
- 31.Snell, F. D., and C. T. Snell. 1949. Colorimetric methods of analysis, 3rd ed. Van Nostrand, New York, NY.
- 32.Tamakoshi, M., T. Yaoi, T. Oshima, and A. Yamagishi. 1999. An efficient gene replacement and deletion system for an extreme thermophile, Thermus thermophilus. FEMS Microbiol. Lett. 173:431-437. [DOI] [PubMed] [Google Scholar]
- 33.Timms, A. R., and B. A. Bridges. 1993. Double, independent mutational events in the rpsL gene of Escherichia coli: an example of hypermutability? Mol. Microbiol. 9:335-342. [DOI] [PubMed] [Google Scholar]
- 34.Vassylyev, D. G., S. Sekine, O. Laptenko, J. Lee, M. N. Vassylyeva, S. Borukhov, and S. Yokoyama. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417:712-719. [DOI] [PubMed] [Google Scholar]
- 35.Vieille, C., and G. J. Zeikus. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56, 29. [DOI] [PubMed] [Google Scholar]
- 37.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, Jr., R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407:327-339. [DOI] [PubMed] [Google Scholar]
- 38.Zafra, O. 2004. Implicación de un citocromo c en la biosíntesis de la nitrato reductasa respiratoria de Thermus thermophilus. Ph.D. dissertation. Autónoma de Madrid, Madrid, Spain.
- 39.Zafra, O., F. Cava, F. Blasco, A. Magalon, and J. Berenguer. 2005. Membrane-associated maturation of the heterotetrameric nitrate reductase of Thermus thermophilus. J. Bacteriol. 187:3990-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zafra, O., S. Ramirez, P. Castan, R. Moreno, F. Cava, C. Valles, E. Caro, and J. Berenguer. 2002. A cytochrome c encoded by the nar operon is required for the synthesis of active respiratory nitrate reductase in Thermus thermophilus. FEBS Lett. 523:99-102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.