Abstract
Transcriptome analysis was used to investigate the global stress response of the gram-positive bacterium Bacillus subtilis caused by overproduction of the well-secreted AmyQ α-amylase from Bacillus amyloliquefaciens. Analyses of the control and overproducing strains were carried out at the end of exponential growth and in stationary phase, when protein secretion from B. subtilis is optimal. Among the genes that showed increased expression were htrA and htrB, which are part of the CssRS regulon, which responds to high-level protein secretion and heat stress. The analysis of the transcriptome profiles of a cssS mutant compared to the wild type, under identical secretion stress conditions, revealed several genes with altered transcription in a CssRS-dependent manner, for example, citM, ylxF, yloA, ykoJ, and several genes of the flgB operon. However, high-affinity CssR binding was observed only for htrA, htrB, and, possibly, citM. In addition, the DNA macroarray approach revealed that several genes of the sporulation pathway are downregulated by AmyQ overexpression and that a group of motility-specific (σD-dependent) transcripts were clearly upregulated. Subsequent flow-cytometric analyses demonstrate that, upon overproduction of AmyQ as well as of a nonsecretable variant of the α-amylase, the process of sporulation is severely inhibited. Similar experiments were performed to investigate the expression levels of the hag promoter, a well-established reporter for σD-dependent gene expression. This approach confirmed the observations based on our DNA macroarray analyses and led us to conclude that expression levels of several genes involved in motility are maintained at high levels under all conditions of α-amylase overproduction.
The gram-positive bacterium Bacillus subtilis is capable of secreting large amounts of endogenous proteins into the extracellular medium (41). Therefore, this bacterium and its relatives are often exploited as hosts for the production and secretion of heterologous industrially interesting enzymes. Secretion of heterologous proteins in large quantities has been shown to lead to the unfavorable condition of protein misfolding and subsequent degradation (36). In B. subtilis, accumulation of misfolded proteins at the membrane-cell wall interface is sensed by the CssRS two-component system, which consists of the membrane-embedded sensor kinase CssS and the response regulator CssR (14). This system responds to high-level protein secretion and heat stress by phosphorylation of CssR, which, in turn, activates transcription of the monocistronic htrA and htrB genes (6). The CssRS system has also been shown to regulate the expression of its own operon (6, 20). HtrA and HtrB are membrane-bound serine proteases whose major functions are degradation of misfolded and aggregated proteins. Expression of both proteases is induced upon excessive expression of secretory proteins or a temperature increase (31). Thus, the CssRS regulon forms a quality control and defense system when cells are confronted with secretion stress. The members of the CssRS quality control system become involved at that stage and take action in two possible manners, either by acting as a chaperone aiding refolding or preventing unfolding or by HtrA/B-dependent proteolysis.
Recently, proteomic studies to investigate the global effects of secretion stress have been performed (3). These studies showed that HtrA is also present in the culture supernatant and, in addition to its protease activity, can act as a molecular chaperone. In addition, global effects of secretion stress in B. subtilis were also investigated by transcriptome analysis during the exponential growth phase. This study, however, revealed differential expression of only relatively few genes, including the genes encoding the HtrA and HtrB proteases (15).
Because most of protein secretion in B. subtilis takes place during the onset of the stationary phase of growth (5, 12), we set out to examine the global transcriptional response of B. subtilis under secretion stress conditions at this specific growth phase. Secretion stress was applied by overproducing the well-secreted AmyQ α-amylase from Bacillus amyloliquefaciens. Besides examining secretion stress in wild-type cells, we compared transcriptome profiles of a cssS mutant strain under conditions of high-level AmyQ production. In this work, we have identified and verified putative novel members of the CssRS regulon and we dissected direct and indirect effects of α-amylase overproduction and protein secretion in stationary-phase cultures. Our study reveals that, upon overproduction of a nonsecreted α-amylase, as well as the secreted wild-type variant, the process of sporulation is severely inhibited. In addition, we show that expression levels of genes involved in motility are maintained at high levels under all conditions of α-amylase overproduction.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are shown in Table 1. B. subtilis 168 with an integrated spectinomycin marker (sp) in the pks locus (B. subtilis 168::sp) (10) and B. subtilis 168 cssS::sp (14) were used for transcriptional analyses. B. subtilis 168::sp was used to avoid possible effects on transcriptional profiling due to the presence of the spectinomycin gene alone, as was shown by Hamoen et al., and to be able to compare the data obtained for this strain with those for the cssS insertion mutant. Both strains were transformed with either the pUB110 (empty vector) (6) or the pKTH10 plasmid (pUB110 derivative containing the amyQ α-amylase gene of B. amyloliquefaciens) (32). B. subtilis strains were grown in TY medium (1% tryptone, 0.5% yeast extract, and 1% NaCl) containing the appropriate antibiotics at 37°C and 250 rpm. Antibiotic concentrations were as follows: kanamycin, 10 μg/ml; spectinomycin, 100 μg/ml; chloramphenicol, 5 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| B. subtilis strain or plasmid | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| 168 | trpC2 | 21 |
| 168::sp | 168, pks::sp Spr | 10 |
| IIA-gfp | 168, PspoIIA-gfp Cmr | 39 |
| hag-gfp | 168, Phag-gfp Cmr | This work |
| IIA/E | 168, PspoIIA-gfp Cmr pUB110 | This work |
| IIA/Q | 168, PspoIIA-gfp Cmr pKTHM10 | This work |
| IIA/QAla | 168, PspoIIA-gfp Cmr pKTHM102 | This work |
| hag/E | 168, Phag-gfp Cmr pUB110 | This work |
| hag/Q | 168, Phag-gfp Cmr pKTHM10 | This work |
| hag/QAla | 168, Phag-gfp Cmr pKTHM102 | This work |
| CB100 | sigD::cat Cmr | 27 |
| Sik243 | spo0A::Em Emr | 16 |
| BV2001 | cssS::sp Spr | 14 |
| Plasmids | ||
| pUB110 | Kmr | |
| pKTH10 | Kmr, pUB110 derivative containing the α-amylase gene (amyQ) of B. amyloliquefaciens | 32 |
| pKTHM10 | Kmr, pKTH10 derivative | 44 |
| pKTHM102 | Kmr, pKTHM10 with the Ala-rich signal peptide of AmyQ | 44 |
Strain construction.
To construct plasmid pGFP-hag, a PCR with primer pair hag-F and hag-R (Table 2) was performed using chromosomal DNA of B. subtilis 168 as a template. The amplified fragment containing the promoter region of the hag gene was subsequently cleaved with HindIII and EcoRI and ligated into the corresponding sites of pSG1151, in that way generating a fusion with the gfpmut1 gene (22). B. subtilis strain hag-gfp was obtained by Campbell-type integration of plasmid pGFP-hag into the chromosome of B. subtilis 168. Transformants were selected on TY agar plates containing chloramphenicol (5 μg/ml), after overnight incubation at 37°C. Correct integration was verified by PCR (data not shown).
TABLE 2.
Primer sequences
| Primer | Sequence (5′→3′) | Target fragment | Restriction site |
|---|---|---|---|
| hag-F | GGGATCAAGTGAAGCTTGGAATTGACG | hag | HindIII |
| hag-R | CGGAATTCCATTTCCTCTCCTCCTTGAATATGTTGTTAAGGCACGTCC | hag | EcoRI |
| QE60-cssR-F | CATGCCATGGCATACACCATTTATCTAGTTGAAGA | cssR | NcoI |
| QE60-cssR-R | CGGGATCCTGATGACATCATCCTGTAGCCGAAACCGTA | cssR | BamHI |
| QE30-cssR-F | CGGGATCCTTGTCATACACCATTTATCTAGTTGAAG | cssR | BamHI |
| QE30-cssR-R | CCGAGCTCTTATCATGATGACATCATCCTGTAGCCGAAA | cssR | SacI |
| htrA-EMSA-F | AACGGATCAGCCGATACGTT | PhtrA | None |
| htrA-EMSA-R | TCATCACGATAGTTATCCAT | PhtrA | None |
| htrB-EMSA-F | CGTCAGCAGTTCATTCAG | PhtrB | None |
| htrB-EMSA-R | CCATCACGTCGATAATCC | PhtrB | None |
| citM-EMSA-F | AGGTCACCTCCTCACCTGAA | PcitM | None |
| citM-EMSA-R | CATGAGAAAGCCTAAGATTGCTAAC | PcitM | None |
| flgB-EMSA-F | TGTATCGTTCCAGAAAATAAGC | PflgB | None |
| flgB-EMSA-R | TCAGGGCGAGAAATGTAGTTC | PflgB | None |
| ykoJ-EMSA-F | TGCCGATCAAATCAGCAG | PykoJ | None |
| ykoJ-EMSA-R | TTGTGAGCCCCTCCTTTGT | PykoJ | None |
| yloA-EMSA-F | CCAGAAGCACAAGCACCATA | PyloA | None |
| yloA-EMSA-R | GTATGTAAACATGCCATCAAACG | PyloA | None |
| pK-F | AATCTATCGACATATCCTGCAA | PcomK | None |
| K-FP-R | GGAATTCTTGCGCCGTTCACTTCATAC | PcomK | None |
RNA isolation, preparation of labeled cDNA, and hybridization.
Cells were grown overnight in 10 ml of TY medium with kanamycin (10 μg/ml) and then diluted to an optical density at 600 nm of 0.1 in 40 ml of TY medium containing kanamycin. Samples for transcriptome analyses were collected at the late-exponential-growth stage (1 hour before the transition point) and 3 h after entry in the stationary-growth phase. Three independent cultures of each strain were used, and cells were sampled for macroarray experiments. RNA was isolated by spinning down cells from 4 ml of culture, with subsequent cell disruption, phenol-chloroform extraction, and then RNA purification with a High Pure RNA isolation kit from Roche, as described previously (10). RNA was eluted with 50 μl of elution buffer and subsequently quantified with GeneQuant (Amersham), and the RNA integrity was checked on agarose gels. Four micrograms of RNA as a template and 1 pmol of open reading frame-specific primers (Eurogentec) were used for the reverse transcriptase reaction with SuperscriptII (Gibco BRL). The detailed protocols of reverse transcription and purification of 33P-labeled cDNA are outlined by Hamoen et al. (10). Labeled cDNA was hybridized to B. subtilis Panorama arrays (Sigma-Genosys), according to the manufacturer's instructions. The membranes were exposed to Cyclone phosphorimager screens (Packard) for approximately 2.5 days, and signals were quantified with Array-Pro Analyzer 4.5 (Media Cybernetics).
Data analysis.
Duplicate spots were averaged in Array-Pro software (Media Cybernetics, Inc.), and the signal was normalized after background subtraction by calculation of the percentage of total signal per gene using Microsoft Excel. Outstandingly high signals (for example, whole genomic DNA control spots, which act as a positive control and always show very strong signals) were excluded from calculations of the total signal. Statistical significance of the obtained gene expression ratios was assessed by the Cyber-T program (23). Genes with a Cyber-T P value lower than 0.05 are discussed in this paper. Raw data and images are available at http://molgen.biol.rug.nl/publication/secstress_data/.
SDS-PAGE and Western blotting.
To detect the precursor and mature forms of AmyQ in cells and growth medium, 1 ml of culture was centrifuged and the cell pellet was washed once with fresh medium. After separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transferred to Immobilon polyvinylidene difluoride membranes. The visualization of the AmyQ protein was carried out with specific anti-AmyQ antibodies, horseradish peroxidase-anti-rabbit immunoglobulin G conjugates, and ECL immunoblotting detection reagents (Amersham).
Amylase activity assay.
Samples from culture supernatants were diluted 100-fold and were subjected to amylase activity quantification with the EnzCheck Ultra amylase assay kit (Molecular Probes), according to the manufacturer's instructions. Assays were carried out in 96-well microtiter plates in a 100-μl total volume using 50 μl diluted supernatants. Reaction mixtures contained 200 μg/ml DQ starch substrate and were carried out in 100 mM MOPS (morpholinepropanesulfonic acid) at pH 6.9. The degradation of the substrate by amylase yields highly fluorescent fragments, and fluorescence was monitored at room temperature every 5 min for 1 h with the microplate reader TECAN (GENios) using standard fluorescein filters. Samples were taken from three independent experimental replicates.
Flow cytometry.
Cells were diluted 100-fold in 0.2 μM of filtered minimal medium (2) and directly measured on an Epics XL-MCL flow cytometer (Beckman Coulter, Mijdrecht, The Netherlands), operating an argon laser (488 nm) as essentially described by Veening et al. (39). For each sample, at least 20,000 cells were analyzed. Data containing the green fluorescent signals were collected with a fluorescein isothiocyanate filter, and the photomultiplier voltage was set between 700 and 800 V. Data were captured using System II software (Beckman Coulter) and further analyzed using WinMDI 2.8 software (http://facs.scripps.edu/software.html). Figures were prepared for publication using WinMDI 2.8 and Corel Graphics Suite 11. To distinguish background fluorescence from green fluorescent protein-specific fluorescence, the parental strain B. subtilis 168 was also analyzed with each flow-cytometric experiment.
Construction and purification of six-His-tagged CssR.
Plasmids encoding CssR protein with a six-His tag were constructed as follows. For C-terminally tagged proteins, the cssR open reading frame was amplified by PCR using primers QE60-cssR-F and QE60-cssR-R (Table 2) and Phusion polymerase (Finnzymes). Chromosomal DNA of B. subtilis strain 168 was used as a template. The product of this reaction was digested with NcoI and BamHI and cloned into the similarly digested pQE30 plasmid (QIAGEN), yielding plasmid pQE60-cssR.
To obtain an N-terminally tagged CssR protein, the cssR open reading frame was amplified by PCR using primers QE30-cssR-F and QE30-cssR-R. The product of this reaction was digested with BamHI and SacI and cloned into the similarly digested pQE30 plasmid (QIAGEN), yielding plasmid pQE30-cssR.
To induce expression, Escherichia coli JM109 carrying pQE60-cssR or pQE30-cssR was diluted 1:100 to fresh medium from an overnight culture. At an optical density at 600 nm of 0.6, expression was induced by the addition of 1 mM of IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested after an additional 2 hours of growth, after which protein was purified as described elsewhere (37a). The purity of the protein was estimated to be >95% on the basis of Coomassie-stained SDS-PAGE gels. After purification, the protein was dialyzed against dialysis buffer (20 mM Tris-HCl, pH 8, 1 mM EDTA, 10 mM MgCl2, 0.2 M NaCl, 1 mM β-mercaptoethanol, 5 mM imidazole, 7% glycerol). The concentration of protein was determined on the basis of A280 using a Nanodrop machine (Nanodrop Technologies), with 0.1% (1 g/liter), giving a value of 1.272 (ExPASy Protparam Tool; http://www.expasy.org/tools/protparam.html).
Electrophoretic mobility shift assays (EMSAs).
To establish direct binding of CssR to putative target promoters, gel shift experiments were carried out as described before (1). In our experiments, we observed no significant difference in affinity between C- and N-terminally His-tagged proteins. Subsequent experiments were therefore carried out using only N-terminally His-tagged proteins. The fragments obtained corresponded to base pairs −339 to +19 (htrA), −335 to +19 (htrB), −333 to +26 (citM), −191 to +25 (flgB), −320 to 0 (ykoJ), and −312 to +32 (yloA) compared to the start of the open reading frame, as annotated in SubtiList R16.1 (http://genolist.pasteur.fr/SubtiList/). As a negative control the promoter of comK (−201 to +47) was amplified. All primer sequences are available in Table 2.
Macroarray data accession number.
The array data have been deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE8014.
RESULTS
Transcriptomics of secretion stress in late-exponential and stationary-phase cells.
AmyQ is the α-amylase from Bacillus amyloliquefaciens and contains a Sec-type signal sequence. This protein has been shown to trigger a specific secretion stress response when overexpressed in B. subtilis (14). DNA macroarray analysis was used to compare transcriptional profiles of B. subtilis 168::sp containing plasmid pUB110 (empty vector) with those of B. subtilis 168::sp containing plasmid pKTH10 (AmyQ overexpression) grown in TY broth. Samples were taken for transcriptome analyses during late exponential (1 hour before the transition point) and stationary (3 h after the transition point) phases of growth. Amylase-dependent starch degradation on TY-agar plates and Western blot analysis of the growth media from the cells used for the macroarray experiment verified the expected AmyQ overproduction for the pKTH10-containing cells (data not shown).
Differentially expressed genes in the late exponential phase are listed in Table S1 in the supplemental material. As expected, the most highly upregulated genes are htrA and htrB, encoding the Htr-like proteases. Also, moderately elevated transcription levels of the cssRS operon, encoding the secretion stress response regulator (CssR) and histidine kinase (CssS), were observed upon AmyQ overproduction, indicating that cells were clearly subjected to secretion stress. In addition, a stimulatory effect was observed for the transcripts of the ribosomal protein RpsB and the genes for the general stress proteins GroEL and DnaK, suggesting that, next to secretion stress, a cytosolic stress, most likely resulting from the presence of misfolded and aggregated proteins, is induced. Moreover, two members of the peroxide stimulon, ahpF and mrgA, next to the genes involved in cell wall homeostasis (dltA, acpA, and accC), showed elevated expression levels. Other genes with known function that were induced upon AmyQ overproduction include pycA and citB of the tricarboxylic acid cycle and sodA and trxA, which play an important role in maintaining the redox balance of the cell. The secretion stress directly or indirectly caused increased mRNA levels of four genes with an unknown or predicted function, including yvfW, encoding a putative iron-sulfur-binding protein, and yvqH, which has similarity to the gene encoding E. coli phage shock protein A, which was shown to be induced under anaerobic growth (42) and ethanol stress conditions (33). Also ykoJ and ydbK showed increased transcript levels. The former codes for a conserved protein of unknown function, and the latter encodes a putative ABC transport system permease protein. Besides the upregulation of a number of genes, some of which have been mentioned above, more than 40 genes were significantly downregulated upon AmyQ overproduction. These downregulated genes are classified within different functional categories including, among others, genes involved in the metabolism of lipids and sporulation and genes encoding transport/binding proteins (see Table S1 in the supplemental material).
Under natural conditions, most of the secretion takes place during the stationary-growth phase. Therefore, transcriptome analyses were also performed on AmyQ-overproducing and nonoverproducing cells in this phase. Again, a strong effect on the transcriptional levels of htrA, htrB, cssS, and cssR was observed (see Table S2 in the supplemental material). Thus, secretion stress also occurs during later stages of growth. From the transcriptional profiling in this growth phase, two main novel findings can be deduced: several genes involved in motility and chemotaxis, including sigD, had increased mRNA levels, in contrast to a group of sporulation-related genes, which appeared to have decreased transcription levels (Fig. 1). The list of downregulated sporulation-related genes is more extensive, although for some of them the obtained expression ratios were slightly below the statistical cutoffs. The protein phosphatase encoded by prpC (yloO) was activated, and the product of this gene, together with the PrkC protein kinase, has been implicated in the sporulation process (25). Two sporulation genes, spo0JA and spo0E, had increased transcription levels under secretion stress, but, interestingly, both of them affect sporulation in a negative manner (34). Expression of several other genes seems also to be affected by AmyQ overproduction. The products of these genes are associated with different cellular processes such as metabolism of amino acids, lipids, and carbohydrates and transport and binding activity (see Table S2 in the supplemental material). For the genes with unknown function, yloA and ykoJ showed the strongest upregulation (11.2-fold and 5.3-fold, respectively). Both genes are well conserved among bacterial species. YloA bears similarity to a fibronectin-binding protein as well as to RNA-binding proteins that show homology to the eukaryotic snRNP's. YkoJ was also induced at the earlier time point. This protein contains a putative signal peptide and two PepSY domains, which, it has been suggested, have peptidase-regulatory activity in proximity to the cell wall (43).
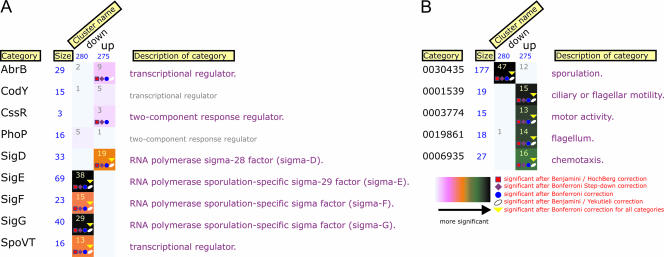
FIG. 1.
FIVA analysis of interaction and GO (gene ontology) categories of the transcriptome data. Genes from the DNA macroarray data set were divided into up- and downregulated clusters [comparison of gene expression ratios between B. subtilis 168(pKTH10)/B. subtilis 168]. “Interaction” (A) and GO (B) categories are shown. The size of each cluster is presented in blue underneath the cluster name. Numbers in rectangles represent how many genes were up- or downregulated per cluster in each category. The colors of square boxes depict the significance of gene enrichment per category as defined in the key. Detailed information on significance analyses is available in Blom et al. (4).
Verification of putative members of the CssRS regulon.
As secretion stress clearly affected the expression of the known members of the CssRS regulon, we attempted to identify novel members of this regulon, with a focus on the upregulated genes resulting from AmyQ overexpression. For this purpose we compared the transcriptome profiles of B. subtilis 168::sp (wild type) and 168 cssS::sp (ΔcssS), both containing the pKTH10 plasmid. Previous research showed that htrA and htrB are regulated by the CssRS system and that disruption of cssS reduces htrA and htrB transcription (6, 14). Our transcriptome data confirmed these observations since both genes showed decreased mRNA levels in the cssS mutant strain. Besides htrA and htrB, similar expression patterns from our macroarray approach were observed for citM (secondary Mg-citrate transporter), the flgB operon (flagellar synthesis and chemotaxis), ylxF, yloA, and ykoJ.
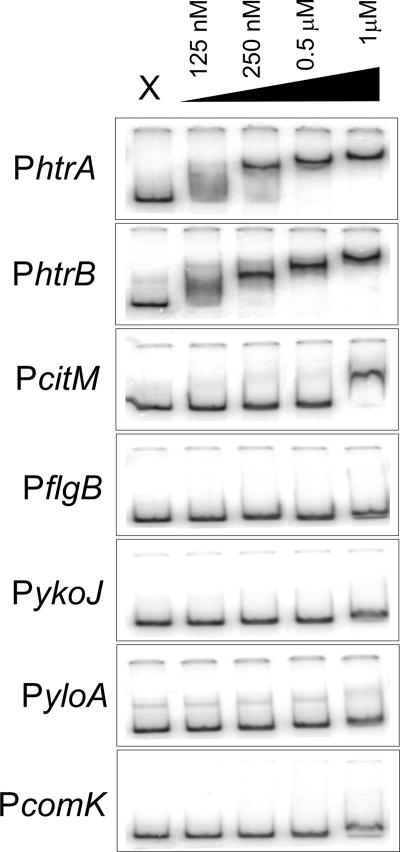
Our analysis revealed several genes with altered transcription levels in response to overproduction of AmyQ, in a CssR-dependent manner. Though CssR is a two-component regulator, it is unknown if the observed effects are due to direct binding of CssR to the target promoters. In fact, the binding of CssR to target promoters has not been reported so far. Therefore, several candidate genes were selected and EMSAs were carried out using purified N-terminally His-tagged CssR protein. Phosphorylation of nontagged CssR by acetyl-phosphate enhances the affinity of the protein for DNA two- to fourfold, depending on the promoter fragment used (E. Darmon, unpublished observations). Figure 2 shows that unphosphorylated, six-histidine-tagged CssR has a high affinity for the DNA fragments containing the promoters of the htrA and htrB genes, with an apparent equilibrium dissociation constant (KD) of ∼125 nM; therefore, the tagged protein likely mimics the phosphorylated form of the protein. These observations are in good agreement with previous reporter studies that demonstrate the CssR-dependent induction of these genes upon protein overproduction (6, 14). In contrast, unphosphorylated, His-tagged CssR does not to appear to bind strongly to any of the other fragments, with the possible exception of citM, for which consistently a reduced mobility was observed at 1 μM of CssR protein. We conclude that, for the fragments used, only htrA and htrB contain a high-affinity CssR-binding site. As for the other genes, the observed transcriptional changes most likely result from secondary effects.
FIG. 2.
Binding of CssR-His6 to the indicated promoter regions. Gel retardation reactions were performed with radiolabeled DNA fragments prepared by PCR and end labeled with 32P. Promoter regions were incubated with increasing concentrations of purified CssR-His6 (see Materials and Methods), ranging from 0.125 μM to 1 μM of the protein. In each panel lane X corresponds to the reaction with no protein added.
Sporulation and motility are affected by AmyQ overproduction and secretion.
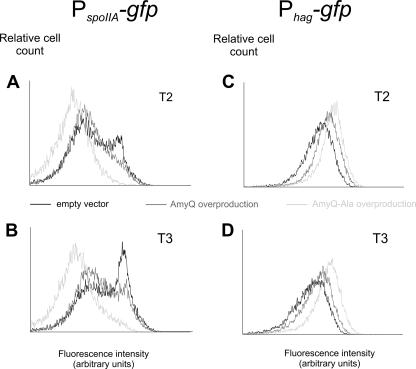
Several genes that were significantly downregulated by AmyQ overexpression belong to the sporulation pathway. This suggests that secretion stress results in inhibition of spore formation. Interestingly, overexpression of a cytoplasmic protein did not affect early sporulation gene expression (18), indicating that secretion stress rather than overproduction stress might be the cause for the observed downregulation. To examine whether the observed downregulation of the sporulation pathway in our transcriptome analysis by AmyQ overproduction was specifically caused by secretion stress or was a result of an indirect effect caused by AmyQ overproduction, we made use of a sporulation-specific reporter strain carrying the spoIIA promoter in front of the gene encoding green fluorescent protein. The spoIIA promoter is directly activated by the key sporulation regulator, Spo0A, and was shown to be a good reporter for cells that initiate sporulation (39). Plasmids pKTHM10 (AmyQ overexpression), pKTHM102 (AmyQ-Ala overproduction), and pUB110 (empty vector) were introduced in the indicator strain, and cells were analyzed for their expression levels by flow cytometry. The pKTHM102 vector is a derivative of pKTHM10 (which is similar to pKTH10) and encodes the AmyQ protein with an Ala-rich signal sequence that renders it inactive in translocation across the cytoplasmic membrane (44). Importantly, this AmyQ variant does not evoke a secretion stress response (40). Strains were grown in TY medium, and samples were taken at hourly intervals and examined by flow cytometry. As depicted in Fig. 3, strain IIA/Q (PspoIIA-gfp pKTHM10), harboring an AmyQ overexpression vector with the wild-type signal peptide, was slightly delayed in the activation of the spoIIA operon (Fig. 3). A more pronounced effect was observed in cells containing the pKTHM102 plasmid (strain IIA/QAla). Overall, these results show that both secretion stress and AmyQ overproduction (IIA/QAla) in particular inhibit the process of sporulation. In addition, this inhibition already occurs at the earliest stages in spore formation, at the level of phosphorylation of Spo0A, as shown by the single-cell analyses.
FIG. 3.
Single-cell analysis of sporulation- and motility-specific gene expression. Strains were grown in TY medium at 37°C with shaking. Samples for flow-cytometric analysis were taken every hour during growth. Two time points are represented, T2 and T3, which correspond to 2 and 3 hours after the entry into stationary phase, respectively. The numbers of cells measured are indicated on the y axis, and their relative fluorescence levels are indicated on the x axis. (A and B) Strains IIA/E (PspoIIA-gfp), IIA/Q (AmyQ overproduction, dark gray line), and IIA/QAla (AmyQ-Ala overproduction). (C and D) Strains hag/E (Phag-gfp), hag/Q (AmyQ overproduction), and hag/QAla (AmyQ-Ala overproduction).
Another adaptive response that B. subtilis utilizes is motility. In this case, cells physically escape from adverse circumstances towards more promising ones to increase their survival chances. Regulation of synthesis of flagellum and motility gene expression is known to be orchestrated by alternative sigma factor σD (11, 27). As described above, we observed a strong upregulation of several genes involved in motility upon AmyQ overproduction. To investigate the increase of flagellar gene expression in B. subtilis as a result of protein overproduction, we constructed a strain in which the σD-dependent hag promoter was fused to the gfp gene (hag-gfp). The hag gene codes for the flagellin protein, and it was shown before that this gene can be used as a good reporter for studying environmental effects on σD-dependent gene expression (28). The AmyQ overproduction plasmids were introduced in this reporter strain, resulting in strains hag/Q (Phag-gfp pKTHM10), hag/QAla ((Phag-gfp pKTHM102), and hag/E (Phag-gfp pUB110). Cells were grown in TY medium, and samples were collected at hourly intervals for flow-cytometric analyses. As shown in Fig. 3, the promoter activity of the flagellin gene is the highest for the strain overproducing AmyQ with the Ala-rich signal peptide. This again suggests that the enhanced expression of motility genes is not a direct effect of secretion stress itself but rather results from AmyQ overproduction.
Effects of sigD and spo0A deletions on AmyQ overproduction and secretion.
The results of the transcriptome analyses have shown that two cellular adaptive processes were clearly affected under secretion stress conditions. This raised the question whether disruption of these processes would affect heterologous protein secretion by B. subtilis.
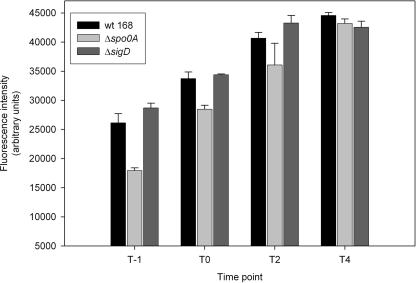
It has been previously investigated how the yields of protein production can be improved by modification of the components engaged in the late stages of protein secretion, especially the ones influencing cell wall-associated protease activity or cell wall composition (41). To further study whether sporulation and/or motility affect protein secretion in B. subtilis, sigD (σD) and spo0A mutations were introduced into a strain containing the pKTHM10 plasmid (AmyQ overproduction). Mutants of spo0A are defective in sporulation since this transcriptional regulator plays a central role in the initiation of this developmental process (34). The null mutant of σD is nonmotile and shows reduced levels of autolysins (27). The levels of production of secreted AmyQ in both mutant strains and the parental strain were determined at different time points of growth by means of the EnzCheck Ultra amylase assay kit as described in Materials and Methods. In all cases, the amount of active amylase in the culture medium increased from exponential phase (1 h after entry into stationary phase [T1]) and reached a maximum in the late stationary phase (T4) (Fig. 4). However, a clear delay was observed in the spo0A mutant, as this strain secreted significantly less amylase than the wild-type strain during exponential phase. In the late stationary phase the difference between the two strains became minimal. More interestingly, under our experimental setup, the sigD mutant reached slightly higher levels of the active enzyme at earlier growth phases compared to the wild-type strain. Again, 4 hours after entry into stationary phase the differences were negligible.
FIG. 4.
Activity of secreted amylase from B. subtilis 168 wild-type (wt) and sporulation- and motility-deficient strains. Amylase activity was quantified in the culture medium of the wild-type strain, the spo0A mutant, and the sigD mutant. All strains were transformed with the pKTHM10 plasmid (AmyQ overproduction), and levels of activity of secreted AmyQ were determined at different time points of growth by using the EnzCheck Ultra amylase assay kit as described in Materials and Methods. T0 corresponds to the transition phase of growth. Error bars represent standard errors over three independent biological replicates.
These results indicate that, when the cells are devoid of the possibility of initiating sporulation or entering the motile phase, the efficiency and timing of protein secretion by B. subtilis are modulated.
DISCUSSION
Protein secretion, like competence development and sporulation, is one of the postexponential processes that B. subtilis employs as a response to altering growth conditions. The secretion activity is rather low during exponential growth and increases substantially at the onset of stationary phase (35). It has been shown before that several components of the Sec machinery in B. subtilis reach maximum expression at the end of exponential growth (12) or in the early postexponential phase (5). Based on these facts, we set out to study the global cellular response of B. subtilis to α-amylase (AmyQ) overproduction and secretion stress during late exponential and stationary phases. In all our macroarray analyses, we observed upregulation of the known targets of the secretion stress regulon, htrA and htrB (Fig. 1; see Tables S1 and S2 in the supplemental material). This validates the quality of the obtained transcriptome data and agrees well with previously published results (6, 14). The comparison of the transcriptional patterns of the control strain and the AmyQ-overproducing strain at the late exponential phase of growth disclosed a rather modest response of B. subtilis to the applied stress. This result is in a good accordance with the studies of Hyyrylaïnen et al., where the induction of only seven genes upon high-level AmyQ secretion was found (15). However, our transcriptome analysis at the late exponential phase revealed the activation of a higher number of genes, which most likely results from differences in the experimental approaches, especially the sampling at the later phase of growth in our case (i.e., late exponential). In addition to the stimulation of the components of the CssRS quality control system, several other genes were weakly induced. Most of the gene products are involved in the stress response evoked by protein misfolding/aggregation events in the cytoplasm, detoxification, and fatty acid metabolism, indicating that cells sensed and tried to counteract the adverse conditions.
Our data revealed the CssRS-dependent regulation of several genes in response to AmyQ overproduction. However, for only two of the (putative) targets (htrA and htrB) could binding of the purified CssR protein to the DNA be demonstrated, indicating that CssR directly binds to and regulates expression of these operons. How can the apparent CssR-dependent expression of these genes then be explained? First, it is possible that some of the promoter regions tested by EMSA require additional cofactors in vivo for a proper binding of CssR. Second, phosphorylated CssR might bind different DNA sequences and with other affinities than those for unphosphorylated CssR. Since our EMSAs were carried out with unphosphorylated protein, we cannot exclude the possibility that in vivo some of the target genes are more strictly dependent on the phosphorylated form of CssR. Another explanation is that the observed effects are indirect and independent of CssR, for instance, via increased levels of HtrA, the activity of which was postulated to influence the regulatory effects of a two-component system in Streptococcus pneumoniae (37). Finally, data obtained from a PykoJ-gfp fusion at its native locus indicate that upregulation of this gene in response to protein overproduction is indeed CssR dependent but that promoter activity could not be demonstrated in an area of 500 bp upstream of the ykoJ open reading frame (30; R. Nijland, unpublished observations). The promoters of htrA and htrB are remarkably similar, making the in silico identification of a CssR binding motif very difficult. However, a Gibbs sampling method (38) identifies a CATTTTTATC motif that is present in the 300-bp region upstream of both genes. In addition, close matches to this motif (GATTTTTTTC and CATTTTTTTC) can be identified 300 bp upstream of citM. The importance of this putative binding sequence remains to be established in future investigations.
The most striking outcome of the transcriptome analysis of stationary-phase cells was the influence of protein overproduction on the processes of sporulation and motility. The effect on sporulation was much more pronounced at the later stages of growth, and reduced expression levels of many SigE, SigF, and SigG regulon members were observed in AmyQ-overproducing cells (Fig. 1). Interestingly, the level of spo0E transcript (phosphatase of Spo0A∼P) in the AmyQ-overexpressing cells is higher than in the wild type (pUB110), because of which the Spo0A∼P pool might be drained. The inhibition of sporulation was confirmed by gfp-reporter experiments, and these analyses showed that sporulation is already blocked at the earliest stages (at the level of Spo0A∼P) of amylase overproduction (Fig. 3). The differences between secretion stress and AmyQ production stress were examined using a nontranslocated AmyQ variant. This approach led us to conclude that the impaired-sporulation phenotype is most likely not an exclusive and direct effect of secretion stress but is rather caused by high-level cellular protein production and accumulation, possibly at the membrane. In fact, overproduction stress seems to inhibit sporulation more efficiently than secretion-specific stress (Fig. 3). It is plausible that the unprocessed AmyQ-Ala variant is still targeted to the Sec translocon, thereby leading to a kind of membrane congestion stress. This type of stress would account for the observed effect on sporulation and motility and would be less severe in the case of the wild-type AmyQ variant, which is translocated across the cytoplasmic membrane.
Sporulation and protein secretion are multistep and energy-consuming processes, and since protein production and secretion take place earlier in the life cycle of B. subtilis than endospore formation, they most likely inhibit the latter due to energy constraints. Our data suggest that B. subtilis cells sense a variety of environmental and intracellular signals under protein overproduction stress, which leads to the decision that commencing energy-consuming spore development is not appropriate since it is highly unlikely that this process can be completed successfully.
Another survival strategy which B. subtilis employs as an adaptive response under unfavorable environmental conditions is motility. Remarkably, our transcriptome profiling revealed that a substantial part of the motility regulon was induced by high-level protein production and secretion. The alternative sigma factor σD orchestrates the transcription of many genes whose products are involved in flagellar assembly, motility, chemotaxis, and autolysis (11, 27). In a complex medium the amount of the σD protein increases during growth, reaching the maximum level at the transition point (28, 29). By single-cell flow-cytometric analysis, we show that AmyQ overproduction prolongs the motile phase of B. subtilis (Fig. 3). One of the explanations for this prolonged motile phase includes the putative replacement and competition of alternative sigma factors for core RNA polymerase (RNAP) during stationary phase, a phenomenon well documented for both E. coli (8, 26) and B. subtilis (13, 17, 24). The consecutive sporulation-specific sigma factors conduct developmental events during endospore formation (7). Since sporulation is blocked upon protein overproduction (Fig. 3), SigD does not have to compete with sporulation-specific sigma factors for free RNAP. However, it has been demonstrated that RNAP is present in excess in B. subtilis cells, and the engagement of anti-sigma factors or other mechanisms, which would give rise to deactivation of σA, could be conceived (9). An extended expression of the motility regulon can provide cells with a rescue machinery under the deficiency of sporulation, but, on the other hand, the members of yet uncharacterized σD-regulated genes could be also involved in helping cells to survive stress circumstances.
Our transcriptome results, combined with the single-cell analyses, predicted that both initiation of sporulation and motility play an important role in combating protein secretion stress. Indeed, when either spo0A or sigD was mutated, protein secretion was significantly affected (Fig. 4). It is demonstrated that the presence of a functional spo0A is required for efficient protein secretion, while the removal of sigD moderately but consistently increases secretion (Fig. 4). It has been shown before that a sigD mutation causes an increased extracellular accumulation of an artificial cell wall-binding lipase by an unknown mechanism (19). Likewise in our experiments, the secretion of enzymatically active AmyQ turned out to be enhanced in the sigD mutant, especially in earlier phases of growth, which renders this mutant strain a good host for efficient extracellular protein production (Fig. 4).
Supplementary Material
Acknowledgments
We thank Holger Jahr and Jan Jongbloed for their contributions in the initial stages of the project, Elise Darmon and Sacha van Hijum for helpful discussions, and Rutger Brouwer for the preparation of the website with supplementary materials.
The work of A.T.L. and G.B. was supported by grant IGE01018 from the Dutch Ministry of Economic Affairs. W.K.S. and J.-W.V. were supported by grants 811.35.002 and ABC-5587 from The Netherlands Organization of Scientific Research, Earth and Lifesciences (NWO-ALW) and Technology Foundation (NWO-STW), respectively. Work performed by E.J.B. was supported by grant 050.50.206 from the NWO-BMI.
Footnotes
Published ahead of print on 22 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Albano, M., W. K. Smits, L. T. Ho, B. Kraigher, I. Mandic-Mulec, O. P. Kuipers, and D. Dubnau. 2005. The Rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions. J. Bacteriol. 187:2010-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., E. Darmon, D. Noone, J.-W. Veening, H. Westers, S. Bron, O. P. Kuipers, K. M. Devine, M. Hecker, and J. M. van Dijl. 2003. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49:143-156. [DOI] [PubMed] [Google Scholar]
- 4.Blom, E. J., D. W. Bosman, S. A. van Hijum, R. Breitling, L. Tijsma, R. Silvis, J. B. Roerdink, and O. P. Kuipers. 2007. FIVA: Functional Information Viewer and Analyzer extracting biological knowledge from transcriptome data of prokaryotes. Bioinformatics 23:1161-1163. [DOI] [PubMed] [Google Scholar]
- 5.Bolhuis, A., C. P. Broekhuizen, A. Sorokin, M. L. van Roosmalen, G. Venema, S. Bron, W. J. Quax, and J. M. van Dijl. 1998. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J. Biol. Chem. 273:21217-21224. [DOI] [PubMed] [Google Scholar]
- 6.Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. van Dijl. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 9.Fujita, M. 2000. Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 5:79-88. [DOI] [PubMed] [Google Scholar]
- 10.Hamoen, L. W., W. K. Smits, A. de Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmann, J. D., L. M. Marquez, and M. J. Chamberlin. 1988. Cloning, sequencing, and disruption of the Bacillus subtilis σ28 gene. J. Bacteriol. 170:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herbort, M., M. Klein, E. H. Manting, A. J. Driessen, and R. Freudl. 1999. Temporal expression of the Bacillus subtilis secA gene, encoding a central component of the preprotein translocase. J. Bacteriol. 181:493-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicks, K. A., and A. D. Grossman. 1996. Altering the level and regulation of the major sigma subunit of RNA polymerase affects gene expression and development in Bacillus subtilis. Mol. Microbiol. 20:201-212. [DOI] [PubMed] [Google Scholar]
- 14.Hyyrylaïnen, H. L., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Pragai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 15.Hyyrylaïnen, H. L., M. Sarvas, and V. P. Kontinen. 2005. Transcriptome analysis of the secretion stress response of Bacillus subtilis. Appl. Microbiol. Biotechnol. 67:389-396. [DOI] [PubMed] [Google Scholar]
- 16.Ireton, K., D. Z. Rudner, K. J. Siranosian, and A. D. Grossman. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 7:283-294. [DOI] [PubMed] [Google Scholar]
- 17.Ju, J., T. Mitchell, H. Peters III, and W. G. Haldenwang. 1999. Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol. 181:4969-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurgen, B., R. Hanschke, M. Sarvas, M. Hecker, and T. Schweder. 2001. Proteome and transcriptome based analysis of Bacillus subtilis cells overproducing an insoluble heterologous protein. Appl. Microbiol. Biotechnol. 55:326-332. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, G., J. Toida, T. Akamatsu, H. Yamamoto, T. Shida, and J. Sekiguchi. 2000. Accumulation of an artificial cell wall-binding lipase by Bacillus subtilis wprA and/or sigD mutants. FEMS Microbiol. Lett. 188:165-169. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, K., M. Ogura, H. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183:7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, and. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, P. J., and A. L. Marston. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101-110. [DOI] [PubMed] [Google Scholar]
- 23.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 24.Lord, M., D. Barilla, and M. D. Yudkin. 1999. Replacement of vegetative σA by sporulation-specific σF as a component of the RNA polymerase holoenzyme in sporulating Bacillus subtilis. J. Bacteriol. 181:2346-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madec, E., A. Laszkiewicz, A. Iwanicki, M. Obuchowski, and S. Seror. 2002. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46:571-586. [DOI] [PubMed] [Google Scholar]
- 26.Malik, S., K. Zalenskaya, and A. Goldfarb. 1987. Competition between sigma factors for core RNA polymerase. Nucleic Acids Res. 15:8521-8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marquez, L. M., J. D. Helmann, E. Ferrari, H. M. Parker, G. W. Ordal, and M. J. Chamberlin. 1990. Studies of σD-dependent functions in Bacillus subtilis. J. Bacteriol. 172:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirel, D. B., and M. J. Chamberlin. 1989. The Bacillus subtilis flagellin gene (hag) is transcribed by the σ28 form of RNA polymerase. J. Bacteriol. 171:3095-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirel, D. B., W. F. Estacio, M. Mathieu, E. Olmsted, J. Ramirez, and L. M. Marquez-Magana. 2000. Environmental regulation of Bacillus subtilis σD-dependent gene expression. J. Bacteriol. 182:3055-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nijland, R., R. Heerlien, L. W. Hamoen, and O. P. Kuipers. 2007. Changing a single amino acid in Clostridium perfringens β-toxin affects the efficiency of heterologous secretion by Bacillus subtilis. Appl. Environ. Microbiol. 73:2390-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noone, D., A. Howell, and K. M. Devine. 2000. Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated. J. Bacteriol. 182:1592-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palva, I. 1982. Molecular cloning of alpha-amylase gene from Bacillus amyloliquefaciens and its expression in B. subtilis. Gene 19:81-87. [DOI] [PubMed] [Google Scholar]
- 33.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579-586. [DOI] [PubMed] [Google Scholar]
- 35.Priest, F. G. 1977. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol. Rev. 41:711-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarvas, M., C. R. Harwood, S. Bron, and J. M. van Dijl. 2004. Post-translocational folding of secretory proteins in gram-positive bacteria. Biochim. Biophys. Acta 1694:311-327. [DOI] [PubMed] [Google Scholar]
- 37.Sebert, M. E., K. P. Patel, M. Plotnick, and J. N. Weiser. 2005. Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J. Bacteriol. 187:3969-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Smits, W. K., T. T. Hoa, L. W. Hamoen, O. P. Kuipers, and D. Dubnau. 2007. Antirepression as a second mechanism of transcriptional activation by a minor groove binding protein. Mol. Microbiol. 64:368-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thijs, G., K. Marchal, M. Lescot, S. Rombauts, B. De Moor, P. Rouze, and Y. Moreau. 2002. A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 9:447-464. [DOI] [PubMed] [Google Scholar]
- 39.Veening, J.-W., L. W. Hamoen, and O. P. Kuipers. 2005. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol. Microbiol. 56:1481-1494. [DOI] [PubMed] [Google Scholar]
- 40.Westers, H., L. Westers, E. Darmon, J. M. van Dijl, W. J. Quax, and G. Zanen. 2006. The CssRS two-component regulatory system controls a general secretion stress response in Bacillus subtilis. FEBS J. 273:3816-3827. [DOI] [PubMed] [Google Scholar]
- 41.Westers, L., H. Westers, and W. J. Quax. 2004. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim. Biophys. Acta 1694:299-310. [DOI] [PubMed] [Google Scholar]
- 42.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeats, C., N. D. Rawlings, and A. Bateman. 2004. The PepSY domain: a regulator of peptidase activity in the microbial environment? Trends Biochem. Sci. 29:169-172. [DOI] [PubMed] [Google Scholar]
- 44.Zanen, G., E. N. Houben, R. Meima, H. Tjalsma, J. D. Jongbloed, H. Westers, B. Oudega, J. Luirink, J. M. van Dijl, and W. J. Quax. 2005. Signal peptide hydrophobicity is critical for early stages in protein export by Bacillus subtilis. FEBS J. 272:4617-4630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.