Abstract
Citrinin, a secondary fungal metabolite of polyketide origin, is moderately nephrotoxic to vertebrates, including humans. From the red-pigment producer Monascus purpureus, a 21-kbp region flanking pksCT, which encodes citrinin polyketide synthase, was cloned. Four open reading frames (ORFs) (orf1, orf2, orf3, and orf4) in the 5′-flanking region and one ORF (orf5) in the 3′-flanking region were identified in the vicinity of pksCT. orf1 to orf5 encode a homolog of a dehydrogenase (similarity, 46%), a regulator (similarity, 38%), an oxygenase (similarity, 41%), an oxidoreductase (similarity, 26%), and a transporter (similarity, 58%), respectively. orf2 (2,006 bp with four introns) encodes a 576-amino-acid protein containing a typical Zn(II)2Cys6 DNA binding motif at the N terminus and was designated ctnA. Although reverse transcriptase PCR analysis revealed that all of these ORFs, except for orf1, were transcribed with pksCT under citrinin production conditions, the disruption of ctnA caused large decreases in the transcription of pksCT and orf5, together with reduction of citrinin production to barely detectable levels, suggesting that these two genes are under control of the ctnA product. Complementation of the ctnA disruptant with intact ctnA on an autonomously replicating plasmid restored both transcription and citrinin production, indicating that CtnA is a major activator of citrinin biosynthesis.
Filamentous fungi have a versatile ability to produce different kinds of secondary metabolites, including antibiotics, anticancer drugs, and pigments (3). During the biosynthesis of these secondary metabolites, many biosynthetic enzymes are required and should function coordinately in the synthesis of these structurally complex metabolites. The genes encoding these enzymes have often been reported to localize in an adjacent region or to form a gene cluster (6, 13), similar to the situation of biosynthetic gene clusters for secondary metabolites in prokaryotic actinomycetes. In addition to such structural genes, each gene cluster may contain a gene encoding a regulator that functions to coordinate expression of the structural genes in the cluster and a gene encoding a transporter that excludes harmful intracellular secondary metabolites as a self-defense mechanism (7, 22), respectively.
Regulators possessing a Zn(II)2Cys6 binuclear motif represent one of the largest classes of transcriptional regulators in filamentous fungi. They generally act as transcriptional activators, as exemplified by GAL4 from Saccharomyces cerevisiae (21). A large number of Zn(II)2Cys6 regulators have been identified exclusively in fungi and characterized as regulators of primary metabolism, secondary metabolism, drug resistance, or meiotic development. Through its DNA binding motif (CX2CX6CX6CX2CX6C [C, cysteinyl residue; X, any amino acid]) located at the N terminus, the Zn(II)2Cys6 regulator binds to a specific binding site (CGGN6/11CCG [N, any nucleotide]) in the promoter regions of target genes. Previous research on secondary metabolites in filamentous fungi revealed that several gene clusters contain this type of activator gene and that the activator plays a major role in synchronizing the expression of several genes in the biosynthetic cluster (22).
The mycotoxin citrinin (CT) was first isolated as a secondary metabolite from Penicillium citrinum (12) and was subsequently identified in many fungal species, such as Penicillium, Aspergillus, and Monascus. CT was first considered an antibiotic against gram-positive bacteria, but toxicity studies have revealed that it should be regarded as a mycotoxin, based on its nephrotoxicity to humans, causing endemic Balkan nephropathy (16).
The fungus Monascus purpureus is highly useful for its ability to produce red pigments (11) as a food colorant, γ-aminobutyric acid as an antihypertensive agent, and monacolin K as an antihypercholesterolemic agent. However, its potential for producing CT limits its wider use. Detailed knowledge of the biosynthetic pathway and regulation mechanism is required to eliminate possible contamination by CT in food products. In our previous study, a polyketide synthase (PKS) gene for CT (pksCT) was cloned from M. purpureus (18). In this study, we cloned the genes in the vicinity of pksCT to obtain new genes involved in CT biosynthesis. An activator gene essential for the efficient production of CT was found in the upstream region of pksCT, and we demonstrate that an extremely low-CT producer can be created by disrupting the gene.
MATERIALS AND METHODS
Strain, growth conditions, and transformation.
A mycelial mat (an area of about 1 cm2) of wild-type Monascus purpureus (strain NBRC30873) was taken from a master plate, and spores on it were spread by daubing onto a new plate (2% agar) of Monascus cultivation (MC) solid medium consisting of glucose (50 g/liter), polypeptone (7.5 g/liter), NH4H2PO4 (2.0 g/liter), MgSO4·7H2O (0.5 g/liter), CaCl2·2H2O (0.1 g/liter), KNO3 (2.0 g/liter), and agar (20.0 g/liter). Plates were incubated for 7 to 10 days at 28°C. For liquid cultivation, a mycelial mat (an area of about 1 cm2) was taken from the agar plate, inoculated into 100 ml of MC liquid medium in a 500-ml baffled Erlenmeyer flask, and incubated at 28°C with reciprocating shaking at 120 strokes per minute. M. purpureus was transformed as described by Shimizu et al. (18, 19).
For the genetic manipulation of Escherichia coli, E. coli XL10-Gold ultracompetent cells (Stratagene, La Jolla, CA) and Library Efficiency DB3.1 competent cells (Invitrogen, Carlsbad, CA) were used as the cloning hosts.
Southern and Northern blot analyses.
Southern and Northern blot analyses were conducted essentially by the method of Sambrook et al. (17). For Southern blot analysis, genomic DNA (20 μg) was digested overnight with restriction enzyme, separated on a 1.0% agarose gel, and transferred to a Hybond N+ membrane (Amersham Biosciences, Piscataway, NJ). For Northern blot analysis, total RNA (8 μg) was separated on a 1% agarose-formaldehyde gel. The probe was labeled with [α-32P]dCTP using the Random Primer DNA labeling kit, version 2 (Takara Bio, Otsu, Japan).
Colony hybridization.
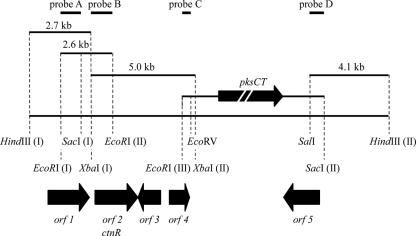
To clone the CT biosynthetic gene cluster, four successive rounds of Southern blot analysis were conducted with four different probes. Probes A, B, C and D were the 1.6-kb EcoRI (I)-SacI (I) fragment, the 960-bp XbaI (I)-EcoRI (II) fragment, the 750-bp EcoRI (III)-EcoRV fragment, and the 820-bp SalI-SacI (II) fragment, respectively, and they hybridized to a 2.7-kb HindIII (I)-XbaI (I) fragment, a 2.6-kb EcoRI (I)-EcoRI (II) fragment, a 5.0-kb XbaI (I)-XbaI (II) fragment, and a 4.1-kb SalI-HindIII (II) fragment, respectively (Fig. 1). (The Roman numbers in parentheses indicate the specific site among several sites for the corresponding restriction enzyme.) Monascus genomic DNA was digested with each restriction enzyme and electrophoresed on a 1% agarose gel. Corresponding regions were extracted and subcloned into the multicloning site of pUC19, resulting in the construction of individual partial genomic libraries. From each library, 2,000 colonies were selected. Positive clones were then identified by colony hybridization with each probe.
FIG. 1.
Restriction enzyme map of cloned fragments. The 5.0-kb XbaI (I)-XbaI (II), 4.1-kb SalI-HindIII (II), 2.6-kb EcoRI (I)-EcoRI (II), and 2.7-kb HindIII (I)-XbaI (I) fragments were obtained by colony hybridization. orf1, orf2, orf3, orf4, and orf5 indicate identified genes encoding a homolog of a dehydrogenase, a regulator, an oxygenase, an oxidoreductase, and a transporter, respectively. pksCT is the CT PKS gene. The thick black arrows indicate position and direction. Probes A, B, C, and D are the 1.6-kb EcoRI (I)-SacI (I), 960-bp XbaI (I)-EcoRI (II), 750-bp EcoRI (III)-EcoRV, and 820-bp SalI-SacI (II) regions, respectively.
Sequence analysis.
Four cloned DNA fragments covering the CT biosynthetic gene cluster (Fig. 1) were sequenced on both strands by an ABI PRISM 3100 sequencer with the BigDye Terminator Cycle Sequencing Ready Reaction, version 3.1, cycle sequencing kit (Applied Biosystems, Foster City, CA) at Hitachi High-Tech Science Systems Corporation (Ibaragi, Japan) by primer walking. The sequence was analyzed with the BLASTP program (2) in the DDBJ homology search system (http://www.ddbj.nig.ac.jp/E-mail/homology-j.html).
RT-PCR.
For reverse transcriptase PCR (RT-PCR), total RNA was extracted from Monascus mycelia cultivated in MC liquid medium every other day from the second to the sixth day of cultivation using the RNeasy Mini Kit (QIAGEN, Tokyo, Japan), according to the manufacturer's protocol, and treated with DNase I to remove the contaminating genomic DNA. The cDNA was synthesized with SuperScript III RNase H− RT (Invitrogen). For PCR, the amplification conditions were 25 cycles of denaturation (95°C for 30 s), annealing (55°C for 30 s), and extension (72°C for 2 min), followed by a single extension at 72°C for 4 min. The primers used in this study are shown in Table 1.
TABLE 1.
Primers used in this study
| Primera | Sequence (5′ to 3′)b |
|---|---|
| act F | GGAATTCTGCAGATTCTACAACGAACTCCG |
| act R | GGAATTCTGCAGTCAGGGAGTTCATAGGAC |
| amp F | GTAGATAACTACGATACGGG |
| amp R | TATGTGGCGCGGTATTATCC |
| hph F | GGGGTACCCTTCTGATCGAAAAGTTCGAC |
| hph R | GGGGTACCCCTCTGATAGAGTTGGTCAAG |
| dehy F | TAGCATCTACTCCAGCGTGA |
| dehy R | TCGGGTAAAGATCTTTGTGC |
| reg F | AAACTACGCTGTGACGGACA |
| reg R | TAACTGCACCAGACGAAACG |
| oxg F | TGCACCTCTACAGGGTTATT |
| oxg R | TCTGCTCATCATATCTCCA |
| oxr F | TAATGCTGCCATCTTCCAAG |
| oxr R | ATACGCAACGAGCGAGACAT |
| pksCT F | CCAGTGTGGCTATTCACC |
| pksCT R | CCTAAGGACATTACTCTT |
| tra F | TGACCATCTGGGTAACCTTC |
| tra R | GAGATGAGTCGACGATGAAG |
| dis F | ACACGGAAGAGATCCAAGCA |
| dis R | AGAGATGAATCACACTCGCC |
The forward (F) and reverse (R) primer pairs act, amp, hph, dehy, reg, oxg, oxr, pksCT, and tra were used in RT-PCR for amplification of the actin gene, ampicillin resistance gene, hygromycin resistance gene, dehydrogenase gene (orf1), regulator gene (ctnA), oxygenase gene (orf3), oxidoreductase gene (orf4), polyketide synthase gene (pksCT), and transporter gene (orf5), respectively.
Underlining indicates EcoRI and PstI restriction sites.
Construction of the pReg-dis vector.
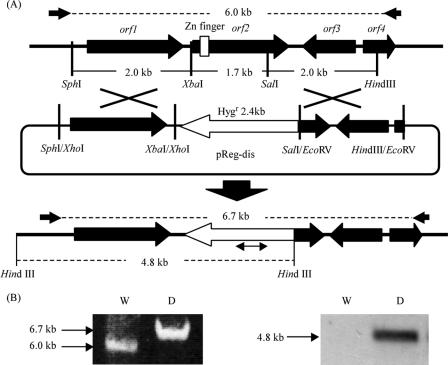
The 2.0-kb SphI-XbaI and 2.0-kb SalI-HindIII fragments (see Fig. 4) were treated with T4 DNA polymerase and inserted into the blunt-ended XhoI and EcoRV sites in pCSN44, respectively, which yields hygromycin B resistance to transformants (Fungal Genetics Stock Center [http://www.fgsc.net/]) (20). The construct contains a hygromycin B resistance gene as a selection marker flanked by the 2.0-kb SphI-XbaI and 2.0-kb SalI-HindIII fragments. This vector, designated pReg-dis, was used for ctnA disruption.
FIG. 4.
orf2 disruption by double crossover via homologous recombination (A) and PCR (left) and Southern blot (right) analyses of the orf2 disruptant (B). The orf2 gene was disrupted by inserting the hygromycin resistance gene (open arrow, Hygr) from pReg-dis (see Materials and Methods). The small arrows indicate the primers, dis F and dis R, used for PCR analysis. A part of the hygromycin resistance gene was used as a probe for Southern blot analysis (the double-headed arrow under Hygr). W, wild-type strain; D, ctnA disruptant.
Construction of the pAG-reg vector.
During the construction of pAG-reg, used for complementing the ctnA disruptant, some restriction enzyme sites were treated with T4 DNA polymerase after enzyme digestion and are indicated below by asterisks. Before SalI digestion, the vector pCSN44 was partially digested with BamHI, followed by T4 DNA polymerase treatment. A 2.1-kb SalI-BamHI* fragment (fragment S0-Bm2; see Fig. S1 in the supplemental material) containing the trpC promoter, terminator, and hygromycin resistance gene from pCSN44, was extracted from the agarose gel after electrophoresis and inserted into the SalI and BamHI* sites of the vector pUC19 (pTRPpt). To replace the hygromycin resistance gene with ctnA, pTRPpt was linearized by digestion with ClaI* and BamHI* (C and Bm1; see Fig. S1 in the supplemental material) and was ligated with a 2.4-kb ApaLI*-NdeI* fragment containing the regulator gene (fragment A-N; see Fig. S1 in the supplemental material), resulting in pTRPpregt. A 3.4-kb HindIII*-SmaI* fragment (fragment H-Sm; see Fig. S1 in the supplemental material) containing the regulator gene flanked by the trpC promoter and terminator) (regulator cassette) was isolated from pTRPpregt and inserted into the EcoRI* site in the entry vector pENTR11 (Invitrogen) (pENTR-TRPpregt). By use of the Gateway system (Invitrogen), pAG-reg was finally constructed through a transfer reaction of the regulator cassette from pENTR-TRPpregt to the destination vector pAG, carrying both the autonomous replicating sequence AMA1 and AurAr, which confers aureobasidin A resistance to M. purpureus (19).
Analysis of pigment and CT production.
Monascus was cultivated in MC liquid medium as described above. The amounts of CT and pigment in the mycelia and in the mycelium-free filtrate were measured every other day from the second to the tenth day of cultivation, as described by Shimizu et al. (18, 19). The mycelia harvested by suction-filtration were dried in an oven at 80°C for 3 days.
Nucleotide sequence accession number.
The sequence surrounding the pksCT gene determined in this study has been deposited in the DDBJ database under accession number AB243687.
RESULTS
Cloning and sequence analysis of the pksCT-flanking region in M. purpureus.
To learn more about the regulation mechanism in CT biosynthesis, a 21,917-bp fragment containing pksCT was cloned by four rounds of genome walking and its sequence was determined (Fig. 1) (DDBJ accession no. AB243687). Sequence analysis and a database search with BLASTP revealed that, in addition to pksCT, this region contains five putative open reading frames (ORFs)—orf1, orf2, orf3, orf4, and orf5—whose products displayed the highest similarity to the aldehyde dehydrogenase from Nocardia farcinica (BAD58112) (similarity, 46%), the binuclear Zn transcription factor from Nectria haematococca (AY218847) (similarity, 38%), the isopenicillin N synthase and related dioxygenases from Aspergillus oryzae (BAE62120) (similarity, 41%), the oxidoreductase from Penicillium citrinum (BAC20563) (similarity, 26%), and the membrane transporter from Aspergillus fumigatus (EAL93985) (similarity, 58%), respectively (Table 2).
TABLE 2.
Sequence analysis of putative ORFs
| ORF | Size (bp/aa)a | Plausible functionb | Protein with highest similarity | Similarity at amino acid level (%) |
|---|---|---|---|---|
| orf1 | 1,506/501 | Dehydrogenase | Nocardia farcinica aldehyde dehydrogenase (BAD58112) | 46 |
| orf2 | 1,731/576 | Transcriptional activator | Nectria haematococca binuclear Zn transcription factor (AY218847) | 38 |
| orf3 | 990/329 | Oxygenase | Aspergillus oryzae isopenicillin N synthase and related dioxygenases (BAE62120) | 41 |
| orf4 | 942/313 | Oxidoreductase | Penicillium citrinum oxidoreductase (BAC20563) | 26 |
| orf5 | 1,566/521 | Transporter | Aspergillus fumigatus membrane transporter (EAL93985) | 58 |
The predicted amino acid sequences for the ORFs were deduced following removal of the putative intron regions. aa, amino acids.
The function of each encoded protein was deduced with its putative functional domains and from the reported function of the protein homolog showing the highest degree of similarity.
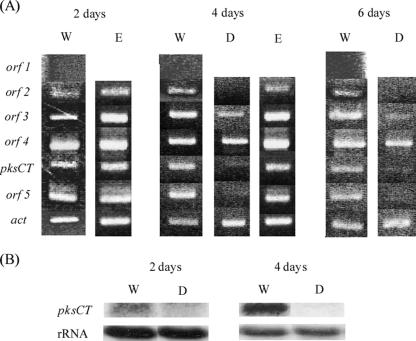
The transcription patterns of the five ORFs were examined by RT-PCR with RNA samples prepared from mycelia cultivated for 2, 4, or 6 days under CT production conditions. pksCT was transcribed from the 2-day cultivation, at which point CT production started in the wild-type strain (Fig. 2A). Similarly, the transcripts of the four other plausible genes (orf2, encoding a regulator; orf3, encoding an oxygenase; orf4, encoding an oxidoreductase; and orf5, encoding a transporter) were detected from the 2-day cultivation, whereas no transcription of orf1 was detected.
FIG. 2.
Transcriptional analysis of putative CT biosynthetic genes. (A) RT-PCR was performed against RNA samples extracted from mycelia harvested from MC liquid medium after the indicated period of cultivation. (B) Northern blot analysis of the pksCT gene. Shown is the fragment amplified by RT-PCR, with the primers pksCT F and pksCT R used as a probe. W, wild-type strain; D, ctnA disruptant; E, ctnA-complemented ctnA disruptant; act (encoding actin), control.
Characterization of the putative regulator Orf2.
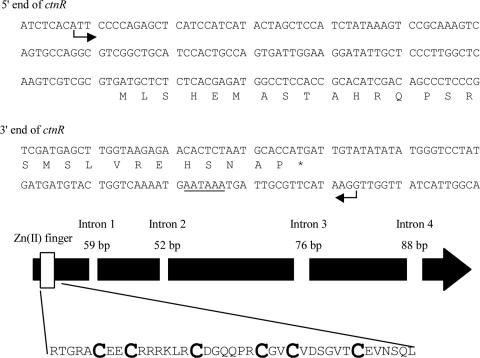
The orf2 gene, located 3.9 kbp upstream of pksCT, encoded a protein that showed significant similarity to the members of the Zn(II)2Cys6 binuclear DNA binding proteins (21). These Zn(II)2Cys6 proteins occur in a number of gene clusters for secondary metabolites, where they serve as transcriptional activators involved in pathway gene regulation (1, 8, 9). This suggests that orf2 may therefore control the CT biosynthesis in Monascus. To deduce the detailed structure of the orf2 transcript, 5′ and 3′ random amplification of cDNA ends was performed against mRNA from mycelia cultivated for 6 days. An A residue 12 nucleotides (nt) downstream from an XbaI (I) site in the XbaI (I)-XbaI (II) fragment was identified as the transcriptional start site (TSS). The start codon, stop codon, and polyadenylation site (AATAAA) were located at 126 nt, 2,005 nt, and 2,048 nt downstream of the TSS, respectively. The presence of four introns was deduced from the presence of a typical splicing site (5′-GT-AG-3′). With RT-PCR followed by sequencing, they were confirmed to be at nt 238 to 296, 571 to 622, 1131 to 1206, and 1696 to 1783 from the TSS (Fig. 3) (DDBJ accession no. AB243687).
FIG. 3.
Structure of ctnA. In the sequences of the 5′ and 3′ termini of ctnA, arrows mark the TSS and termination site. The underlined sequence is a putative polyadenylation site. The thick black arrow indicates the ctnA gene, of 2,006 bp, encoding a 576-amino-acid protein with four introns shown by empty rectangles (with intron sizes shown). The rectangle on the far left indicates the position of the Zn(II)2Cys6 DNA binding motif. The large, bold letters in the highlighted sequence are cysteinyl residues constituting the Zn(II)2Cys6 DNA binding motif at the N terminus.
Gene disruption of orf2.
To analyze the in vivo function of the Orf2 protein, orf2 disruptants were created by homologous recombination with pReg-dis (Fig. 4A), by which a major part of orf2, including the DNA binding Zn(II)2Cys6 motif, was lost. The remaining portion of the orf2 coding region corresponded to the C-terminal 107 amino acids. The predicted recombination product at the orf2 locus was confirmed by PCR and Southern blot analyses (Fig. 4B).
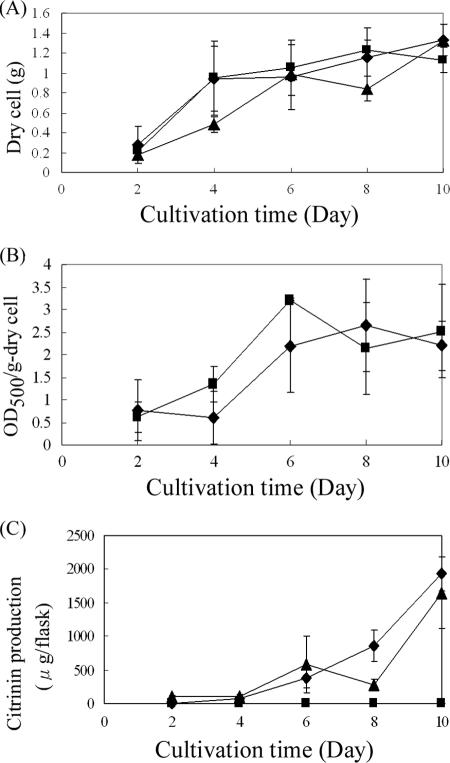
The orf2 disruptant did not show any significant differences from the wild-type strain in either growth (Fig. 5A) or pigment production (Fig. 5B). However, in sharp contrast to the progressive production of CT by the wild-type strain (1,930 μg per flask at 10 days' cultivation), CT production in the orf2 disruptant was kept very low (less than 0.1 μg per flask) throughout the cultivation period (Fig. 5C), suggesting that the orf2 product is important for the efficient production of CT. Regarding PksCT, a central player in CT biosynthesis as the PKS for CT, RT-PCR revealed a drastic reduction in its transcription in the orf2 disruptant (Fig. 2A). Northern blot analysis confirmed that pksCT transcription was almost negligible in the orf2 disruptant (Fig. 2B), indicating that the orf2 product is necessary for transcriptional activation of pksCT. In addition to pksCT, RT-PCR also suggested that the orf2 product would be required for the efficient transcription of orf5, though the weak effect was observed for orf3 transcription. These results suggested that, as in the case of AflR in the aflatoxin biosynthetic cluster (9), the orf2 product (designated CtnA for being the first factor related to CT biosynthesis other than PksCT) acts as an activator at least on pksCT and orf5 transcription. This occurs probably through binding to specific DNA sequences in the upstream region of each of the two genes, although no definite conserved sequence was discovered in the promoter regions of pksCT and orf5.
FIG. 5.
Phenotypic analysis of the ctnA disruptant and the ctnA-complemented ctnA disruptant. (A) Growth in liquid culture. Growth was expressed by the weight of dry mycelia for the wild-type strain (diamonds), the ctnA disruptant (squares), and the ctnA-complemented ctnA disruptant (triangles). (B) Pigment production. Pigments were extracted from dry mycelia with 70% ethanol, and the optical density at 500 nm (OD500) was measured for the wild-type strain (diamonds) and the ctnA disruptant (squares). (C) Comparison of CT production among the wild-type strain (diamonds), the ctnA disruptant (squares), and the ctnA-complemented ctnA disruptant (triangles). The CT contents in both the mycelia and mycelium-free filtrate were measured by high-performance liquid chromatography and enzyme-linked immunosorbent assay, and the sum is shown. These experiments were performed at least three times independently.
ctnA gene expression in the ctnA disruptant.
To establish that CtnA acts as an activator in CT biosynthesis, an intact copy of ctnA under the constitutive trpC promoter was introduced into the ctnA disruptant by means of an autonomously replicating vector, pAG-Reg (see Fig. S1 in the supplemental material), and transformants harboring the intact ctnA gene on the replicating plasmid (strains E3 and E4 [Fig. 6]) were obtained. The ctnA-complemented strains regained the ability to produce CT, although to an extent slightly lower (1,649 μg/flask at 10 days' cultivation) than that of the wild-type strain (1,930 μg/flask at 10 days' cultivation). In addition, in the ctnA-complemented strain, transcription of the orf5, pksCT, and orf3 genes became detectable by RT-PCR, with levels comparable to those of the wild-type strain (Fig. 2A). Based on these results, it can be concluded that CtnA is a major activator of the CT pathway in M. purpureus.
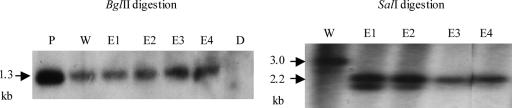
FIG. 6.
Complementation of the ctnA disruptant by intact ctnA, as determined by Southern blot analysis of the ctnA-complemented strain. Part of the regulator gene was used as a probe (see the double-headed arrow above ctnA in Fig. S1 of the supplemental material). By BglI digestion, any strains containing ctnA show a 1.3-kb band (corresponding to fragment B1-B2 in Fig. S1 in the supplemental material). By SalI digestion, any strains containing pAG-reg-derived ctnA show a 2.2-kb band (fragment S0-S2 in Fig. S1 in the supplemental material), while the wild-type strain shows a 3.0-kb band (fragment S1-S2 in Fig. S1 in the supplemental material). The band smaller than 2.2 kbp in strains E1 and E2 may indicate the presence of genome-integrated pAG-reg in addition to the autonomously existing pAG-reg. E4 was selected for phenotypic analysis. P, pAG-reg (positive control); W, wild-type strain; E1 to E4, ctnA-complemented ctnA disruptants; D, ctnA disruptant.
DISCUSSION
In addition to PKS and post-PKS-modifying enzymes, the biosynthesis of fungal polyketides often requires several proteins, such as transcriptional regulators and a transporter(s) encompassing resistance, and most of the genes encoding these proteins form a gene cluster in an adjacent region on a chromosome. Our previous study found that the pksCT gene was essential for CT biosynthesis in M. purpureus, based on the fact that pksCT encodes a homolog of fungal iterative type I PKS and on the fact that its disruption resulted in the complete loss of CT production. However, no regulatory gene(s) is known for CT biosynthesis despite the likely importance of this gene product(s) in manipulating CT production in fungi.
In this study, we investigated the presence of the genes involved in CT production in the flanking regions of pksCT and discovered five putative ORFs (orf1, ctnA, orf3, and orf4 in the 5′-flanking region and orf5 in the 3′-flanking region of pksCT). A database search revealed the plausible functions of the encoded proteins as a dehydrogenase for orf1, a transcriptional activator for orf2 (ctnA), an oxygenase for orf3, an oxidoreductase for orf4, and a transporter for orf5 (Table 2). The presence of an oxygenase (orf3) and an oxidoreductase (orf4) corresponds well to the proposed pathway of CT biosynthesis (10), which, together with their positions adjacent to pksCT, raises the possibility that ctnA, orf3, orf4, pksCT, and orf5 may be involved in CT biosynthesis. Based on BLASTP alignments, the product of ctnA was found to be most similar (27% identity and 38% similarity) to a transcriptional regulator for pisatin demethylase (AY218847) from Nectria haematococca, which contains a Zn(II)2Cys6 DNA binding motif (14). Based on the results that (i) ctnA disruption drastically decreased CT production (to 0.0016% of that of the wild-type strain) and (ii) the disruptant regained CT production by the complementation of intact ctnA, ctnA is predicted to be a required factor for efficient CT production. Transcriptional analysis of the ctnA disruptant revealed that ctnA disruption affected the transcription of pksCT and orf5, though the effect on orf3 was not clear. Again, complementation of the intact ctnA restored the transcription of pksCT and orf5 to wild-type levels. These findings, taken together with the recovery of CT production, suggest that the ctnA gene encodes a transcriptional activator for the CT biosynthetic cluster, including pksCT. In the ctnA-complemented strain, however, cellular growth did not reach the wild-type level throughout cultivation and the production of CT was higher from the second to the sixth day and lower from the eighth to the tenth day than in the wild-type strain. This might be attributed to the uncontrolled expression of ctnA under the constitutive trpC promoter, which could result in the production of CT to levels harmful to cellular growth in the early phase of cultivation.
Zn(II)2Cys6-type regulators have been reported to activate target genes by binding to consensus binding sequences consisting of conserved terminal trinucleotides (CGGN6CCG or CGGN11CCG) in their promoter regions (21). Searches for such conserved sequences revealed none in the promoter region of either pksCT or orf5, whose transcripts were dramatically decreased in the ctnA disruptant. Therefore, different terminal trinucleotides might be recognized by CtnA. In the future, in vitro analysis using recombinant CtnA, such as a gel shift assay or DNase I footprinting, will clarify the actual CtnA binding site in the CT biosynthetic cluster.
CT was first discovered in Penicillium citrinum (12) and was considered an antibiotic, but it was later clarified to be a nephrotoxin to animals, including humans (16). Subsequent research discovered that several species of Penicillium, Aspergillus (15), and Monascus (5) produce CT. Moreover, CT contamination has been detected in wheat, oats, rye, corn, barley, and rice in many countries (4). This paper is the first report of the presence of an activator of CT biosynthesis among CT-producing fungi. The identification of an activator and an apparent CT biosynthetic gene cluster in Monascus may aid in the identification of corresponding gene clusters in different fungal species and in the development of strategies for suppressing CT production through the application of detailed knowledge of the regulation of CT biosynthesis.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from YAEGAKI Bio-Industry, Inc.; by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT); and by Special Coordination Funds for the Promotion of Science and Technology from the JST-NRCT joint program of the Japan Science and Technology Agency (JST) and the National Research Council of Thailand (NRCT).
This paper is part of the Ph.D. dissertation of T.S.
Footnotes
Published ahead of print on 22 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abe, Y., C. Ono, M. Hosobuchi, and H. Yoshikawa. 2002. Functional analysis of mlcR, a regulatory gene for ML-236B (compactin) biosynthesis in Penicillium citrinum. Mol. Genet. Genomics 268:352-361. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, J. W. 1998. Mycotechnology: the role of fungi in biotechnology. J. Biotechnol. 66:101-107. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, J. W., and M. Klich. 2003. Mycotoxins. Clin. Microbiol. Rev. 16:497-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanc, P. J., M. O. Loret, and G. Goma. 1995. Production of citrinin by various species of Monascus. Biotechnol. Lett. 17:291-294. [Google Scholar]
- 6.Brown, M. P., C. S. Brown-Jenco, and G. A. Payne. 1999. Genetic and molecular analysis of aflatoxin biosynthesis. Fungal Genet. Biol. 26:81-98. [DOI] [PubMed] [Google Scholar]
- 7.Calabrese, D., J. Bille, and D. Sanglard. 2000. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 146:2743-2754. [DOI] [PubMed] [Google Scholar]
- 8.Chang, P. K., K. C. Ehrlich, J. Yu, D. Bhatnagar, and T. E. Cleveland. 1995. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl. Environ. Microbiol. 61:2372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrlich, K. C., B. G. Montalbano, and J. W. Cary. 1999. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 230:249-257. [DOI] [PubMed] [Google Scholar]
- 10.Hajjaj, H., A. Klaébé, M. O. Loret, G. Goma, P. J. Blanc, and J. François. 1999. Biosynthetic pathway of citrinin in the filamentous fungus Monascus ruber as revealed by 13C nuclear magnetic resonance. Appl. Environ. Microbiol. 65:311-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajjaj, H., A. Klaébé, G. Goma, P. J. Blanc, E. Barbier, and J. François. 2000. Medium-chain fatty acids affect citrinin production in the filamentous fungus Monascus ruber. Appl. Environ. Microbiol. 66:1120-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hetherington, A. C., and H. Raistrick. 1931. Studies in biochemistry of microorganism. XI. On the production and chemical constitution of a new yellow colouring matter, citrinin, produced from glucose by Penicillium citrinum. Thom. Philos. Trans. R. Soc. Lond. B 220:269-297. [Google Scholar]
- 13.Kennedy, J., K. Auclair, S. G. Kendrew, C. Park, J. C. Vederas, and C. R. Hutchinson. 1999. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science 284:1368-1372. [DOI] [PubMed] [Google Scholar]
- 14.Khan, R., R. Tan, A. G. Mariscal, and D. Straney. 2003. A binuclear zinc transcription factor binds the host isoflavonoid-responsive element in a fungal cytochrome p450 gene responsible for detoxification. Mol. Microbiol. 49:117-130. [DOI] [PubMed] [Google Scholar]
- 15.Manabe, M. 2001. Fermented foods and mycotoxins. Mycotoxins 51:25-28. [Google Scholar]
- 16.Sabater-Vilar, M., R. F. M. Maas, and J. Fink-Gremmels. 1999. Mutagenicity of commercial Monascus fermentation products and the role of citrinin contamination. Mutat. Res. 444:7-16. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Shimizu, T., H. Kinoshita, S. Ishihara, K. Sakai, S. Nagai, and T. Nihira. 2005. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl. Environ. Microbiol. 71:3453-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu, T., H. Kinoshita, and T. Nihira. 2006. Development of transformation system in Monascus purpureus using an autonomous replication vector with aureobasidin A resistance gene. Biotechnol. Lett. 28:115-120. [DOI] [PubMed] [Google Scholar]
- 20.Staben, C., B. Jensen, M. Singer, J. Pollock, M. Schechtman, J. Kinsey, and E. Selker. 1989. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal. Genet. Newsl. 36:79-81. [Google Scholar]
- 21.Todd, R. B., and A. Andrianopoulos. 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21:388-405. [DOI] [PubMed] [Google Scholar]
- 22.Walton, J. D. 2000. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet. Biol. 30:167-171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.