Abstract
d- and l-amino acids were produced from l- and d-amino acid amides by d-aminopeptidase from Ochrobactrum anthropi C1-38 and l-amino acid amidase from Pseudomonas azotoformans IAM 1603, respectively, in the presence of α-amino-ɛ-caprolactam racemase from Achromobacter obae as the catalyst by dynamic kinetic resolution of amino acid amides.
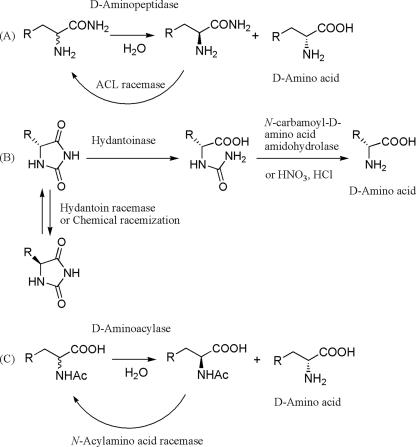
Several dynamic kinetic enzymatic resolutions of synthetic substrates have been developed for the industrial production of chiral amino acids. The use of l-α-amino-ɛ-caprolactam (ACL)-hydrolase in the presence of ACL racemase is a typical example of l-lysine production (1, 2, 12). l-cysteine is produced by hydrolases acting on amino-Δ2-thiazoline-4-carboxylic acid in the presence of a racemase (23). Used in combination with d-specific hydantoinase (22, 26), either with spontaneous racemization of hydantoin or with hydantoin racemase (21, 24), N-carbamoyl-d-amino acid amidohydrolase catalyzes the hydrolysis of N-carbamoyl-d-amino acids to form d-amino acids (3, 16-20). N-acyl-d-amino acid amidohydrolase (27) and N-acyl amino acid racemase (25) are utilized together to form d-amino acids. Fig. 1 compares several dynamic kinetic enzymatic resolutions of synthetic substrates. The optically active amino acids can be synthesized in three steps with amidases from aldehydes via aminonitrile and amino acid amide, while the d-amino acylase route requires four steps via aminonitrile, amino acid, and N-acyl amino acid.
FIG. 1.
Dynamic kinetic enzymatic resolutions of synthetic substrates to form d-amino acids. (A) Dynamic kinetic resolution of amino acid amide by DAP in the presence of ACL racemase (this study). (B) Dynamic kinetic resolution of hydantoinase substituted for hydantoin by chemical racemization or in the presence of hydantoin racemase. (C) Dynamic kinetic resolution of N-acyl-amino acid by d-aminoacylase in the presence of N-acylamino acid racemase.
We isolated and characterized several d-stereospecific amino acid amidases, such as d-aminopeptidase (DAP), d-amino acid amidase, alkaline d-peptidase, and r-amidase, and utilized them in the kinetic resolution of amino acid amides (4-9, 14). l-Amino acid amide hydrolases such as LaaA and LaaABd have been characterized previously (15).
In this report, the effect of a high concentration of the substrate on the dynamic kinetic resolution of amino acid amide, together with the optimum pH, was investigated using DAP or LaaA in the presence of ACL racemase (10, 11).
ACL racemase was purified from Escherichia coli JM109/pACL60, and its activity was assayed as described previously (10). The expression level of ACL racemase was about 300 U/liter culture (5% of the total soluble protein). The amount of inclusion body was not measured. DAP expressed in E. coli JM109 was purified by a similar procedure, and its activity was assayed as described previously (8). About 8,000 U/liter of DAP could be produced maximally in the cell extract (about 6% of the total soluble protein). LaaA activity toward l-alanine amide was determined by measuring the amount of l-alanine formed. The reaction mixture (1.0 ml) contained 0.1 M potassium phosphate buffer (KPB; pH 7.0), 0.1 M l-alanine amide, and an appropriate amount of LaaA. The reaction was performed at 30°C for 10 min and stopped by adding 200 μl 2 N HClO4. The amount of l-alanine formed was determined by the method used for d-ACL, and the enzyme activity was measured by the same method as described previously (10). Recombinant E. coli JM109/pSTB20 with the LaaA gene of Pseudomonas azotoformans IAM 1603 was cultured and LaaA was purified by the same procedure as described previously (13), and 5,360 U/liter could be produced in the cell extract (about 7% of the total soluble protein).
The effect of the concentration of l-alanine amide on the racemization reaction catalyzed by ACL racemase was investigated. The reaction mixture (2.0 ml), containing 2 μM pyridoxal 5′-phosphate (PLP), 0.1 M KPB (pH 7.0), 0.6 M or 1.2 M l-alanine amide, and ACL racemase (2.8 U, 0.3 mg), was incubated at 30°C for 3 h. Formation of d-alanine amide increased when the concentration of l-alanine amide was increased from 0.6 M to 1.2 M, indicating that the enzyme activity was not inhibited by the high substrate concentration. ACL racemase and DAP can be used for efficient chiral d-amino acid synthesis, since DAP was shown to catalyze the synthesis of d-alanine, even in a 5-M concentration of the substrate (6).
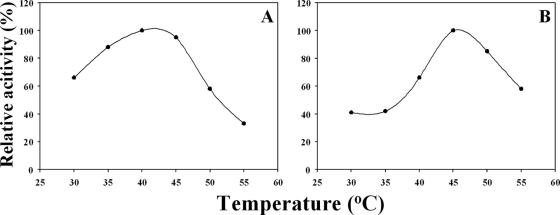
The effect of pH on d- or l-alanine production from l- or d-alanine amide with DAP or LaaA in the presence of ACL racemase was investigated. The reaction mixture (0.5 ml), containing 0.2 M KPB, 2 μM PLP, 50 mM l-alanine amide, and ACL racemase (0.15 U, 16 μg) with DAP (0.15 U, 0.36 μg) or LaaA (0.15 U, 8 μg), was incubated at 30°C for 30 min. The maximum conversion rate was observed at pH 7.3 (data not shown). The conversion of l-alanine amide to d-alanine was carried out at the temperatures indicated for 90 min (Fig. 2A). The maximum rate of conversion to d-alanine was observed at between 40 and 45°C. However, at 50°C, the rate of conversion to d-alanine was decreased because of the instability of DAP at the higher temperature. It was previously reported that ACL racemase is stable at up to 65°C for 20 min (1), whereas DAP loses 50% of its activity in incubation at 50°C for 10 min (9). Racemization was performed by ACL racemase at pH values of 6.5 to 9.2, and the maximum rate of conversion was found at pH 7.5 (data not shown) and at a temperature of around 45°C at 90 min (Fig. 2B). Above 45°C, the rate of conversion decreased. It was previously reported that LaaA retained only 25% of its activity after being incubated at 50°C for 5 min (13).
FIG. 2.
Effect of temperature on the conversion of alanine amide to alanine. (A) The conversion of d-alanine from l-alanine amide by DAP in the presence of ACL racemase. (B) The conversion of l-alanine from d-alanine amide by LaaA in the presence of ACL racemase.
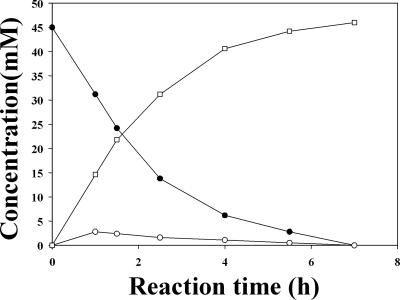
The time course of d-alanine synthesis from l-alanine amide, using DAP in the presence of ACL racemase, was measured in a reaction mixture (2.0 ml) containing 2 μM PLP, 0.1 M KPB (pH 7.0), 45 mM l-alanine amide, and ACL racemase (0.5 U, 68 μg) with DAP (2.2 U, 5.4 μg). The reaction mixture, containing 50 mM of the amino acid amide substrates listed in Tables 1 and 2, 1 M KPB (pH 7.5), 2 μM PLP, and DAP or LaaA, was incubated with ACL racemase at 30°C for 20 h. The time course of d-alanine synthesis from l-alanine amide, using DAP and ACL racemase, was measured. After 7 h, l-alanine amide was completely converted to d-alanine with an optical purity of 99.7% enantiomeric excess (ee) (Fig. 3). The synthesized d-alanine was purified and identified with nuclear magnetic resonance as described previously (8, 11).
TABLE 1.
Synthesis of various d-amino acids from the corresponding l-amino acid amides by DAP in the presence of ACL racemase
| Substrate | Product | Yield (%)a | ee (%) |
|---|---|---|---|
| l-Alanine amide | d-Alanine | 100b | >99 |
| l-2-Aminobutyric amide | d-2-Aminobutyric acid | 100b | >99 |
| l-Serine amide | d-Serine | 94c | >99 |
| l-Methionine amide | d-Methionine | 100c | >99 |
Concentration in the reaction mixture.
Synthesized for 12 h using DAP (0.21 mg) and ACL racemase (10 mg).
Synthesized for 20 h using DAP (2.1 mg) and ACL racemase (0.13 mg).
TABLE 2.
Synthesis of various l-amino acids from the corresponding d-amino acid amides by LaaA in the presence of ACL racemase
| Substrate | Product | Yield (%)a | ee (%) |
|---|---|---|---|
| d-Alanine amide | l-Alanine | 100b | >99 |
| d-Leucine amide | l-Leucine | 100c | >99 |
| d-Methionine amide | l-Methionine | 100c | >99 |
Concentration in the reaction mixture.
Synthesized for 12 h using LaaA (0.45 mg) and ACL racemase (10 mg).
Synthesized for 20 h using LaaA (0.45 mg) and ACL racemase (0.13 mg).
FIG. 3.
Production of d-alanine from l-alanine amide by DAP in the presence of ACL racemase. (○), d-alanine amide; (•), l-alanine amide; (□), d-alanine.
We extended the conditions to the production of d-amino acids from different l-amino acid amides (Table 1). l-Alanine amide, l-2-aminobutyric acid amide, and l-methionine amide were completely converted to the corresponding d-amino acids, using ACL racemase with DAP. d-Alanine and d-2-aminobutyric acid were synthesized in 12 h, using ACL racemase (10 μg) and DAP (0.21 mg). Conversion of various d-amino acid amides to the corresponding l-amino acids was also examined by using LaaA in the presence of ACL racemase (Table 2). d-Alanine amide, d-leucine amide, and d-methionine amide were completely converted to l-alanine, l-leucine, and l-methionine, respectively.
Acknowledgments
This research was supported in part by grants from the Japan Society for the Promotion of Sciences to Y. Asano (Ministry of Education, Culture, Sports, Science and Technology, Japan).
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Ahmed, S. A., N. Esaki, H. Tanaka, and K. Soda. 1983. Properties of α-amino-ɛ-caprolactam racemase from Achromobacter obae. Agric. Biol. Chem. 47:1887-1893. [Google Scholar]
- 2.Ahmed, S. A., N. Esaki, H. Tanaka, and K. Soda. 1986. Mechanism of α-amino-ɛ-caprolactam racemase reaction. Biochemistry 25:385-388. [DOI] [PubMed] [Google Scholar]
- 3.Altenbuchner, J., M. Siemann-Herzberg, and C. Syldatk. 2001. Hydantoinases and related enzymes as biocatalysts for the synthesis of unnatural chiral amino acids. Curr. Opin. Biotechnol. 12:559-563. [DOI] [PubMed] [Google Scholar]
- 4.Asano, Y. 2000. New enzyme acting on peptides containing d-amino acids: their properties and application. J. Microbiol. Biotechnol. 10:573-579. [Google Scholar]
- 5.Asano, Y., H. Ito, T. Dairi, and Y. Kato. 1996. An alkaline d-stereospecific endopeptidase with β-lactamase activity from Bacillus cereus. J. Biol. Chem. 271:30256-30262. [DOI] [PubMed] [Google Scholar]
- 6.Asano, Y., K. Kishino, A. Yamada, S. Hanamoto, and K. Kondo. 1991. Plasmid-based, d-aminopeptidase-catalysed synthesis of (R)-amino acids. Recl. Trav. Chim. Pays-Bas Belg. 110:206-208. [Google Scholar]
- 7.Asano, Y., T. Mori, S. Hanamoto, Y. Kato, and A. Nakazawa. 1989. A new d-stereospecific amino acid amidase from Ochrobactrum anthropi. Biochem. Biophys. Res. Commun. 162:470-474. [DOI] [PubMed] [Google Scholar]
- 8.Asano, Y., A. Nakazawa, Y. Kato, and K. Kondo. 1989. Properties of a novel d-stereospecific aminopeptidase from Ochrobactrum anthropi. J. Biol. Chem. 264:14233-14239. [PubMed] [Google Scholar]
- 9.Asano, Y., and K. Yamaguchi. 1995. Mutants of d-aminopeptidase with increased thermal stability. J. Ferment. Bioeng. 79:614-616. [Google Scholar]
- 10.Asano, Y., and S. Yamaguchi. 2005. Discovery of amino acid amides as new substrate for α-amino-ɛ-caprolactam racemase from Achromobacter obae. J. Mol. Catal. B Enzym. 36:22-29. [Google Scholar]
- 11.Asano, Y., and S. Yamaguchi. 2005. Dynamic kinetic resolution of amino acid amide catalyzed by d-aminopeptidase and α-amino-ɛ-caprolactam racemase. J. Am. Chem. Soc. 127:7696-7697. [DOI] [PubMed] [Google Scholar]
- 12.Fukumura, T. 1977. Conversion of d- and dl-α-amino-ɛ-caprolactam into l-lysine using both yeast cells and bacterial cells. Agric. Biol. Chem. 41:1327-1330. [Google Scholar]
- 13.Komeda, H., H. Harada, S. Washika, T. Sakamoto, M. Ueda, and Y. Asano. 2004. S-Stereoselective piperazine-2-tert-butylcaroxamide hydrolase from Pseudomonas azotoformans IAM 1603 is a novel l-amino acid amidase. Eur. J. Biochem. 271:1465-1475. [DOI] [PubMed] [Google Scholar]
- 14.Komeda, H., H. Harada, S. Washika, T. Sakamoto, M. Ueda, and Y. Asano. 2004. A novel r-stereoselective amidase from Pseudomonas sp. MCI3434 acting on piperazine-2-tert-butylcaroxamide. Eur. J. Biochem. 271:1580-1590. [DOI] [PubMed] [Google Scholar]
- 15.Komeda, H., N. Hariyama, and Y. Asano. 2006. l-Stereoselective amino acid amidase with broad substrate specificity from Brevundimonas diminuta: characterization of a new member of the leucine aminopeptidase family. Appl. Microbiol. Biotechnol. 70:412-421. [DOI] [PubMed] [Google Scholar]
- 16.Leuchtenberger, W., K. Huthmacher, and K. Drauz. 2005. Biotechnological production of amino acids and derivatives: current status and prospects, Appl. Microbiol. Biotechnol. 69:1-8. [DOI] [PubMed] [Google Scholar]
- 17.May, O., P. T. Nguyen, and F. H. Arnold. 2000. Inverting enantioselectivity by directed evolution of hydantoinase for improved production of l-methionine, Nat. Biotechnol. 18:317-320. [DOI] [PubMed] [Google Scholar]
- 18.Nakai, T., T. Hasegawa, E. Yamashita, M. Yamamoto, T. Kumasaka, T. Ueki, H. Nanba, Y. Ikenaka, S. Takahashi, and M. Sato. 2000. Crystal structure of N-carbamyl-d-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure 8:729-739. [DOI] [PubMed] [Google Scholar]
- 19.Nanba, H., Y. Ikenaka, Y. Yamada, K. Yajima, M. Takano, and S. Takahashi. 1998. Isolation of Agrobacterium sp. strain KNK712 that produces N-carbamyl-d-amino acid amidohydrolase, cloning of the gene for this enzyme, and properties of the enzyme. Biosci. Biotechnol. Biochem. 62:875-881. [DOI] [PubMed] [Google Scholar]
- 20.Nozaki, H., I. Kira, K. Watanabe, and K. Yokozeki. 2005. Purification and properties of d-hydantoin hydrolase and N-carbamoyl-d-amino acid amidohydrolase from Flavobacterium sp. AJ1199 and Pasteurella sp. AJ11221. J. Mol. Catal. B 32:205-211. [Google Scholar]
- 21.Nozaki, H., Y. Takenaka, I. Kira, K. Watanabe, and K. Yokozeki. 2005. d-amino acid production by E. coli co-expressed three genes encoding hydantoin racemase, d-hydantoinase and N-carbamoyl-d-amino acid amidohydrolase. J. Mol. Catal. B 32:213-218. [Google Scholar]
- 22.Ogawa, J., and S. Shimizu. 1997. Diversity and versatility of microbial hydantoin-transforming enzymes. J. Mol. Catal. B 2:163-176. [Google Scholar]
- 23.Sano, K., and K. Mitsugi. 1978. Enzymatic production of l-cysteine from dl-2-amino-Δ2-thiazoline-4-carboxylic acid by Pseudomonas thiazolinophilum: optimal conditions for enzyme formation and enzymatic reaction. Agric. Biol. Chem. 41:2315-2321. [Google Scholar]
- 24.Suzuki, S., N. Onishi, and K. Yokozeki. 2005. Purification and characterization of hydantoin racemase from Microbacterium liquefaciens AJ 3912. Biosci. Biotechnol. Biochem. 69:530-536. [DOI] [PubMed] [Google Scholar]
- 25.Tokuyama, S., and K. Hatano. 1996. Overexpression of the gene for N-acylamino acids racemase from Amycolatopsis sp. TS-1-60 in Escherichia coli and continuous production of optically active methionine by a bioreactor. Appl. Microbiol. Biotechnol. 44:744-777. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi, S., T. Ohashi, Y. Kii, H. Kumagai, and H. Yamada. 1979. Microbial transformation of hydantoins to N-carbamyl-d-amino acids. J. Ferment. Technol. 57:328-332. [Google Scholar]
- 27.Wakayama, M., K. Yoshimune, Y. Hirose, and M. Moriguchi. 2003. Production of d-amino acids by N-acyl-d-amino acid amidohydrolase and its structure and function. J. Mol. Catal. B 23:71-85. [Google Scholar]