Abstract
Pediocin PA-1 is a member of the class IIa bacteriocins, which show antimicrobial effects against lactic acid bacteria. To develop an improved version of pediocin PA-1, reciprocal chimeras between pediocin PA-1 and enterocin A, another class IIa bacteriocin, were constructed. Chimera EP, which consisted of the C-terminal half of pediocin PA-1 fused to the N-terminal half of enterocin A, showed increased activity against a strain of Leuconostoc lactis isolated from a sour-spoiled dairy product. To develop an even more effective version of this chimera, a DNA-shuffling library was constructed, wherein four specific regions within the N-terminal half of pediocin PA-1 were shuffled with the corresponding sequences from 10 other class IIa bacteriocins. Activity screening indicated that 63 out of 280 shuffled mutants had antimicrobial activity. A colony overlay activity assay showed that one of the mutants (designated B1) produced a >7.8-mm growth inhibition circle on L. lactis, whereas the parent pediocin PA-1 did not produce any circle. Furthermore, the active shuffled mutants showed increased activity against various species of Lactobacillus, Pediococcus, and Carnobacterium. Sequence analysis revealed that the active mutants had novel N-terminal sequences; in active mutant B1, for example, the parental pediocin PA-1 sequence (KYYGNGVTCGKHSC) was changed to TKYYGNGVSCTKSGC. These new and improved DNA-shuffled bacteriocins could prove useful as food additives for inhibiting sour spoilage of dairy products.
Many food industries, such as those involved in producing cooked meat and fish, suffer losses due to sour spoilage (37, 44, 47). This type of spoilage is mainly caused by lactic acid bacteria (LAB), such as Leuconostoc spp., Lactobacillus spp., and Carnobacterium spp., which produce abundant lactic acid via sugar metabolism (37, 44, 47). In recent years, researchers have examined the possibility of using bacteriocins, a group of LAB-directed antimicrobial peptides, to guard against sour spoilage (37). Among the bacteriocins, the class IIa (pediocin-like family) bacteriocins have been widely studied because they are digestible and seemingly safe for human use and show activity against both LAB and the food-borne pathogen Listeria monocytogenes (1, 3, 10, 14, 16, 41). The class IIa bacteriocins are composed of 37 to 48 residues and have a characteristic YGNGV/L consensus sequence. The more than 20 class IIa bacteriocins identified to date (18) may be structurally and functionally divided into two independent domains, a highly homologous N-terminal domain and a variable C-terminal domain (20, 27). Three-dimensional structural analysis has indicated that the N-terminal domain forms a three-stranded β-sheet while the C-terminal domain is folded into an amphiphilic α-helical structure (23, 25, 46, 48). The bacteriocins exert their antimicrobial activity by associating with target bacteria via an electrostatic interaction at the N-terminal domain (7, 8, 29). The amphiphilic α-helical domain then inserts into the bacterial membrane via a presumed receptor interaction (39), leading to the formation of a membrane pore on the bacterial cell (9, 26).
Although the class IIa bacteriocins have proven effective as food additives in some cases, their usefulness has been limited by differences in efficacy between and sometimes even within bacterial species (15). Thus, it would be useful to improve the class IIa bacteriocins by making them capable of acting against a wide variety of sour-spoilage-causing LAB species. A number of mutants have been generated by site-directed mutagenesis (17, 19, 21, 22, 28, 29) and error-prone PCR (31, 35, 38), but few showed higher activities than the parent peptides. This may suggest that the parent bacteriocins isolated from nature had been functionally optimized through natural selection processes. This would explain why the generated point mutations resulted in decreased activity and might suggest that it will be necessary to simultaneously substitute multiple residues in order to generate an innovative molecule.
Recently, an in vitro strategy for mimicking natural DNA recombination processes has been applied for the functional improvement of various proteins (12, 32, 33, 40). In this technique, called DNA shuffling, multiple homologous genes are randomly fragmented by DNase I and then reassembled by PCRs in which the fragments are utilized as primers for each other. The resulting mutant library consists of homologous recombined molecules that differ dramatically from their parents in terms of sequence and function.
Here, DNA shuffling among 10 class IIa bacteriocins was used to generate new peptides from pediocin PA-1. The N-terminal domain was selected for shuffling because this region contains several conserved sequences that could be used as shuffling scaffolds. Within this domain, four regions were selected for shuffling based on multiple alignment and data from a previous study examining which residues were essential or variable for pediocin PA-1 activity (45). The antimicrobial activities of the resulting shuffled peptides against Leuconostoc lactis YKLAB10, which had been isolated from a sour-spoiled prepared food product, were examined. This screening revealed that 63 out of 280 shuffled molecules had higher activities than the parental molecule. Moreover, several mutants with activity against L. lactis YKLAB10 were also active against various species of Lactobacillus, Pediococcus, and Carnobacterium, suggesting that the DNA-shuffled bacteriocins could prove useful as food additives for inhibiting sour spoilage of dairy products.
MATERIALS AND METHODS
Bacterial strains and culture.
Escherichia coli DH5α and E. coli JE5505 were used as the cloning host strain and the bacteriocin leaky host strain, respectively. E. coli cells were cultured at 37°C in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg ml−1) as needed. Indicator strains are shown in Table 1. They were all cultivated in MRS (Difco Laboratories) broth or agar.
TABLE 1.
Bacterial strains used as indicators in this study
| Organism | Strain(s)a |
|---|---|
| Leuconostoc lactis | JCM 6123T, JCM 11052, NBRC 12455 |
| Leuconostoc mesenteroices subsp. | |
| mesenteroides | IAM 13004T, NBRC 3832, NBRC 12060 |
| Leuconostoc carnosum | JCM 9695T |
| Leuconostoc citreum | JCM 9698T |
| Leuconostoc fallax | JCM 9694T |
| Leuconostoc ficulneum | JCM 12225T |
| Leuconostoc fructosum | NBRC 3516T |
| Leuconostoc gasicomitatum | JCM 12535T |
| Leuconostoc gelidum | JCM 9697T |
| Leuconostoc pseudomesenteroides | JCM 9696T |
| Carnobacterium maltaromaticum | NBRC 15684 |
| Lactobacillus curvatus | NBRC 15884T |
| Lactobacillus plantarum | NBRC 3075, NBRC 14711, NBRC 14713 |
| Pediococcus parvulus | NBRC 100673T |
| Pediococcus pentosaceus | JCM 2026 |
| Weisella confusa | NBRC 3955 |
| Weisella hellenica | NBRC 15553T |
JCM, Japan Collection of Microorganisms; NBRC, NITE Biological Resource Center.
Isolation and identification.
BCP plate count agar (Nissui) was used to isolate bacterial strain L. lactis YKLAB10 from a sour-spoiled prepared food product. L. lactis YKLAB10 was identified through morphological, cultural, biochemical (e.g., API 50CHL medium and API 50CH strips; bioMérieux), and genetic analyses. The 16S rRNA gene and the intergenic transcribed spacer region between the 16S and 23S rRNA genes were sequenced as described previously (2, 30, 34).
Construction of the pFLAG-enterocin A, -divercin V41, -carnobacteriocin BM1, -EP, and -PE plasmids and their derivative plasmids.
The presently used oligonucleotide and primer sequences are listed in the supplemental material. The enterocin A, divercin V41, and carnobacteriocin BM1-encoding fragment was obtained essentially as described previously (45), using EntF and EntR, DivF and DivR, and CarF and CarR, respectively, as oligonucleotides and EntAExtF and EntACR, DivExtF and DivExtR, and CarExtF and CarExtR, respectively, as primers. The resultant fragment was cloned into the pFLAG-ATS expression vector (Sigma). The chimeric constructs using pediocin PA-1 and enterocin A (chimeras EP and PE) (Fig. 1) were generated as follows. First, pFLAG-enterocin A (0.5 μl) was subjected to PCR amplification (first PCR) using, in a final volume of 50 μl, 25 pmol each of primers PA plus Ent and EntACR, 2 mM of Mg2+, 0.2 mM of mixed deoxynucleoside triphosphates, and 2.5 units of ExTaq DNA polymerase. The cycling conditions used in this protocol were as follows: 3 min at 95°C followed by 25 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C, followed by a final soak for 10 min at 72°C. Cycling was performed in an iCycler (Bio-Rad). A second PCR was performed using pFLAG-pediocin PA-1 as the template, along with 2.5 μl of the products of the first PCR and 25 pmol of pUCHinF as primers. The third PCR was performed using 0.5 μl of the products of the second PCR as the template, along with 25 pmol each of primers EntACR and pUCHinF. The product of the third PCR was cloned into pFLAG-ATS, yielding pFLAG-PE. pFLAG-EP was constructed using the same procedure as that described for pFLAG-PE, with the first PCR using pFLAG-pediocin PA-1 as the template and primers Ent plus PA and pUCEcoR, the second PCR using pFLAG-enterocin A as the template and primer EntAExtF, and the third PCR using primers EntAExtF and pUCEcoR. PE +TTHSG, EP −TTHSG, Ped +TTHSG, and Ent −TTHSG, chimeric construct variants (Fig. 1), were generated by PCR amplification using pFLAG-PE, pFLAG-EP, pFLAG-pediocin PA-1, and pFLAG-enterocin A, respectively, as the templates and TTHSGadd and pUCEcoR, TTHSGdel and pUCEcoR, TTHSGadd and pUCEcoR, and TTHSGdel and pUCEcoR, respectively, as the primers, followed by cloning into pFLAG-ATS.
FIG. 1.
Amino acid sequences of pediocin PA-1 (Ped), enterocin A (Ent), and the constructed chimeric bacteriocins. Residues in common between pediocin PA-1 and enterocin A are in black boxes. Regarding the chimeric bacteriocins, the fragments derived from pediocin PA-1 are in black boxes. The peptide sequences were derived from the NCBI protein database under the following accession numbers: pediocin PA-1, AAA25559, and enterocin A, CAA63890.
DNA shuffling.
The strategy for constructing the DNA-shuffling library is depicted schematically in Fig. 2B. The recombinant molecules were produced by a synthetic method in which four rounds of PCR were used to sequentially extend the products. The first round of PCR consisted of two independent amplifications of pFLAG-pediocin PA-1 with 15 pmol either of primer CSVDWGKA or CSVNWGKA, along with 15 pmol of primer pUCEcoR, in a final volume of 50 μl containing 1 mM of MgSO4, 0.2 mM of mixed deoxynucleoside triphosphates, and 1 unit of KOD-Plus (Toyobo). The PCR amplification conditions for all steps of library construction were as follows: 2 min at 94°C followed by 25 cycles of 15 s at 94°C, 30 s at 50°C, and 30 s at 68°C, followed by a final soak for 10 min at 68°C. The resulting products (1 ng each) were mixed and used as templates for the second PCRs, wherein the CSV scaffold introduced during the first PCR was used as the scaffold for the addition of the second layer of sequences. Individual second PCRs were performed using forward primers LeuA1st, Mund1st, Pis1st, Bav1st, EntP1st, CarBM1st, SakA1st, CarB2-1st, Ped1st, or EntA1st and reverse primer pUCEcoR. The resulting products (1 ng each) were mixed and used as the template for the third PCRs, wherein the NGV introduced in the second PCRs was used as a scaffold for continued extension. The third PCRs were performed separately, using forward primer NGVHC, NGVSC, NGVTC, or NGVYC, along with reverse primer pUCEcoR. The resulting products (1 ng each) were mixed and used as the templates for the fourth PCRs, wherein the NGV generated by the third PCRs was used as a scaffold for continued extension. The fourth PCRs were performed separately, using forward primer Bav2nd, CarBM1-2nd, EntA2nd, or Ped2nd, along with reverse primer pUCEcoR. The resulting PCR products, designated, respectively, groups B (from the Bav2nd forward primer in the fourth PCR), C (CarBM1-2nd), E (EntA2nd), and P (Ped2nd), were separately purified and ligated into pFLAG-ATS.
FIG. 2.
(A) Multiple sequence alignment of class IIa bacteriocins used in the DNA-shuffling experiment. The common residues in all bacteriocins are in black boxes. The peptide sequences were derived from the NCBI protein database under the following accession numbers: pediocin PA-1, AAA25559; enterocin A, CAA63890; leucocin A, AAA68003; mundticin KS, BAB88211; piscicolin 126, P80569; bavaricin MN, P80493; divercin V41, CAA11804; enterocin P, AAC45870; carnobacteriocin BM1, AAA23014; sakacin A, CAA86942; and carnobacteriocin B2, AAA72431. (B) DNA-shuffling strategy. In the first PCR, CSVDWGKA or CSVNWGKA was used as a forward primer. In the second PCR, Ped1st, Ent1st, LeuA1st, Mund1st, Pis1st, Bav1st, EntP1st, CarBM1st, SakA1st, or CarB2-1st was used as a forward primer. In the third PCR, NGVHC, NGVSC, NGVTC, or NGVYC was used as a forward primer. In the fourth PCR, Bav2nd, EntA2nd, Ped2nd, or CarBM1-2nd was used as a forward primer. All primers used in this study are listed in the supplemental material. There were no degenerate primers.
Determination of antimicrobial activity.
Colony overlay assays were performed as described previously (45). Briefly, single E. coli JE5505 colonies containing the desired expression vector were picked with toothpicks and touched lightly onto the surface of 2.5% LB agar plates. The plates were incubated at 37°C for 4 h and then overlain with 5 ml of 1.2% melted MRS soft agar containing 1 mM isopropyl-β-d-thiogalactopyranoside and indicator culture corresponding to an optical density at 600 nm of 0.03 (∼107 CFU). After the agar solidified, 5 ml of 1.2% melted soft agar was poured onto each plate, and the plates were incubated at 30°C for 21 h for the development of growth inhibition zones. On the other hand, Leuconostoc gelidum was cultured at 25°C for 45 h and Leuconostoc gasicomitatum was cultured at 22°C for 45 h when both bacteria were used as indicators. The peptide activities were assessed by measuring the diameter of each inhibition zone.
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA gene and the intergenic transcribed spacer region between the 16S and 23S rRNA genes of L. lactis YKLAB10 were deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers AB295116 and AB295117. The sequences of the synthetic enterocin A, divercin V41, and carnobacteriocin BM1 were deposited in DDBJ under accession numbers AB270898, AB306976, and AB306977.
RESULTS
Isolation and identification of sour spoilage LAB.
BCP plate count agar was used to isolate a bacterial strain from a sour-spoiled prepared food product. The strain appeared to be acid-producing, based on its ability to change the color of nearby agar from purple to yellow. Morphological, cultural, and biochemical analyses were used to identify the strain as being either Leuconostoc mesenteroides subsp. mesenteroides or Leuconostoc lactis. Genetic analysis of 16S rRNA gene sequences revealed that the 16S rRNA gene of strain YKLAB10 was 97.8% identical to that of L. mesenteroides subsp. mesenteroides but was 99.9% identical with that of L. lactis, indicating that strain YKLAB10 was L. lactis (2). In agreement with this conclusion, a partial sequence from the intergenic transcribed spacer region of strain YKLAB10 was only 89.3% identical to that of L. mesenteroides subsp. mesenteroides but showed 98.4% identity with that of L. lactis. Therefore, strain YKLAB10 was identified as L. lactis and was duly designated L. lactis YKLAB10.
Activities of the chimeric bacteriocins.
Pediocin PA-1 and enterocin A show relatively high activities compared to those of the other class IIa bacteriocins, and their target specificities frequently differ (15). To examine whether pediocin PA-1 and/or enterocin A showed any activity against L. lactis YKLAB10, pediocin PA-1 (45) and enterocin A were constructed and their activities were examined. Neither of the bacteriocins showed antimicrobial activity against the test strain (data not shown). Some previous studies have reported increased activity in chimeric bacteriocins in which the N- or C-terminal domain of one bacteriocin was replaced with that of another (27). Based on these previous findings, two reciprocal chimeras between pediocin PA-1 and enterocin A (chimeras EP and PE) (Fig. 1) were constructed and evaluated for activity against L. lactis YKLAB10. PE showed no detectable activity against the test microbe. In contrast, EP-producing E. coli showed antibacterial activity in a colony overlay assay (data not shown). The N terminus of EP contained a TTHSG sequence, which is characteristic of enterocin A (Fig. 1). To examine whether EP activity depended on this motif, four other bacteriocin variants containing additions or deletions of TTHSG were constructed (Fig. 1). Activity assays revealed that only EP showed detectable antimicrobial activity, yielding an inhibition circle 3.1 ± 0.24 mm in diameter (n = 7; data not shown). This suggests that the TTHSG motif is necessary but not sufficient for the activity.
Activities of DNA-shuffled bacteriocins.
The findings that EP was more active than either parent peptide and that the TTHSG motif was necessary but not sufficient for activity suggested that the increased activity was due to the exchange of multiple residues within the N-terminal domains of pediocin PA-1 and enterocin A. Multiple sequence alignment of representative class IIa bacteriocins (Fig. 2A) compared to pediocin PA-1 showed that the other family members varied at some N-terminal residues, including the extreme N terminus, as well as T8, G10 to S13, and D17 (the amino acid numbers correspond to those of pediocin PA-1). Consistent with this, a previous study showed that all of these candidate residues, with the exception of D17, could be varied without affecting the antimicrobial activity of pediocin PA-1 (45). Based on these findings, a DNA-shuffling library in which the N terminus, T8, G10 to S13, and D17 of pediocin PA-1 were shuffled with the corresponding residues from 10 other class IIa bacteriocins was constructed. Then, to examine the feasibility of the strategy, the activities of 70 mutants in each of the four shuffled groups (designated groups B, E, P, and C, based on their N-terminal ends; refer to the fourth-PCR primers in Fig. 2B), which were estimated to cover more than 50% of all possible mutants, were examined (data not shown). No group C mutant showed antimicrobial activity, but 16 active mutants were obtained from group P, 17 from group E, and 30 from group B, for a total of 63 active mutants isolated from 280 DNA-shuffled mutants. Notably, some of these mutants showed enhanced activities compared to that of EP (Fig. 3A).
FIG. 3.
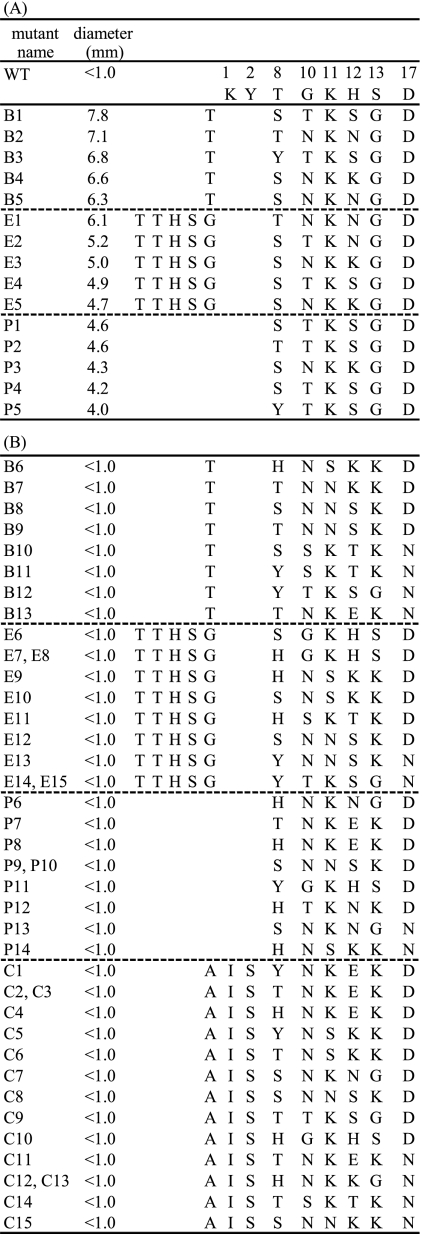
Sequence analysis of highly active (A) and inactive (B) mutants against L. lactis YKLAB10. Only the N-terminal domain of pediocin PA-1 is shown. Mutant names correspond to group B, E, P, or C. For example, B1 belongs to mutants of group B. The diameters of the inhibition circles of the inactive mutants, whose activity was under the detection level, are given as <1.0 mm. WT indicates parental pediocin PA-1. The numbers above the columns on the right correspond to the amino acid numbers of pediocin PA-1.
Sequence analysis of active and inactive DNA-shuffled mutants.
To examine whether the active mutants shared any sequence characteristics, 15 highly active mutants from groups B, E, and P were sequenced (Fig. 3A). All of the sequenced mutants had an aspartic acid at position D17, suggesting that aspartic acid was likely to be essential for activity. At the G10-to-S13 region, although 1 unpredicted sequence was observed (E2; TKNG), probably due to PCR error, only 3 out of 10 possible sequences were observed (TKSG, NKNG, and NKKG), suggesting that these sequences may be associated with antimicrobial activity. As for T8, every possible residue was observed except histidine, suggesting that histidine may block antimicrobial activity at this position.
To examine whether these apparent trends were the result of PCR bias during library construction or reflected essentiality for the activity, 42 inactive mutants of groups B, E, P, and C were sequenced and the sequences compared with the above-described results (Fig. 3B). At position D17, both aspartic acid and asparagine were seen in the inactive mutants; however, approximately two-thirds of the sequences contained aspartic acid at this position, suggesting some PCR preference. At G10 to S13, all 10 possible sequences were found in the inactive mutants, indicating that the DNA shuffling occurred randomly at this site. At T8, all four possible residues were found in the inactive mutants, again arguing against the influence of PCR bias on the present results.
Antimicrobial spectrum of DNA-shuffled mutants.
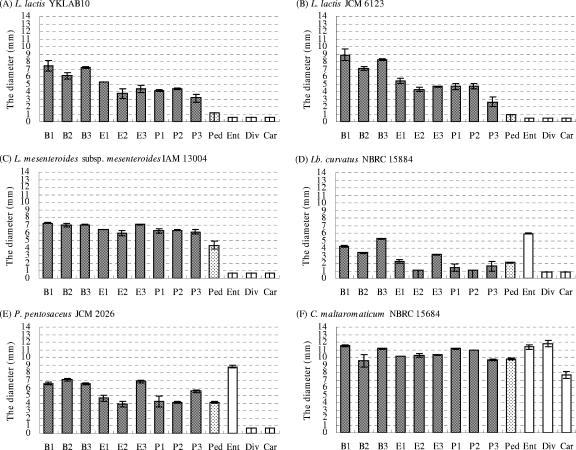
To examine whether shuffled mutants with activity against L. lactis YKLAB10 were also active against other L. lactis strains, representative active mutants from each group (B1, B2, B3, E1, E2, E3, P1, P2, and P3) were tested for antimicrobial activity against L. lactis. Consistent with their activity against L. lactis YKLAB10 (Fig. 4A), all nine mutants showed higher activities than the parental pediocin PA-1 when tested against L. lactis JCM 6123 (type strain) (Fig. 4B). Similar results were obtained for activity against L. lactis JCM 11052 and NBRC12455 (data not shown). Thus, all of the tested mutants having activity against L. lactis YKLAB10 also showed activity against other L. lactis strains, whereas the parental pediocin PA-1 showed no detectable activity against these four strains.
FIG. 4.
The antimicrobial activities of DNA-shuffled mutants and pediocin PA-1 (Ped), enterocin A (Ent), divercin V41 (Div), and carnobacteriocin BM1 (Car) against L. lactis YKLAB10 (A), L. lactis JCM 6123 (B), L. mesenteroides subsp. mesenteroides IAM 13004 (C), L. curvatus NBRC 15884 (D), P. pentosaceus JCM 2026 (E), and C. maltaromaticum NBRC 15684 (F) as indicated by the diameters of their inhibition circles. Bavaricin MN was not sequenced completely, so divercin V41, which had almost the same sequence as bavaricin MN (Fig. 2), was synthesized instead. The mutant names, as shown also in Fig. 3, are shown under the horizontal axis. The data represent the average values from two independent experiments, and the error bars show standard deviations.
Several Leuconostoc bacteria were then used in activity assays in order to examine whether the activity of the shuffled mutants extended to other species. Mutants B1, B2, B3, and E3 were active against L. gelidum JCM 9697 (data not shown) and Leuconostoc carnosum JCM 9695, while only B1 and B3 showed activity against L. gasicomitatum JCM 12535. In contrast, parental pediocin PA-1 and all of the shuffled mutants showed activity against L. mesenteroides subsp. mesenteroides IAM 13004 (Fig. 4C), NBRC 3832, NBRC 12060, and Leuconostoc pseudomesenteroides JCM 9696 (data not shown), with the shuffled mutants showing consistently higher activity than the parental pediocin PA-1. No activity against Leuconostoc citreum JCM 9698, Leuconostoc fallax JCM 9694, Leuconostoc ficulneum JCM 12225, or Leuconostoc fructosum NBRC 3516 (data not shown) was detected.
The active mutants always showed higher activities than the parental pediocin PA-1 against members of the genus Leuconostoc. Then, non-Leuconostoc LAB strains were used as indicators to compare activity levels among the mutants and parental pediocin PA-1. The activities against Lactobacillus plantarum NBRC 14711, NBRC 14713, and NBRC 3075 were similar to those observed for members of Leuconostoc (data not shown). However, mutants E2 and P2 showed lower activities than the parental pediocin PA-1 against Lactobacillus curvatus NBRC 15884, whereas mutants B1, B2, B3, and E3 showed higher activities than the parental pediocin PA-1 (Fig. 4D). The latter four mutants also showed higher activities than the parental pediocin PA-1 against Pediococcus pentosaceus JCM 2026 (Fig. 4E) and Pediococcus parvulus NBRC 100673. However, out of these four mutants, only B1 and B3 showed distinctly higher activities than the parental pediocin PA-1 against Carnobacterium maltaromaticum NBRC 15684 (Fig. 4F). These observations suggest that shuffled mutants showing activity against members of genus Leuconostoc did not always show higher activity than the parental pediocin PA-1 against other LAB genera. However, B1 and B3 were generally more active than the parental pediocin PA-1 against the indicator strains used in this study. On the other hand, the levels of activity of B1 and B3 did not reach the level of activity of enterocin A, which has a C-terminal half different from that of pediocin PA-1 and all present shuffled mutants. Finally, no detectable activity against Weissella confusa NBRC 3955 or Weissella hellenica NBRC 15553 from either mutant or parental pediocin PA-1 was observed.
DISCUSSION
In the present study, four regions of the N-terminal domain of pediocin PA-1 were shuffled with the corresponding residues from 10 other class IIa bacteriocins, and some of the novel molecules generated showed increased antimicrobial activity against sour spoilage LAB.
In considering which bacteriocin residues would be appropriate for shuffling, the relatively well-conserved N-terminal domain was chosen as the starting scaffold. A previous study examining essential and variable residues of pediocin PA-1 using saturation mutagenesis (NNK scanning) (45) showed that residues K1, T8, and G10 to S13 of the N-terminal domain could be replaced without affecting the antimicrobial activity of pediocin PA-1. In addition, multiple alignment of various bacteriocins suggested that D17 might be substituted for N17. Accordingly, these four regions of the N-terminal domain of pediocin PA-1 were shuffled with the corresponding residues from 10 other class IIa bacteriocins, yielding 63 active mutants in a partial screen of the libraries.
In general, DNA shuffling utilizes DNase I for the fragmentation of parent proteins. In the case of class IIa bacteriocins, however, the parent peptides are so short in length that it was feared that DNase I digestion might yield fragments which could be too small to effectively ligate in subsequent steps. Recently, a synthetic PCR-based method not using DNase I was reported for DNA shuffling of growth factors and subtilisin (11, 36), which used synthetic oligonucleotides as fragmented DNA and linked them by annealing the homologous scaffold and subsequently performing PCR. Thus, the synthetic method was applied for the class IIa bacteriocins, with modifications to allow DNA shuffling by sequential PCR extension reactions, since the bacteriocins were too short to anneal the homologous region adequately in comparison with the annealing achieved in the previous studies. To avoid the generation of biased products, separate PCRs were performed during each step and equal amounts of each product were mixed for the following step. The sequence analysis revealed that the resulting DNA-shuffled bacteriocins had low PCR bias, confirming the validity of the present modified method.
Sequence analysis of the active and inactive mutants suggests the existence of a consensus-like sequence governing antimicrobial activity in these peptides, although we did not evaluate the activities of all possible mutants. Although there appeared to be some PCR bias at D17, all active mutants had aspartic acid at this residue, whereas the inactive mutants had either aspartic acid or asparagine. Our previous study suggested that few substitutions at this residue could retain activity (45). In addition, substitution of D17 for N17 in sakacin P resulted in 3- to 20-fold reductions in potency (29). Moreover, it has been speculated that the negatively charged D17 residue is required to interact with the C-terminal hairpin domain (27). These studies and the present finding collectively suggest that D17 is important for antimicrobial activity in class IIa bacteriocins whose C-terminal hairpin domain is derived from pediocin PA-1. At residues G10 to S13, 3 out of 10 possible sequence types were observed in the active mutants. Notably, K11 and G13 were found in all active mutants, suggesting that these two residues may be important for increased activity. The G10-to-S13 sequence is a region between the C9-C14 disulfide bonds (23, 25, 46, 48) and is considered to be flexible. The positive charge of K11 is considered essential to the initial binding of bacteriocins to target bacteria (29), while glycine is generally thought to confer structural flexibility. Thus, G13 may make the G10-to-S13 region more flexible and allow K11 to contact bacteria more easily and/or strengthen the binding, leading to increased activity. To confirm this hypothesis, the binding affinity for lipid vesicles needs to be compared (8) among mutants in a future study.
The relatively conserved N-terminal domain of class IIa bacteriocins is thought to be involved in initial binding to the bacterial membrane, whereas the more variable C-terminal domain is likely to be involved in recognizing putative receptors that may vary widely among bacterial strains. Therefore, it seems reasonable to hypothesize that functional mutations within the N-terminal domain might be expected to improve bacteriocin function against a wide variety of bacteria, whereas a similar change in the C-terminal domain might improve the function against only a limited number of species. This expectation was largely borne out by the results of the present study. None of the active shuffled mutants showed lower activity than the parental pediocin PA-1 against Leuconostoc species, while some of the mutants showed higher activity than the parental pediocin PA-1 even against members of other genera, including Lactobacillus, Pediococcus, and Carnobacterium, all of which are important bacteria in sour spoilage. For example, L. lactis has been found in spoiled Spanish blood sausage (44), while L. mesenteroides subsp. mesenteroides, L. carnosum, L. gelidum, and L. gasicomitatum have been found in spoiled cooked meat products, vacuum-packaged sausages and ham, and tomato-marinated broiler meat (4, 5, 6, 13, 24, 43), and L. curvatus is a spoilage bacterium of cooked meat (5, 42). However, the activities of mutants were lower than that of enterocin A against L. curvatus, P. pentosaceus, and C. maltaromaticum. This was probably because the C-terminal half of enterocin A might be more effective against these bacterial strains than that of pediocin PA-1. Thus, the future use as food additives of active shuffled mutants with the enterocin A C-terminal half in addition to shuffled mutants with the pediocin PA-1 C-terminal half could help prevent the spoilage of various food products.
None of the tested peptides showed activity against L. citreum or W. confusa, which also cause meat spoilage (24, 44). However, even though 320 (2 × 10 × 4 × 4) types of DNA-shuffled mutants could theoretically be generated in the system utilized, only 280 mutants were screened in the present study. As the sequencing results revealed that the mutants were likely to include redundancy, it is highly probable that additional active shuffled mutants could have been excluded from the present study. The present screening size might cover only ∼60% of the probable mutants, so more than 960 mutants should have been screened to evaluate more than 95% of the probable mutants. In addition, only 10 out of 24 class IIa bacteriocins were used to construct the DNA-shuffling library, meaning that additional effective mutants were likely to have been generated if all class IIa bacteriocins were used as parent molecules. On the other hand, in the present work, DNA shuffling was performed at only four regions within the N-terminal domain. The addition of more regions could lead to the generation of new effective mutants. Lastly, the present method of evaluating activity was only rough, with the assumption that the rates of bacteriocin synthesis and export were the same for all variants. Then, there was the possibility of missing the active mutants by underestimation. A more precise method, like liquid dilution, would overcome this problem. Therefore, future studies using L. citreum or W. confusa as the target, screening with larger pools, employing different parent molecules, and/or including additional shuffled regions, evaluated using the liquid dilution method, may yield active DNA-shuffled bacteriocins against other spoilage-causing LAB.
In nature, DNA shuffling can act to generate novel proteins during the evolutionary process. Here, this natural process was mimicked in vitro, rapidly yielding DNA-shuffled class IIa bacteriocins having different antimicrobial spectra. In the future, it may be possible to use DNA-shuffled bacteriocins to benefit food industries by prolonging the shelf lives of various products.
Supplementary Material
Acknowledgments
We thank Y. Husimi, K. Nishigaki, H. Gotoh, K. Hanada, M. Suzuki, N. Nemoto, M. Nakayama, T. Aita, O. Takei, L. Futatsugi, M. Biyani, Y. Ishijima, Y. Honda, H. Nakajima, and M. Sekine for useful discussions and suggestions and the National Institute of Genetics and IAM culture collection for providing us with bacterial strains. We also thank S. Butler for critical reading of the manuscript.
This work was performed as a part of the Rational Evolutionary Design of Advanced Biomolecules (REDS) Project, Saitama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, supported by JST.
Footnotes
Published ahead of print on 25 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abee, T., L. Krockel, and C. Hill. 1995. Bacteriocins: modes of action and potentials in food preservation and control of food poisoning. Int. J. Food Microbiol. 28:169-185. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou, R., S. Yoon, F. Breidt, Jr., H. P. Fleming, and T. R. Klaenhammer. 2002. Identification and characterization of Leuconostoc fallax strains isolated from an industrial Sauerkraut fermentation. Appl. Environ. Microbiol. 68:2877-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhunia, A. K., M. C. Johnson, and B. Ray. 1988. Purification, characterization and antimicrobial spectrum of a bacteriocin produced by Pediococcus acidilactici. J. Appl. Bacteriol. 65:261-268. [DOI] [PubMed] [Google Scholar]
- 4.Björkroth, K. J., P. Vandamme, and H. J. Korkeala. 1998. Identification and characterization of Leuconostoc carnosum, associated with production and spoilage of vacuum-packaged, sliced, cooked ham. Appl. Environ. Microbiol. 64:3313-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Björkroth, K. J., R. Geisen, U. Schillinger, N. Weiss, P. De Vos, W. H. Holzapfel, H. J. Korkeala, and P. Vandamme. 2000. Characterization of Leuconostoc gasicomitatum sp. nov., associated with spoiled raw tomato-marinated broiler meat strips packaged under modified-atmosphere conditions. Appl. Environ. Microbiol. 66:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, Y., Y. Benno, A. Takeda, T. Yoshida, T. Itaya, and T. Nakase. 1998. Characterization of Leuconostoc species isolated from vacuum-packaged ham. J. Gen. Appl. Microbiol. 44:153-159. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., R. Shapira, M. Eisensterin, and T. J. Montville. 1997. Functional characterization of pediocin PA-1 binding to liposomes in the absence of a protein receptor and its relationship to a predicted tertiary structure. Appl. Environ. Microbiol. 63:524-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., R. D. Ludescher, and T. J. Montville. 1997. Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phospholipid vesicles. Appl. Environ. Microbiol. 63:4770-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chikindas, M. L. M. J. García-Garcerá, A. J. M. Driessen, A. M. Ledeboer, J. Nissen-Meyer, I. F. Nes, T. Abee, W. N. Konings, and G. Venema. 1993. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl. Environ. Microbiol. 59:3577-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 11.Coco, W. M., L. P. Encell, W. E. Levinson, M. J. Crist, A. K. Loomis, L. L. Licato, J. J. Arensdorf, N. Sica, P. T. Pienkos, and D. J. Monticello. 2002. Growth factor engineering by degenerate homoduplex gene family recombination. Nat. Biotechnol. 20:1246-1250. [DOI] [PubMed] [Google Scholar]
- 12.Crameri, A., S. Raillard, E. Bermudez, and W. P. C. Stemmer. 1998. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391:288-291. [DOI] [PubMed] [Google Scholar]
- 13.Dykes, G. A., T. E. Cloete, and A. von Holy. 1994. Identification of Leuconostoc species associated with the spoilage of vacuum-packaged Vienna sausages by DNA-DNA hybridization. Food Microbiol. 11:271-274. [Google Scholar]
- 14.Eijsink, V. G. H., L. Axelsson, D. B. Diep, L. S. Håvarstein, H. Holo, and I. F. Nes. 2002. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Leeuwenhoek 81:639-654. [DOI] [PubMed] [Google Scholar]
- 15.Eijsink, V. G. H., M. Skeie, P. H. Middelhoven, M. B. Brurberg, and I. F. Nes. 1998. Comparative studies of class IIa bacteriocions of lactic acid bacteria. Appl. Environ. Microbiol. 64:3275-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ennahar, S., K. Sonomoto, and A. Ishizaki. 1999. Class IIa bacteriocins from lactic acid bacteria: antibacterial activity and food preservation. J. Biosci. Bioeng. 87:705-716. [DOI] [PubMed] [Google Scholar]
- 17.Fimland, G., J. Pirneskoski, J. Kaewsrichan, A. Jutila, P. E. Kristiansen, P. K. J. Kinnunen, and J. Nissen-Meyer. 2006. Mutational analysis and membrane-interactions of the β-sheet-like antimicrobial peptide sakacin P. Biochim. Biophys. Acta 1764:1132-1140. [DOI] [PubMed] [Google Scholar]
- 18.Fimland, G., L. Johnsen, B. Dalhus, and J. Nissen-Meyer. 2005. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J. Peptide Sci. 11:688-696. [DOI] [PubMed] [Google Scholar]
- 19.Fimland, G., L. Johnsen, L. Axelsson, M. B. Brurberg, I. F. Nes, V. G. H. Eijsink, and J. Nissen-Meyer. 2000. A C-terminal disulfide bridge in pediocin-like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J. Bacteriol. 182:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fimland, G., O. R. Blingsmo, K. Sletten, G. Jung, I. F. Nes, and J. Nissen-Meyer. 1996. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl. Environ. Microbiol. 62:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fimland, G., V. G. H. Eijsink, and J. Nissen-Meyer. 2002. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 41:9508-9515. [DOI] [PubMed] [Google Scholar]
- 22.Fleury, Y., M. A. Dayem, J. J. Montagne, E. Chaboisseau, J. P. L. Caer, P. Nicolas, and A. Delfour. 1996. Covalent structure, synthesis, and structure-function studies of mesentericin Y 10537, a defensive peptide from gram-positive bacteria Leuconostoc mesenteroides. J. Biol. Chem. 271:14421-14429. [DOI] [PubMed] [Google Scholar]
- 23.Fregeau Gallagher, N. L., M. Sailer, W. P. Niemczura, T. T. Nakashima, M. E. Stiles, and J. C. Vederas. 1997. Three-dimensional structure of Leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry 36:15062-15072. [DOI] [PubMed] [Google Scholar]
- 24.Hamasaki, Y., M. Ayaki, H. Fuchu, M. Sugiyama, and H. Morita. 2003. Behavior of psychotrophic lactic acid bacteria isolated from spoiling cooked meat products. Appl. Environ. Microbiol. 69:3668-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haugen, H. S., G. Fimland, J. Nissen-Meyer, and P. E. Kristiansen. 2005. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide curvacin A. Biochemistry 44:16149-16157. [DOI] [PubMed] [Google Scholar]
- 26.Héchard, Y., and H. G. Sahl. 2002. Mode of action of modified and unmodified bacteriocins from gram-positive bacteria. Biochimie 84:545-557. [DOI] [PubMed] [Google Scholar]
- 27.Johnsen, L., G. Fimland, and J. Nissen-Meyer. 2005. The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J. Biol. Chem. 280:9243-9250. [DOI] [PubMed] [Google Scholar]
- 28.Johnsen, L., G. Fimland, V. Eijsink, and J. Nissen-Meyer. 2000. Engineering increased stability in the antimicrobial peptide pediocin PA-1. Appl. Environ. Microbiol. 66:4798-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazazic, M., J. Nissen-Meyer, and G. Fimland. 2002. Mutational analysis of the role of charged residues in target-cell binding, potency and specificity of the pediocin-like bacteriocin sakacin P. Microbiology 148:2019-2027. [DOI] [PubMed] [Google Scholar]
- 30.Klijn, N., A. H. Weerkamp, and W. M. de Vos. 1991. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl. Environ. Microbiol. 57:3390-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, K. W., R. Schamber, O. Osmanagaoglu, and B. Ray. 1998. Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl. Environ. Microbiol. 64:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minshull, J., and W. P. Stemmer. 1999. Protein evolution by molecular breeding. Curr. Opin. Chem. Biol. 3:284-290. [DOI] [PubMed] [Google Scholar]
- 33.Miyazaki, K., M. Takenouchi, H. Kondo, N. Noro, M. Suzuki, and S. Tsuda. 2006. Thermal stabilization of Bacillus subtilis family-11 xylanase by directed evolution. J. Biol. Chem. 281:10236-10242. [DOI] [PubMed] [Google Scholar]
- 34.Mori, K., K. Yamazaki, T. Ishiyama, M. Katsumata, K. Kobayashi, Y. Kawai, N. Inoue, and H. Shinano. 1997. Comparative sequence analyses of the genes coding for 16S rRNA of Lactobacillus casei-related taxa. Int. J. Syst. Bacteriol. 47:54-57. [DOI] [PubMed] [Google Scholar]
- 35.Morisset, D., J.-M. Berjeaud, D. Marion, C. Lacombe, and J. Frère. 2004. Mutational analysis of mesentericin Y105, an anti-Listeria bacteriocin, for determination of impact on bactericidal activity, in vitro secondary structure, and membrane interaction. Appl. Environ. Microbiol. 70:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ness, J. E., S. Kim, A. Gottman, R. Pak, A. Krebber, T. V. Borchert, S. Govindarajan, E. C. Mundorff, and J. Minshull. 2002. Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine independently. Nat. Biotechnol. 20:1251-1255. [DOI] [PubMed] [Google Scholar]
- 37.Paludan-Müller, C., P. Dalgaard, H. H. Huss, and L. Gram. 1998. Evaluation of the role of Carnobacterium piscicola in spoilage of vacuum- and modified-atmosphere-packed cold-smoked salmon stored at 5°C. Int. J. Food Microbiol. 39:155-166. [DOI] [PubMed] [Google Scholar]
- 38.Quadri, L. E. N., L. Z. Yan, M. E. Stiles, and J. C. Vederas. 1997. Effect of amino acid substitutions on the activity of carnobacteriocin B2. J. Biol. Chem. 272:3384-3388. [DOI] [PubMed] [Google Scholar]
- 39.Ramnath, M., S. Arous, A. Gravesen, J. W. Hastings, and Y. Héchard. 2004. Expression of mptC of Listeria monocytogenes induces sensitivity to class IIa bacteriocins in Lactococcus lactis. Microbiology 150:2663-2668. [DOI] [PubMed] [Google Scholar]
- 40.Reetz, M. T. 2004. Controlling the enantioselectivity of enzymes by directed evolution: practical and theoretical ramifications. Proc. Natl. Acad. Sci. USA 101:5716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodríguez, J. M., M. I. Martínez, and J. Kok. 2002. Pediocin PA-1, a wide-spectrum bacteriocin from lactic acid bacteria. Crit. Rev. Food Sci. Nutr. 42:91-121. [DOI] [PubMed] [Google Scholar]
- 42.Samelis, J., A. Kakouri, and J. Rementzis. 2000. Selective effect of the product type and the packaging conditions on the species of lactic acid bacteria dominating the spoilage microbial association of cooked meats at 4 °C. Food Microbiol. 17:329-340. [Google Scholar]
- 43.Samelis, J., A. Kakouri, K. G. Georgiadou, and J. Metaxopoulos. 1998. Evaluation of the extent and type of bacterial contamination at different stages of processing of cooked ham. J. Appl. Microbiol. 84:649-660. [DOI] [PubMed] [Google Scholar]
- 44.Santos, E. M., I. Jaime, J. Rovira, U. Lyhs, H. Korkeala, and J. Björkroth. 2005. Characterization and identification of lactic acid bacteria in “morcilla de Burgos”. Int. J. Food Microbiol. 97:285-296. [DOI] [PubMed] [Google Scholar]
- 45.Tominaga, T., and Y. Hatakeyama. 2006. Determination of essential and variable residues in pediocin PA-1 by NNK scanning. Appl. Environ. Microbiol. 72:1141-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uteng, M., H. H. Hauge, P. R. L. Markwick, G. Fimland, D. Mantzilas, J. Nissen-Meyer, and C. Muhle-Goll. 2003. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P and sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. Biochemistry 42:11417-11426. [DOI] [PubMed] [Google Scholar]
- 47.Vermeiren, L., F. Devlieghere, V. De Graef, and J. Debevere. 2005. In vitro and in situ growth characteristics and behaviour of spoilage organisms associated with anaerobically stored cooked meat products. J. Appl. Microbiol. 98:33-42. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Y., M. E. Henz, N. L. Fregeau Gallagher, S. Chai, A. C. Gibbs, L. Z. Yan, M. E. Stiles, D. S. Wishart, and J. C. Vederas. 1999. Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria. Biochemistry 38:15438-15447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.