Abstract
In tropical ecosystems, termite mound soils constitute an important soil compartment covering around 10% of African soils. Previous studies have shown (S. Fall, S. Nazaret, J. L. Chotte, and A. Brauman, Microb. Ecol. 28:191-199, 2004) that the bacterial genetic structure of the mounds of soil-feeding termites (Cubitermes niokoloensis) is different from that of their surrounding soil. The aim of this study was to characterize the specificity of bacterial communities within mounds with respect to the digestive and soil origins of the mound. We have compared the bacterial community structures of a termite mound, termite gut sections, and surrounding soil using PCR-denaturing gradient gel electrophoresis (DGGE) analysis and cloning and sequencing of PCR-amplified 16S rRNA gene fragments. DGGE analysis revealed a drastic difference between the genetic structures of the bacterial communities of the termite gut and the mound. Analysis of 266 clones, including 54 from excised bands, revealed a high level of diversity in each biota investigated. The soil-feeding termite mound was dominated by the Actinobacteria phylum, whereas the Firmicutes and Proteobacteria phyla dominate the gut sections of termites and the surrounding soil, respectively. Phylogenetic analyses revealed a distinct clustering of Actinobacteria phylotypes between the mound and the surrounding soil. The Actinobacteria clones of the termite mound were diverse, distributed among 10 distinct families, and like those in the termite gut environment lightly dominated by the Nocardioidaceae family. Our findings confirmed that the soil-feeding termite mound (C. niokoloensis) represents a specific bacterial habitat in the tropics.
Termites are recognized as one of the major ecosystem engineers in tropical soils (18, 23). Their effects on soil are caused mostly by their major construction activities, of which their mounds are the most complex type. A termite mound is usually built from a mineral matrix mixed with feces or saliva, depending upon the termite species (16). The construction of these mounds causes both physical changes (in water-holding capacity, bulk density, and structural stability) and chemical changes (in cation-exchange capacity and organic matter content) in the surrounding environment for microorganisms (5, 18, 27). For this reason, termite mounds are a major functional compartment in tropical soils (2), comparable to the drilosphere created by earthworms, which can modify the activity and diversity of microbial soil communities (25). However, although the relationship between earthworm casts and microorganism structures in soil has been well characterized (10), this relationship has not yet been determined for bacterial communities in termite constructions.
The effect of termites on soil is closely linked to their feeding habits and the type of constructions they build (18). Of the six feeding groups described (8), we have chosen to study specifically the microbial compartment of a mound formed by a species of soil-feeding termite (Cubitermes niokoloensis). This choice was based on both the ecological importance of this feeding group (60% of the 2,600 termite species described [11]) and the way in which its mounds are built. This feeding group is the only group to build mounds from a fine mixture of soil and feces containing a dense and specific microbial community. Soil-feeding termite mounds, therefore, provide a useful model for studying the relationship between macrofauna and the soil microbial compartment, which is the principal aim of this study.
Soil-feeding termite mounds (such as those of C. niokoloensis) have very specific properties arising from the combination of materials of two distinct origins, feces and soil (reviewed in reference 5). This creates an increased richness in clay (5 times more), minerals (2 to 3 times more P and Ca and up to 50 times more NH4 [32]), and organic matter (5 to 7 times more C and N [12]) with respect to neighboring soil. The environment for microorganisms derived from soil and feces is modified not just by an increase in available organic compounds but also by a change in their qualities (C/N, humic acid/sugar content [12, 36]) and their availability by the formation of stable clay-humus complexes (14). This richness in organic matter appears to be the reason for the increase in microbial density in termite mounds (3 to 24 times [13]). However, this increase in density is not accompanied by a significant increase in bacterial activity (mineralization) with respect to neighboring soil (31). C. niokoloensis mounds, therefore, provide a site where organic matter is protected from the strong mineralization that is characteristic of the tropical savanna ecosystem. Apart from the physicochemical differences, the termite mounds also have bacterial (13, 17) and fungal (35) communities that are very different from those in neighboring soil.
The aim of this study was to characterize both the structure of the dominant bacterial communities in soil-feeding termite mounds and the specificity of bacterial communities within mounds with respect to the digestive and soil origins of the mound. Using denaturing gradient gel electrophoresis (DGGE) coupled with clonal analysis, the microbial communities in the mound were compared with those of the three main gut segments (anterior gut, midgut, and posterior gut) and with fresh materials used for building mounds (feces and galleries). We were able to demonstrate that the soil-feeding termite mound harbors a bacterial community that differs in terms of structure and diversity from those of its building materials, i.e., termite gut feces and surrounding savanna soil particles, and that the termite mound bacterial community is characterized by the domination of Actinobacteria families. These Actinobacteria clones of soil-feeding termite mound were diverse, distributed among 10 distinct families, and lightly dominated by the Nocardioidaceae family.
MATERIALS AND METHODS
Mound samples.
A whole mound constructed by Cubitermes niokoloensis was collected from a 10-year-old fallow located in the village of Sare Yorobana (12°49′N, 14°53′W) in the east of Casamance (Senegal) and was brought to the laboratory in a vat filled with the soil from the original collection site. The surrounding soil (SS) sample was collected at about 10 m from the mound and was comprised of a pool of 10 subsamples collected from depths of 0 to 10 cm. In the laboratory, the mound was kept in a conservatory at room temperature (27 to 33°C), and the soil under the mound was kept saturated. Measurements were taken within 1 to 15 days after collection.
Termite dissection.
Just after the mound was collected in the field, approximately 50 termites were dissected, and the total guts were pooled in 100 μl sodium dodecyl sulfate (4%). This sample was frozen during transport to the laboratory and before DNA extraction. Between 1 and 2 weeks after the mound had been brought to the laboratory, sampling of additional termite guts was completed as follows. Under a hood, individual worker caste termites were dissected in aseptic conditions. By use of two pairs of tweezers, the whole gut of each termite was extracted by pulling the anus with one set of tweezers and securing the head with the other. The whole guts were divided into three segments (Fig. 1): anterior gut, midgut, and posterior gut. About 150 samples of each gut segment were pooled in 2-ml Eppendorf tubes containing 100 μl sodium dodecyl sulfate (4% wt/vol) solution and stored at −20°C before analysis.
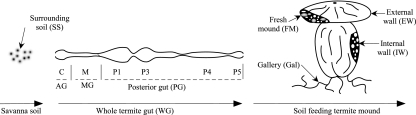
FIG. 1.
Schematics of gut and mound of soil-feeding termites showing the various environments sampled. AG, anterior gut; MG, midgut. Anatomic gut segments: C, crop; M, midgut; P1 to P5, proctodeal segments 1 to 5, respectively.
Soil samples.
When the termites had been dissected, the mound was separated into two different compartments: the external mound wall (EW) and the internal mound wall (IW) (avoiding the termites) (Fig. 1). These mound compartments represent different patterns of soil organic matter evolution and associated microbial communities, as previously described by Fall et al. (12). About 1 kilogram of soil from each compartment was sampled and homogenized separately, using mortar and pestle, and an aliquot of about 100 g of soil was frozen at −20°C for DNA analysis. To compare the mound material with the termite gut contents, portions of around 100 g of homogenized soil from two other samples were collected: fresh mound soil and gallery samples. Fresh mound soil comprised fresh feces (less than 2 h old) from termite workers which had been used to repair the mound wall after it had been partially destroyed by hand. Galleries, which are built using fecal products (15), were sampled 24 h after they had been built in the vat containing the mound.
DNA extraction.
Total DNA was extracted from soil samples and from the various gut segments by use of a direct lysis extraction procedure described previously (13). A subsample (0.5 g for soil samples and 0.25 g for gut samples) was prepared using 0.5 g of glass beads (0.1-mm diameter; Biospec Products, Inc., Bartlesville, OK) and 925 μl of sodium dodecyl sulfate buffer (4% wt/vol) in a 2-ml Eppendorf tube (polypropylene; Poly Labo, France). The subsamples were shaken at maximum speed for 5 min using a Biospec 8TM Mini-BeadBeater. After incubation for 1 h at 68°C, 300 μl of 5 M NaCl was added, and the mixture was vortexed and stored for 5 min on ice. This step precipitated the clay to provide optimum DNA recovery. The DNA was precipitated using 40% polyethylene glycol and 2.5 M ammonium acetate successively. The crude DNA was purified using an S400 HR spin column fast DNA purification kit (Pharmacia Amersham, Freiburg, Germany) and then a QIAGEN mini column (QIAGEN, France) using the procedure described by Ranjard et al. (34).
PCR amplification of 16S rRNA gene and DGGE.
PCR amplifications were performed using the forward primer EUB338 (22) with a GC clamp (29) and reverse primer UNIV518 (33). The total reaction mixture (50 μl) contained 2.6 U of Expand Fidelity PCR Taq (Boehringer Mannheim, Mannheim, Germany), 5 μl PCR buffer (10×), 1.5 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, 500 nM of each primer, 0.25 μl of T4 gene 32 protein (Boehringer Mannheim, Mannheim, Germany), sterile water, and about 50 ng of sample DNA. Three replicates were performed for each sample. A Perkin-Elmer GenAmp PCR system 2400 (Perkin-Elmer, Corporation Norwalk, CT) thermocycler was used for PCR amplification with 5 min at 94°C followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. The first 20 cycles had an annealing temperature of 65°C, which decreased 0.5°C every second cycle until a touchdown at 55°C. The primer extension was carried out at 72°C for 10 min. The PCR products were checked for size on an agarose gel (1.2% for the DGGE product) stained with ethidium bromide. An 8% polyacrylamide gel was run using a D-Gene system (Bio-Rad Laboratories, Hercules, CA) with a denaturing gradient between 30% and 55%. In addition, one gel including all 10 samples was made so that DGGE patterns for the different compartments could be run under identical electrophoresis conditions. A total of 15 μl of PCR product (mixture of three replicates) was loaded. The gel was run for 5 h at 150 V in Tris-acetate-EDTA buffer (0.5×) at 60°C with a prerun of 10 min at 25 V. After migration, the DGGE profiles were stained with SYBR Green (1:10,000 dilutions) for 20 min and then scanned under UV. The gel images were captured using Bio-capt software (Ets Vilber Lourmat, France).
Gel analysis.
The DGGE band patterns were recovered using Gel Compar software (Applied Maths, Belgium), and the matrices of the relative intensities and distances of migration of the bands were obtained. The various DGGE profiles were compared with the Bray-Curtis distance, and the unweighted-pair group method using average linkages was used for hierarchical cluster analysis to produce the dendrogram by use of R 2.0.1 software (R Development Core Team 2004).
Band extraction and purification.
Because the microbial community profiles of the anterior gut and midgut segments were very similar (about 20%) (see Fig. 2, lanes 1 and 2, respectively; cluster analysis) and the profiles of the posterior gut and whole gut were also very similar (see Fig. 2, lanes 3, 4, and 5; cluster analysis), their respective clone libraries were pooled. A clone library originating from one DGGE profile was set up for each cluster (anterior gut and midgut cluster and posterior gut and whole gut cluster). The DGGE bands were extracted from the gel as described by Jensen et al. (22). These extracts were reamplified and reanalyzed by DGGE to check the electrophoretic mobility. The DGGE bands with the expected mobility were excised from the various samples for sequencing.
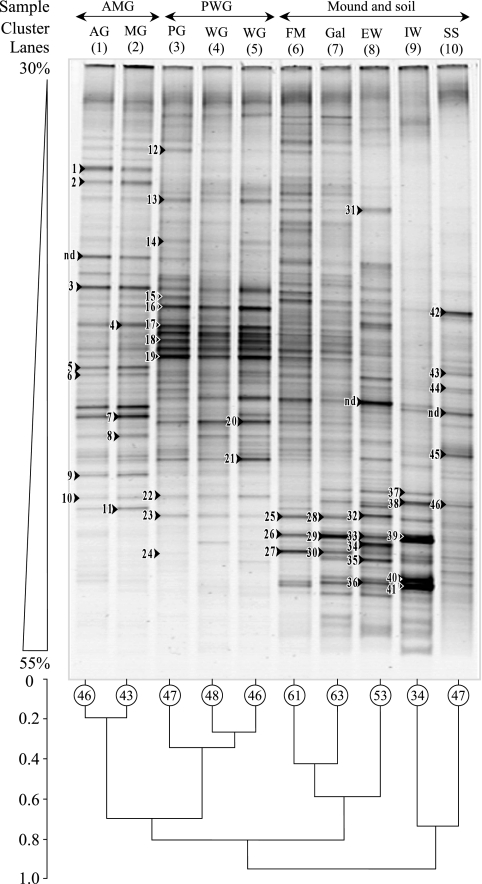
FIG. 2.
DGGE profiles of bacterial 16S rRNA genes amplified from gut and mound environments of soil-feeding termites (Cubitermes niokoloensis) and dendrogram of DGGE profile similarities. The numbers on profiles indicate the bands that were cloned and sequenced. Band richness is marked in the circles. AG, anterior gut; MG, midgut; PG, posterior gut; FM, fresh mound; Gal, gallery; AMG, cluster of anterior and midgut profiles; PWG, cluster of posterior and whole gut profiles.
Sequencing of DGGE bands.
For the DGGE bands, the PCR products were purified with a Nucleotrap PCR purification kit (Macherey-Nagel, GmbH, Düren, Germany). Portions of about 15 ng were cloned into competent Escherichia coli cells (XL1) by use of Promega pGEM-T vector (Promega Corporation, Madison, WI) according to the manufacturer's instructions.
The recombinants were screened by PCR with T7 and SP6 primers to check the size of the inserts, and these PCR products were restricted using HaeIII endonuclease. For each DGGE band, one to three distinct clones with the correct insert size were sequenced.
Construction of 16S rRNA clone library.
A 16S rRNA clone library was constructed from the three compartments: SS, the IW, and the total termite gut. PCR amplifications were performed using the pA (forward) and pH (reverse) (9) primers. The amplification reaction mixture used was the same as that used to amplify DNA for DGGE analysis, as described above. The PCR amplification with a Perkin-Elmer GenAmp PCR system 2400 was as follows: 5 min at 94°C followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. The amplicons were purified in 0.8% agarose gel containing ethidium bromide at a concentration of 0.5 μg ml−1 and were recovered from the gel using GFX PCR DNA and gel purification kits (Amersham Biosciences, Germany); then they were cloned into the pCR 2.1 vector (Invitrogen). Competent cells of E. coli DH5α cells were transformed with the ligations, and white colonies were randomly picked and screened directly for inserts by performing colony PCR with M13r and M13f primers.
Sequencing the 16S rRNA clone library.
Plasmid DNA was prepared from the positive clones with the Nucleotrap PCR purification kit (Macherey-Nagel, GmbH, Düren, Germany). Plasmid DNA was then sequenced using an ABI model 3730xl capillary DNA sequencer (Applied Biosystems, Foster City, CA) and a BigDye v3.1 Terminator cycle sequencing kit (Applied Biosystems).
Diversity measurements.
The Shannon diversity index (H′) (39) and the Simpson index (D) (40) were calculated according to the following equations:
 |
where n equals the total number of clones sequenced for the three compartments (SS, IW, and gut) and pi corresponds to the relative frequency of each phylotype based on 97% similarity between sequences. The nonparametric estimators of coverage and of phylotype richness, CACE (26) and SChao 1 (6), respectively, were calculated for the molecular inventories with the EstimateS software package (version 8.00, R. K. Colwell; http://viceroy.eeb.uconn.edu/estimates).
Phylogenetic analysis.
All of the sequences were compared with similar sequences from the reference organisms by performing BLAST searches (1). Phylogenetic Actinobacteria trees were constructed with the complete 16S rRNA gene sequences by use of the alignment programs in the ARB package (28), which are based on the rRNA secondary structure. To load our sequences, the nearest neighboring species for each 16S rRNA sequence was found using a BLAST search of the NCBI database. The neighboring sequences were considered to be the references and all were downloaded. The sequences were then aligned using ClustalX. The aligned sequences were imported into the ARB database, with T's already substituted by U's. Consensus-aligned sequences were used to align our sequences with the ARB database. The quick add marked species feature of ARB (parsimony) was used to position our sequences, and the position was manually corrected using the neighboring species found with NCBI BLAST.
Nucleotide sequence accession numbers.
The nucleotide sequence data have been deposited in the GenBank database under accession numbers from AY100703 to AY100745, from AY293289 to AY293299, and from DQ347842 to DQ347944.
RESULTS
DGGE fingerprinting.
The bacterial community structure in the mound compartments and gut of the soil-feeding termites (Cubitermes niokoloensis) and in the SS was determined using DGGE (Fig. 2). Visual examination of the DGGE gels (Fig. 2) showed that the termite mound compartments (lanes 8 and 9) had profiles very different from those for the termite gut (lanes 1 to 5) and the SS (lane 10). The differences were particularly noticeable in the profiles for the IW (lane 9). There were several differences between the other environments: a significant reduction in the number of bands was observed (34 bands) with respect to those for the SS and the whole gut (47 and 48 bands, respectively), and the densest bands were exclusively in the lower part of the gel with the strongest denaturing conditions. Eighty-five percent of the bands were situated between 40% and 55% denaturing conditions (Fig. 2), whereas most of the bands (85% to 95%) for the whole gut and different sections of the gut were located in the weak and medium denaturing conditions (30% to 45%). The profiles of the whole gut (lanes 4 and 5) were very close to the profiles of the posterior gut (lane 3) and were characterized by six strong bands situated in the top of the gel, with weak denaturing conditions. In order to determine whether the length of time that termites were kept affected the detectable community structure of bacterial communities in the termite gut, the guts were sampled just after the mounds had been collected (lane 4) and after the termites had been kept for 2 weeks (lane 5). The closeness of these profiles (Fig. 2, lanes 4 and 5) showed that the length of time had little effect on bacterial communities in the gut. Fresh feces, and to a lesser extent the galleries (Fig. 2, lanes 6 and 7), had intermediate DGGE profiles between those of the posterior gut and those of the IW. The SS (Fig. 2, lane 10) was characterized by a fairly large number of weak bands (47 bands).
Cluster analysis.
A cluster analysis of the DGGE profiles (Fig. 2) was carried out to determine similarities between bacterial communities in the different environments studied. For this analysis, the profiles were categorized into three groups. In the first level, the IW and the SS (lanes 9 and 10) were separated from the other environments (digestive tract, fresh feces, and galleries). Although the bacterial communities of the IW and the SS were clustered in this level, there was less than 18% similarity between these communities. In the next level (about 20% similarity) the bacterial communities in the various segments of the gut (lanes 1, 2, and 3) were separated from the soil reworked by the termites (fresh mound, galleries, and EW). Finally, in the third level (i) the bacterial communities in the gut cluster were split, with the bacterial communities in the anterior gut and midgut (lanes 1 and 2) separated from the communities in the posterior gut and whole gut (lanes 3, 4, and 5), and (ii) the bacterial communities in the reworked soil cluster were split between the freshly reworked soil (fresh feces and galleries; lanes 6 and 7, respectively) and the EW (lane 8).
Sequencing and identification of DGGE fragments.
To determine the affiliation of the dominant bacterial communities in the termite mounds and the other environments, the 46 strongest bands representative of the DGGE profiles were excised, cloned, and sequenced (Table 1). In the anterior gut and midgut (Fig. 2), 14 clones obtained from 11 bands were assigned to five distinct phylogenetic groups (Table 1). Nine clone sequences belonged to the group of Firmicutes. The other groups were spread over four phylogenetic groups.
TABLE 1.
Phylogenetic affiliation and sequence similarities of DNA recovered from DGGE gel (Fig. 2)
| Sample typea | Band no.b | Accession no. | Phylogenetic affiliation or origin | Closest relative organism | Accession no. | Similarity (%) | Nearest 16S rRNA clone library | Similarity (%) |
|---|---|---|---|---|---|---|---|---|
| AG and MG | 1 | AY100703 | Bacteroidetes/Chlorobi group | Uncultured Bacteroidales | AB191999 | 89 | DQ347922 | 66 |
| 2 | AY100707 | Firmicutes | Unidentified rumen bacterium | AF018555 | 94 | DQ347938 | 72 | |
| 3-1 | AY100708 | Firmicutes | Paenibacillus sp. | AJ582393 | 94 | DQ347934 | 71 | |
| 3-2 | AY100709 | Firmicutes | Uncultured Mollicutes bacterium | AB234515 | 88 | DQ347871 | 75 | |
| 4 | AY100710 | Firmicutes | Clostridium oroticum | M59109 | 99 | DQ347941 | 93 | |
| 5-1 | AY100711 | Firmicutes | Clostridium sp. | AF157052 | 91 | DQ347941 | 86 | |
| 5-2 | AY100712 | Bacteroidetes/Chlorobi group | Cytophaga sp. | AY238333 | 96 | DQ347859 | 76 | |
| 6 | AY100713 | Alphaproteobacteria | Sphingomonas alaska | AF145754 | 99 | DQ347872 | 100 | |
| DQ347910 | 100 | |||||||
| 7 | AY100714 | Deltaproteobacteria | Desulfobulbus rhabdoformis | U12253 | 93 | DQ347934 | 73 | |
| 8 | AY100715 | Firmicutes | Uncultured Clostridiales | AB088977 | 97 | DQ347938 | 72 | |
| 9 | AY100716 | Firmicutes | Bacillus fastidiosus | X60615 | 94 | DQ347934 | 86 | |
| 10-1 | AY100704 | Firmicutes | Uncultured Eubacteriaceae | AB100462 | 88 | DQ347934 | 84 | |
| 10-2 | AY100705 | Firmicutes | Uncultured Eubacteriaceae | AB192223 | 94 | DQ347934 | 91 | |
| 11 | AY100706 | Actinobacteria | Tsukamurella tyrosinosolvens | AY26287 | 99 | DQ347886 | 92 | |
| PG and WG | 12 | AY100717 | Bacteroidetes/Chlorobi group | Prevotella ruminicola | AY699286 | 96 | DQ347859 | 72 |
| 13 | AY100724 | Bacteroidetes/Chlorobi group | Uncultured bacterium | AB277981 | 91 | DQ347859 | 70 | |
| 14 | AY100725 | Spirochaetes | Uncultured spirochete | DQ307690 | 91 | DQ347917 | 74 | |
| 15 | AY100726 | Firmicutes | Clostridium sp. | DQ347920 | 93 | DQ347941 | 82 | |
| 16 | AY100727 | Firmicutes | Ruminococcus sp. | X94964 | 95 | DQ347941 | 93 | |
| 17 | AY100728 | Firmicutes | Dethiosulfovibrio acidaminovorans | AY005466 | 89 | DQ347917 | 82 | |
| 18-1 | AY100729 | Firmicutes | Clostridium sp. | AY330126 | 96 | DQ347941 | 91 | |
| 18-2 | AY100730 | Soil | Uncultured bacterium | DQ509993 | 88 | DQ347860 | 74 | |
| 19 | AY100731 | Betaproteobacteria | Acidovorax avenae | AY512827 | 96 | DQ347922 | 85 | |
| 20-1 | AY100718 | Alphaproteobacteria | Sphingomonas phyllosphaerae | AY453855 | 97 | DQ347872 | 97 | |
| DQ347910 | 97 | |||||||
| DQ347860 | 97 | |||||||
| 20-2 | AY100719 | Firmicutes | Uncultured Clostridiaceae | AB100480 | 95 | DQ347917 | 92 | |
| 21 | AY100720 | Firmicutes | Uncultured Clostridiaceae | AB100483 | 92 | DQ347938 | 91 | |
| 22 | AY100721 | Actinobacteria | Nocardia lactamdurans | AF154128 | 93 | DQ347886 | 86 | |
| 23 | AY100722 | Actinobacteria | Nocardioides aquiterrae | AF529063 | 100 | DQ347886 | 98 | |
| 24 | AY100723 | Actinobacteria | Cellulomonas flavigena | AF140036 | 96 | DQ347886 | 90 | |
| DQ347928 | 90 | |||||||
| FM and Gal | 25-1 | AY293292 | Uncultured rumen bacterium | AB034016 | 96 | DQ347886 | 80 | |
| 25-2 | AY293291 | Actinobacteria | Streptomyces spiralis | EF178683 | 99 | DQ347900 | 92 | |
| 26/39 | AY293298 | Chloroflexi | Uncultured Chloroflexi | AY921935 | 93 | DQ347864 | 100 | |
| 27 | AY293293 | Actinobacteria | Propionibacterineae bacterium | AB271050 | 99 | DQ347928 | 99 | |
| 28 | AY293294 | Actinobacteria | Uncultured bacterium | DQ347885 | 98 | DQ347928 | 98 | |
| 29 | AY293295 | Chloroflexi | Sphaerobacter thermophilus | AJ871227 | 92 | DQ347864 | 100 | |
| 30-1 | AY293296 | Actinobacteria | Uncultured bacterium | DQ347843 | 98 | DQ347843 | 96 | |
| 30-2 | AY293297 | Actinobacteria | Mycobacterium sp. | AY312273 | 92 | DQ347865 | 98 | |
| IW and EW | 31 | AY100732 | Chloroflexi | Uncultured Chloroflexi | AY186850 | 96 | DQ347859 | 85 |
| 32 | AY100734 | Actinobacteria | Uncultured actinobacterium | 96 | DQ347876 | 95 | ||
| 33 | AY100735 | Actinobacteria | Rhodococcus sp. | AJ007003 | 95 | DQ347846 | 94 | |
| 34 | AY293289 | Actinobacteria | Streptomyces yerevanensis | AB184099 | 98 | DQ347876 | 92 | |
| 35 | AY100733 | Actinobacteria | Uncultured actinobacterium | AF544363 | 99 | DQ347876 | 92 | |
| 36 | AY293290 | Actinobacteria | Uncultured bacterium | DQ347843 | 98 | DQ347843 | 97 | |
| 37 | AY100739 | Chloroflexi | Bacteroides forsythus | L16495 | 92 | DQ347860 | 72 | |
| 38 | AY100738 | Firmicutes | Uncultured Clostridiacea | AB089020 | 92 | DQ347875 | 82 | |
| 40-1 | AY293299 | Actinobacteria | Uncultured bacterium | DQ001685 | 97 | DQ347865 | 99 | |
| 40-2 | AY100737 | Actinobacteria | Rhodococcus opacus | AF095715 | 96 | DQ347876 | 94 | |
| 41 | AY100736 | Planctomycetes | Uncultured Planctomycetacia | EF074758 | 99 | DQ347886 | 65 | |
| SS | 42 | AY100740 | Firmicutes | Bacillus benzoevorans | AY043085 | 100 | DQ347938 | 76 |
| 43 | AY100741 | Alphaproteobacteria. | Nitrobacter winogradskyi | AY055796 | 99 | DQ347872 | 87 | |
| DQ347910 | 87 | |||||||
| DQ347860 | 87 | |||||||
| 44 | AY100745 | Bacteroidetes/Chlorobi group | Uncultured Flavobacteria | EF073338 | 90 | DQ347859 | 84 | |
| 45-1 | AY100742 | Deltaproteobacteria | Angiococcus disciformis | AJ233910 | 96 | DQ347872 | 80 | |
| DQ347910 | 80 | |||||||
| 45-2 | AY100743 | Deltaproteobacteria | Uncultured proteobacterium | AJ532714 | 92 | DQ347908 | 75 | |
| 46 | AY100744 | Chloroflexi | Uncultured Chloroflexi | DQ828843 | 97 | DQ347859 | 84 |
AG, anterior gut; MG, midgut; PG, posterior gut; WG, whole gut; FM, fresh mound; Gal, gallery.
Certain bands consisted of multiple sequences, and representative sequences in each clone library are shown.
From the posterior and whole gut profiles (Fig. 2), the 15 clones that were analyzed originating from 13 bands and were assigned to seven distinct phylogenetic groups (Table 1). Once again, the Firmicutes clones formed the dominant group found in this part of the gut, accounting for slightly less than half of the clones identified (six clones). Three clones were assigned to phylogenetic groups which seemed to be more characteristic of this part of the gut: three clones belonged to Actinobacteria in the genera Nocardioides and Cellulomonas. Two clones were affiliated with the Bacteroidetes/Chlorobi group, and one clone was fairly similar (91% homologous) to the Spirochaetes phylum. The other clones were assigned to uncultured bacteria (one clone) and to Proteobacteria (two clones).
In the mound compartments (EW and IW), 7 out of 12 clones (Table 1) originating from 11 excised bands were affiliated with Actinobacteria. To determine whether Actinobacteria species were also present in freshly reworked soil (galleries and feces), six DGGE bands (Fig. 2, lanes 6 and 7) from the lower part of the gel were sequenced. As for the IW, most of the clones (five out of eight) were strongly affiliated to various Actinobacteria families. Of all the clones sequenced, only one (AY293298) was found in both fresh construction material and the IW.
Because most of the bands belonging to the SS profiles were faint, we were able to excise only six dominant bands of the SS samples. Four of the six clones belong to Proteobacteria (Table 1).
Bacterial community structure as determined by clone library analysis.
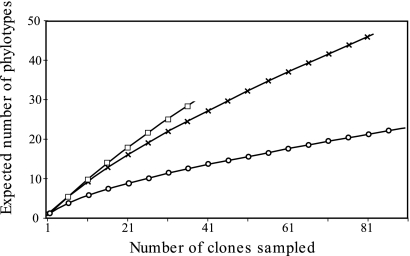
To complete the DGGE fingerprinting analysis, a 16S rRNA clone library was constructed. A total of 212 clones originating from the mound IW (83 clones), the SS (92 clones), and the termite gut (37 clones) were sequenced (Table 2), using primers that targeted a 650-bp portion of the gene. Rarefaction analysis (Fig. 3) together with coverage estimation (CACE; Table 2) revealed that the sampling depth of the individual clone library was sufficient to cover most of the bacterial diversity of the SS (62%) and, to a lesser extent, that of the termite mound (55%). For the termite gut, the relatively weak coverage estimation (35%) and the slope of the rarefaction curve (Fig. 3) indicated that further sampling may be needed; however, this environment has been already extensively inventoried by previous cloning analysis for a neighboring termite species (38, 41).
TABLE 2.
Clone library sizes, numbers of phylotypes, and diversity indices of Bacteria and Actinobacteria from three compartments investigated
| Phylum | Sample | Library size (no. of clones) | No. of phylotypesa | Value for indicated index of diversity
|
|||
|---|---|---|---|---|---|---|---|
| Coverage (CACE) | Richness (SChao 1) | Shannon (H′) | Simpson (D) | ||||
| Bacteria | Whole gut | 37 | 28 | 0.35 | 120 | 3.16 | 0.05 |
| Mound IW | 83 | 49 | 0.55 | 144 | 3.60 | 0.04 | |
| SS | 92 | 26 | 0.62 | 111 | 2.24 | 0.22 | |
| Total | 212 | 101b | |||||
| Actinobacteria | Whole gut | 5 | 5 | NDc | NDc | NDc | NDc |
| Mound IW | 42 | 22 | 0.64 | 50 | 2.76 | 0.09 | |
| SS | 28 | 13 | 0.69 | 39 | 1.97 | 0.19 | |
| Total | 75 | 39b | |||||
FIG. 3.
Rarefaction curve of bacterial 16S rRNA gene clones recovered from the gut, mound of soil-feeding termites (Cubitermes niokoloensis), and surrounding savanna soil. The expected number of phylotypes was calculated from the number of clones, with inclusion in the same species based on 97% sequence similarity. The three compartments studied are represented as follows: squares, whole gut; crosses, IW; and circles, surrounding savanna soil.
The presence of more than 101 different phylotypes (Table 2; also see Fig. S1 in the supplemental material) showed high diversity in the bacterial communities in the three compartments investigated in this study. The diversity indices (Table 2) reflect both the richness and the relative abundance of phylotypes in each environment. First, the SChao 1 species richness estimation increased from gut (111) to SS (120) to mound (144) inventories. The highest value (3.16) of the Shannon indices (H′) for the termite gut indicated a relatively even phylotype distribution, whereas the low Shannon (2.24) and high Simpson (0.22) diversity indices in the SS are due to the presence of a few dominant phylotypes. In this compartment, one clone (AF094766) belonging to the Gammaproteobacteria group represented about 45% of the total SS clone library.
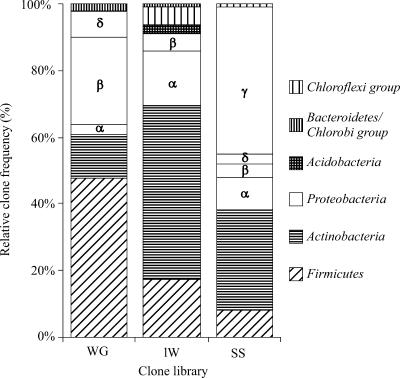
The bacterial compositions at the phylum and phylotype levels differed between the termite gut, mound, and savanna soil (Fig. 4; also see Fig. S1 in the supplemental material). Εach compartment was dominated by a particular phylum. The termite mound library was dominated by the Actinobacteria phylum (∼50% of all assigned clones), whereas Proteobacteria (mainly the γ subgroup) dominated the SS library (∼45% of the clones) (Fig. 4). The Actinobacteria phylum represents another important phylum (∼30% of the clones) in the SS. The termite gut library was dominated by clones affiliated with Firmicutes (Fig. 4; also see Fig. S1 in the supplemental material). The Bacteroidetes/Chlorobi phylum, dominated by bacteria considered to be fermentative, was found as expected in the termite gut library, but interestingly, it was also found to a lesser extent in the mound.
FIG. 4.
Relative clone frequencies in major phylogenetic groups of the clone libraries from the gut of soil-feeding termites (Cubitermes niokoloensis), mound of soil-feeding termites, and SS. The various subphyla of Proteobacteria are indicated by Greek letters. WG, whole termite gut.
Phylogenetic tree of the Actinobacteria phylum.
The diversity of phylotypes belonging to the Actinobacteria phylum was estimated only for the mound and the SS (Table 2) but not for the gut environment, where this phylum was not sufficiently represented. The whole 16S rRNA gene from one representative clone of each phylotype was completely sequenced. A total of 37 phylotypes representing 72 different clones from the Actinobacteria phylum were obtained in this study (Table 2). These clone libraries cover nearly two-thirds of the Actinobacteria diversity of these two soil compartments (Table 2). Compared to the SS (SChao 1 = 39; H′ = 1.97), the termite mound harbored a more diverse (SChao 1 = 50) and evenly distributed (H′ = 2.76) Actinobacteria community.
The 37 phylotypes were classified into 16 different Actinobacteria families (Fig. 5). However, for the termite mound and the SS, there was a clear separation of the clones, depending on their origin. Within the 15 Actinobacteria families, only two families (Nocardioidaceae and Coriobacteriaceae) contained phylotypes from both compartments (Fig. 5). Although the termite gut and the SS did not have any common Actinobacteria phylotypes, nearly all of the Actinobacteria phylotypes (four out of five) from the termite gut were also found in the termite mound.
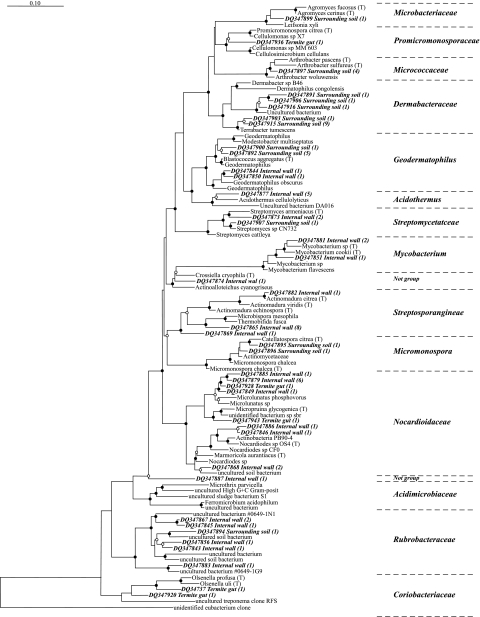
FIG. 5.
Subtree of actinomycetes from the complete tree (1,578 species; 2005 database) of the 16S rRNA database in the ARB package showing the phylogenetic positions of 40 phylotypes from a soil-feeding termite mound (IW), termite gut, and SS. The numbers of clones in each phylotype (97% sequence similarity) are given in parentheses. The quick add marked species feature of ARB (parsimony) was used to position our sequences, and the position was manually corrected using the neighboring species found with NCBI BLAST. Nodes with bootstrap values of >70% and between 50 and 69% are marked with black and empty circles, respectively. The scale bar indicates an approximately 10% difference in nucleotide sequence. posit, positive.
Within the termite mound, the 22 Actinobacteria phylotypes representing 42 clones were distributed among 10 distinct families (including 2 unknowns). Most of the Actinobacteria clones were affiliated to four dominant families: the Nocardioidaceae family had the most with six phylotypes (representing 12 clones), the Rubrobacteraceae family had five phylotypes (representing 6 clones), the Streptosporangineae family had three phylotypes (representing 10 clones), and finally the Acidothermus family had one phylotype (representing 5 clones).
For the SS, the 13 phylotypes were distributed in only seven Actinobacteria families, thereby showing a diversity lower than that of the mound environment. Above all, this environment was characterized by the dominance of one family, the Dermabacteraceae, characteristic of the savanna soil, which covered nearly half of all the clones in the SS. The two other important groups were the Geodermatophilus and Micromonospora genera, represented by six and two clones, respectively. The former was not specific to the SS, as it included two clones found in the termite mound.
The five termite gut Actinobacteria clones were affiliated with three families: two specific families, the Coriobacteriaceae (two clones) and the Promicromonosporaeae (one clone), and one nonspecific family, the Nocardioidaceae (two clones), also found in the termite mound.
DISCUSSION
This study is the first comprehensive analysis using molecular characterization based on the 16S rRNA gene of the bacterial community of a soil-feeding termite mound (C. niokoloensis). We were able to demonstrate that the termite mound harbors a bacterial community particular in terms of structure and diversity and characterized by the domination of Actinobacteria phylotypes, probably originating from the termite gut, that was distinct from the building materials, termite gut feces, and particles from the surrounding savanna soil.
Shift in the bacterial community structure between the termite gut and the mound.
Because of the dual origin (gut feces and soil particles) of the soil-feeding termite mound, we investigated the bacterial community structures of the various gut sections, feces, mound, and surrounding savanna soil using DNA fingerprinting (DGGE). The cluster analysis of the DGGE profiles (Fig. 2) showed unequivocally that the three compartments investigated (gut, mound, and soil) each harbored different bacterial communities (Fig. 2). There was a major shift in the structure of the bacterial communities between the various segments of the gut and the various mound compartments. This shift is seen in the different distributions of the dominant bands along the DGGE profiles. The strong bands in the gut profiles were situated mainly at the top of the gel (weak denaturing conditions), while the profiles for the mound compartments were all in the lower part of the gel (strong denaturing conditions). This change may indicate that the bacterial community naturally present in the posterior gut is replaced in the mound by a new bacterial community after excretion in the form of feces. This change seems to be very rapid, as fresh feces sampled less than an hour after being deposited in the mound already had a profile that was intermediate between the posterior gut and the mound. This almost rapid change is probably caused by the nature of the environment in the soil-feeding termite gut, which is characterized by several physicochemical (pH, O2, H2) gradients both axially and longitudinally (37). On the other hand, the mound is characterized by a very compact structure rich in organic matter resulting from the fine mixture of feces and soil particles (35). Within the mound, various different bacterial communities coexist in microenvironments (aggregates) created by the termites (13). Passing from an environment that is almost anoxic, alkaline, at micron scale, and half-liquid (gut) to an environment that is oxic, slightly acidic, solid, and rich in available organic matter (mound) may be the cause of the shift observed between the bacterial communities.
The DGGE profiles of the IW is more related (Fig. 2) to the surrounding reference soil than to the mound EW, a mound compartment closer to the fresh termite structures (feces and galleries). This study thus confirmed the differences in terms of bacterial diversity (13) and bacterial activity (32) in termite mounds related to the pedological differences between these two structures (12).
Another interesting finding highlighted by this fingerprinting analysis is the difference between the compositions of the bacterial communities in the reference soil and those in the anterior gut, which should presumably hold soil freshly ingested by the termite. This shows that the anterior gut and midgut have a specific microbial community, even though the physicochemical conditions are not particularly specific (neutral pH and oxic conditions [37]). This study, therefore, confirms the hypothesis, proposed by Schmitt-Wagner et al. (38), that the shift observed from the structure of the digestive bacterial communities to the soil bacterial communities was not caused solely by the extreme pH conditions but also by the lysis of soil bacteria on entry into the gut.
Bacterial composition.
The bacterial community structure in the IW of the mound was strongly dominated (∼50% of clones) by bacterial sequences affiliated with Actinobacteria families. This result agreed with the DGGE band cloning analysis, where two-thirds of the clones were affiliated with this group. Besides the Actinobacteria, the Firmicutes clones represented another important component of the termite mounds, with nearly ∼17% of all assigned clones in the library. Nearly half of the mound's Firmicutes clones were from Clostridiales genera closely affiliated with previously retrieved sequences from termite guts (see Fig. S1 in the supplemental material) (38, 41, 42). This indicates that the change in the bacterial community structure between the gut and the mound shown by the DGGE analysis was not complete. The presence of typical anaerobic gut organisms in the mound is not surprising, as the dominance of fine aggregate soil fractions (87% of soil weight [12]) might favor the preservation of conditions for anaerobic and microaerophilic organisms in the mound.
Our investigation of the microbial diversity of soil-feeding termite guts did not completely describe this astonishingly diverse bacterial environment (Table 1), where more than 300 different bacterial phylotypes, representing more than 700 species per termite, have been reported for the gut of the termite Reticulitermes speratus (20, 21). However, our results were in agreement with those obtained by Schmitt-Wagner et al. (38) for a closely related termite species (C. ugandensis) and provided convincing evidence that the bacterial community compositions identified in this study were representative of those of termite gut of this genus (see Fig. S1 in the supplemental material). The gut bacterial community of C. niokoloensis (Senegal) showed that more than 50% of the gut clone libraries were affiliated with the Firmicutes phylum and mostly with the Clostridiales genus (about 50% and 45% of the clones, respectively, by use of DGGE and cloning analysis). Moreover, the majority of the clones were closely affiliated with sequences found also in termite guts (see Fig. S1 in the supplemental material). These results are in good agreement with a study by Hongoh et al. (19), who showed that the majority of termite gut bacteria represent true symbionts intimately linked with termites during their speciation.
A cloning analysis was undertaken from the main DGGE bands and from clone libraries of the main compartments. Comparison of both libraries shows that large proportions (19% and 41% obtained with 97% and 90% of sequence identities, respectively) of DGGE clones (Table 1) were found in 16S rRNA gene clone libraries, indicating a reasonable coverage between the two libraries. The DGGE cloning analysis also confirmed the compartmentalization of the bacterial community in the gut sections investigated. While the Firmicutes, mostly Clostridiales, dominated the anterior gut and midgut communities, the hindgut bacterial community was more diverse, including phyla more characteristic of this part of the gut, such as Spirochaetes, Actinobacteria, and Betaproteobacteria (38). Analysis of the gut clone libraries in this study revealed the presence of Actinobacteria clones (seven clones from all clone libraries), whereas sequences belonging to the Actinobacteria phylum have not been reported in clone libraries from similar termite species like C. orthognathus and C. ugandensis (38). However, the same study (38) reported the detection of Actinobacteria by direct counts using in situ hybridization in the gut. Other reports identified Actinobacteria-related phylotypes specific to the termite gut (belonging to termite clusters) in different termite species (20, 21), including soil feeders (41). Nakajima et al. (30) found that Actinobacteria was one of the dominant phyla colonizing the gut wall of a xylophagous species (R. speratus). Because of their gut wall localization, Actinobacteria species could have been underestimated in these authors' termite gut studies, where they represented less than 1.1% of the total clone libraries in the lumen of the gut (30).
Phylogenetic affiliation of mound Actinobacteria population.
The dominance of the Actinobacteria phylum in the termite mound (Fig. 2) represents the main results of this study. A phylogenetic tree (Fig. 4) clearly demonstrated both the high level of diversity of this Actinobacteria population (41 phylotypes in 16 different Actinobacteria families) and its specificity (70% of families had clones from only one environment).
In spite of its mixed origin, the termite mound had Actinobacteria populations dominated by the Nocardioidaceae family. Only one previous study has reported on the presence of Actinobacteria in soil-feeding termite mounds based on conventional culturing analysis (3). Contrary to our results, the isolates were not significantly different from those in the reference soil, which is not surprising considering that some of the Actinobacteria families (such as Rubrobacteraceae and Coriobacteriaceae) contain phylotypes that have not yet been cultured. Is the dominance of Actinobacteria species due simply to the physicochemical characteristics of the mound being favorable to the development of such a community, or is the community maintained or helped by the termites? Our hypothesis is that C. niokoloensis may cause or help to maintain the dominance of Actinobacteria in its mounds. It has been well established that termites maintain endosymbiotic relationships (digestive bacteria) and exosymbiotic relationships (fungus-growing termites) with microbial communities (4). A symbiotic relationship between insects and Actinobacteria has already been shown in the case of fungus-growing ants (7). This mechanism would imply the reingestion of the IW by the termites as a form of coprophagy, which is widespread among other ecosystem engineers such as earthworms or macroarthropods such as diplopods and isopods (24). Microscopic and ethologic studies are currently in progress to validate this hypothesis.
Although this study was carried out for only one termite mound and its associated population, the results obtained may be considered as representative for this species and even for the genus Cubitermes. Recent studies have shown remarkable stability and uniformity in the bacterial communities of soil-feeding termite mounds of the same species (13) and in different genera of soil feeders (17). This specificity also seems true for fungal populations (35).
Supplementary Material
Acknowledgments
This study was supported financially by the CNRS French Biodiversity Institute Environmental Life and Society program and by the French Institute for Research and Development (IRD, ex-ORSTOM).
We thank Amadou Lamine Ndieng for his technical assistance, two anonymous reviewers for comments that greatly improved the manuscript, and Amy R. Sapkota for proofreading this paper.
Footnotes
Published ahead of print on 15 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Beare, M. H., D. C. Coleman, D. A. Crossley, Jr., P. F. Hendrix, and E. P. Odum. 1995. A hierarchical approach to evaluating the significance of soil biodiversity to biogeochemical cycling. Plant Soil 170:5-22. [Google Scholar]
- 3.Bignell, D. E., J. M. Anderson, and R. Crosse. 1991. Isolation of facultatively aerobic actinomycetes from the gut, parent soil and mound materials of the termites Procubitermes aburiensis and Cubitermes severus. FEMS Microb. Ecol. 85:151-160. [Google Scholar]
- 4.Bignell, D. E., and P. Eggleton. 2000. Termites in ecosystems, p. 363-387. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbioses, ecology, vol. 1. Kluwer Academic, Dordrecht, The Netherlands. [Google Scholar]
- 5.Brauman, A. 2000. Effect of gut transit and mound deposit on soil organic matter transformations in the soil feeding termite: a review. Eur. J. Soil Biol. 36:117-125. [Google Scholar]
- 6.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 7.Currie, C. R., J. A. Scott, R. C. Summerbell, and D. Malloch. 1999. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701-704. [Google Scholar]
- 8.Donovan, S., P. Eggleton, and D. E. Bignell. 2001. Gut content analysis and a new feeding group classification of termites. Ecol. Entomol. 26:356-366. [Google Scholar]
- 9.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egert, M., S. Marhan, B. Wagner, S. Scheu, and M. W. Friedrich. 2004. Molecular profiling of 16S rRNA genes reveals diet-related differences of microbial communities in soil, gut, and casts of Lumbricus terrestris L. (Oligochaeta: Lumbricidae). FEMS Microbiol. Ecol. 48:187-197. [DOI] [PubMed] [Google Scholar]
- 11.Eggleton, P. 2000. Global patterns of termite diversity, p. 25-51. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbioses, ecology, vol. 1. Kluwer Academic, Dordrecht, The Netherlands. [Google Scholar]
- 12.Fall, S., A. Brauman, and J. L. Chotte. 2001. Comparative distribution of organic matter in particle and aggregate size fraction in the mounds of with different feeding habitats in Senegal: Cubitermes niokoloensis and Macrotermes bellicosus. Appl. Soil Ecol. 17:131-140. [Google Scholar]
- 13.Fall, S., S. Nazaret, J. L. Chotte, and A. Brauman. 2004. Bacterial density and community structure associated with aggregate size fractions of soil-feeding termite mounds. Microb. Ecol. 28:191-199. [DOI] [PubMed] [Google Scholar]
- 14.Garnier-Sillam, E., and M. Harry. 1995. Distribution of humic compounds in mounds of soil-feeding termites: its influence on soil structural stability. Insect Soc. 42:167-185. [Google Scholar]
- 15.Grassé, P. P. 1986. Termitologia. Comportement, socialité, ecologie, evolution, systématique, vol. III. Masson, Paris, France.
- 16.Grassé, P. P. 1984. Termitologia. Fondation des sociétes—constructions, vol. II. Masson, Paris, France.
- 17.Harry, M., N. Jusseaume, B. Gambier, and E. Garnier-Sillam. 2001. Use of RAPD markers for the study of microbial community similarity from termite mounds and tropical soils. Soil Biol. Biochem. 33:417-427. [Google Scholar]
- 18.Holt, J. A., and M. Lepage. 2000. Termites and soil properties, p. 389-407. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbioses, ecology, vol. 1. Kluwer Academic, Dordrecht, The Netherlands. [Google Scholar]
- 19.Hongoh, Y., P. Deevong, T. Inoue, S. Moriya, S. Trakulnaleamsai, M. Ohkuma, C. Vongkaluang, N. Noparatnaraporn, and T. Kudo. 2005. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microbiol. 71:6590-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hongoh, Y., L. Ekpornprasit, T. Inoue, S. Moriya, S. Trakulnaleamsai, M. Ohkuma, N. Noparatnaraporn, and T. Kudo. 2006. Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol. Ecol. 15:505-516. [DOI] [PubMed] [Google Scholar]
- 21.Hongoh, Y., M. Ohkuma, and T. Kudo. 2003. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol. Ecol. 44:231-242. [DOI] [PubMed] [Google Scholar]
- 22.Jensen, S., L. Ovreas, F. L. Daae, and V. Torsvik. 1998. Diversity in methane enrichments from agricultural soil revealed by DGGE separation of PCR amplified 16S rDNA fragments. FEMS Microbiol. Ecol. 26:17-26. [Google Scholar]
- 23.Jones, C. G., J. H. Lawton, and M. Shachak. 1994. Organisms as ecosystem engineers. Oikos 69:373-386. [Google Scholar]
- 24.Lavelle, P. 1997. Faunal activities and soil processes: adaptative strategies that determine ecosystem function. Adv. Ecol. Res. 27:93-130. [Google Scholar]
- 25.Lavelle, P., D. Bignell, M. Lepage, V. Wolters, P. Roger, P. Ineson, O. W. Heal, and S. Dhillion. 1997. Soil function in a changing world: the role of invertebrate ecosystem engineers. Eur. J. Soil Biol. 33:159-193. [Google Scholar]
- 26.Lee, H. S., and A. Chao. 1994. Estimating population size via sample coverage for closed capture-recapture models. Biometrics 50:88-97. [PubMed] [Google Scholar]
- 27.Lobry de Bruyn, L. A., and A. J. Conacher. 1990. The role of termites and ants in soil modification: a review. Aust. J. Soil Res. 28:55-93. [Google Scholar]
- 28.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima, H., Y. Hongoh, R. Usami, T. Kudo, and M. Ohkuma. 2005. Spatial distribution of bacterial phylotypes in the gut of the termite Reticulitermes speratus and the bacterial community colonizing the gut epithelium. FEMS Microbiol. Ecol. 54:247-255. [DOI] [PubMed] [Google Scholar]
- 31.Ndiaye, R. L. D., M. Lepage, and A. Brauman. 2004. The effect of the soil feeding termite Cubitermes niokoloensis on microbial activity in a semi-arid savanna in West Africa. Plant Soil 259:277-286. [Google Scholar]
- 32.Ndiaye, R. L. D., M. Lepage, C. E. Sall, and A. Brauman. 2004. Nitrogen transformations associated with termite biogenic structure in a dry savanna ecosystem. Plant Soil 265:189-196. [Google Scholar]
- 33.Ovreas, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Sælenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranjard, L., F. Poly, J. Combrisson, A. Richaume, F. Gourbiere, J. Thioulouse, and S. Nazaret. 2000. Heterogeneous cell density and genetic structure of bacterial pools associated with various soil microenvironments as determined by enumeration and DNA fingerprinting approach (RISA). Microb. Ecol. 39:263-272. [PubMed] [Google Scholar]
- 35.Roose-Amsaleg, C., Y. Brygoo, and M. Harry. 2004. Ascomycete diversity in soil-feeding termite nests and soils from a tropical rainforest. Environ. Microbiol. 6:462-469. [DOI] [PubMed] [Google Scholar]
- 36.Sall, S. N., A. Brauman, S. Fall, C. Rouland, E. Miambi, and J. L. Chotte. 2002. Variation in the distribution of monosaccharides in soil fractions in the mounds of termites with different feeding habits (Senegal). Biol. Fertil. Soils 36:232-239. [Google Scholar]
- 37.Schmitt-Wagner, D., and A. Brune. 1999. Hydrogen profiles and localization of methanogenic activities in the highly compartmentalized hindgut of soil-feeding higher termites (Cubitermes spp.). Appl. Environ. Microbiol. 65:4490-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6007-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon, C., and W. Weaver. 1963. The mathematical theory of communication. University of Illinois Press, Urbana.
- 40.Simpson, E. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 41.Thongaram, T., Y. Hongoh, S. Kosono, M. Ohkuma, S. Trakulnaleamsai, N. Noparatnaraporn, and T. Kudo. 2005. Comparison of bacterial communities in the alkaline gut segment among various species of higher termites. Extremophiles 9:229-238. [DOI] [PubMed] [Google Scholar]
- 42.Yang, H., D. Schmitt-Wagner, U. Stingl, and A. Brune. 2005. Niche heterogeneity determines bacterial community structure in the termite gut (Reticulitermes santonensis). Environ. Microbiol. 7:916-932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.