Abstract
Salinity effects on microbial community structure and on potential rates of arsenate reduction, arsenite oxidation, sulfate reduction, denitrification, and methanogenesis were examined in sediment slurries from two California soda lakes. We conducted experiments with Mono Lake and Searles Lake sediments over a wide range of salt concentrations (25 to 346 g liter−1). With the exception of sulfate reduction, rates of all processes demonstrated an inverse relationship to total salinity. However, each of these processes persisted at low but detectable rates at salt saturation. Denaturing gradient gel electrophoresis analysis of partial 16S rRNA genes amplified from As(V) reduction slurries revealed that distinct microbial populations grew at low (25 to 50 g liter−1), intermediate (100 to 200 g liter−1), and high (>300 g liter−1) salinity. At intermediate and high salinities, a close relative of a cultivated As-respiring halophile was present. These results suggest that organisms adapted to more dilute conditions can remain viable at high salinity and rapidly repopulate the lake during periods of rising lake level. In contrast to As reduction, sulfate reduction in Mono Lake slurries was undetectable at salt saturation. Furthermore, sulfate reduction was excluded from Searles Lake sediments at any salinity despite the presence of abundant sulfate. Sulfate reduction occurred in Searles Lake sediment slurries only following inoculation with Mono Lake sediment, indicating the absence of sulfate-reducing flora. Experiments with borate-amended Mono Lake slurries suggest that the notably high (0.46 molal) concentration of borate in the Searles Lake brine was responsible for the exclusion of sulfate reducers from that ecosystem.
Hypersaline lakes are common physiographic features in many of the Earth's arid regions. These lakes typically occupy closed-basin settings, where their size is controlled by the balance between evaporation and freshwater inputs to their drainage basins. As a result, the salinity of a given lake may oscillate dramatically over both long and short time intervals in response to regional changes in climate, topography, or hydrologic conditions. To date, relatively little research has been conducted to investigate the effect of changing salinity on the resident populations of microorganisms that mediate various biogeochemical reactions in these environments. A better understanding of the response of microbial communities to changing salinity and of the relative importance of particular electron acceptors at high salt concentrations may offer insight into the development of life in hypersaline conditions on this planet (4, 9), as well as the potential for the existence of life in evaporated brines from extraterrestrial settings (24). In addition, research on this topic may help us to better understand the broader ecological effects that may accompany salinity changes brought about by anthropogenic causes, including the diversion of tributary streams for irrigation or drinking water uses (5).
As evaporative lakes become increasingly hypersaline, the microorganisms that inhabit them come to occupy a highly specialized environmental niche. Life at high salt concentrations is bioenergetically taxing because organisms must maintain an osmotic balance between their cytoplasm and the surrounding medium while excluding sodium ions from the cell interior. The metabolic pathways utilized by halophilic microorganisms must therefore supply sufficient energy to meet the cellular requirements for osmoadaptation at high salinity (21). In addition, evaporation may increase the concentration of dissolved toxic constituents (e.g., arsenic and boron) that serve to inhibit particular metabolic processes in the residual brine. As a result, physiological groups that are ubiquitous in freshwater or marine settings may become strongly inhibited or even excluded from evaporative environments that approach salt saturation if the energetic cost of adaptation exceeds the bioenergetic yield of their respective dissimilatory processes.
Mono Lake (ML) and Searles Lake (SL) are closed-basin terminal lakes located in the rain shadow of the Sierra Nevada Range in California. These hypersaline and alkaline soda lakes represent two remnants of an extensive system of interconnected freshwater lakes and rivers that drained the eastern side of the central Sierra Nevada during the late Pleistocene, when regional climatic conditions were much wetter. As regional climatic conditions became increasingly arid during the Holocene, these lakes became increasingly evaporative and hypersaline. Benson et al. (2) estimates that the surface areas of ML and SL have decreased in size by factors of 4.2 and 5.5, respectively, relative to the surface areas of these lakes during the last Pleistocene highstand. Currently, ML is a moderately hypersaline soda lake that contains ∼5.5% of its Pleistocene water volume (19), while SL is a partially dry and salt-saturated residual playa. Both of these lakes are notably enriched in dissolved arsenic as well as containing abundant dissolved sulfate and borate (see “Site description and sample collection” below). Previously, we reported that dissimilatory arsenate reduction (DAsR) and dissimilatory sulfate reduction (DSR) occur in the anoxic water column and sediments of ML (10, 16). We also reported that while DAsR was active in SL sediments, DSR was not detectable and furthermore could not be elicited by various manipulative experiments (e.g., addition of electron donors, removal of competitive electron acceptors, and lowering salinity) with sediment slurries (10, 17). We attributed the absence of DSR in SL to Oren's theory (21) that anaerobic processes, such as sulfate reduction and methanogenesis, yield insufficient metabolic energy to support the metabolism of these microbes under salt-saturated conditions. In this contribution, we further explore the effect of salinity on the metabolic processes and community structure of natural populations in the sediments of ML and SL. We focused on the processes of DAsR and DSR, with particular emphasis aimed at addressing why the latter process appears to be absent from SL sediments. We also describe salinity-induced changes in the microbial community structure of arsenate-reducing sediment microcosms as an example of the response of anaerobic populations to changing salinity conditions.
MATERIALS AND METHODS
Site description and sample collection.
Mono Lake and Searles Lake are alkaline (pH 9.8) evaporative soda lakes that occupy arid, closed-basin settings east of the Sierra Nevada mountain range in eastern California. ML is a large moderately hypersaline (total salinity of 90 g liter−1) soda lake with a mean depth of approximately 17 m. During most of the year, SL is a partially dry residual playa and saltern containing a shallow (∼15-cm-deep) salt-saturated (total salinity of >300 g liter−1) brine that is overlain by a 5-cm-thick salt crust. These lakes are naturally enriched in numerous dissolved constituents that are largely derived from hydrothermal sources and concentrated in the lakes by evaporation. In particular, both lakes contain abundant dissolved sulfate (total SO42− of 0.1 molal [m] for ML and 0.8 m for SL) and arsenate [total As(V) of 0.2 mM for ML and 3.4 mM for SL]. Dissolved borate also occurs at high concentrations (0.03 m) in ML and reaches commercially significant concentrations (0.46 m) in SL, which has been mined as an industrial source of borax since the late 1800s. We collected surficial sediments (upper 6- to 8-cm sediment depth) from the shallow littoral zone of ML (10, 14) (38°04′27″N, 119°00′46″W) and from below the salt crust and brine of the SL saltern (10, 17) (35°42′38″N, 117°20′12″W). ML sediments were collected in May 2005, while SL sediments were collected from the same location in the SL saltern during two separate sampling trips (April 2004 and February 2005). All sediment was collected in Mason jars that were completely filled prior to sealing and transported on ice to the laboratory where they were stored at 8°C for up to 6 months prior to use in the sediment slurry experiments.
Sediment slurry preparation.
All sediment slurries were prepared using ML or SL sediment mixed with artificial media that were designed to mimic the water chemistry of the corresponding lake. Artificial ML lake water prepared at the ambient ML salinity of 90 g liter−1 was composed of the following ingredients (concentrations are grams liter−1 and are shown in the parentheses): NaCl (75), (NH4)2SO4 (0.1), KH2PO4 (0.08), K2HPO4 (0.15), MgSO4·7H2O (0.025), Na2CO3 (10.6), NaHCO3 (4.2), Na2WO4 (0.00001), and Widdel et al. (25) trace elements solution (5 ml). ML media of higher or lower total salinity were achieved by altering the amount of NaCl added. The artificial SL lake water, when prepared at the ambient SL condition of salt saturation (346 g liter−1), was composed of the following ingredients (concentrations are grams liter−1 and are shown in the parentheses): NaCl (180), Na2SO4 (100), K2SO4 (30), (NH4)2SO4 (0.05), KH2PO4 (0.08), K2HPO4 (0.15), MgSO4·7H2O (0.025), Na2WO4 (0.075), H3BO4 (4.0), Na2SeO4 (0.00001), Na2CO3 (27), NaHCO3 (5), and Widdel et al. (25) trace elements solution (5 ml). SL media of lower salinity concentrations were prepared by altering the amount of NaCl, Na2SO4, and K2SO4 added, while maintaining a constant ratio between the concentrations of these three salts. No borate was added to the ML media, and the borate concentration of the SL media (51 mM) was considerably lower than the ambient borate concentration of the natural SL brine. All media were adjusted to pH 9.8 with 0.1 M NaOH prior to use in sediment slurries. Sediments were slurried (1:4) with artificial lake water under nitrogen flow using an electric blender, and the resulting homogenates (10 ml) were transferred into 57-ml serum bottles containing an additional 20 ml of artificial brine (final dilution of sediment in medium of 1:12). Aerobic slurries were capped with a permeable foam stopper that allowed free exchange with the atmosphere during incubation. For anaerobic slurries, all preparations and manipulations were conducted under a flow of O2-free N2, and the serum bottles were subsequently crimp sealed and flushed for 10 min with O2-free N2. In some cases, H2 gas was substituted for N2 in order to serve as an electron donor. Heat-sterilized control slurries were autoclaved twice (121°C, 250 kPa for 60 min each time) prior to receiving further amendments. Amendments of electron acceptors, electron donors, and inhibitors were made from sterile, anaerobic stock solutions as described below for each experiment. Unless otherwise noted, the slurries were incubated in the dark at 25°C on a rotary shaking table (150 rpm).
Bioassays.
Anaerobic sediment slurries to assay DAsR activity and aerobic slurries to assay As(III) oxidation were set up at the following salinities for ML: 25, 50, 100, 150, 200, 250, 300, and 346 g liter−1. For SL, the same salinity concentrations were used except that the 25 g liter−1 condition was excluded and a 275 g liter−1 condition was added. Slurries for all salinity conditions were prepared in triplicate, and duplicate autoclaved controls were prepared for three representative salinities (50, 150, and 346 g liter−1) in each experiment. Arsenate reduction slurries were amended with 1 mM As(V) added as Na2HAsO4, and As(III) oxidation slurries received 1 mM As(III) added as NaH2AsO3. Over the course of 2- to 3-week incubations, slurry subsamples were collected periodically with a syringe (0.3 ml), and the subsamples were filter centrifuged (Spin-X centrifuge tube filters; pore size, 0.2 μM; Corning, Inc., Corning, NY) to separate the liquid phase from the sediment. An aliquot of the liquid phase was diluted in deionized water prior to determination of As(III) and As(V) concentrations using high-performance liquid chromatography with UV wavelength (210-nm) detection (6). We monitored methane production in the headspace of the anaerobic As(V) reduction slurries that were incubated with ambient electron donors. The gas phase was periodically sampled with a syringe (0.1 ml), and the methane concentration was measured using flame ionization gas chromatography (17).
We conducted sediment slurry experiments with radiolabeled [35S]sulfate to measure potential rates of DSR in the sediments as a function of salinity. Anaerobic sediment slurries were prepared using lower-sulfate (50 mM) versions of the ML and SL media (see above) in order to minimize isotopic dilution of the radiolabeled tracer. We prepared slurries (total volume of 20 ml) of sediments from both lakes in triplicate at 25, 90, and 325 g liter−1 total salinity. Two sets of slurries were prepared for each lake under N2: the first set received no additional electron donor amendment, while the second was amended with 10 mM sodium lactate. A third set of slurries for each lake was prepared under a 100% H2 atmosphere. Autoclaved control slurries were prepared at 25 and 325 g liter−1 for each condition. Each slurry was injected with 1.85 MBq Na235SO4 (50 μl; carrier free; American Radiolabeled Chemicals, Inc., St. Louis, MO) and incubated at 30°C (to encourage microbial activity) with constant rotary shaking (150 rpm). After 7 days, the slurries were injected with 2 ml of a 10% zinc acetate solution and stored at −70°C to terminate activity prior to distillation processing, trapping, and quantification of [35S]sulfide that had been produced by dissimilatory reduction of 35SO4 (10, 18). The rate of sulfate reduction in the slurries was calculated from the rate of radiotracer turnover and the ambient sulfate concentration of the medium.
Manipulation experiments were also conducted with regard to sulfate reduction in these sediment slurries. First, we tested the possibility that sulfate reduction could be restored to SL by providing a small inoculum of ML sediments. Two triplicate sets of SL sediment slurries were prepared at 25, 90, and 325 g liter−1 salinity. One set of SL slurries was inoculated with a small volume (0.6 ml) of live ML sediment mixed with SL media (1:12 dilution). The second set of SL slurries was inoculated with an equivalent volume of ML sediment that had first been heat sterilized by autoclaving twice at 121°C and 250 kPa for 60 min each time. A third set of slurries was prepared with SL sediments in ML media at 25, 90, and 325 g liter−1 salinity. We also tested for a possible inhibitory influence of borate ions upon sulfate reduction in ML sediments. Three sets of ML sediment slurries were prepared in triplicate at 50 g liter−1 salinity and amended with 0, 40, or 200 mM H3BO3. Each of the sediment slurries in these manipulation experiments was prepared using low-sulfate media, amended with 10 mM sodium lactate, injected with [35S]sulfate, and incubated for 10 days prior to quantification of [35S]sulfide production as described above.
Denitrification was assayed using the acetylene block method (20, 22). Anaerobic slurries were prepared at the same salinity conditions used for As(V) reduction and As(III) oxidation bioassays described above. The sediment slurries were amended with 2 mM NaNO3 and 2 mM glucose, followed by injection with C2H2 (initial pressure of 0.15 atm [14.8 kPa]). The production of N2O in the headspace of the slurry bottles was monitored over the course of 2- to 4-week incubations using gas chromatography with a 63Ni electron capture detector (12).
Extraction and DGGE sequencing of DNA from As(V) reduction slurries.
Following termination of the As(V) reduction bioassays (described above), slurries representing low, intermediate, and high salinities from each lake were chosen for extraction and denaturing gradient gel electrophoresis (DGGE) analysis of partial 16S rRNA genes from the microbial population that grew during incubation. To minimize the biases from DNA extraction and PCR amplification, we extracted DNA with triplicate, amplified 16S rRNA genes, and combined the PCR products for DGGE analysis. For ML, we extracted DNA from sediment slurries that had been incubated at 25, 100, 200, and 325 g liter−1 total salinity, and for SL, we extracted slurries representing 50, 200, and 346 g liter−1 salinity. The 100 g liter−1 condition was included for ML in order to approximate the ambient salinity of ML water. The contents of one replicate slurry representing each condition were transferred to a Nalgene centrifuge tube (50 ml) and centrifuged (500 × g for 25 min) to separate the sediment from the liquid phase. The supernatant was decanted, and the sediment was collected for DNA extraction. DNA was extracted from 0.5 g (wet weight) of sediment using a FastDNA Soil Spin kit (Qbiogene, Irvine, CA). Twenty nanograms of environmental DNA was used as a template for amplification of partial 16S rRNA genes with bacterium-specific primers 341F-gc (5′CCTACGGGAGGCAGCAG3′ with 40-bp GC clamp) and 518R (5′ATTACCGCGGCTGCTGG3′) (10). DGGE was carried out with a D-Code universal mutation detection system (Bio-Rad Laboratories, Inc., Hercules, CA) using 8% polyacrylamide gel with a 40% to 60% gradient of denaturing reagent as described previously (10). DGGE was performed with 800 ng of PCR products, and the gel was run at 100 V for 14 h at 60°C in 1× Tris-acetate-EDTA (TAE) buffer. The gel was stained with ethidium bromide for 15 min, rinsed with nucleic acid-free water, and photographed using a Geldoc XR system (Bio-Rad Laboratories, Inc., Hercules, CA). Major DGGE bands were chosen based on band intensity and/or uniqueness, excised by using a Spot picker (Gelcompany, CA), and transferred into 20 μl of nucleic acid-free water. The bands were frozen and thawed two times to elute the DNA from the polyacrylamide gel pieces. Two microliters of the supernatant was used for reamplification with the original primer set (without a GC clamp). PCR products were purified with a QIAquick PCR purification kit (QIAGEN) and sequenced with primer 357F (5′CCTACGGGAGGCAGCAG3′ without a GC clamp). Partial sequences were determined, and the affiliations at the phylum or class level were established by RDP Classifier (http://rdp.cme.msu.edu/classifier/) (3) and BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). DGGE sequences containing ambiguities were omitted. Digital images of the gel were captured with Quantity-One software (version 4.3.1; Bio-Rad) for use in comparative image analyses. In order to analyze bacterial community structure, cluster analysis was performed with Gelcomopar II software (version 4.5; Applied Maths, Kortrijk, Belgium) using the unweighted-pair group method with arithmetic mean based on Dice correlation coefficients that accounted for band position. The number of bands observed after DGGE separation represented the number of bacterial species in each sample, and band intensity was used to represent the relative abundance of each species (13).
PCR amplification and sequencing of arrA genes.
DNA extracts from the sediment incubations were analyzed by PCR for the presence of the arrA gene using methods developed by Kulp et al. (10). The reactions contained 300 nM of each primer (HAArrA-D1F [5′-CCGCTACTACACCGAGGGCWWYTGGGRNTA-3′] and HAArrA-G2R [5′-CGTGCGGTCCTTGAGCTCNWDRTTCCACC-3′]), 20 ng of DNA extract, and 25 μl of a 2× PCR Taq mix (Promega), and nuclease-free water added to a total volume of 50 μl. The PCR conditions included incubation at 95°C for 5 min, followed by 40 cycles of 95°C for 30 seconds, 53.5°C for 30 seconds, and 72°C for 30 seconds. The PCR products were visualized by UV fluorescence following electrophoresis in a 1.5% agarose gel in 1× TAE buffer and staining with 1 μg ml−1 ethidium bromide solution. Methods for cloning and sequencing the partial arrA PCR products and using HAArrA primers were reported previously (10).
Phylogenetic analysis of the arrA gene.
The partial arrA sequences obtained from the sediment incubation experiments were aligned using ClustalW and MacVector. Phylogenetic analyses were performed using ARB and the neighbor-joining method, Jukes-Cantor DNA substitution correction, and bootstrap analysis with 1,000 replications. Also included in the phylogeny were overlapping regions of known arrA sequences from Shewanella sp. strain ANA-3 and Bacillus arseniciselenatis E1H and several arrA sequences obtained from an environmental DNA survey for arrA in ML and SL sediment performed by Kulp et al. (10).
Nucleotide sequence accession numbers.
The 16S rRNA and arrA gene sequences have been deposited in the GenBank database under the accession numbers EF710661 to EF710720.
RESULTS
Arsenate reduction rates.
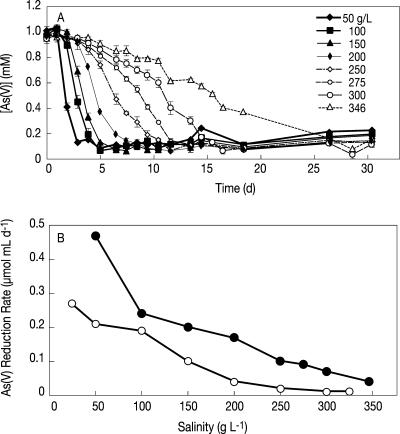
We observed dissimilatory As(V) reduction in active anaerobic slurries of both Mono and Searles Lake sediments at all salinities up to and including salt saturation. Live slurries reduced 1 mM concentrations of amended As(V) to As(III). No activity was observed in autoclaved controls (data not shown). In slurries from both lakes, the potential rate of DAsR in sediment slurries exhibited an inverse relationship to total salinity. An example is provided in Fig. 1A, which presents time course data of As(V) reduction in SL slurries at each salinity condition. Figure 1B shows the maximum rates of DAsR observed at the various salinity levels in each lake using naturally present electron donors in the sediment. Arsenate reduction rates were similar in slurries from both lakes and decreased between the lowest and highest applied salinities (Fig. 1B).
FIG. 1.
(A) Potential arsenate reduction in SL sediment slurries at various salinities (in grams/liter). (B) Arsenate reduction rates as a function of salinity in ML (open symbols) and SL (closed symbols) sediment slurries. The incubation times were 20 days (d) for ML sediments and 30 days for SL sediments.
DGGE comparison of 16S rRNA gene sequences from As(V)-reducing populations grown at various salinities.
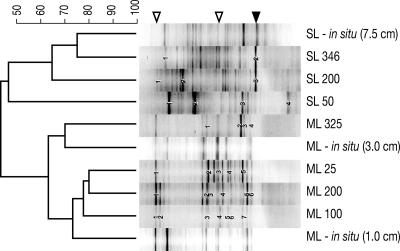
DGGE analysis of 16S rRNA genes amplified from the As(V)-reducing sediment slurries following incubation demonstrated that distinct microbial communities developed at low, intermediate, and high salinities in both lakes (Fig. 2). Cluster analysis of DGGE banding patterns showed strong clustering within the lakes; no clustering was observed for equivalent salinities from different lakes. For SL, the two higher-salinity samples clustered together, while the lowest-salinity sample was quite distinct (<55% similarity), whereas for ML, the three low-salinity samples were quite similar while the highest salinity was distinct (Fig. 2). For SL, the highest-salinity sediment slurry showed highest similarity (75%) to a sediment core section previously recovered from the lake (10). For ML, the three lowest-salinity sediments showed high similarity (∼73%) to previously recovered surface sediment samples (10), whereas the highest-salinity sediment slurry showed strong similarity to subsurface sediments (10). The number of bands was higher for SL (mean number of bands, 25.7) than ML (mean number of bands, 16.3), indicating a more diverse community in SL sediments. No specific trend was observed regarding the overall complexity of the bacterial community with salinity except that diversity dropped drastically in ML at the highest salinity; no equivalent change was observed in SL samples. The highest number of bands was observed at 200 g liter−1 total salinity for both lakes. Sequence analysis of selected DGGE bands indicated that a diverse and distinct microbial community grew in sediments from both lakes at each salinity condition studied (see Table S1 in the supplemental material). The majority of clones sequenced closely matched gene sequences that had been previously retrieved from other hypersaline and alkaline environments (see Table S1 in the supplemental material and see the arrowheads in Fig. 2). Selected bands from ML slurries incubated at 25 and 200 g liter−1 salinity were closely related (100% and 94%, respectively) to uncultured sequences obtained directly from the ML water column (8) or from sulfide-enriched incubated ML water (7). Similarly, SL slurries incubated at 200 and 346 g liter−1 salinity exhibited a prominent band that matched (100% 16S rRNA gene sequence identity) strain SLAS-3, a clone obtained from natural SL sediment samples (10) that is most closely related (96.1% rRNA gene sequence similarity) to an obligately halophilic As(V) respirer previously isolated from SL sediments (strain SLAS-1 [17]). Strain SLAS-3 was not observed in SL slurries incubated at 50 g liter−1 salinity, consistent with the observation that strain SLAS-1 does not grow in culture at salinities below 200 g liter−1 (17).
FIG. 2.
DGGE of 16S rRNA obtained from SL and ML sediment slurries following incubation with added arsenate at various salinities and compared to natural sediment samples. DGGE profiles of natural sediments are profiles of samples collected from sediment hand cores at the labeled depth intervals (in centimeters). The salinities range from 25 to 346 g liter−1 and are shown after the lake. The similarities between samples are shown in an unweighted-pair group method with arithmetic mean dendrogram of Dice similarity coefficients based on band position. The scale bar indicates levels of percent similarity. The numbered bands refer to the sequences in Table S1 in the supplemental material. Open arrowheads indicate bands that match 16S rRNA gene sequences from uncultivated organisms previously discovered in ML water (see text for details). The band indicated by the solid arrowhead matches strain SLAS-3, a clone that is closely related to the halophilic As(V)-respiring strain SLAS-1 isolated previously from SL sediments (16). (DGGE profiles of natural sediments are reprinted from reference 10.)
Phylogenetic analysis of arrA.
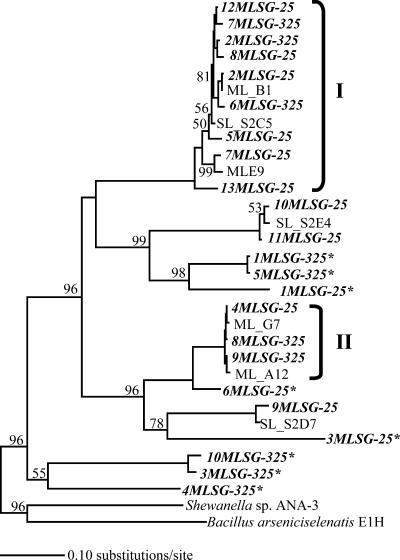
Figure 3 shows the phylogenetic relationships of arrA sequences obtained in this study (n = 23), including selected arrA sequences obtained from DNA extracts of ML and SL environmental samples (10). More than half of the partial arrA clones (n = 15) were very similar (>95%) to ML and SL environmental clones. Specifically, group I (n = 9), group II (n = 3), and several other arrA clones (n = 3) from the incubations were greater than 95% identical to several SL and ML environmental arrA sequences. However, there was a significant number (n = 8) of single divergent phylotypes (indicated with an asterisk in Fig. 3) that did not cluster well with arrA phylotypes retrieved from ML sediments. These sequences were considerably different (more than 10% nucleotide changes) compared to the other partial arrA sequences shown in Fig. 3. The nearest neighbors to most of these divergent arrA sequences were several SL and ML environmental clones, which shared about 65 to 72% nucleotide identities. These results indicate that salt amendments of ML sediments caused shifts in the arrA phylotypes. However, it is unclear from the limited number of clones whether there are associations of specific arrA phylotypes with low- or high-salinity conditions.
FIG. 3.
Phylogenetic relationship of partial arrA DNA sequences retrieved from Mono Lake sediments incubated at low (25 g/liter NaCl; MLSG-25) and high salinities (325 g/liter NaCl; MLSG-325). SL and ML refer to sequences originating from Searles Lake and Mono Lake sediments, respectively. Partial arrA sequences (n = 23) from the clone libraries obtained in this study (italicized, boldface type) were aligned with various known sequences using ClustalW in MacVector (v. 8). The tree was constructed using ARB and neighbor-joining analysis with Jukes-Cantor correction for transitions and transversions. Bootstrap values of at least 50% from 1,000 replications are labeled on the corresponding nodes. Clones marked with asterisks indicate unique arrA phylotypes. The scale bar represents 0.1 nucleotide substitution per site. Strains and GenBank accession numbers used in the tree are as follows: Bacillus arseniciselenatis strain E1H, AY660885; Shewanella sp. strain ANA-3, AAQ01672; and other SL and ML environmental sequences previously reported by Kulp et al. (10).
Arsenite oxidation and denitrification.
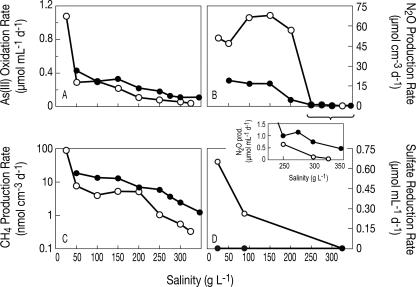
Rates of aerobic As(III) oxidation decreased linearly above 50 g liter−1 salinity in slurries from both lakes (Fig. 4A). Amendments of 1 mM As(III) were completely oxidized to As(V) in active slurries from both lakes at all salinity concentrations (average recovery, 97.6%), while sterilized control slurries did not oxidize As(III) over the duration of the incubation (data not shown). Arsenite oxidation was markedly stimulated in ML sediments incubated at a salinity of 25 g liter−1, which exhibited As(III) oxidation rates that were fourfold higher than at a salinity of 50 g liter−1.
FIG. 4.
Potential rates of As(III) oxidation (A), denitrification (measured as N2O production) (B), methanogenesis (C), and sulfate reduction (D) versus total salinity in ML (open symbols) and SL (closed symbols) sediment slurries. The inset in panel D shows N2O production at salinities above 200 g liter−1 on a finer scale. Arsenite oxidation slurries were incubated 20 days for ML and 14 days for SL. Denitrification slurries were incubated 14 days for ML and 27 days for SL. Methanogenesis was measured in As(V) reduction slurries incubated 20 days for ML and 30 days for SL. Sulfate reduction slurries were incubated 7 days for both lakes.
Production of N2O occurred at all salinity levels in nitrate- plus glucose-amended slurries that were incubated in the presence of C2H2 (Fig. 4B). Denitrification was markedly inhibited at salinity concentrations above 200 g liter−1 in sediments from both lakes. Nevertheless, modest N2O production (0.04 to 0.4 μmol cm−3 day−1) persisted even at salt-saturated conditions in sediments from both lakes. Maximum rates of N2O production in the ML slurries were fourfold higher than those in the SL slurries. The maximum rates of N2O production in the SL sediment slurries (15.7 to 18.0 μmol cm−3 day−1) occurred at salinities of 50 to 100 g liter−1, whereas ML sediments exhibited maximum rates (53.0 to 63.2 μmol cm−3 day−1) at 100 to 200 g liter−1 total salinity. No N2O production occurred in autoclaved control slurries (data not shown). No accumulation of N2O occurred in slurries that were not amended with C2H2, although we did observe a complete removal of the added NO3−, presumably via reduction to N2 because no NH4+ production was observed at any salinity condition (detection limit of 100 μM; T. Kulp, unpublished data).
Methanogenesis.
The anaerobic slurries utilized in the As(V) reduction experiments also produced methane at a rate that decreased with increasing salinity (Fig. 4C). Low but measurable rates of methane production (0.3 nmol cm−3 day−1 for ML; 1.2 nmol cm−3 day−1 for SL) were observed at salt saturation. The generally inverse linear relationship between methanogenic activity and total salinity was observed in sediment slurries from both lakes; however, ML sediments incubated at 25 g liter−1 salinity produced methane at a rate that was an order of magnitude greater than those incubated at 50 g liter−1.
Sulfate reduction.
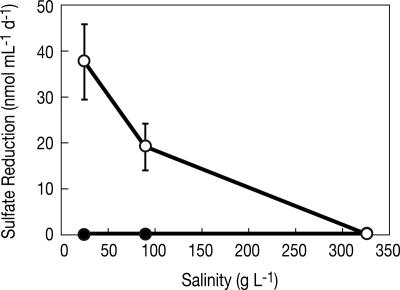
Dissimilatory sulfate reduction was observed in ML sediment slurries prepared at low (25 g liter−1) and intermediate (90 g liter−1) total salinities. In lactate-amended ML sediment, sulfate reduction rates of 0.66 μmol ml−1 day−1 and 0.26 μmol ml−1 day−1 were measured at low and intermediate salinity, respectively (Fig. 4D). Similar rates of sulfate reduction were observed at low (0.63 μmol ml−1 day−1) and intermediate (0.21 μmol ml−1 day−1) salinity in ML slurries when a H2 atmosphere was provided in lieu of lactate (data not shown). ML slurries that did not receive electron donor amendments reduced sulfate at a rate of 0.03 μmol ml−1 day−1 at 25 g liter−1 salinity and 0.02 μmol ml−1 day−1 at 90 g liter−1 (data not shown).
No sulfate reduction activity was detected in ML sediments incubated at a salinity of 325 g liter−1 or in SL sediments incubated at any salinity tested (Fig. 4D). Tracer (35S) activity in these samples was statistically identical to that from autoclaved controls and sample blanks (∼100 cpm), and DSR could not be elicited under these conditions with either lactate or H2. Substitution of ML media for SL media also did not elicit DSR from SL sediment slurries (data not shown). SL sediment slurries that were inoculated with live ML sediment, however, were able to reduce sulfate at the low and intermediate salinities. This effect was not observed when a sterilized ML inoculum was used (Fig. 5).
FIG. 5.
Sulfate reduction activity in SL sediment slurries inoculated with ML sediment. Sulfate reduction was stimulated at 25 and 90 g liter−1 salinity in SL sediments that received 3% (by volume) active ML sediment as an inoculum (open symbols). No activity was observed in SL slurries where sterilized ML sediment was used as the inoculum (closed symbols). Incubation time was 10 days.
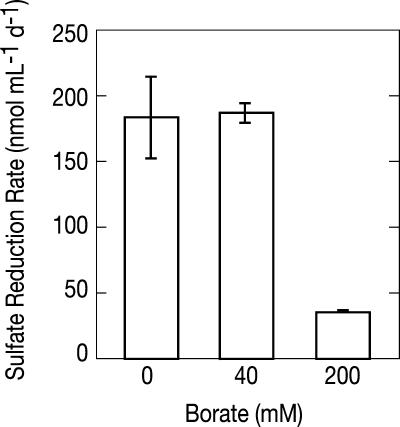
The effect of borate on sulfate reduction rates in ML sediment slurries at 50 g liter−1 salinity is shown in Fig. 6. No inhibition of DSR was observed in slurries that contained 40 mM borate compared with borate-free slurries. In slurries that contained 200 mM borate, however, DSR was inhibited by 80% compared to equivalent slurries without borate.
FIG. 6.
Sulfate reduction rates in ML sediment slurries showing the effect of borate additions. Slurries were incubated at a salinity of 50 g liter−1 for 10 days.
DISCUSSION
Sedimentary communities from moderately hypersaline Mono Lake and salt-saturated Searles Lake utilized a variety of active biogeochemical pathways as indicated by apparent As(V) reduction, As(III) oxidation, denitrification, and methanogenesis in sediment slurries that were tested across a wide range of total salinities up to and including salt saturation. With the exception of denitrification in ML, the fastest potential rates for each of these microbial processes were observed at the lowest experimental salinities (25 or 50 g liter−1), and these rates decreased incrementally as salinity increased. The fact that the fastest potential rates for these processes occurred at salt concentrations that were considerably lower than the ambient salinity of each lake suggests the presence of a salt-stressed microbial community at the in situ lake conditions or that distinct populations of microorganisms rapidly develop in these sediments as a response to changing salinity. The latter explanation is supported by our DGGE comparison of sedimentary 16S rRNA partial gene sequences amplified from As(V) reduction slurries, which revealed distinct variations in the microbial community that developed at low, intermediate, and high salinities after 20 to 30 days of incubation. We suggest that organisms adapted to lower salinities remain viable but comprise a relatively small (or inactive) component of the sedimentary microflora during periods of high salinity. The overall community structure in the lake sediments may shift rapidly in favor of these organisms during wetter climatic periods, when lake water conditions become less saline. Conversely, the fact that these biological processes were observed when moderately hypersaline ML sediments were incubated at concentrations near or at salt saturation demonstrates that a component of the microbial community is able to function at much higher salinities, as can occur in arid climatic periods.
In addition to shifts in the microbial community composition as determined by 16S rRNA gene DGGE, we observed changes in the composition of arrA-containing microorganisms in ML sediment samples incubated at lower salinities (25 g liter−1) and at salt saturation (325 g liter−1), respectively. Many of these sequences were closely related to those previously found in the sediments recovered directly from the lake (10). However, a number of unique arrA phylotypes were detected in the sediment incubations. This observation indicates that an enrichment effect had occurred in response to the incubation conditions. DGGE analysis of the 16S rRNA gene provides additional evidence that the microbial community in the sediment incubations was altered in response to the salinity. Because of the small number (n = 23) of partial arrA sequences in the clone library from both lakes, it is unclear whether the arrA phylotypes are associated with low or high salinity.
Interestingly, both low- and higher-salinity sediment incubations contained a high percentage of arrA phylotypes that were previously observed in these two lakes (10). These sequences are most likely part of the background or native arrA-containing microorganisms within the sediment samples. Because these sequences are found in both ML and SL sediment, it is possible that they are associated with arsenate-respiring prokaryotes that are metabolically active at both moderate and extreme hypersalinity. Of the four known halophilic arsenate-respiring bacteria held in culture, Bacillus selenitireducens appears to be the best adapted for growth over a wide range of salinities (20 to 220 g liter−1) (23). However, we did not observe environmental arrA phylotypes that were similar to the arrA of B. selenitireducens (1). In contrast, two other existing cultures of moderate halophiles, namely, B. arseniciselenatis (23) and strain MLMS-1 (6) exhibit maximum growth at salinities lower than 140 g liter−1. Strain SLAS-1 is extremely halophilic and does not grow at salinities less than 200 g liter−1 (17). A second explanation for why we observed the group I/II arrA phylotypes in both low- and higher-salinity sediment conditions could be that horizontal gene transfer events resulted in the spread of certain arrA phylotypes within the microbial communities. In natural sediments of ML and SL, we observed that certain arrA phylotypes were present in both environments despite the geographic separation (10). Whether this observation is due to horizontal gene transfer events or to being native to the environment due to adaptation to high or lower salinities will require additional molecular and culture-dependent work aimed at identifying the links between the arrA sequences and specific microorganisms and investigating the diversity of arrA in various environments.
Our sulfate reduction experiments, however, yielded anomalous results compared to the other metabolic processes that were investigated in this study. The lack of sulfate reduction in SL sediment slurries at any salinity confirms observations that we reported previously (10) regarding the absence of DSR in SL sediments that support active DAsR. SL sediment slurries were able to reduce sulfate after the introduction of sulfate reducers with a small inoculum from the ML sediments (Fig. 5). This result suggests that the absence of DSR in SL sediment slurries was caused by the lack of viable sulfate-reducing flora within the microbial community of SL, rather than by inhibition of sulfate reduction by some constituent of the SL sediment or media. This interpretation is supported by a lack of DSR activity in SL slurries that received a sterilized ML inoculum (Fig. 5) and also by the fact that SL sediments did not reduce sulfate when slurried with ML media rather than SL media (data not shown).
The lack of detectable DSR activity in salt-saturated ML slurries demonstrates the inhibition of this process at the highest salinities (Fig. 4D). This result is consistent with the hypothesis of Oren (21), which postulates that DSR supplies insufficient metabolic energy for bacterial osmoadaptation as conditions approach salt saturation. However, the fact that the relatively less bioenergetically favorable process of methanogenesis (CO2/CH4) (E0′ = −244 mV) persisted at salt saturation while DSR (SO42−/HS−) (E0′ = −220 mV) was excluded was an unexpected finding (Fig. 4C). Our experiments with ML sediments demonstrated that DSR was strongly inhibited by high (200 mM) borate concentrations (Fig. 6); however, DSR was not inhibited in ML slurries amended with 40 mM borate or when ML sediments were incubated in SL medium that contained 51 mM borate (Fig. 5). We infer that the notably high in situ borate content of the concentrated SL brine (0.46 m) may act as a specific inhibitor of DSR in a manner analogous to that previously described for molybdate (15), thus excluding sulfate reducers, but not methanogens, from the SL ecosystem. Further work is needed to establish the tolerance of sulfate-reducing bacteria from other settings to borate and to investigate the physiological mechanism by which high borate concentrations may inhibit DSR.
Thus, as conditions approach salt saturation, the relative importance of DSR may become diminished (or absent) relative to less abundant but bioenergetically more favorable electron acceptors like arsenate or nitrate. Nonetheless, the ability of methanogenesis to persist at the extreme salinity condition while DSR did not is a curiosity perhaps dependent upon the availability of a suitable methanogenic precursor. For example, Marvin-DiPasquale et al. (11) demonstrated the ability of salt-saturated Dead Sea sediments to produce [14C]methane from [14C]methanol but not from other potential methanogenic substrates (e.g., 14C-labeled acetate, trimethylamine, dimethylsulfide, and methionine). We have noted that methanogenesis in low-salinity SL sediment slurries was significantly stimulated by amendment with millimolar levels of methionine, trimethylamine, and methanol, and yet these substrates did not enhance the minimal amount of activity observable at salt saturation (T. R. Kulp, unpublished data). Clearly, the specific case of methanogenetic activity occurring in salt-saturated ecosystems also needs further investigation.
In summary, it appears that distinct members of the resident microbial population may either gain or lose predominance in the overall community structure of these hypersaline soda lakes as broad salinity changes occur, possibly as a response to climatic events. As regional climatic conditions become more arid or as tributary streams are diverted by human activity, the microbial population of an increasingly saline lake can be expected to shift in favor of organisms that utilize higher energy-yielding metabolic processes (e.g., denitrification and arsenate reduction), although the processes themselves will occur at suboptimal rates. Of the processes we examined, it is evident that sulfate reduction is the most sensitive to both salt saturation and the presence of abundant naturally occurring minerals like borate. In the specific instance of soda lakes, but perhaps not for other high-density brines of different composition (24), it appears that borate ions can actually eliminate sulfate reducers from these ecosystems.
Supplementary Material
Acknowledgments
We are grateful to J. Stolz and M. Voytek for constructive criticism of earlier versions of the manuscript. We also thank L. Miller and S. Hoeft for their valuable assistance in the field and for their thoughtful comments.
This work was supported in part by the USGS National Research Program and by a grant to R.S.O. from the NASA Exobiology Research Program.
Footnotes
Published ahead of print on 29 June 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Afkar, E., J. Lisak, C. Saltikov, P. Basu, R. S. Oremland, and J. F. Stolz. 2003. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol. Lett. 226:107-112. [DOI] [PubMed] [Google Scholar]
- 2.Benson, L. V., D. R. Currey, R. I. Dorn, K. R. Lajoie, C. G. Oviatt, S. W. Robinson, G. I. Smith, and S. Stine. 1990. Chronology of expansion and contraction of four Great Basin lake systems during the past 35,000 years. Palaeogeogr. Palaeoclimatol. Palaeoecol. 78:241-286. [Google Scholar]
- 3.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:294-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dundas, I. 1998. Was the environment for primordial life hypersaline? Extremophiles 2:375-377. [DOI] [PubMed] [Google Scholar]
- 5.Herbst, D. B., and D. W. Blinn. 1998. Experimental mesocosm studies of salinity effects on the benthic algal community of a saline lake. J. Phycol. 34:772-778. [Google Scholar]
- 6.Hoeft, S. E., T. R. Kulp, J. F. Stolz, J. T. Hollibaugh, and R. S. Oremland. 2004. Dissimilatory arsenate reduction with sulfide as an electron donor: experiments with Mono Lake water and isolation of strain MLMS-1, a chemoautotrophic arsenate respirer. Appl. Environ. Microbiol. 70:2741-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollibaugh, J. T., C. Budinoff, R. A. Hollibaugh, B. Ransom, and N. Bano. 2006. Sulfide oxidation coupled to arsenate reduction by a diverse microbial community in a soda lake. Appl. Environ. Microbiol. 72:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knauth, L. P. 1998. Salinity history of the Earth's early ocean. Nature 395:554-555. [DOI] [PubMed] [Google Scholar]
- 10.Kulp, T. R., S. E. Hoeft, L. G. Miller, C. Saltikov, J. Murphy, S. Han, B. Lanoil, and R. S. Oremland. 2006. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl. Environ. Microbiol. 72:6514-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marvin-DiPasquale, M., A. Oren, Y. Cohen, and R. S. Oremland. 1999. Radiotracer studies of bacterial methanogenesis in sediments from the Dead Sea and Solar Lake (Sinai), p. 149-159. In A. Oren (ed.) Microbiology and biogeochemistry of hypersaline environments. CRC Press, Boca Raton, FL.
- 12.Miller, L. G., M. D. Coutlakis, R. S. Oremland, and B. B. Ward. 1993. Selective inhibition of ammonium oxidation and nitrification-linked N2O formation by methyl fluoride and dimethyl ether. Appl. Environ. Microbiol. 59:2457-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muylaert, K., K. Van der Gucht, N. Vloemans, L. De Meester, M. Gillis, and W. Vyverman. 2002. Relationship between bacterial community composition and bottom-up versus top-down variables in four eutrophic shallow lakes. Appl. Environ. Microbiol. 68:4740-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oremland, R. S. 1990. Nitrogen fixation dynamics of two diazotrophic communities in Mono Lake, California. Appl. Environ. Microbiol. 56:614-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oremland, R. S., and D. G. Capone 1988. Use of “specific” inhibitors in biogeochemistry and microbial ecology. Adv. Microb. Ecol. 10:285-383. [Google Scholar]
- 16.Oremland, R. S., P. R. Dowdle, S. Hoeft, J. O. Sharp, J. K. Schaefer, L. G. Miller, J. Switzer-Blum, R. L. Smith, N. S. Bloom, and D. Wallschlaeger. 2000. Bacterial dissimilatory reduction of arsenate and sulfate in meromictic Mono Lake, California. Geochim. Cosmochim. Acta 64:3073-3084. [Google Scholar]
- 17.Oremland, R. S., T. R. Kulp, J. Switzer-Blum, S. E. Hoeft, S. Baesman, L. G. Miller, and J. Stolz. 2005. A microbial arsenic cycle in a salt-saturated, extreme environment. Science 308:1305-1308. [DOI] [PubMed] [Google Scholar]
- 18.Oremland, R. S., and L. G. Miller. 1993. Biogeochemistry of natural gases in three alkaline, permanently stratified (meromictic) lakes, p. 439-452. In D. G. Howell (ed.), The future of energy gases. U.S. Geological Survey, Menlo Park, CA.
- 19.Oremland, R. S., L. G. Miller, C. W. Culbertson, S. W. Robinson, R. L. Smith, D. Lovley, M. J. Whiticar, G. M. King, R. P. Kiene, N. Iversen, and M. Sargent. 1993. Aspects of the biogeochemistry of methane in Mono Lake and the Mono Basin of California, p. 704-741. In. R. S. Oremland (ed.), Biogeochemistry of global change: radiatively active trace gases. Chapman and Hall, Inc., New York, NY.
- 20.Oremland, R. S., C. Umberger, C. W. Culbertson, and R. L. Smith. 1984. Denitrification in San Francisco Bay intertidal sediments. Appl. Environ. Microbiol. 47:1106-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oren, A. 1999. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63:334-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sørensen, J. 1978. Denitrification rates in a marine sediment as measured by the acetylene inhibition technique. Appl. Environ. Microbiol. 36:139-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Switzer Blum, J., A. Burns Bindi, J. Buzzelli, J. F. Stolz, and R. S. Oremland. 1998. Bacillus arsenicoselenatis sp. nov., and Bacillus selenitireducens sp. nov.: two haloalkaliphiles from Mono Lake, California which respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19-30. [DOI] [PubMed] [Google Scholar]
- 24.van der Wielen, P. W. J. J., H. Bolhuis, S. Borin, D. Daffonchio, C. Corselli, L. Giuliano, G. D'Auria, G. J. de Lange, A. Huebner, S. P. Varnavas, J. Thomson, C. Tamburini, D. Marty, T. J. McGenity, K. N. Timmis, and BioDeep Scientific Party. 2005. The enigma of prokaryotic life in deep hypersaline anoxic basins. Science 307:121-123. [DOI] [PubMed] [Google Scholar]
- 25.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.