Abstract
Corynebacterium glutamicum is able to accumulate up to 600 mM cytosolic phosphorus in the form of polyphosphate (poly P). Granular poly P (volutin) can make up to 37% of the internal cell volume. This bacterium lacks the classic enzyme of poly P synthesis, class I polyphosphate kinase (PPK1), but it possesses two genes, ppk2A (corresponds to NCgl0880) and ppk2B (corresponds to NCgl2620), for putative class II (PPK2) PPKs. Deletion of ppk2B decreased PPK activity and cellular poly P content, while overexpression of ppk2B increased both PPK activity and cellular poly P content. Neither deletion nor overexpression of ppk2A changed specific activity of PPK or cellular poly P content significantly. Purified PPK2B of C. glutamicum is active as a homotetramer and formed poly P with an average chain length of about 125, as determined with 31P nuclear magnetic resonance. The catalytic efficiency of C. glutamicum PPK2B was higher in the poly P-forming direction than for nucleoside triphosphate formation from poly P. The ppk2B deletion mutant, which accumulated very little poly P and grew as C. glutamicum wild type under phosphate-sufficient conditions, showed a growth defect under phosphate-limiting conditions.
Inorganic polyphosphate (poly P), a linear polymer consisting of three to hundreds of orthophosphate residues linked by phosphoanhydride bonds (20, 37), has been found in all living cells examined: archaea, bacteria, and eukarya (7, 20, 37). Poly P is synthesized by polyphosphate kinases (PPKs), using the terminal phosphate of ATP as the substrate, and is degraded to inorganic phosphate (Pi) by both endo- and exopolyphosphatases (19, 20, 37). Among the many functions poly P performs, the most prominent are the responses to many stresses. A variety of defects in responses to environmental stresses and/or virulence were observed in PPK gene mutants of Escherichia coli, Pseudomonas aeruginosa, Shigella, Salmonella, Vibrio cholerae, and Helicobacter pylori (5, 14, 17, 32). In E. coli, it could be demonstrated that under conditions of amino acid starvation, poly P accumulates and is bound by Lon protease (21). The Lon-poly P complex degrades ribosomal proteins supplying amino acids in response to starvation (21).
PPKs, which can be grouped into two classes, catalyze the reversible transfer of the gamma phosphoryl group of ATP or GTP to poly P. Genetic and biochemical studies have established the role of PPKs for biosynthesis of poly P, e.g., in the gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa (2, 3, 44). The archetypes of PPKs are class I PPK, PPK1, from E. coli, and class II PPK, PPK2, from P. aeruginosa. PPK2 of P. aeruginosa differs from ATP-specific PPK1, as it accepts GTP as well as ATP (12). Moreover, while the rate of GTP synthesis catalyzed by P. aeruginosa PPK2 is 75-fold higher than the rate of poly P synthesis, E. coli PPK1 shows a preference for poly P synthesis (12). Based on the presence or absence of PPK1 and/or PPK2 genes within complete genome sequences, Zhang et al. (44) distinguished four groups of microorganisms containing neither PPK1 nor PPK2 (e.g., Bacillus subtilis), only PPK1 (e.g., E. coli), PPK1 and PPK2 (e.g., P. aeruginosa), or only PPK2 (e.g., Corynebacterium glutamicum). The characteristics of PPK from the group of microorganisms containing only PPK2, i.e., Magnetococcus MC-1, Plectonema boryanum, or C. glutamicum, have not yet been reported.
C. glutamicum is a gram-positive rod belonging to the suborder Corynebacterineae together with its pathogenic relative Corynebacterium diphtheriae, of the family Corynebacteriaceae, and Mycobacterium tuberculosis, of the family Mycobacteriaceae (39). C. glutamicum is widely used for the biotechnological production of >1,200,000 tons of l-glutamate per year, of >550,000 tons of l-lysine per year, and of several other amino acids (8).
C. glutamicum is able to produce high amounts of soluble cytoplasmic poly P when cells are aerated and supplied with substrate and Pi, and intracellular poly P granules (volutin) are formed preferentially in the early exponential phase of growth (6, 18, 22, 28). Moreover, the poly P granules of C. glutamicum contain an enzyme predicted to be involved in poly P metabolism, i.e., a putative poly P glucokinase (6, 18, 22, 28). To date, the enzymes involved in synthesis, conversion, and degradation of poly P have not been characterized in C. glutamicum. Genes encoding PPK1s, such as E. coli ppk1, are absent from the genome of C. glutamicum or other corynebacteria but are present in M. tuberculosis and other mycobacteria (42, 44). However, C. glutamicum possesses two genes for putative PPK2s: ppk2A (corresponds to NCgl0880 and cg1046) and ppk2B (corresponds to NCgl2620 and cg3007) (42, 44). These genes encode proteins that share 57% and 64% sequence identity with P. aeruginosa PPK2, the only characterized PPK2 so far (44), and 51% sequence identity with each other. The gram-negative P. aeruginosa also possesses a PPK1. However, to date, characteristics of a PPK2 from a gram-positive bacterium are not known. Moreover, neither the role nor the characteristics of a PPK2 from a bacterium that lacks PPK1 and only possesses PPK2 have been determined so far. The objectives of the present work were the determination of the roles of the ppk2A and ppk2B gene products for polyphosphate biosynthesis in C. glutamicum and the biochemical characterization of PPK2 from a gram-positive bacterium.
MATERIALS AND METHODS
Microorganisms and cultivation conditions.
E. coli DH5α (11) was used for cloning and BL21(DE3) (41) for protein production. The wild-type strain of Corynebacterium glutamicum ATCC 13032 (1), named WT in the following text, the lysine producer MH20-22B (38), and the respective ppk2A and ppk2B deletion mutants C. glutamicum WTΔppk2A, WTΔppk2B, MH20-22BΔppk2A, and MH20-22BΔppk2B described in this work, were used and are listed in Table 1. Plasmids pVWEx1-ppk2A and pVWEx1-ppk2B for IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible overexpression of the genes ppk2A and ppk2B, respectively, were constructed on the basis of pVWEx1 (29). All C. glutamicum strains were precultured on Luria-Bertani complex medium (35) with kanamycin (50 μg/ml) or chloramphenicol (10 μg/ml) added when appropriate. Exponentially growing cells were harvested by centrifugation (5,000 × g, 5 min, 4°C), washed twice in 200 mM potassium phosphate buffer, pH 7, and used to inoculate CgXII minimal medium (8), which contained 0.3 g liter−1 leucine. For overexpression of ppk2A and ppk2B, 1 mM IPTG was added to minimal medium cultures of C. glutamicum WT(pVWEx1-ppk2A), WT(pVWEx1-ppk2B), and WT(pVWEx1) at an optical density at 600 nm (OD600) of 0.5. For protein purification, E. coli BL21(DE3) was used and cultured on LB medium with kanamycin (50 μg/ml) and 10 g of glucose per liter.
TABLE 1.
C. glutamicum strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| WT | ATCC 13032, wild-type strain auxotrophic for biotin | 1 |
| WTΔppk2A | In-frame deletion of the ppk2A gene in WT | This study |
| WTΔppk2B | In-frame deletion of the ppk2B gene in WT | This study |
| MH20-22B | Lysine producer, resistant against S-2-aminoethyl-l-cysteine, leucine auxotrophic | 38 |
| MH20-22BΔppk2A | In-frame deletion of the ppk2A gene in MH20-22B | This study |
| MH20-22BΔppk2B | In-frame deletion of the ppk2B gene in MH20-22B | This study |
| Plasmids | ||
| pGEM-T | Ampr, PCR cloning vector (PT7 PSP6) | Promega, WI |
| pK19mobsacB | Kanr, E. coli-C. glutamicum shuttle vector for the construction of insertion and deletion mutants in C. glutamicum (pK18 oriVE. coli sacB lacZα) | 36 |
| pK19mobsacBΔppk2B | Kanr, pk19mobsacB with the deletion construct of the gene ppk2B | This study |
| pK19mobsacBΔppk2A | Kanr, pk19mobsacB with the deletion construct of the gene ppk2A | This study |
| pVWEx1 | Kanr PtaclacIq | 29 |
| pVWEx1-ppk2A | Kanr, pVWEx1 with the ORF of the ppk2A gene and an artificial RBS | This study |
| pVWEx1-ppk2B | Kanr, pVWEx1 with the ORF of the ppk2B gene and an artificial RBS | This study |
| pET16b | Ampr, overproduction of proteins with an N-terminal decahistidine tag in E. coli (pBR322 oriVE. coli PT7 lacI) | Novagen |
| pET16b-ppk2B | Ampr, pET16b with the ORF of the ppk2B gene | This study |
ORF, open reading frame; RBS, ribosome binding site.
For growth comparisons of C. glutamicum WT and the ppk2B deletion mutant (WTΔppk2B), cells were plated on LB media and incubated for 24 h. Single colonies were picked and grown overnight at 30°C in 70 ml LB media. Cells were pelleted by centrifugation for 5 min at 4,120 × g, washed in phosphate-free CgXII medium, centrifuged again, and resuspended in 5 ml phosphate-free CgXII media. Cultures of 60 ml CgXII media with 13 mM, 0.13 mM, or no phosphate added and 4% (wt/vol) glucose were inoculated to a final OD600 of 1 and incubated for 26 h at 30°C. The biomass concentration was calculated from the OD600 values using an experimentally determined correlation factor of 0.25 g dry weight for an OD600 of 1 (43).
For lysine production, 4% (wt/vol) CgXII minimal medium was used supplemented with 0.3 g liter−1 leucine. When appropriate, kanamycin (50 μg/ml) and IPTG (1 mM) were added. Supernatants (10 min, 5,000 × g) of the cultures were analyzed 120 h after inoculation by high-performance liquid chromatography as described previously (16).
Homologous overexpression of ppk2A and ppk2B from C. glutamicum.
For overexpression of ppk2A, the gene was amplified from genomic DNA of C. glutamicum WT, which was prepared as described previously (9), by PCR using primers with the following sequences: 5′-GCGTCGACAAGGAGATATAGATATGCGAAAGAAAAAAGACGG-3′ (nucleotides [nt] 970864 to 970883 in the sequence under GenBank gi 23308765; the start codon is underlined, the ribosome binding site is given in italics, and the SalI restriction site is highlighted in bold) and 5′-GCTCTAGACTATTTCTTGGACTTCTTCTTGCC-3′ (nt 971803 to 971826 in the sequence under GenBank gi 23308765; the stop codon is underlined, and the XbaI restriction site is highlighted in bold). Similarly, for amplification of ppk2B, primers with the following sequences were used: 5′-GCGTCGACAAGGAGATATAGATATGGTGGGTAAACTTCCCATC-3′ (nt 2887814 to 2887834 in the sequence under GenBank gi 23308765; the start codon is underlined, the ribosome binding site is given in italics, and the SalI restriction site is highlighted in bold) and 5′-GCTCTAGACTAGTCACCGATCTGGTCGCGCCC-3′ (nt 2886912 to 2886937 in the sequence under GenBank gi 23308765; the stop codon is underlined, and the XbaI restriction site is highlighted in bold). The amplification products were subcloned into vector pGEM-T (Promega, Mannheim, Germany), resulting in vector pGEM-T-ppk2A and pGEM-T-ppk2B. Afterwards, SalI-XbaI fragments of pGEM-T-ppk2A and pGEM-T-ppk2B were cloned into SalI, and XbaI restricted pVWEx1 to yield pVWEx1-ppk2A and pVWEx1-ppk2B, respectively, which allow IPTG-inducible expression of ppk2A and ppk2B in C. glutamicum.
Construction of the deletion mutants C. glutamicum WTΔppk2A, WTΔppk2B, MH20-22BΔppk2A, and MH20-22BΔppk2B.
In-frame deletion mutants of C. glutamicum were constructed via a two-step homologous recombination procedure as described previously (8). To amplify flanking regions of ppk2A, pairs Δppk2A-1 (5′-TGGTCTAGATGCGACGGAACATAAGAAG-3′; nt 970597 to 970615 in the sequence under GenBank gi 23308765; the XbaI restriction site is highlighted in bold) and Δppk2A-2 (5′-CCCATCCACTAAACTTAAACAGTCTTTTTTCTTTCG-3′; nt 970867 to 970881 in the sequence under GenBank gi 23308765; the linker sequence is given in italics), as well as Δppk2A-3 (5′-TGTTTAAGTTTAGTGGATGGGTCCGGAGGAAAAGGC-3′; nt 971791 to 971805 in the sequence under GenBank gi 23308765; the linker sequence is given in italics) and Δppk2A-4 (5′-CAGTAAGCTTTCCGCCCCCGGACCGCAG-3′; nt 972074 to 972091 in the sequence under GenBank gi 23308765; the HindIII restriction site is highlighted in bold), were used. A crossover PCR product, which was generated using primers Δppk2A-1 and Δppk2A-4 and both of the generated PCR fragments as templates, was cloned into pK19mobsacB (36) via its primer-attached XbaI and HindIII sites. Similarly, a crossover PCR product of the flanking regions of ppk2B was generated using primer pairs Δppk2B-1 (5′-TGGTCTAGACTCAACTCTTAATAGGAG-3′; nt 2888088 to 2888105 in the sequence under GenBank gi 23308765; the XbaI restriction site is highlighted in bold) and Δppk2B-2 (5′-CCCATCCACTAAACTTAAACAGGGAAGTTTACCCAC-3′; nt 2887817 to 2887831 in the sequence under GenBank gi 23308765; the linker sequence is given in italics), as well as Δppk2B-3 (5′-TGTTTAAGTTTAGTGGATGGGGTTGTGCTTCGTGGG-3′; nt 2886935 to 2886950 in the sequence under GenBank gi 23308765; the linker sequence is given in italics) and Δppk2B-4 (5′-CAGTAAGCTTGGCAACTAACCCAAAGTG-3′; nt 2886656 to 2886674 in the sequene under GenBank gi 23308765; the HindIII restriction site is highlighted in bold) and cloned into pK19mobsacB. Gene-deletion mutagenesis with pK19mobsacBΔppk2A or pK19mobsacBΔppk2B was carried out as described previously (33). The correct genotype of the deletion mutants was verified by PCR analysis using the primer pair ppk2A-con1 (5′-ATTCGCTTCAAGCCCAAGGC-3′, nt 970433 to 970452 in the sequence under GenBank gi 23308765) and ppk2A-con2 (5′-AGAGGGCGTTATTGTGGTGC-3′; nt 972264 to 972283 in the sequence under GenBank gi 23308765), as well as the primer pair ppk2B-con1 (5′-TACCGCTCTTCAGAGCATCC-3′; nt 2888255 to 2888274 in the sequence under GenBank gi 23308765) and ppk2B-con2 (5′-TTAGGCAGCGGACCTAAAGG-3′; nt 2886464 to 2886483 in the sequence under GenBank gi 23308765), and the mutants were designated WTΔppk2A and WTΔppk2B. Accordingly, the mutants MH20-22BΔppk2A and MH20-22BΔppk2B were constructed.
Heterologous expression of ppk2B from C. glutamicum and protein purification.
For the expression of the PPK gene, ppk2B was amplified via PCR from genomic DNA of C. glutamicum WT. The primers used had the following sequences: 5′-GGAACCCATATGGTGGGTAAACTT-3′ (nt 2886914 to 2886928 in the sequence under GenBank gi 1020661 are italicized, the start codon is underlined, and the NdeI recognition site is shown in bold) and 5′-GGTGGATCCTAGTCACCGATCTGG-3′ (nt 2887819 to 2887834 in the sequence under GenBank gi 1020661 are italicized, the stop codon is underlined, and the BamHI restriction side is shown in bold). The amplification product was cloned into vector pGEM-T (Promega, Mannheim, Germany), resulting in vector pGEM-T-ppk2B. After restriction with NdeI and BamHI, the ∼1-kb fragment from pGEM-T-ppk2b was ligated to NdeI and BamHI-restricted pET16b (Novagen, Madison, WI). The vector, pET16b-ppk2B, allows IPTG-inducible expression of the gene encoding PPK2B with an N-terminal decahistidyl tag and a factor Xa cleavage site in E. coli BL21(DE3). The induction with 0.5 mM IPTG was started at an OD600 of 0.5 to 0.6 in LB. The cells (500 ml) were harvested 4 h after induction and washed in 20 mM Tris, 300 mM NaCl, 5 mM imidazol, and 5% (vol/vol) glycerol. Pelleted cells were stored at −20°C until protein purification. Prior to lysis by French press, cells were resuspended in 20 mM Tris, 300 mM NaCl, 5 mM imidazol, and 5% (vol/vol) glycerol, and protease activity was inhibited by addition of 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM diisopropylfluorophosphate (DFP). The extract was cleared by centrifugation for 1.5 h at 140,000 × g. Peak fractions of Ni-nitrilotriacetic acid (Ni-NTA) agarose affinity chromatography eluted with 20 mM Tris, 300 mM NaCl, 100, 200, or 400 mM imidazol, and 5% (vol/vol) glycerol were pooled, and the pooled fractions were desalted using Sephadex G25 gel filtration (Amersham Bioscience, Uppsala) and buffered in 50 mM triethylamine (TEA), pH 7.2. After purification, the His tag was cleaved by factor Xa (Novagen, San Diego) according to the manufacturer's recommendations.
PPK assay.
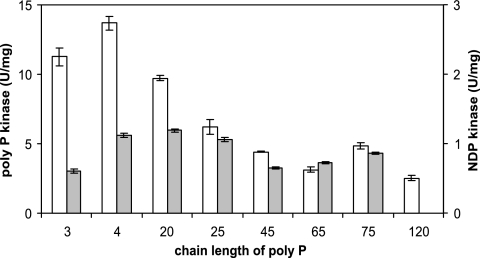
The activity of PPK2B as a PPK was measured in a spectrophotometric assay containing 10 mM MnCl2, 80 mM (NH4)2SO4, 50 mM TEA, pH 7.2, 0.25 mM NADH, 0.2 mM phosphoenolpyruvate, and rabbit muscle pyruvate kinase-l-lactate dehydrogenase (4.5 U). At least two experiments with at least three replicates (variation <10%) were performed at 30°C, the optimal temperature for C. glutamicum growth. For optimization, concentrations of 0 to 100 mM MnCl2, 0 to 100 mM MgCl2, and 0 to 500 mM (NH4)2SO4, temperatures from 11°C to 60°C, and a pH range from 5.5 to 8.9 were used. Nucleoside triphosphate (NTP) concentrations were varied up to 5 mM and poly P (P68 [28] with an average chain length of 20) concentrations up to 50 mM (in Pi residues) were added. The assay was started by the addition of poly P. NTP was regenerated from nucleoside diphosphate (NDP) and phosphoenolpyruvate by pyruvate kinase, and the formed pyruvate was reduced to lactate by NADH-dependent lactate dehydrogenase. The decrease of NADH was measured spectrophotometrically at 340 nm (ɛ340 nm = 6.22 mM−1 cm−1). To determine the influence of poly P chain length on the activity of poly P kinase (and NDP kinase; see next paragraph), the following standards were used (see Fig. 4): for a poly P chain length of 3, pentasodiumtripolyphosphate (Sigma-Aldrich, Taufkirchen, Germany); for a poly P chain length of 15, P15 (Sigma-Aldrich, Taufkirchen, Germany; gift of U. Selig, Rostock); for poly P chain lengths of 4 and 20, natriumtetrapolyphosphate and natriumhexametaphosphate (gifts of F. Wahl and B. K. Giulini, Ladenburg); for poly P chain lengths of 25, 45, 65, and 75, phosphate glasses (gifts of W. E. G. Müller, Mainz); and for a poly P chain length of 120, sodiumdihydrogenphosphate heated to constant weight at 700°C by F. Michulitz (Zentralabteilung für Chemische Analysen, Forschungszentrum Jülich). Chain length was checked with 31P nuclear magnetic resonance (NMR).
FIG. 4.
Influence of poly P chain length on the activities of the poly P and NDP kinase reactions of PPK2B. Open columns, PPK; shaded columns, NDP kinase. Bars represent standard errors of two to three determinations. For poly P standards, see Materials and Methods.
NDP kinase assay.
The NDP kinase activity of PPK2B was measured with ADP as the substrate in a spectrophotometric assay containing 10 mM MnCl2, 80 mM (NH4)2SO4, 50 mM TEA, pH 7.2, 1.5 mM NADP+, 1 mM glucose, yeast hexokinase (3 U), and Leuconostoc mesenteroides glucose-6-phosphate dehydrogenase (1.5 U). At least two experiments with at least three replicates (variation <10%) were performed at 30°C. ADP concentrations used were varied up to 1 mM, and P68 concentrations up to 40 mM (in Pi residues) were added. The assay was started by the addition of poly P. ATP formed by PPK2B was converted with glucose to ADP and glucose-6-phosphate by hexokinase. NADP+-dependent glucose-6-phosphate dehydrogenase converted glucose-6-phosphate to 6-phosphogluconate, and the NADPH formed was measured spectrophotometrically at 340 nm (ɛ340 nm = 6.22 mM−1 cm−1).
Molecular weight determination.
The molecular weight of purified PPK2B was determined by gel filtration using a Superdex column (HiLoad 26/60 Superdex 200 prep grade; Amersham). Therefore, 2 mg PPK2B dissolved in 2 ml of 50 mM TEA, pH 7.2, containing 150 mM NaCl and 5% glycerol (vol/vol) was applied to the column. The calibration was carried out using gel filtration molecular weight markers (MW-GF-200; Sigma) containing the proteins cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), and β-amylase (200 kDa). The void volume was determined with blue dextran.
Quantification of poly P by toluidine blue.
Poly P was quantified by measurement of a 10-μl sample in 1 ml toluidine blue solution (6 mg/liter) containing 40 mM acetic acid (25). The ratio of 530 to 630 nm was measured spectrophotometrically to quantify poly P using appropriate concentrations of P68 as a standard (see next paragraph, “31P NMR spectroscopy”).
31P NMR spectroscopy.
Poly P standards and samples from the PPK2B reaction were prepared for NMR spectroscopy with EDTA buffer and D2O as described previously (22, 28), except that 700-μl aliquots in 5-mm NMR tubes were analyzed. During preparation, all samples were kept on ice or were kept frozen until further use.
For cellular poly P, cultures at the late exponential phase of growth were harvested by centrifugation, and 1 g of wet cell pellet weight was suspended in 4 ml absolute ethanol, mixed for 1 min, and centrifuged for 10 min at 4,400 × g and 4°C. The supernatant was discarded, and EDTA, D2O, and H2O were added as described previously (28).
The 31P NMR spectra were measured at 5°C on a Varian Inova 400 MHz spectrometer. An amount of D2O sufficient to obtain a stable lock signal was added to each sample prior to measurement. The following parameters were used: frequency, 161,985 MHz; excitation pulse width, 9.25 μs; pulse repetition delay, 1 s; and spectral width, 18.35 kHz. Routine spectra were acquired with 4,096 scans. When indicated, trace signals were detected using 122,880 scans, divided into blocks of 12,288 each. Chemical shifts were referenced to 85% orthophosphoric acid (0 ppm). Orthophosphate and poly P P68 and P100 with an average chain length of 150 (10 mM P each) (18) served as standards for quantification. Spectra were Fourier transformed and processed with MestReC v4.9.9.6 (Mestrelab Research, Santiago de Compostela, Spain).
Estimation of the average chain length of poly P.
All commercially available poly Ps with chain lengths greater than 3 are mixtures of low- through high-molecular species (23, 40). The average chain length, n̄, can be determined from the signal intensities of the terminal and core phosphate groups according to the following equation, which was modified from one described previously (30):
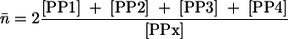 |
where x is either 1, 2, or 3, whichever signal is best resolved (PP1, phosphate endgroups; PP2 and PP3, groups next to PP4, which denotes the core phosphate groups).
It proved impossible to perform 31P NMR with the PPK2B reaction mixture directly, as poly P was too dilute for 31P NMR, and the concentration of Mn2+ ions was too high when the sample was concentrated by evaporation. Therefore, poly P was concentrated as described previously (4), yet by replacing Glassmilk with QIAprep columns (QIAprep Spin Miniprep; QIAGEN, Valencia, CA), which were eluted with 200 μl H2O after incubation for 2 min at 95°C. In tests, we could show that extraction with QIAprep yielded a near-quantitative recovery of long-chain poly P by comparing 31P NMR spectra of the poly P standards P68 and P100 before and after treatment with QIAprep (spectra not shown). With poly P P68, the sum of the poly P signals was only slightly reduced from 10 to 9.5 mM P. This was due to a decrease of low-molecular-weight poly P trace signals from the PP1, PP2, and PP3 regions. Thus, the average polymerization grade n̄ (calculated as described above) increased from 19 ± 2 before to 30 ± 1 after treatment with QIAprep. Cyclic poly Ps with ring sizes of 3 through 7 P units were also reduced. They appeared as singulet signals around −21 to −23 ppm and were assigned as described previously (10). Applying 122,880 scans, it could be shown that, e.g., cyclic poly P4 was reduced from 150 μM P to 5 μM P by the QIAprep treatment.
RESULTS
Overexpression and deletion of genes coding for putative PPKs.
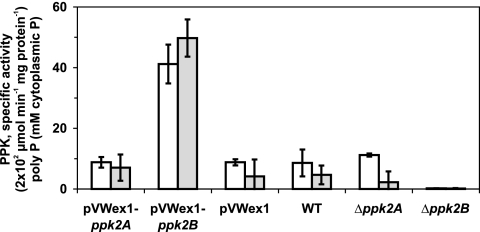
First, it was tested whether the deletion or overexpression of either ppk2A or ppk2B (predicted to code for PPK2s) (42) showed any effect on the metabolism of C. glutamicum. Each gene was deleted and overexpressed, respectively, in C. glutamicum WT (see Materials and Methods for details) and growth, enzyme activities in crude extracts, and cellular poly P content were analyzed in these strains and the respective controls. C. glutamicum WT, the deletion mutants WTΔppk2A and WTΔppk2B, as well as WT(pVWEx1) and the overexpression strains WT(pVWEx1-ppk2A) and WT(pVWEx1-ppk2B) grew similarly on minimal medium with 200 mM glucose (data not shown). Crude cell extracts prepared from these cultures contained about a fourfold increase in PPK activity when ppk2B was overexpressed compared to strains WT(pVWEx1) and WT(pVWEx1-ppk2A) (Fig. 1).
FIG. 1.
PPK activity and poly P accumulation in various strains of C. glutamicum. Open columns, PPK activity; shaded columns, poly P content. Average values and standard deviations of at least three independent determinations are given.
Moreover, a strain lacking ppk2B showed at least a threefold reduction in PPK activity compared to the parent strain. Deletion or overexpression of ppk2A did not change PPK activity notably.
When the cellular poly P content was compared in these strains by 31P NMR, the strain overexpressing ppk2B showed about sixfold higher intracellular poly P concentrations than the strain overexpressing ppk2A or the empty vector control, and the strain lacking ppk2B showed hardly any intracellular poly P, while poly P could easily be detected in the wild type or the ppk2A deletion mutant. These data indicate that ppk2B encodes a major PPK of C. glutamicum.
Overexpression or deletion of ppk2A or ppk2B had little effect on lysine production by C. glutamicum MH20-22B (53 ± 21 mM), MH20-22BΔppk2A (55 ± 8 mM), MH20-22BΔppk2B (42 ± 7 mM), MH20-22B(pVWEx1) (65 ± 2 mM), MH20-22B(pVWEx1-ppk2A) (59 ± 6 mM), and MH20-22B(pVWEx1-ppk2B) (58 ± 9 mM) on 4% (wt/vol) glucose minimal medium.
Biochemical characterization of the PPK encoded by ppk2B. (i) Overproduction, purification, and molecular mass.
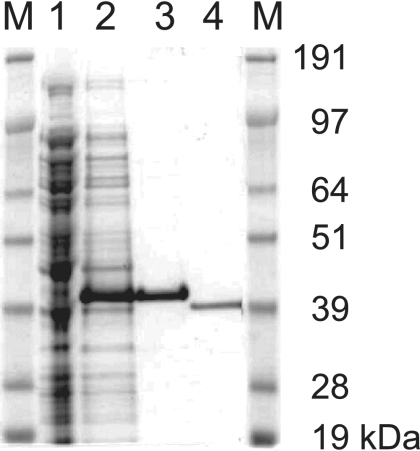
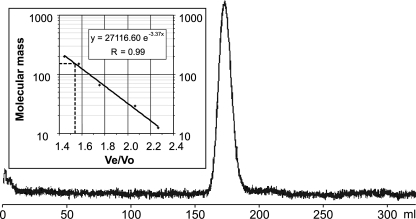
To characterize the enzymatic properties of the gene product of ppk2B, the protein carrying a His tag was overproduced in E. coli (Fig. 2). After Ni-NTA chromatography and factor Xa cleavage, a single band of approximately 39 kDa was detected, indicating purification of PPK2B to apparent homogeneity. Gel filtration of this protein and activity assays showed that PPK2B is active as a homotetramer with a calculated molecular mass of 151.4 kDa (Fig. 3).
FIG. 2.
Purification of PPK2B of Corynebacterium glutamicum from Escherichia coli BL21(DE3) (pET16b-ppk2B). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of protein from extracts before (lane 1) and after (lane 2) induction with 0.5 mM IPTG as well as from Ni-NTA chromatography purified (lane 3) and factor Xa-cleaved protein (lane 4) is shown. Lane M, SeeBlue Plus2 prestained standard (Invitrogen).
FIG. 3.
Molecular weight determination of PPK2B of C. glutamicum by gel filtration resulted in a single peak, as shown. The ratio of Ve (elution volume) and Vo (void volume; measured by blue dextran) of the standard proteins (see Materials and Methods) was plotted against the molecular weight, as seen in the inset. From this plot, the molecular weight of PPK2b was calculated.
(ii) Optimum conditions for activity.
C. glutamicum PPK2B showed a narrow pH optimum at pH 7.2 in 50 mM TEA. The optimal concentrations of MnCl2 and MgCl2 were 10 mM, but the PPK2B activity with magnesium was only 7% of the optimum activity with manganese. The optimum concentration of (NH4)2SO4 was 80 mM.
To determine the stability against irreversible thermal denaturation, PPK2B was preheated in the absence or presence of 40 mM poly P before assaying for activity at 30°C, the optimal temperature for C. glutamicum growth. After preincubation for 30 min at 20°C or 25°C, more than 50% of the activity remained (data not shown). The stability of PPK2B towards higher temperatures was low, as the enzyme almost completely lost its activity after 5 min of preheating at 45°C (data not shown). However, when 40 mM poly P was added during preheating, the activity attained the same stability at 45°C as was found in the controls without poly P at 20°C and 25°C, respectively (data not shown).
(iii) Kinetic parameters.
Kinetic parameters were determined both in the direction of synthesis of poly P and in the direction of NTP formation from poly P with various nucleotides as substrates. The chain length preference of PPK2B was similar in the poly P-forming and in the NTP-forming directions (Fig. 4), i.e., around 3 to 20 and about 4 to 25, respectively. In both directions, about 15 mM poly P (in Pi units) supported half-maximal activity (Table 2).
TABLE 2.
Kinetic parameters of PPK2B from C. glutamicum
| Reaction and substrate | Kinetic parameter
|
|||
|---|---|---|---|---|
| Km (mM) | Vmax (μmol min−1 mg−1 of protein) | kcat (s−1) | kcat/Km (s−1 mM−1) | |
| Poly P forming | ||||
| ATP | 0.17 | 31 | 74 | 438 |
| GTP | 0.14 | 14 | 34 | 240 |
| ITP | 1 | 18 | 43 | 43 |
| Poly Pa | 15 | 32 | 77 | 5 |
| NTP forming | ||||
| Poly P | 16 | 3 | 7 | 0.45 |
| ADP | 0.04 | 4 | 10 | 250 |
Kinetic parameters of the reaction with ATP as the substrate.
In the poly P-forming direction, the maximal activity was twofold higher with ATP (Vmax of 31 μmol min−1 mg−1) than with GTP and ITP (Table 2). While the Km values were 0.17 and 0.14 mM for ATP and GTP, respectively, the Km value was six- to sevenfold higher with ITP (Table 2), and hence the highest kcat/Km value was for ATP, followed by GTP (∼1/2) and ITP (∼1/10). The maximal activity in the NTP-forming direction was about tenfold lower than in the poly P-forming direction (Table 2). While the Km value for poly P was about 15 mM in both directions, the Km value for ADP in the NTP-forming direction was about fourfold lower than that for ATP in the poly P-forming direction (Table 2). Based on the Vmax and kcat/Km values for ATP, ADP, and poly P, PPK2B is more active in the poly P-forming direction.
Poly P chain elongation by PPK2B was inhibited by mononucleotides with Ki values of 3.9 mM for GMP, 6.6 mM for IMP, and 7.9 mM for AMP, while UMP and CMP had no influence on this reaction.
(iv) Primer-independent formation of poly P from ATP by PPK2B.
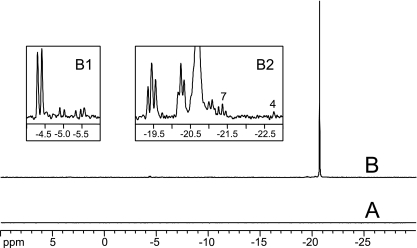
C. glutamicum PPK2B was able to synthesize poly P from ATP without the need for added poly P as a primer (Fig. 5). The product of the PPK2B reaction with ATP as the only added substrate was poly P with an average polymerization of n̄ = 125 ± 15 (for calculation of n̄, see Materials and Methods). Insets B1 and B2 in Fig. 5 show that traces of ATP, ADP, and PPi were present after the PPK2B reaction, as were traces of cyclic poly Ps (about 0.1% P, i.e., about 10 μM P each). Not even traces of poly P signals could be detected in the control with heat-inactivated (preheated for 1 h at 95°C) enzyme (Fig. 5A). In a separate experiment, the reaction of PPK2B with ATP was stopped after 5 min. Already, about 30% of the main poly P signal had been formed.
FIG. 5.
31P NMR analysis of the poly P product from the PPK2B reaction with ATP. (A) Heat-inactivated enzyme. (B) Reaction product after 24-h incubation in reaction buffer. B1 and B2 show selected regions of spectrum B. Major signals are doublets near −4.5 ppm (PP1); triplets around −19.5 and −20.3 (PP2 and PP3); and the core phosphates signal around −20.6 ppm (PP4); see Materials and Methods. In inset B1, the small signals at −5 and −5.5 ppm are due to traces of ATP, ADP, and PPi. In inset B2, the small signals from about 20.75 to 20.7 ppm are due to traces of cyclic polyphosphates (true metaphosphates; for assignment of tetra- and heptametaphosphate [4 and 7 in inset B2], indicated by numerals, see Materials and Methods); the other trace signals are unassigned metaphosphates. The spectra depicted in insets B1 and B2 were acquired with 122,880 scans to enhance details.
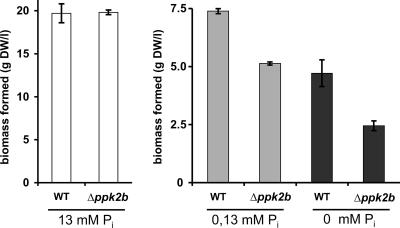
Growth of C. glutamicum WT and WTΔppk2B affected by phosphate availability.
As the ppk2B deletion mutant contained only traces of cytoplasmic poly P, we investigated how C. glutamicum copes with phosphate starvation. C. glutamicum WT and the ppk2B deletion mutant showed comparable growth under phosphate-sufficient conditions (13 mM) (Fig. 6). However, the ppk2B deletion mutant exhibited a growth disadvantage when transferred from phosphate-sufficient to phosphate-limiting conditions (Fig. 6) and formed about 30% and 50% less biomass than C. glutamicum WT in medium with 0.13 and 0 mM added phosphate (Fig. 6). Thus, the presence of PPK2B and the ability to store poly P enable C. glutamicum to better strive under conditions when phosphate becomes scarce.
FIG. 6.
Biomass formation of C. glutamicum WT and Δppk2B after transfer to minimal medium with different phosphate concentrations. Cells cultured overnight on LB medium were used to inoculate CgXII minimal medium with 4% (wt/vol) glucose and either 13 mM, 0.13 mM, or 0 mM Pi. Biomass formation was determined after 26 h of cultivation, and average values and errors of at least two independent cultivations are given.
DISCUSSION
ppk2B was shown to encode PPK2 of C. glutamicum.
PPK2B catalyzes poly P synthesis from ATP or GTP with higher catalytic efficiency than the reverse poly P-dependent NDP kinase reaction. With respect to pH and temperature, PPK2B showed optimal activity under physiologically relevant conditions, i.e., at neutral pH and at the optimal temperature for growth. So far, only one example of PPK2, the enzyme from Pseudomonas aeruginosa PAOM5, has been characterized (12). Both enzymes share a preference for manganese and ATP, GTP, and short-chain poly Ps as substrates. However, when comparing PPK2 of P. aeruginosa (12) with PPK2B of C. glutamicum, there is a striking difference: the former prefers the direction of NTP synthesis (∼75-fold); the latter is more effective in the poly P-forming direction (∼10-fold). While C. glutamicum lacks PPK1, PPK1 is the major poly P-synthesizing enzyme in P. aeruginosa, as the ppk1 mutant PAOM5 possesses five times less cellular poly P than the parent strain, P. aeruginosa PAO1 (31). Here, we have shown that the C. glutamicum ppk2B mutant accumulated at least 25-fold less poly P than C. glutamicum WT (Fig. 1). Thus, in accordance with its catalytic properties, PPK2 is a major poly P-synthesizing enzyme of C. glutamicum.
Sequence homologues of PPK2 from P. aeruginosa and C. glutamicum found in many other microorganisms (44) may therefore show preference either for poly P synthesis from ATP/GTP or for NTP formation from poly P and may have evolved to serve different functions. In P. aeruginosa, PPK2 provides GTP from poly P for the synthesis of alginate, and PPK2 levels are highest in the late-exponential-growth phase, when alginate production occurs. By contrast, C. glutamicum lacks an exopolysaccharide coat, and we could show that the ppk2B deletion mutant exhibited a growth disadvantage under phosphate-limiting conditions (Fig. 6); thus, C. glutamicum PPK2B serves a function in formation of poly P stores. It has to be noted that the genomes of P. aeruginosa and C. glutamicum encode two PPK2 sequence homologues, but for both bacteria, the characteristics of only one of them are known. The biochemical characteristics and functional roles of C. glutamicum PPK2A and P. aeruginosa PA2428 remain to be studied.
PPK2B of C. glutamicum yielded poly P of an average chain length of 125 ± 15. This is considerably shorter than poly P formed by PPK1 of E. coli, exhibiting a chain length of about 750 residues (4). Both PPK1 and PPK2 from Pseudomonas aeruginosa produce a broader range of poly P species, i.e., about 500 to 800 for PPK1 and 200 to 800 for PPK2. At present, we cannot completely rule out that some longer chain poly P species, possibly produced by the action of C. glutamicum PPK2B, have been retained during silica-based poly P purification. As the chain length of poly P extracted from C. glutamicum cells was 800 to 1,000 (C. Lambert and S. M. Schoberth, unpublished), it is tempting to speculate that PPK2A might play a role in elongating poly P formed by PPK2B.
Poly P may have many functions, e.g., as a substitute for ATP in kinase reactions, as a reservoir of phosphorus, as a chelator of metal ions, and in adaptation to environmental stress. Especially in pathogenic bacteria like P. aeruginosa, Shigella, Salmonella, Vibrio cholerae, Neisseria meningitidis, and Helicobacter pylori, PPK gene mutants were shown to have defects in responses to environmental stresses and were less virulent than the parent strains (5, 14, 17, 26, 31, 32). In Vibrio cholerae, large poly P stores enhance the ability of this gram-negative bacterium to adapt to high calcium ion concentrations (26) and to survive in low Pi environments, i.e., outside the human host, e.g., in estuarine and coastal waters (14). The C. glutamicum ppk2B deletion mutant exhibited a growth disadvantage in a shift from Pi-sufficient to Pi-starvation conditions (Fig. 6), indicating that poly P serves as a reservoir of phosphorus in this bacterium.
Lysine production by the model strain MH20-22B was not influenced by deletion or overexpression of either ppk2A or ppk2B. It remains to be studied whether PPK2B is important under conditions employed in large-scale fermentation, which may be characterized by stresses such as high osmolarity or low oxygen availability. Aside from amino acids, corynebacteria are also used for the biotechnological production of phosphorus-containing compounds, e.g., Corynebacterium ammoniagenes produces the flavor-enhancing purine nucleotides inosine 5′-monophosphate (IMP) and xanthosine 5′-monophosphate (XMP) (24). It is likely, but not yet experimentally shown, that engineering polyphosphate accumulation in C. ammoniagenes by overexpression or deletion of the ppk2B homolog(s) affects overproduction of IMP and XMP.
The determination of the phosphate starvation stimulon revealed the genetic program of C. glutamicum to cope with phosphate starvation (13). When Pi becomes scarce, genes (i) for the high-affinity Pi uptake system PstSCAB, (ii) for the glycerol-3-phosphate uptake system UgpAEBC and glycerophosphoryl diester phosphodiesterase GlpQ1, and (iii) for the 5′-nucleotidase UshA (34), the putative nuclease NucH, and other systems for uptake of phosphorous compounds or for mobilization of extracellular Pi, are induced (13). However, evidence for Pi-dependent transcriptional control of the genes ppk2A and ppk2B or of the putative exopolyphosphatase genes NCgl0396 and NCgl0938 was not obtained (13), which indicates that synthesis and degradation of poly P in C. glutamicum appear to be regulated at another level, e.g., by allosteric control. Synthesis of poly P by PPK2B was inhibited by AMP, GMP, and IMP, with inhibition constants in the millimolar range (3.9 to 7.9 mM). AMP in the same concentration range (2 mM) activated C. glutamicum pyruvate kinase fourfold (15), while the inhibition constant of AMP for C. glutamicum fructose-1,6-bisphosphatase was much lower (0.09 mM) (33). Thus, at least to some extent, poly P synthesis by PPK2B appears to be regulated by the “energy charge” of the cell.
In conclusion, we have shown that PPK2B is a major PPK of C. glutamicum and is responsible for the accumulation of high concentrations of soluble and granular poly P (18, 22, 27) in this gram-positive bacterium. The PPK2 PPK2B from C. glutamicum shows a preference for poly P synthesis from ATP or GTP over the reverse NDP kinase reaction and, thus, differs from the only other characterized PPK2, i.e., PPK2 from P. aeruginosa (44), which preferably catalyzes the NDP kinase reaction providing GTP from poly P for the synthesis of alginate. While growth under Pi-sufficient conditions as well as lysine production were not affected by the absence or overproduction of PPK2B, the presence of PPK2B confers a growth advantage to C. glutamicum when shifted to Pi-starvation conditions.
Acknowledgments
We thank F. Wahl for helpful discussions on chemistry and analysis of poly P throughout this work and for gifts of sodiumhexametaphosphate P 68 (poly P P68) and sodiumtetrapolyphosphate P 60 (poly P P60). We thank Werner E. G. Müller and Uwe Seelig for gifts of various types of sodium phosphate glasses used as standards in this study and Hermann Sahm for support.
Footnotes
Published ahead of print on 1 June 2007.
REFERENCES
- 1.Abe, S. G.-G., K. I. Takayama, and S. Kinoshita. 1967. Taxonomical studies on glutamic acid-producing bacteria. J. Gen. Appl. Microbiol. 13:279-285. [Google Scholar]
- 2.Ahn, K., and A. Kornberg. 1990. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 265:11734-11739. [PubMed] [Google Scholar]
- 3.Akiyama, M., E. Crooke, and A. Kornberg. 1992. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J. Biol. Chem. 267:22556-22561. [PubMed] [Google Scholar]
- 4.Ault-Riché, D., C. D. Fraley, C. M. Tzeng, and A. Kornberg. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayraud, S., B. Janvier, A. Labigne, C. Ecobichon, C. Burucoa, and J. L. Fauchere. 2005. Polyphosphate kinase: a new colonization factor of Helicobacter pylori. FEMS Microbiol. Lett. 243:45-50. [DOI] [PubMed] [Google Scholar]
- 6.Coello, N., J. G. Pan, and J. M. Lebeault. 1992. Corynebacterium glutamicum: morphological and ultrastructural changes of l-lysine producing cells in continuous culture. Appl. Microbiol. Biotechnol. 38:34-38. [Google Scholar]
- 7.Docampo, R. 2006. Acidocalcisomes and polyphosphate granules, p. 53-70. In J. M. Shively (ed.), Inclusions in prokaryotes, vol. 1. Springer, Berlin, Germany. [Google Scholar]
- 8.Eggeling, L., and M. Bott (ed.). 2005. Handbook of Corynebacterium glutamicum. CRC Press LLC, Boca Raton, FL.
- 9.Eikmanns, B. J., D. Rittmann, and H. Sahm. 1995. Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J. Bacteriol. 177:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glonek, T., J. R. van Wazer, M. Mudgett, and T. C. Myers. 1972. Cyclic metaphosphates from hydrolysis of the products from phosphoric acid condensation with carbodiimides. Inorg. Chem. 11:567-570. [Google Scholar]
- 11.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning: a practical approach, vol. 1. IRL Press, Oxford, United Kingdom. [Google Scholar]
- 12.Ishige, K., H. Zhang, and A. Kornberg. 2002. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. USA 99:16684-16688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahid, I. K., A. J. Silva, and J. A. Benitez. 2006. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl. Environ. Microbiol. 72:7043-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jetten, M. S., M. E. Gubler, S. H. Lee, and A. J. Sinskey. 1994. Structural and functional analysis of pyruvate kinase from Corynebacterium glutamicum. Appl. Environ. Microbiol. 60:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabus, A., T. Georgi, V. F. Wendisch, and M. Bott. 2007. Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves L-lysine formation. Appl. Microbiol. Biotechnol. 75:47-53. [DOI] [PubMed] [Google Scholar]
- 17.Kim, K. S., N. N. Rao, C. D. Fraley, and A. Kornberg. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. USA 99:7675-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klauth, P., S. R. Pallerla, D. Vidaurre, C. Ralfs, V. F. Wendisch, and S. M. Schoberth. 2006. Determination of soluble and granular inorganic polyphosphate in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 72:1099-1106. [DOI] [PubMed] [Google Scholar]
- 19.Kornberg, A. 1999. Inorganic polyphosphate: a molecule of many functions, p. 1-26. In H. Schröder and W. Müller (ed.), Inorganic polyphosphates: biochemistry, biology, biotechnology. Progress in molecular and subcellular biology, vol. 23. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 20.Kulaev, I. S., V. M. Vagabov, and T. V. Kulakovskaya. 2004. The biochemistry of inorganic polyphosphates, 2nd ed. John Wiley & Sons, Chichester, United Kingdom.
- 21.Kuroda, A., K. Nomura, R. Ohtomo, J. Kato, T. Ikeda, N. Takiguchi, H. Ohtake, and A. Kornberg. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293:705-708. [DOI] [PubMed] [Google Scholar]
- 22.Lambert, C., D. Weuster-Botz, R. Weichenhain, E. W. Kreutz, A. A. de Graaf, and S. M. Schoberth. 2002. Monitoring of inorganic polyphosphate dynamics in Corynebacterium glutamicum using a novel oxygen sparger for real time 31P in vivo NMR. Acta Biotechnol. 22:245-260. [Google Scholar]
- 23.Lorenz, B., and H. C. Schröder. 1999. Methods for investigation of inoganic polyphosphates and polyphosphate-metabolizing enzymes. Prog. Mol. Subcell. Biol. 23:217-239. [DOI] [PubMed] [Google Scholar]
- 24.Mori, H., A. Iida, T. Fujio, and S. Teshiba. 1997. A novel process of inosine 5′-monophosphate production using overexpressed guanosine/inosine kinase. Appl. Microbiol. Biotechnol. 48:693-698. [DOI] [PubMed] [Google Scholar]
- 25.Mullan, A., J. P. Quinn, and J. W. McGrath. 2002. A nonradioactive method for the assay of polyphosphate kinase activity and its application in the study of polyphosphate metabolism in Burkholderia cepacia. Anal. Biochem. 308:294-299. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa, N., C. M. Tzeng, C. D. Fraley, and A. Kornberg. 2000. Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J. Bacteriol. 182:6687-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallerla, S. R. 2004. Characterisation of the polyphosphate metabolism in Corynebacterium glutamicum using newly developed fluorescence microscopic and photometric methods. M. S. thesis. Fachhochschule Reutlingen, Reutlingen, Germany.
- 28.Pallerla, S. R., S. Knebel, T. Polen, P. Klauth, J. Hollender, V. F. Wendisch, and S. M. Schoberth. 2005. Formation of volutin granules in Corynebacterium glutamicum. FEMS Microbiol. Lett. 243:133-140. [DOI] [PubMed] [Google Scholar]
- 29.Peters-Wendisch, P. G., B. Schiel, V. F. Wendisch, E. Katsoulidis, B. Mockel, H. Sahm, and B. J. Eikmanns. 2001. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3:295-300. [PubMed] [Google Scholar]
- 30.Pilatus, U., A. Mayer, and A. Hildebrandt. 1989. Nuclear polyphosphate as a possible source of energy during the sporulation of Physarum polycephalum. Arch. Biochem. Biophys. 275:215-223. [DOI] [PubMed] [Google Scholar]
- 31.Rashid, M. H., N. N. Rao, and A. Kornberg. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182:225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rittmann, D., S. Schaffer, V. F. Wendisch, and H. Sahm. 2003. Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and biochemical characterization of the enzyme. Arch. Microbiol. 180:285-292. [DOI] [PubMed] [Google Scholar]
- 34.Rittmann, D., U. Sorger-Herrmann, and V. F. Wendisch. 2005. Phosphate starvation-inducible gene ushA encodes a 5′ nucleotidase required for growth of Corynebacterium glutamicum on media with nucleotides as the phosphorus source. Appl. Environ. Microbiol. 71:4339-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 37.Schröder, H. C., and W. E. G. Müller (ed.). 1999. Inorganic polyphosphates: biochemistry, biology, biotechnology. Progress in molecular and subcellular biology, vol. 23. Springer, Berlin, Germany.
- 38.Schrumpf, B., L. Eggeling, and H. Sahm. 1992. Isolation and prominent characteristics of an L-lysine hyperproducing strain of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 37:566-571. [Google Scholar]
- 39.Stackebrandt, E., F. Rainey, and N. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 40.Staffel, T., and T. Klein. 1999. Phosphoric acid and phosphates, p. 127. In Ullmann's encyclopedia of industrial chemistry, 6th ed. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- 41.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 42.Wendisch, V. F., and M. Bott. 2005. Phosphorus metabolism of Corynebacterium glutamicum, p. 379-398. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press LLC, Boca Raton, FL.
- 43.Wendisch, V. F., A. A. de Graaf, H. Sahm, and B. J. Eikmanns. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 182:3088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, H., K. Ishige, and A. Kornberg. 2002. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. USA 99:16678-16683. [DOI] [PMC free article] [PubMed] [Google Scholar]