Abstract
Four laboratory sourdough fermentations, initiated with wheat or spelt flour and without the addition of a starter culture, were prepared over a period of 10 days with daily back-slopping. Samples taken at all refreshment steps were used for determination of the present microbiota. Furthermore, an extensive metabolite target analysis of more than 100 different compounds was performed through a combination of various chromatographic methods including liquid chromatography-mass spectrometry and gas chromatography-mass spectrometry. The establishment of a stable microbial ecosystem occurred through a three-phase evolution within a week, as revealed by both microbiological and metabolite analyses. Strains of Lactobacillus plantarum, Lactobacillus fermentum, Lactobacillus rossiae, Lactobacillus brevis, and Lactobacillus paraplantarum were dominating some of the sourdough ecosystems. Although the heterofermentative L. fermentum was dominating one of the wheat sourdoughs, all other sourdoughs were dominated by a combination of obligate and facultative heterofermentative taxa. Strains of homofermentative species were not retrieved in the stable sourdough ecosystems. Concentrations of sugar and amino acid metabolites hardly changed during the last days of fermentation. Besides lactic acid, ethanol, and mannitol, the production of succinic acid, erythritol, and various amino acid metabolites, such as phenyllactic acid, hydroxyphenyllactic acid, and indolelactic acid, was shown during fermentation. Physiologically, they contributed to the equilibration of the redox balance. The biphasic approach of the present study allowed us to map some of the interactions taking place during sourdough fermentation and helped us to understand the fine-tuned metabolism of lactic acid bacteria, which allows them to dominate a food ecosystem.
Sourdough is a mixture of ground cereals (e.g., wheat or rye) and water that is spontaneously fermented. Sourdough fermentations improve dough properties, enhance both bread texture and bread flavor, and delay bread spoilage (28). Lactic acid bacteria (LAB) and yeasts play a key role in sourdough fermentation processes (9, 21, 26, 28, 29, 55). Sourdough LAB have been intensively studied with respect to their carbohydrate metabolism (16, 24, 60), proteolysis and amino acid metabolism (16, 23, 58, 69), lipid metabolism (16), and production of volatile compounds (7, 31, 32). Besides these general metabolic traits, specific metabolic properties have been recognized in sourdough LAB, such as the use of alternative electron acceptors (59, 61), the production of antifungal compounds (38, 45, 57), the biosynthesis of exopolysaccharides (36, 63, 64), and arginine catabolism (8, 53). These metabolic traits of sourdough LAB highlight their adaptation to the sourdough environment. For instance, fructose-to-mannitol and arginine-to-ornithine conversion favor ATP generation and/or acid stress (16). Also, interactions between sourdough LAB and yeasts have been studied in detail (9, 21).
The microbial growth and activity of LAB in sourdough are influenced by endogenous factors (e.g., flour carbohydrates and enzymes); process parameters (e.g., water content and temperature); and interactions between LAB, yeasts, and other bacteria (21, 28, 44, 71). Despite changes in raw material or the bakery environment, the sourdough microbiota is often remarkably stable, even for several years (17, 54). However, the metabolism of certain sourdough LAB may be influenced by process factors to obtain higher-quality end products. An example of the latter is the stimulated release or addition of alternative electron acceptors (e.g., fructose) or pentosans (e.g., through pentosanases) to stimulate the production of acetic acid (25, 37, 52). In this way, an optimal fermentation quotient (molar ratio of lactic acid to acetic acid) of about 2.5 can be attained (52).
The population dynamics of microbial food ecosystems have been studied mainly through microbiological analysis (20). In recent years, culture-independent methods have been developed to circumvent the limitations of conventional cultivation for the analysis of microbial communities in fermented foods (11). In this regard, denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA fragments (16S rRNA PCR-DGGE) is frequently used as a relatively rapid and reliable cultivation-independent method to study the biodiversity and population dynamics of microbial communities (11). A few studies have already described the use of PCR-DGGE to monitor the diversity and dynamics of LAB and yeast populations during sourdough fermentation processes (18, 42, 44, 51).
In the last couple of years, metabolomics has found its place among the “-omics” technologies, such as genomics, transcriptomics, and proteomics (48). Unfortunately, due to differences in physical and chemical properties of metabolites, a comprehensive approach is lacking (12). Therefore, metabolite target analysis and metabolite profiling are the most widespread techniques to study the metabolism of organisms (13). Metabolite target analysis involves a combination of techniques to quantify a limited number of metabolites. Metabolite profiling aims at generating metabolic profiles of certain classes of compounds and usually generates only semiquantitative data (13, 14). Recently, a combined analysis of acidity, amino acids, and selected volatiles has been performed for wheat sourdoughs to elucidate possible relationships between process parameters and formation of metabolites (34). However, the last study did not report on population dynamics during sourdough fermentation, because well-defined starter cultures were used that dominated the entire fermentation process.
The aim of the present study was an extended biphasic approach of spontaneously fermented laboratory sourdoughs. The population dynamics of yeasts and LAB were assessed by microbiological analysis (both yeasts and LAB) and PCR-DGGE (only LAB). In parallel, an extensive metabolite target analysis of more than 100 compounds was performed on the whole sourdough ecosystem to document the in situ metabolic potential of sourdough LAB and to explain why certain sourdough LAB species became dominant during spontaneous sourdough fermentation.
MATERIALS AND METHODS
Flours.
Wheat and spelt flours were provided by two local flour mills (A and B). Two types of each flour were used to prepare spontaneous sourdoughs by means of back-slopping (Table 1). Flour protein content was determined using near-infrared spectroscopy, and amylase activity was assessed by the Hagberg method (6).
TABLE 1.
Characteristics of flours from two mills (A and B) used for the production of laboratory sourdoughs
| Flour | Moisture (%) | Ash (% of dry matter) | Protein (% of dry matter) | Amylase activity (Hagberg method [6]; s) |
|---|---|---|---|---|
| Wheat A | 13.8 | 0.51 | 14.3 | 412 |
| Spelt A | 13.9 | 0.62 | 12.2 | 369 |
| Wheat B | 14.1 | 0.51 | 12.6 | 384 |
| Spelt B | 13.6 | 0.61 | 13.5 | 281 |
Laboratory sourdough preparation.
The production of sourdough (8 kg) was carried out in two Biostat C fermentors (Sartorius AG, Goettingen, Germany) and started by mixing 6 liters of sterile water with 2 kg of flour in one fermentor with a resulting dough yield [(dough mass/flour mass) × 100] of 400. This dough was incubated at 30°C for 24 h, during which the mixture was kept homogeneous through stirring (300 rpm). After 24 h of incubation, 800 g of ripe sourdough was collected in a sterile bottle to inoculate a fresh water-flour mixture (5.2 liters of water and 2 kg of flour) in the second fermentor on day 2. This dough was again incubated under the same conditions as described above. The back-slopping was carried out during a period of 10 days. At each refreshment step (every 24 h), samples were withdrawn from the ripe sourdough for culture-dependent microbiological analysis and immediate measurement of pH and total titratable acidity (TTA). The latter was done by suspending 10 g of sourdough in 100 ml of ultrapure water. The TTA value is expressed as the amount (in milliliters) of 0.1 M NaOH needed to achieve a final pH of 8.5. Cell counts were determined by mixing 10 g of fresh sourdough with 90 ml of sterile 0.85% (wt/vol) NaCl solution and homogenizing the mixture for 5 min (Stomacher 400; Seward, Worthington, United Kingdom). A 10-fold dilution series was made and plated on de Man-Rogosa-Sharpe-5 (MRS-5) agar (44) or yeast extract-glucose-chloramphenicol agar (43) to determine LAB and yeast counts, respectively, after incubation of the agar plates at 30°C for 48 h. A given amount of sourdough (approximately 100 g) was centrifuged (8,041 × g for 20 min at 4°C) to remove solids, and the supernatant was stored at −20°C for metabolite target analysis. In addition, part of the ripe sourdough was stored at −20°C for culture-independent microbiological analysis through PCR-DGGE.
Bacterial population dynamics of back-slopped sourdoughs analyzed through a culture-dependent approach.
On average 10 to 15 purified gram-positive and catalase-negative isolates, recovered from MRS-5 agar plates based on colony morphology, were subjected to repetitive DNA element PCR (rep-PCR) analysis to dereplicate and classify potential LAB. Therefore, a small portion of one colony was resuspended in 20 μl lysis buffer (2.5 ml of 10% [wt/vol] sodium dodecyl sulfate, 5.0 ml of 1 N NaOH, and 92.5 ml of ultrapure water). The mixture was heated at 95°C for 15 min and cooled immediately on ice. After a short centrifugation step at high speed, 180 μl of sterile ultrapure water was added. Subsequently, the mixture was centrifuged (13,000 × g for 5 min) and stored at −20°C. If this alkaline lysis method was not able to produce high-quality rep-PCR profiles, the phenol-chloroform method was used, as described by Gevers et al. (19). The rep-PCR oligonucleotide primer pair used was (GTG)5 (5′-GTGGTGGTGGTGGTG-3′) with the following temperature program: initial denaturation (95°C, 7 min); 30 cycles of denaturation (94°C, 1 min), annealing (40°C, 1 min), and extension (65°C, 8 min); and a single final extension step (65°C, 16 min) (70). The PCR products were separated in a 1.5% (wt/vol) agarose gel at a constant voltage of 55 V in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0) at 4°C for 16 h. The rep-PCR profiles were visualized after staining with ethidium bromide under UV light, followed by digital image capturing using a charge-coupled device camera. The resulting (GTG)5-PCR fingerprints were analyzed using the BioNumerics v4.0 software package (Applied Maths, Sint-Martens-Latem, Belgium). To obtain a collection of unique strains, the resulting (GTG)5-PCR patterns were clustered together with an in-house reference framework. rep-PCR clusters were delineated at 50% Pearson similarity, and clustering was performed with the unweighted pair group method using arithmetic averages. Isolates were tentatively assigned to a given species when they belonged to a cluster that contained one or more type or reference strains. To verify the tentative identifications obtained by (GTG)5-PCR clustering and to identify the remaining unknown isolates, representatives of each cluster were subjected to a polyphasic taxonomic approach. This included identification through amplified fragment length polymorphism (62), sodium dodecyl sulfate-polyacrylamide gel electrophoresis of whole-cell proteins (50), 16S rRNA gene sequence analysis (65), pheS gene sequence analysis (47), and/or DNA-DNA hybridizations (56).
Bacterial population dynamics of back-slopped sourdoughs analyzed through PCR-DGGE.
For the extraction of total DNA from sourdough samples, 90 ml of peptone-physiological solution (0.1% [wt/vol] bacteriological peptone [Oxoid, Basingstoke, Hampshire, United Kingdom] and 0.85% [wt/vol] NaCl) was added to 10 g of sourdough and homogenized for 5 min. Fifty milliliters of this solution was centrifuged (1,000 × g at 4°C for 5 min). To harvest the cells, the supernatant was centrifuged for a second time (5,000 × g at 4°C for 15 min), and the cell pellet was stored at −20°C. The extraction of total DNA was performed as described by Pitcher et al. (49), except that an extra lysis step with lysozyme (7.5 mg lysozyme powder in 150 μl Tris-EDTA buffer; Serva, Heidelberg, Germany) and mutanolysine (40 μl mutanolysine at 5,000 U/ml; Sigma-Aldrich, St. Louis, MO), as well as an additional chloroform-isoamyl alcohol extraction step, was added.
Amplification of 16S rRNA fragments was carried out with universal primers, amplifying the V3 region of the 16S rRNA gene, by using a thermocycler (MyCycler; Bio-Rad, Hercules, CA). The forward primer F357 possessed the sequence 5′-ATTACCGCGGCTGCTGG-3′ and contained a GC clamp (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGG-3′). The reverse primer 518R possessed the sequence 5′-ATTACCGCGGCTGCTGG-3′. PCR volumes of 50 μl contained 6 μl of 10× PCR buffer (containing 15 mM MgCl2), 2.5 μl bovine serum albumin, 2.5 μl deoxynucleoside triphosphates (2 mM each), 2 μl of each primer (5 μM), 0.25 μl Taq polymerase (5 U μl−1), 32.75 μl ultrapure water, and 2 μl of DNA solution. The following PCR program was used: initial denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 20 s, annealing at 55°C for 45 s, and extension at 72°C for 1 min; and final extension at 72°C for 7 min, followed by cooling to 4°C. PCR products were checked by mixing 8 μl of the PCR product with 2 μl of loading dye, followed by electrophoresis on a 1.5% (wt/vol) agarose gel at 100 V for 30 min, flanked by the EZ Load 100-bp Molecular Ruler (Bio-Rad).
PCR products were analyzed on DGGE gels based on the protocol of Muyzer et al. (46). Polyacrylamide gels (160 × 160 × 1 mm) consisted of 8% (vol/vol) polyacrylamide (National Diagnostics, Atlanta, GA) in 1× TAE buffer (Bio-Rad), using a 35 to 70% denaturant gradient (100% denaturing polyacrylamide solution, corresponding to 7 M urea [National Diagnostics] and 40% [vol/vol] formamide [Sigma-Aldrich]). Electrophoresis was performed for 16.5 h at 70 V in 1× TAE buffer at a constant temperature of 60°C using the Dcode System (Bio-Rad). Gels were stained with CyberGold (solution of 50 μl in 500 ml 1× TAE buffer) for 1 h, followed by visualization of DGGE band profiles under UV light and digital capturing with a charge-coupled device camera.
A reference pattern was designed consisting of V3 16S rRNA amplicons from 13 different bacterial type strains. By inclusion of this reference pattern three to four times on each DGGE gel, resulting band profiles could be digitally normalized by comparison with a standard reference, using the BioNumerics v4.0 software package (Applied Maths).
For sequencing of DGGE bands, bands of interest were excised from the gels, resuspended in 50 μl of sterile water, and incubated overnight at 4°C to allow diffusion of DNA from the gel cuts. Five microliters of this aqueous solution was used to reamplify the PCR products with the same primers, including the GC clamp. The amplicons were checked for purity by another DGGE run under the conditions described above with amplified DNA of the original sample as a control. Only reamplified PCR products migrating as a single band and at the same position as the control were amplified with the primer pair (without GC clamp) and sequenced using an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) with the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems), applied according to the manufacturer's instructions. Searches in the international nucleotide sequence library (EMBL) database were performed using the BLAST program (1) to determine the closest known relatives of the partial 16S rRNA sequences obtained.
Metabolite target analysis.
Concentrations of glucose, fructose, xylose, arabinose, sucrose, maltose, erythritol, and mannitol were determined by high-performance anion-exchange chromatography with pulsed amperometric detection (Dionex, Sunnyvale, CA) using a CarboPacPA10 column. The mobile phase, at a flow rate of 1.0 ml min−1, consisted of ultrapure water (0.015 μS cm−1; eluent A), 167 mM NaOH (eluent B), and 500 mM NaOH (eluent C) with the following gradient: 0.0 min, 87% A and 13% B; 20.0 min, 87% A and 13% B; 40.0 min, 15% A and 85% B; 41.0 min, 100% C; 49.0 min, 100% C; 50.0 min, 87% A and 13% B; 65.0 min, 87% A and 13% B. To remove proteins, sourdough supernatant was treated with Carrez A and B reagent. Therefore, 350 μl of Carrez A reagent [3.6% (wt/vol) K4Fe(CN)6·0.3H2O] and 350 μl of Carrez B reagent (7.2% [wt/vol] ZnSO4·7H2O) were added to 700 μl of sourdough supernatant. After centrifugation (16,060 × g for 15 min) the supernatant was filtered (0.2 μm; Minisart; Sartorius AG) prior to injection.
Concentrations of lactic acid and formic acid were determined by high-performance liquid chromatography analysis with a Waters chromatograph (Waters Corp., Milford, MA), equipped with a 2414 differential refractometer, a 600S controller, a column oven, and a 717plus autosampler. An ICSep ICE ORH-801 column (Interchim, Montluçon, France) was used with 10 mN H2SO4 as mobile phase at a flow rate of 0.4 ml min−1. The column temperature was kept at 35°C. Sample preparation was performed as described above.
Concentrations of citrulline, ornithine, and amino acids were determined using a Waters 2695 liquid chromatograph coupled to a Quattro Micro mass spectrometer (Waters). The column (Atlantis; Waters) was kept at 35°C. The mobile phase, starting at a flow rate of 0.2 ml min−1 and linearly increasing to 0.5 ml min−1 over a period of 45 min with a flow rate of 0.2 ml min−1 afterwards, was composed of 0.1% (vol/vol) formic acid in ultrapure water (eluent A) and 0.1% (vol/vol) formic acid in acetonitrile (eluent B). The following gradient was used (vol/vol): 0.0 min, 90% A and 10% B; 45.0 min, 10% A and 90% B; 46.0 min, 90% A and 10% B; 60.0 min, 90% A and 10% B. Sourdough supernatant was diluted 10-fold with ultrapure water and stirred for 15 s. Then, 100 μl of internal standard (IS) (0.002% [wt/vol] of 2-aminobutyric acid in ultrapure water) was added to 500 μl of the diluted sample. The amino acids were derivatized using an AccQ Fluor reagent kit (Waters). Therefore, 70 μl of borate buffer was added to 15 μl of the sample-IS mixture. The latter was stirred for 15 s, after which 30 μl of derivatization reagent was added. After being stirred for another 15 s, the mixture was heated at 55°C for 10 min, cooled, transferred to a vial, and injected.
Metabolites of the aromatic amino acids (phenylpyruvic acid, phenyllactic acid, phenylacetic acid, phenylpropionic acid, hydroxyphenylpyruvic acid, hydroxyphenyllactic acid, hydroxyphenylacetic acid, hydroxyphenylpropionic acid, indolelactic acid, indoleacetic acid, and indolepropionic acid) and succinic acid were determined using the same liquid chromatography-mass spectrometry apparatus as described above. The mobile phase, at a flow rate of 0.2 ml min−1, was composed of ultrapure water (eluent A), acetonitrile (eluent B), and 10 mM ammonium acetate (pH 6.5; eluent C). The following gradient was used (vol/vol): 0.0 min, 85% A, 5% B, and 10% C; 15.0 min, 40% A, 50% B, and 10% C; 15.1 min, 10% A, 80% B, and 10% C; 23.0 min, 10% A, 80% B, and 10% C; 23.1 min, 85% A, 5% B, and 10% C; 30.0 min, 85% A, 5% B, and 10% C. One hundred microliters of IS (0.25% [wt/vol] of 3,4-dihydroxybenzoic acid in ultrapure water) was added to 500 μl of sourdough supernatant. After mixing, 600 μl of acetonitrile was added and the samples were centrifuged (16,060 × g for 15 min). The supernatant was filtered (Minisart RC4) and injected.
Short-chain fatty acids (SCFA; acetic acid, propionic acid, butyric acid, valeric acid, and caproic acid), branched SCFA (iso-butyric acid, iso-valeric acid, and 2-methylbutyric acid), ethanol, and other volatile compounds (diacetyl, acetaldehyde, acetoin, indole, skatole, cresol, and phenol) were measured by gas chromatography-mass spectrometry as described before (66). Sourdough supernatant was treated with Carrez A and B reagent as described above prior to injection.
Volatile compounds were determined by static headspace analysis on an Agilent 6890 gas chromatograph coupled to an Agilent 5973N mass spectrometer (Agilent Technologies, Palo Alto, CA). A capillary column (DB-WAXetr; Agilent Technologies) was used with a starting temperature of 40°C that increased linearly to 230°C over a period of 30 min, after which the temperature was kept at 230°C for 10 min. Data were acquired in scan mode at 5.2 spectra per s in the mass range m/z of 2 to 300. For sample preparation, 200 mg of NaCl and 1 g of sourdough supernatant were weighed in a 20-ml headspace vial and sealed with a crimp cap with a silicone rubber septum. Samples were shaken for 30 min at 85°C prior to injection of the volatile fraction. Standard solutions of 57 volatile compounds in total were used to determine their retention time, and 2-octanol was used as IS. The AMDIS deconvolution software (NIST, http://www.nist.gov/), used in simple analysis mode and coupled to the NIST98 Mass Spectral Database (NIST), was used for analysis of the chromatograms. Identification of target compounds was performed by combining retention time and mass spectral database information.
All samples were analyzed in triplicate, and the mean values ± standard deviations are represented, except for the results of the headspace analysis, which were expressed as [100 × peak area compound (peak area IS × g sample)−1].
RESULTS
Characteristics of wheat and spelt laboratory sourdough fermentations.
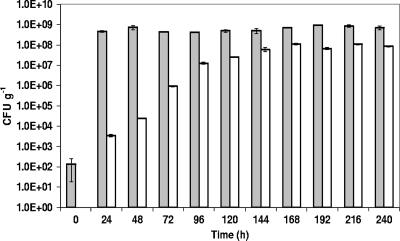
At the start of the fermentations, low colony counts (<104 CFU g−1) were found for both LAB and yeast populations. However, LAB numbers rapidly increased after the first day of fermentation in all sourdoughs and did not change much during the following days (Fig. 1). In general, LAB counts reached up to 108 to 109 CFU g−1. For the yeast counts, more variation was observed. Yeasts developed more slowly than LAB and usually reached high amounts (107 CFU g−1) after 4 to 5 days of back-slopping (Fig. 1). However, during the wheat A sourdough fermentation, the yeast population started to develop only after 8 days of back-slopping, resulting in a low yeast population at the end of the 10-day cycle (Table 2).
FIG. 1.
Population dynamics of LAB (gray bars) and yeast (white bars) counts during a 10-day spelt sourdough fermentation with daily back-slopping. Counts were performed in triplicate, and mean values ± standard deviations are represented.
TABLE 2.
Characteristics of the four laboratory sourdoughs after 10 days of back-slopping
| Flour | pH | TTA | LAB (CFU g−1) | Yeast (CFU g−1) |
|---|---|---|---|---|
| Wheat A | 3.34 ± 0.01 | 9.1 ± 0.1 | 1.0 × 109 ± 0.2 × 109 | 9.1 × 105 ± 1.6 × 105 |
| Spelt A | 3.34 ± 0.01 | 9.0 ± 0.1 | 6.9 × 108 ± 1.3 × 108 | 8.5 × 107 ± 0.7 × 107 |
| Wheat B | 3.33 ± 0.02 | 7.7 ± 0.1 | 3.8 × 108 ± 1.2 × 108 | 3.4 × 107 ± 0.2 × 107 |
| Spelt B | 3.35 ± 0.02 | 9.6 ± 0.2 | 1.5 × 109 ± 0.2 × 109 | 2.7 × 107 ± 0.7 × 107 |
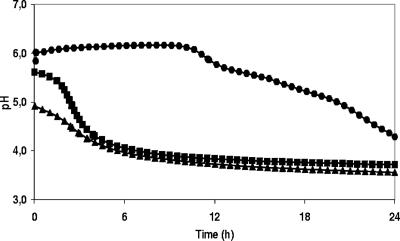
The pH did not change during the first 12 h of fermentation for all sourdoughs (Fig. 2). During the next 12 h, the pH started to drop slowly, ending at pH 4 to 5 after the first day of fermentation. After the first back-slopping, an immediate decrease of the pH was observed during fermentation. As the fresh dough was inoculated with ripe sourdough from the day before, the pH at the start was lower on day 2, and the final pH value was also lower. After day 2 a pH value below 4.0 was reached during all sourdough fermentations. During the following days (days 3 to 10), the pH profile was similar to that for day 2; only the pH at the start and at the end slightly decreased (Table 2). An inverse relation between pH and TTA values was observed. Final TTA values ranged between 9.0 and 9.5, except for the wheat B sourdough, which showed a lower final TTA value (Table 2).
FIG. 2.
Changes in pH during a 10-day spelt sourdough fermentation with daily back-slopping. Symbols: •, changes in pH during the first day of fermentation (0 to 24 h); ▪, changes in pH during the second day of fermentation (24 to 48 h); ▴, changes in pH during the last day of fermentation (216 to 240 h).
Bacterial population dynamics of back-slopped laboratory sourdoughs analyzed through a culture-dependent approach.
Identification of presumptive LAB isolates based on (GTG)5-PCR analysis and/or taxonomic polyphasic characterization revealed several changes in the LAB populations (Table 3). The major changes occurred during the first 4 to 5 days of back-slopping. After the first day of fermentation, LAB atypical for sourdough, such as species belonging to the genera Enterococcus and Lactococcus, were detected. They usually decreased below the detection limit after the first or second day of back-slopping. One exception was the persistence of Lactococcus lactis in the wheat B sourdough; it was still retrieved after 4 days of fermentation. From day 2 to day 4, other LAB species mainly belonging to Lactobacillus or Leuconostoc, and sometimes Pediococcus or Weissella, became dominant in the sourdough ecosystem. From day 5 on, only minor changes were observed in all sourdoughs, indicating achievement of a stable sourdough ecosystem. Lactobacillus plantarum was present in the stable wheat B, spelt A, and spelt B sourdough ecosystems, whereas Lactobacillus fermentum dominated the wheat A sourdough ecosystem and was found in the stable wheat B sourdough. Interestingly, L. fermentum was never retrieved in any of the spelt sourdoughs. Finally, Lactobacillus rossiae was found in the stable spelt A sourdough, whereas Lactobacillus brevis and Lactobacillus paraplantarum were found in the stable spelt B sourdough, in both cases together with L. plantarum.
TABLE 3.
Bacterial population dynamics during four laboratory sourdough fermentations, analyzed by a culture-dependent approach (plating, isolation, and subsequent polyphasic identification)
| Flour and microorganism identified | Identification at time (h):
|
|||||
|---|---|---|---|---|---|---|
| 0 | 24 | 48 | 72 | 96 | 240 | |
| Spelt A | ||||||
| Enterobacter cloacae | X | |||||
| Enterococcus faecium | X | |||||
| Escherichia coli | X | |||||
| Lactococcus lactis subsp. lactis | X | |||||
| Enterococcus hirae | X | X | ||||
| Leuconostoc citreum | X | X | ||||
| Lactobacillus curvatus | X | X | X | |||
| Pediococcus pentosaceus | X | X | X | |||
| Lactobacillus plantarum | X | X | ||||
| Lactobacillus rossiae | X | |||||
| Wheat A | ||||||
| Aerococcus viridans | X | |||||
| Lactobacillus fermentum | X | X | X | X | ||
| Leuconostoc citreum | X | X | X | |||
| Lactobacillus curvatus | X | X | X | X | ||
| Lactococcus lactis | X | |||||
| Weissella cibaria | X | X | ||||
| Wheat B | ||||||
| Aerococcus viridans | X | |||||
| Lactococcus lactis | X | X | X | X | ||
| Leuconostoc citreum | X | X | X | X | ||
| Lactobacillus curvatus | X | X | ||||
| Pediococcus pentosaceus | X | X | ||||
| Lactobacillus fermentum | X | X | X | X | ||
| Lactobacillus plantarum | X | X | X | |||
| Spelt B | ||||||
| Enterococcus mundtii | X | |||||
| Leuconostoc pseudomesenteroides | X | X | X | |||
| Lactobacillus curvatus | X | X | X | X | ||
| Lactococcus lactis subsp. lactis | X | X | ||||
| Leuconostoc citreum | X | X | X | X | ||
| Lactobacillus helveticus | X | |||||
| Pediococcus pentosaceus | X | |||||
| Lactobacillus brevis | X | X | ||||
| Lactobacillus paraplantarum | X | X | ||||
| Lactobacillus plantarum | X | X | ||||
Bacterial population dynamics of laboratory sourdoughs analyzed through a culture-independent approach.
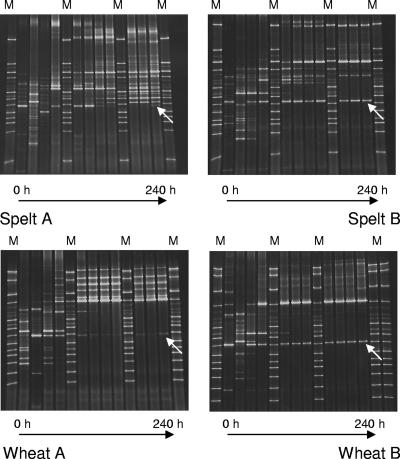
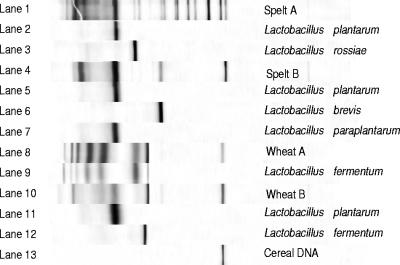
16S rRNA PCR-DGGE was carried out mainly to follow the development of the flour microbiota from an unstable, initial phase to a stable, final sourdough ecosystem. This community fingerprinting was reflected by DGGE band number and position changing over time (Fig. 3). Indeed, the PCR-DGGE band patterns showed major changes in the LAB population during the first 4 to 5 days and only minor changes afterwards. To have an idea about which bands corresponded to which microorganisms in the stable sourdough ecosystem, an additional experiment was performed during which DNA isolated from the stable sourdough at day 10 and DNA isolated from pure cultures isolated from the same sourdough sample at day 10 were included in a single PCR and DGGE run (Fig. 4). By this additional PCR-DGGE experiment, the DGGE band positions of these single isolates could be retrieved in the community DGGE profile of the stable sourdough from which the isolates originated. No comigration of DGGE bands was observed among digitally normalized DGGE band positions of multiple isolates of all bacterial species isolated during this study from day 0 to day 10. In fact, there was only one band, present in all four sourdoughs, that could not be linked to a certain microorganism in this way. Sequencing of this band indicated that it probably corresponded to mitochondrial cereal DNA, indicating that a certain region within the plant genome can be amplified with the universal V3 primer pair (2).
FIG. 3.
Bacterial population dynamics shown by 16S rRNA PCR-DGGE analysis of two spelt (top) and two wheat (bottom) sourdough fermentations, daily back-slopped, with flour from two different mills (A and B), after each day (0, 24, 48, 72, 96, 120, 144, 168, 192, 216, and 240 h). M, reference marker. The indicated bands (arrows) correspond with cereal DNA.
FIG. 4.
Results of the additional 16S rRNA PCR-DGGE run, including the normalized PCR-DGGE profiles of stable sourdoughs after 10 days of back-slopping followed by the PCR-DGGE profiles of strains isolated and purified from the corresponding sourdoughs. Lane 1, profile of the dominant population in the spelt A sourdough; lanes 2 and 3, profiles of purified cultures isolated from the stable spelt A sourdough; lane 4, profile of the dominant population in the spelt B sourdough; lanes 5 to 7, profiles of purified cultures isolated from the stable spelt B sourdough; lane 8, profile of the dominant population in the wheat A sourdough; lane 9, profile of the purified culture isolated from the stable wheat A sourdough; lane 10, profile of the dominant population in the wheat B sourdough; lanes 11 and 12, profiles of purified cultures isolated from the stable wheat B sourdough; lane 13, profile of mitochondrial cereal DNA.
Metabolite target analysis.
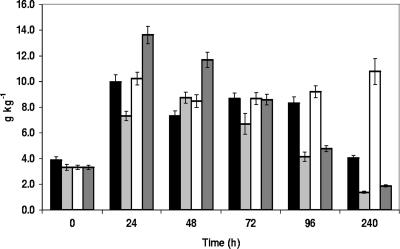
Maltose was abundant in all sourdough fermentations. In unfermented dough mixtures, maltose concentrations varied between 3.3 ± 0.2 and 3.9 ± 0.3 g kg−1. Sucrose, glucose, and fructose were found in concentrations of 0.6 ± 0.1 to 0.8 ± 0.1 g kg−1, 0.2 ± 0.1 to 0.3 ± 0.1 g kg−1, and 0.1 ± 0.1 to 0.2 ± 0.1 g kg−1, respectively. From day 2 to day 4, concentrations of maltose and glucose were changing (Fig. 5), whereas sucrose and fructose disappeared. Maximum concentrations measured for maltose and glucose were 13.6 ± 0.7 g kg−1 (the wheat B sourdough after 24 h) and 2.4 ± 0.3 g kg−1 (the spelt B sourdough after 48 h), respectively. During the last days of fermentation, their concentrations remained constant in the dough. The highest final maltose concentration (10.8 ± 1.0 g kg−1) was measured in the spelt B sourdough, which corresponded to the flour with the highest amylase activity as revealed by the Hagberg method (Table 1). In contrast, the lowest final maltose concentration (1.4 ± 0.1 g kg−1) was measured in the wheat A sourdough, which corresponded to the flour with the lowest amylase activity (Table 1). Other sugars such as arabinose or xylose were hardly found (<50 mg kg−1).
FIG. 5.
Evolution of the maltose concentration during a 10-day daily back-slopped sourdough fermentation with spelt A (black bars), wheat A (light gray bars), spelt B (white bars), and wheat B (dark gray bars).
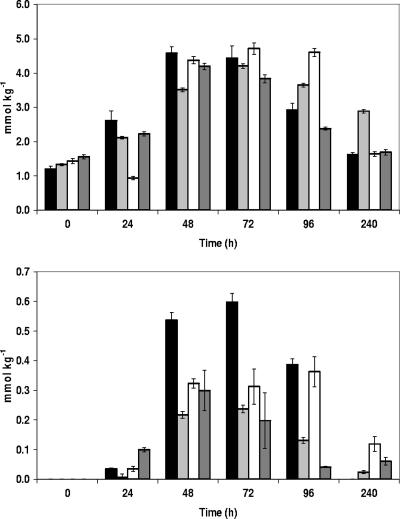
Total amino acid concentrations increased after sourdough fermentation compared to unfermented doughs (Fig. 6). The largest changes were observed from day 2 to day 4, when total amino acid concentrations peaked with a maximum of 4.7 mmol kg−1, as observed in the spelt B sourdough after 3 days. Afterwards, total amino acid concentrations decreased, and after 10 days of sourdough fermentation, differences from unfermented doughs were less clear. One exception was the wheat A sourdough, the total amino acid concentration of which was more than twofold higher than that of the unfermented wheat A dough after 10 days of back-slopping. Interestingly, this wheat A flour showed the highest protein content (Table 1). Asparagine and aspartate were the only amino acids the concentrations of which were higher in unfermented doughs than in 10-day back-slopped sourdoughs. Ornithine was found during all sourdough fermentations. Its concentration was highest from day 2 to day 5, after which it disappeared completely in the spelt A sourdough. In the other three sourdoughs, ornithine was still found in low concentrations (less than 0.15 mmol kg−1) after 10 days.
FIG. 6.
Evolution of total amino acid concentration (top) and ornithine concentration (bottom) during a 10-day daily back-slopped sourdough fermentation with spelt A (black bars), wheat A (light gray bars), spelt B (white bars), or wheat B (dark gray bars).
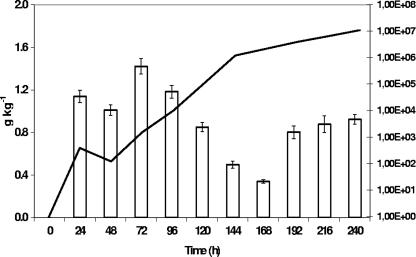
Lactic acid was the major metabolite produced during all sourdough fermentations. Its concentration increased during the first 4 to 5 days of fermentation and remained constant afterwards. Final concentrations ranged between 4.7 ± 0.2 g kg−1 (the wheat B sourdough) and 7.3 ± 0.4 g kg−1 (the spelt B sourdough), which corresponded to the lowest TTA (7.7 ± 0.1) and highest TTA (9.6 ± 0.2), respectively. Ethanol was the second most important metabolite retrieved and correlated in most cases with changes in the yeast population. In some cases, ethanol production could be attributed to heterofermentative LAB, as ethanol concentrations increased when yeast counts were low or decreasing (Fig. 7). Acetic acid was found in all sourdough samples (except in unfermented doughs), but concentrations hardly exceeded 1.0 ± 0.1 g kg−1. Mannitol was produced mainly during the first 4 to 5 days of fermentation. Later on, mannitol was not produced, except in the wheat A sourdough, where it was retrieved till day 10. Concentrations of mannitol were never higher than 1.0 ± 0.1 g kg−1. Besides these more common metabolites, erythritol and succinic acid were found in all sourdoughs (not in unfermented doughs). Concentrations of erythritol and succinic acid were maximally 0.14 ± 0.02 g kg−1 (after day 1 in the spelt B sourdough) and 0.49 ± 0.02 g kg−1 (after day 8 in the spelt B sourdough), respectively.
FIG. 7.
Evolution of ethanol concentration (white bars, left axis) and yeast counts (solid line, right axis) during the 10-day daily back-slopped spelt B sourdough fermentation.
Phenyllactic acid was found in 35 out of 40 sourdough samples (never in unfermented doughs) in concentrations usually lower than 0.3 mmol kg−1. In fact, phenyllactic acid was the only aromatic amino acid metabolite found in the wheat A sourdough. Besides phenyllactic acid, indolelactic acid, hydroxyphenyllactic acid, and hydroxyphenylacetic acid were found but only in a few sourdough samples. Hydroxyphenylacetic acid was found in the wheat B sourdough in concentrations of about 1.0 mmol kg−1. Concentrations of indolelactic acid and hydroxyphenyllactic acid never exceeded 0.05 and 0.15 mmol kg−1, respectively.
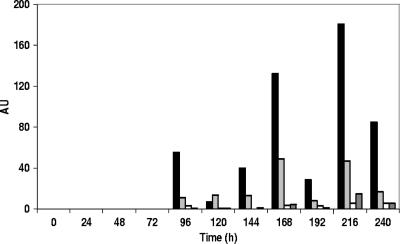
Headspace analysis revealed that ethanol and ethyl acetate were present in all sourdough samples. The smallest amount of volatile compounds was measured in the wheat A sourdough samples. After day 1, acetoin and diacetyl were present in the wheat A sourdough, but they were not retrieved later on. The only volatiles found in the wheat A sourdough were, besides ethanol and ethyl acetate, propyl lactate, 2-pentyl furan, and hexanal. The last compound was found in all sourdoughs prepared and even in unfermented doughs. In the wheat B, spelt A, and spelt B sourdoughs, a larger amount of volatile compounds was found, including compounds such as 1-propanol, 3-methyl-1-butanol, 2-methyl-1-butanol, and 2-methyl-1-propanol. Furthermore, in sourdoughs prepared with spelt flour, additional alcohols such as 1-pentanol, 1-penten-3-ol, and 1-hexanol were found. Other compounds retrieved in several sourdough samples included 3-methyl-1-butanol acetate, pentanol, and acetaldehyde. In general, the amounts of volatile compounds were low during the first days and increased later on (Fig. 8).
FIG. 8.
Changes in the production of 3-methyl-1-butanol (black bars), 2-methyl-1-propanol (light gray bars), 1-propanol (white bars), and 3-methyl-1-butanol acetate (dark gray bars) during the 10-day daily back-slopped spelt A sourdough fermentation. AU, arbitrary units [100 × peak area compound (peak area internal standard × g sample)−1].
Although a lot of other compounds were measured, most of them were not found in the sourdough samples analyzed, either because they were produced in amounts lower than the detection limit or because they were not produced at all. Examples of such compounds were propionic acid and butyric acid; branched SCFA; and amino acid metabolites such as phenol, skatole, indole, phenylacetic acid, and indoleacetic acid.
DISCUSSION
Spontaneous fermentations of raw materials usually lead to stable end products. In the present study, both population dynamics (through a culture-dependent and culture-independent approach) and metabolite target analysis revealed that all sourdough ecosystems, daily back-slopped starting from a water-flour mixture without a starter culture, were stable within 1 week. Not only the composition but also the metabolic activity of the present microbiota remained unchanged once a stable ecosystem was achieved. It could be shown that microbial adaptation resulted in both succession of microbial populations (and strains) and altered metabolic patterns.
Succession of microbial populations could be correlated with acid stress and carbohydrate and amino acid metabolism. In general, a three-phase evolution could be observed during development of a stable sourdough ecosystem, as revealed by culture-dependent analysis. In a first phase (from the start until day 2), the cereal ecosystem was dominated by LAB species that were not specific for sourdough, such as those belonging to the genera Enterococcus, Lactococcus, and Leuconostoc. Although strains belonging to the last genus are often encountered in sourdough ecosystems (10), they decreased below the detection limit within a few days, together with enterococci and lactococci. A decrease of the last two genera can be explained by a lack of adaptation to the sourdough environment, as they are almost never retrieved in stable sourdough ecosystems (9). For leuconostocs, it has been shown that they are more sensitive to acid stress than are some lactobacilli, and hence their numbers decrease upon further acidification of the sourdough ecosystem (15, 41). For instance, succession of Leuconostoc mesenteroides and L. plantarum is seen in the production of sauerkraut (33). During the second phase of sourdough fermentation (days 2 to 5), sourdough-specific LAB dominated the process, such as species belonging to the genera Lactobacillus, Pediococcus, and Weissella. This can be explained by a better acid tolerance and a metabolism adapted to the cereal environment (16). As an example, the use of external electron acceptors, such as fructose present in the sourdough ecosystem, enhances their competitiveness (16). Finally, during a third and last phase (days 5 to 7), well-adapted sourdough strains, such as strains of L. plantarum and L. fermentum, became dominant under the applied laboratory conditions of temperature, dough yield, etc. This observation was confirmed by 16S rRNA PCR-DGGE analysis, yielding stable band patterns upon prolonged fermentation. The three-phase establishment of a stable ecosystem has also been observed during other cereal fermentations such as traditional cassava flour fermentation (3). During the present study, a different ecosystem was found for the flours used, as revealed by both a culture-dependent and a culture-independent approach, indicating that the type of flour has a certain effect on the establishing ecosystem.
In the present study, L. plantarum was found in three out of the four stable sourdoughs. L. plantarum is often encountered in sourdough ecosystems (9), and its dominance in cereal fermentations is attributed to a highly adapted carbohydrate metabolism, possibly reflecting its natural habitat (35). Besides L. plantarum, L. fermentum was found in stable wheat sourdough ecosystems, which can be explained by the extreme acid tolerance and adapted metabolism of the latter species (4, 5). Moreover, L. fermentum dominated the stable wheat A sourdough ecosystem, as revealed by PCR-DGGE. The other three sourdough ecosystems were always dominated by a consortium of obligate and facultative heterofermentative LAB species, whereas obligate homofermentative LAB species were outcompeted in the sourdough ecosystems studied. The observation that L. fermentum was found in stable wheat sourdough ecosystems and was never encountered in spelt sourdoughs is interesting but needs further investigation before valid conclusions can be drawn.
In general, maltose release (mediated by flour amylases and perhaps amylolytic sourdough LAB) and maltose consumption (by sourdough LAB) as well as proteolysis (mainly ascribed to the action of flour proteinases enhanced by acidic conditions) occur during sourdough fermentation. This is reflected by the considerable levels of maltose retrieved after each fermentation step and an increase of total amino acids upon fermentation (27, 39, 40, 69). The present study showed that the competitiveness of sourdough LAB is enhanced by both the use of the arginine deiminase pathway (in an early stage) and the use of alternative electron acceptors.
The arginine deiminase pathway is considered to yield a competitive advantage compared to strains lacking it (8, 16). However, in the present study, ornithine was found only in low concentrations (or not at all) after 10 days of back-slopping. Hence, the dominating strains in the present sourdough ecosystems hardly made use of this metabolic trait. In contrast, it seemed that the strains competing during the first days of fermentation made more use of this specific metabolic capacity to compete in the sourdough ecosystem, which is shown here for the first time, as ornithine concentrations were highest during the first days of back-slopping. However, it appears that counteracting acid stress solely due to ornithine production is not sufficient.
External electron acceptors such as fructose or citrate are preferred (73). The production of mannitol from fructose, which may lead to an efficient equilibration of the redox balance, enhanced energy generation, and, therefore, increased competitiveness (37, 72, 73), was observed during the present study. However, mannitol was produced mainly during the first 2 to 5 days of back-slopping and disappeared afterwards, indicating that the dominating species in the stable ecosystem did not make full use of this metabolic trait. The only exception was the wheat A sourdough, dominated by L. fermentum, where mannitol was also found after 10 days of back-slopping. This mannitol had to be produced by L. fermentum, as it was the only species detected through PCR-DGGE in the stable sourdough. Interestingly, mannitol was not found in the wheat B sourdough, in spite of the presence of L. fermentum. An explanation might be that the precursor, fructose, was not generated in one sourdough whereas it was in the other. Besides the production of lactic acid, ethanol, and mannitol, also other pathways to equilibrate the redox balance were observed. Two examples of this were the production of succinic acid and that of erythritol during fermentation. Succinic acid can be produced through metabolism of citrate, which is usually present in flour in low concentrations (22, 29, 61). Erythritol production from glucose has previously been observed for pure cultures of several LAB strains (59, 67). This study showed for the first time the in situ production of these two compounds during sourdough fermentation. The need for other ways to regenerate NAD(P)+ might be explained by a low activity of the acetaldehyde dehydrogenase enzyme (67, 73).
Besides the equilibration of the redox balance through sugar metabolism, amino acid metabolism may contribute to this. The in situ production of phenyllactic acid, hydroxyphenyllactic acid, indolelactic acid, and hydroxyphenylacetic acid during sourdough fermentation was shown during the present study. It was striking that the aromatic amino acids were mostly converted to their hydroxy acids and that besides hydroxyphenylacetic acid, no other metabolite was found. The equilibration of the redox balance through the conversion of the α-keto acid (e.g., phenylpyruvic acid) to the corresponding hydroxy acid (e.g., phenyllactic acid) might explain this observation. Therefore, these fine-tuning reactions might give an extra competitive advantage to the strains possessing this capability. This hypothesis is strengthened by the observation that these aromatic amino acid metabolites increased towards the end in three out of four sourdough fermentations and were at their highest concentrations in the stable sourdough ecosystems.
Other amino acid catabolic pathways probably contributed to the equilibration of the redox balance. Analysis of volatile compounds revealed that, besides ethanol and ethyl acetate, degradation products of valine, leucine, and iso-leucine in their alcohol form were the most important volatiles. In general, the levels of volatiles increased upon fermentation and alcohols were the most important compounds retrieved, confirming previous results (30, 32). In a similar way, one can assume that they play a role in maintaining the redox balance. However, it is not easy to discriminate between volatiles produced by the LAB populations and those produced by the yeast populations. The importance of redox balance, not only for LAB metabolism but also concerning dough structure, has recently been highlighted by Vermeulen et al., who found an interaction between LAB-catalyzed redox reactions and the gluten structure of the dough (68).
To conclude, it was shown that the establishment of a stable sourdough ecosystem, starting from a water-flour mixture, occurred through a three-phase evolution. LAB atypical for sourdough were outcompeted by sourdough-specific LAB during a first transition phase after 1 or 2 days of back-slopping. After 4 to 5 days of back-slopping, these LAB are in turn outcompeted by sourdough-specific LAB that were highly adapted to the employed conditions of temperature, dough yield, etc. Furthermore, the in situ production of certain metabolites, such as succinic acid and erythritol, during sourdough fermentation was shown for the first time. The use of fructose as an external electron acceptor and arginine-to-ornithine conversion contribute to competitiveness during the first days of back-slopped sourdough fermentation. Together with the production of several amino acid metabolites, especially metabolites of aromatic amino acids, such as phenyllactic acid, their production can contribute to the equilibration of the redox balance to ensure fast growth of the dominating strains. An adapted metabolism and efficient redox balancing strengthen the stable existence of specific LAB strains in a sourdough ecosystem. Stable wheat and spelt sourdoughs did not differ significantly.
Acknowledgments
This work was supported by the Research Council of the Vrije Universiteit Brussel, the Fund for Scientific Research-Flanders, and the Flemish Institute for the Encouragement of Scientific and Technological Research in the Industry (SBO Project 030263). G.H. is a postdoctoral fellow of the Fund for Scientific Research-Flanders (F.W.O.-Vlaanderen, Belgium).
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ampe, F., N. Ben Omar, C. Moizan, C. Wacher, and J. P. Guyot. 1999. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65:5464-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brauman, A., S. Keleke, M. Malonga, E. Miambi, and F. Ampe. 1996. Microbiological and biochemical characterization of cassava retting, a traditional lactic acid fermentation for foo-foo (cassava flour) production. Appl. Environ. Microbiol. 62:2854-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderon-Santoyo, M., G. Loiseau, and J. P. Guyot. 2003. Fermentation by Lactobacillus fermentum Ogi E1 of different combinations of carbohydrates occurring naturally in cereals: consequences on growth energetics and α-amylase production. Int. J. Food Microbiol. 80:161-169. [DOI] [PubMed] [Google Scholar]
- 5.Calderon-Santoyo, M., G. Loiseau, R. R. Sanoja, and J. P. Guyot. 2003. Study of starch fermentation at low pH by Lactobacillus fermentum Ogi E1 reveals uncoupling between growth and α-amylase production at pH 4.0. Int. J. Food Microbiol. 80:77-87. [DOI] [PubMed] [Google Scholar]
- 6.Catterall, P. 1998. Flour milling, p. 296-329. In S. P. Cauvain and L. S. Young (ed.), Technology of breadmaking. Blackie Academic and Professional, London, United Kingdom.
- 7.Czerny, M., and P. Schieberle. 2002. Important aroma compounds in freshly ground wholemeal and white wheat flour—identification and quantitative changes during sourdough fermentation. J. Agric. Food Chem. 50:6835-6840. [DOI] [PubMed] [Google Scholar]
- 8.De Angelis, M., L. Mariotti, J. Rossi, M. Servili, P. F. Fox, G. Rollán, and M. Gobbetti. 2002. Arginine catabolism by sourdough lactic acid bacteria: purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl. Environ. Microbiol. 68:6193-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Vuyst, L., and P. Neysens. 2005. The sourdough microflora: biodiversity and metabolic interactions. Trends Food Sci. Technol. 16:43-56. [Google Scholar]
- 10.De Vuyst, L., and M. Vancanneyt. 2007. Biodiversity and identification of sourdough lactic acid bacteria. Food Microbiol. 24:120-127. [DOI] [PubMed] [Google Scholar]
- 11.Ercolini, D. 2004. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56:297-314. [DOI] [PubMed] [Google Scholar]
- 12.Fernie, A. R., R. N. Trethewey, A. J. Krotzky, and L. Willmitzer. 2004. Innovation—metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5:763-769. [DOI] [PubMed] [Google Scholar]
- 13.Fiehn, O. 2002. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 48:155-171. [PubMed] [Google Scholar]
- 14.Fiehn, O., J. Kopka, P. Dormann, T. Altmann, R. N. Trethewey, and L. Willmitzer. 2000. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18:1157-1161. [DOI] [PubMed] [Google Scholar]
- 15.Gabriel, V., D. Lefebvre, Y. Vayssier, and C. Faucher. 1999. Characterization of the microflora of natural sourdoughs. Microbiol. Aliment. Nutr. 17:171-179. [Google Scholar]
- 16.Gänzle, M. G., N. Vermeulen, and R. F. Vogel. 2007. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough. Food Microbiol. 24:128-138. [DOI] [PubMed] [Google Scholar]
- 17.Gänzle, M. G., and R. F. Vogel. 2003. Contribution of reutericyclin production to the stable persistence of Lactobacillus reuteri in an industrial sourdough fermentation. Int. J. Food Microbiol. 80:31-45. [DOI] [PubMed] [Google Scholar]
- 18.Gatto, V., and S. Torriani. 2004. Microbial population changes during sourdough fermentation monitored by DGGE analysis of 16S and 26S rRNA gene fragments. Ann. Microbiol. 54:31-42. [Google Scholar]
- 19.Gevers, D., G. Huys, and J. Swings. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205:31-36. [DOI] [PubMed] [Google Scholar]
- 20.Giraffa, G. 2004. Studying the dynamics of microbial populations during food fermentation. FEMS Microbiol. Rev. 28:251-260. [DOI] [PubMed] [Google Scholar]
- 21.Gobbetti, M. 1998. The sourdough microflora: interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 9:267-274. [Google Scholar]
- 22.Gobbetti, M., and A. Corsetti. 1996. Co-metabolism of citrate and maltose by Lactobacillus brevis subsp lindneri CB1 citrate-negative strain: effect on growth, end-products and sourdough fermentation. Z. Lebensm. Unters. Forsch. 203:82-87. [Google Scholar]
- 23.Gobbetti, M., A. Corsetti, and J. Rossi. 1994. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: metabolism of amino acids. World J. Microbiol. Biotechnol. 10:275-279. [DOI] [PubMed] [Google Scholar]
- 24.Gobbetti, M., A. Corsetti, and J. Rossi. 1994. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: metabolism of carbohydrates. Appl. Microbiol. Biotechnol. 41:456-460. [DOI] [PubMed] [Google Scholar]
- 25.Gobbetti, M., M. De Angelis, P. Arnaut, P. Tossut, A. Corsetti, and P. Lavermicocca. 1999. Added pentosans in breadmaking: fermentations of derived pentoses by sourdough lactic acid bacteria. Food Microbiol. 16:409-418. [Google Scholar]
- 26.Gobbetti, M., M. De Angelis, A. Corsetti, and R. Di Cagno. 2005. Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci. Technol. 16:57-69. [Google Scholar]
- 27.Gobbetti, M., M. S. Simonetti, J. Rossi, L. Cossignani, A. Corsetti, and P. Damiani. 1994. Free D-amino acid and L-amino acid evolution during sourdough fermentation and baking. J. Food Sci. 59:881-884. [Google Scholar]
- 28.Hammes, W. P., and M. G. Gänzle. 1998. Sourdough breads and related products, p. 199-216. In B. J. B. Wood (ed.), Microbiology of fermented foods, 2nd ed. Blackie Academic and Professional, London, United Kingdom.
- 29.Hammes, W. P., P. Stolz, and M. G. Gänzle. 1996. Metabolism of lactobacilli in traditional sourdoughs. Adv. Food Sci. 18:176-184. [Google Scholar]
- 30.Hansen, A., and B. Hansen. 1994. Influence of wheat flour type on the production of flavour compounds in wheat sourdoughs. J. Cereal Sci. 19:185-190. [Google Scholar]
- 31.Hansen, A., B. Lund, and M. J. Lewis. 1989. Flavour production and acidification of sourdoughs in relation to starter culture and fermentation temperature. Food Sci. Technol. 22:145-149. [Google Scholar]
- 32.Hansen, A., and P. Schieberle. 2005. Generation of aroma compounds during sourdough fermentation: applied and fundamental aspects. Trends Food Sci. Technol. 16:85-94. [Google Scholar]
- 33.Harris, L. J. 1998. The microbiology of vegetable fermentations, p. 45-72. In B. J. B. Wood (ed.), Microbiology of fermented foods, 2nd ed. Blackie Academic and Professional, London, United Kingdom.
- 34.Katina, K., K. Poutanen, and K. Autio. 2004. Influence and interactions of processing conditions and starter culture on formation of acids, volatile compounds, and amino acids in wheat sourdoughs. Cereal Chem. 81:598-610. [Google Scholar]
- 35.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. Fiers, W. Stiekema, R. M. K. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korakli, M., M. Pavlovic, M. G. Gänzle, and R. F. Vogel. 2003. Exopolysaccharide and kestose production by Lactobacillus sanfranciscensis LTH2590. Appl. Environ. Microbiol. 69:2073-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korakli, M., E. Schwarz, G. Wolf, and W. P. Hammes. 2000. Production of mannitol by Lactobacillus sanfranciscensis. Adv. Food Sci. 22:1-4. [Google Scholar]
- 38.Lavermicocca, P., F. Valerio, A. Evidente, S. Lazzaroni, A. Corsetti, and M. Gobbetti. 2000. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl. Environ. Microbiol. 66:4084-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loponen, J., M. Mikola, K. Katina, T. Sontag-Strohm, and H. Salovaara. 2004. Degradation of HMW glutenins during wheat sourdough fermentations. Cereal Chem. 81:87-93. [Google Scholar]
- 40.Martinez-Anaya, M. A. 1996. Carbohydrate and nitrogen related components in wheat sourdough processes: a review. Adv. Food Sci. 18:185-200. [Google Scholar]
- 41.McDonald, L. C., H. P. Fleming, and H. M. Hassan. 1990. Acid tolerance of Leuconostoc mesenteroides and Lactobacillus plantarum. Appl. Environ. Microbiol. 56:2120-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meroth, C. B., W. P. Hammes, and C. Hertel. 2004. Characterisation of the microbiota of rice sourdoughs and description of Lactobacillus spicheri sp. nov. Syst. Appl. Microbiol. 27:151-159. [DOI] [PubMed] [Google Scholar]
- 43.Meroth, C. B., W. P. Hammes, and C. Hertel. 2003. Identification and population dynamics of yeasts in sourdough fermentation processes by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:7453-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meroth, C. B., J. Walter, C. Hertel, M. J. Brandt, and W. P. Hammes. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Messens, W., and L. De Vuyst. 2002. Inhibitory substances produced by lactobacilli isolated from sourdoughs—a review. Int. J. Food Microbiol. 72:31-43. [DOI] [PubMed] [Google Scholar]
- 46.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naser, S. M., F. L. Thompson, B. Hoste, D. Gevers, P. Dawyndt, M. Vancanneyt, and J. Swings. 2005. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 151:2141-2150. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen, J., and S. Oliver. 2005. The next wave in metabolome analysis. Trends Biotechnol. 23:544-546. [DOI] [PubMed] [Google Scholar]
- 49.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 50.Pot, B., P. Vandamme, and K. Kersters. 1994. Analysis of electrophoretic whole-organism protein fingerprints, p. 493-521. In M. Goodfellow and A. G. O'Donnell (ed.), Chemical methods in prokaryotic systematics. J. Wiley & Sons, Chichester, United Kingdom.
- 51.Randazzo, C. L., H. Heilig, C. Restuccia, P. Giudici, and C. Caggia. 2005. Bacterial population in traditional sourdough evaluated by molecular methods. J. Appl. Microbiol. 99:251-258. [DOI] [PubMed] [Google Scholar]
- 52.Röcken, W. 1996. Applied aspects of sourdough fermentation. Adv. Food Sci. 18:212-216. [Google Scholar]
- 53.Rollán, G., G. L. Lorca, and G. F. de Valdez. 2003. Arginine catabolism and acid tolerance response in Lactobacillus reuteri isolated from sourdough. Food Microbiol. 20:313-319. [Google Scholar]
- 54.Rosenquist, H., and A. Hansen. 2000. The microbial stability of two bakery sourdoughs made from conventionally and organically grown rye. Food Microbiol. 17:241-250. [Google Scholar]
- 55.Rossi, J. 1996. The yeasts in sourdough. Adv. Food Sci. 18:201-211. [Google Scholar]
- 56.Scheirlinck, I., R. Van der Meulen, A. Van Schoor, I. Cleenwerck, G. Huys, P. Vandamme, L. De Vuyst, and M. Vancanneyt. 2007. Lactobacillus namurensis sp. nov., isolated from a traditional Belgian sourdough. Int. J. Syst. Evol. Microbiol. 57:223-227. [DOI] [PubMed] [Google Scholar]
- 57.Schnürer, J., and J. Magnusson. 2005. Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 16:70-78. [Google Scholar]
- 58.Spicher, G., and W. Nierle. 1988. Proteolytic activity of sourdough bacteria. Appl. Microbiol. Biotechnol. 28:487-492. [Google Scholar]
- 59.Stolz, P., G. Böcker, W. P. Hammes, and R. F. Vogel. 1995. Utilization of electron acceptors by lactobacilli isolated from sourdough. 1. Lactobacillus sanfrancisco. Food Sci. Technol. 201:91-96. [Google Scholar]
- 60.Stolz, P., W. P. Hammes, and R. F. Vogel. 1996. Maltose-phosphorylase and hexokinase activity in lactobacilli from traditionally prepared sourdoughs. Adv. Food Sci. 18:1-6. [Google Scholar]
- 61.Stolz, P., R. F. Vogel, and W. P. Hammes. 1995. Utilization of electron acceptors by lactobacilli isolated from sourdough. 2. Lactobacillus pontis, L. reuteri, L. amylovorus, and L. fermentum. Z. Lebensm. Unters. Forsch. 201:402-410. [Google Scholar]
- 62.Thompson, F. L., B. Hoste, K. Vandemeulebroecke, and J. Swings. 2001. Genomic diversity amongst Vibrio isolates from different sources determined by fluorescent amplified fragment length polymorphism. Syst. Appl. Microbiol. 24:520-538. [DOI] [PubMed] [Google Scholar]
- 63.Tieking, M., and M. G. Gänzle. 2005. Exopolysaccharides from cereal-associated lactobacilli. Trends Food Sci. Technol. 16:79-84. [Google Scholar]
- 64.Tieking, M., M. Korakli, M. A. Ehrmann, M. G. Gänzle, and R. F. Vogel. 2003. In situ production of exopolysaccharides during sourdough fermentation by cereal and intestinal isolates of lactic acid bacteria. Appl. Environ. Microbiol. 69:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vancanneyt, M., S. M. Naser, K. Engelbeen, M. De Wachter, R. Van der Meulen, I. Cleenwerck, B. Hoste, L. De Vuyst, and J. Swings. 2006. Reclassification of Lactobacillus brevis strains LMG 11494 and LMG 11984 as Lactobacillus parabrevis sp. nov. Int. J. Syst. Evol. Microbiol. 56:1553-1557. [DOI] [PubMed] [Google Scholar]
- 66.Van der Meulen, R., T. Adriany, K. Verbrugghe, and L. De Vuyst. 2006. Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD+ through its growth-associated production. Appl. Environ. Microbiol. 72:5204-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Veiga-da-Cunha, M., H. Santos, and E. Van Schaftingen. 1993. Pathway and regulation of erythritol formation in Leuconostoc oenos. J. Bacteriol. 175:3941-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vermeulen, N., J. Kretzer, H. Machalitza, R. F. Vogel, and M. G. Gänzle. 2006. Influence of redox reactions catalysed by homo- and heterofermentative lactobacilli on gluten in wheat sourdoughs. J. Cereal Sci. 43:137-143. [Google Scholar]
- 69.Vermeulen, N., M. Pavlovic, M. A. Ehrmann, M. G. Gänzle, and R. F. Vogel. 2005. Functional characterization of the proteolytic system of Lactobacillus sanfranciscensis DSM 20451T during growth in sourdough. Appl. Environ. Microbiol. 71:6260-6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Versalovic, J., M. Schneider, F. J. De Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 71.Wick, M., P. Stolz, G. Böcker, and J. M. Lebault. 2003. Influence of several process parameters on sourdough fermentation. Acta Biotechnol. 23:51-61. [Google Scholar]
- 72.Wisselink, H. W., R. A. Weusthuis, G. Eggink, J. Hugenholtz, and G. J. Grobben. 2002. Mannitol production by lactic acid bacteria: a review. Int. Dairy J. 12:151-161. [Google Scholar]
- 73.Zaunmüller, T., M. Eichert, H. Richter, and G. Unden. 2006. Variations in the energy metabolism of biotechnologically relevant heterofermentative lactic acid bacteria during growth on sugars and organic acids. Appl. Microbiol. Biotechnol. 72:421-429. [DOI] [PubMed] [Google Scholar]