Abstract
We examined 219 Shiga toxin-producing Escherichia coli (STEC) strains from meat, milk, and cheese samples collected in Germany between 2005 and 2006. All strains were investigated for their serotypes and for genetic variants of Shiga toxins 1 and 2 (Stx1 and Stx2). stx1 or variant genes were detected in 88 (40.2%) strains and stx2 and variants in 177 (80.8%) strains. Typing of stx genes was performed by stx-specific PCRs and by analysis of restriction fragment length polymorphisms (RFLP) of PCR products. Major genotypes of the Stx1 (stx1, stx1c, and stx1d) and the Stx2 (stx2, stx2d, stx2-O118, stx2e, and stx2g) families were detected, and multiple types of stx genes coexisted frequently in STEC strains. Only 1.8% of the STEC strains from food belonged to the classical enterohemorrhagic E. coli (EHEC) types O26:H11, O103:H2, and O157:H7, and only 5.0% of the STEC strains from food were positive for the eae gene, which is a virulence trait of classical EHEC. In contrast, 95 (43.4%) of the food-borne STEC strains carried stx2 and/or mucus-activatable stx2d genes, an indicator for potential high virulence of STEC for humans. Most of these strains belonged to serotypes associated with severe illness in humans, such as O22:H8, O91:H21, O113:H21, O174:H2, and O174:H21. stx2 and stx2d STEC strains were found frequently in milk and beef products. Other stx types were associated more frequently with pork (stx2e), lamb, and wildlife meat (stx1c). The combination of serotyping and stx genotyping was found useful for identification and for assignment of food-borne STEC to groups with potential lower and higher levels of virulence for humans.
Shiga toxin-producing Escherichia coli (STEC) strains have been known as food-borne disease agents in humans since 1982 (38). STEC strains, also called verotoxin-producing E. coli strains, are characterized by production of one or more types of cytotoxins causing tissue damage in humans and animals (34, 46). Humans infected with STEC show symptoms, such as abdominal pain and watery diarrhea, and a number of patients develop a life-threatening disease, such as hemorrhagic colitis (HC) and hemolytic-uremic syndrome (HUS) (46). The natural reservoirs of STEC are domestic and wild ruminant animals, which shed the bacteria with their feces into the environment (10). STEC-infected animals normally do not show signs of disease and can be included in food production. As a consequence, products of animal origin, such as meat and milk, are at risk of contamination with STEC originating from animals (19). Consumption of food containing STEC was identified as a major route of human infections with these pathogens in different countries (10, 19, 29). STEC strains can be divided into more than 200 E. coli serotypes. The phenotypes of most STEC strains resemble those of the normal fecal E. coli flora, which causes problems with detection (2). Some properties of STEC were shown to facilitate spread into the environment and to animals and humans. STEC strains are highly resistant to physicochemical stresses, such as acidity and dryness, and can survive for long time periods in soil, manure, and water (27, 28). Relatively few organisms are needed to cause disease, and routes of direct transmission from animals, infected humans, and the environment have been identified as important routes for STEC infections in humans (22, 37, 46).
For STEC, two major types of Shiga toxins, called Stx1 and Stx2, which share 56% homology to each other, have been described previously (34). Genetic variants were detected within members of the Stx1 and Stx2 families, and a growing number of toxin types were defined according to differences in toxicity, toxin receptor, and amino acid composition of StxA and StxB subunits (34, 39). Some Stx types, such as Stx2 and the elastase (mucus)-activatable Stx2d type, are associated with the high virulence of STEC and with HC and HUS (5, 6, 15, 21). Other toxin types, such as Stx1c, Stx2-O118 (formerly Stx2d-Ount), Stx2e, and Stx2f, were found to be associated mainly with uncomplicated diarrhea or asymptomatic excreters (4, 15, 16). STEC strains producing Stx1c and/or Stx2d-O118 were associated with sheep (8, 45), Stx2e with pigs (14), and Stx2f with pigeons (42) as natural reservoirs.
Highly human-virulent STEC strains able to cause HC and HUS are also called enterohemorrhagic E. coli (EHEC) strains. Typical EHEC strains possess additional virulence functions encoded by genes of the locus of enterocyte effacement (LEE) pathogenicity island and of a large virulence plasmid. The LEE function enables the bacteria to colonize efficiently the intestine of the host by binding between intimin, the product of the eae gene, and its translocated receptor (Tir). Binding of LEE-positive bacteria to the colonic cells results in the attaching and effacing lesion (A/E phenotype), with final destruction of colonic enterocytes (12, 34). The EHEC virulence plasmid carries the genes for EHEC hemolysin (hly) and other functions which may contribute to the disease process (34). Typical EHEC strains, such as O26, O103, O111, O145, and O157, were shown to express one or more types of Stx, the A/E phenotype, and the plasmid-encoded EHEC hemolysin (10, 31). In humans, typical EHEC strains are isolated frequently from young children, whereas atypical EHEC strains are predominant in older patients (4, 5, 15, 47). Atypical EHEC strains express Stx2 and/or Stx2d but do not carry the LEE and belong to serotypes other than those of typical EHEC strains (31, 33, 36, 43).
Since contaminated foodstuff is one of the major vehicles of STEC transmission to humans, we became interested in investigating STEC strains from food samples for their possible role as human pathogens. In recent years, much data on the epidemiology of human infections with STEC in Germany has become available (4, 5, 15, 16, 47). In this study, we investigated 219 STEC strains isolated from food samples in different parts of Germany for their toxin types, other virulence genes, and serotypes. Typical EHEC strains were found rarely among STEC isolates from food, whereas atypical EHEC strains belonging to known human-pathogenic types were detected frequently.
MATERIALS AND METHODS
Bacteria.
A total of 219 STEC strains originating from food samples collected in Germany in 2005 and 2006 were investigated. The strains were isolated in 15 governmental food inspection laboratories situated in different parts of Germany and were sent to the National Reference Laboratory for Escherichia coli at the Federal Institute for Risk Assessment (BfR) in Berlin, Germany, for examination. Only one isolate per food sample was included in the study in order to avoid investigation of double isolates. The strains originated from beef, lamb, pork, or wildlife meat (n = 180), raw milk (n = 32), and raw-milk cheese (n = 5). Apart from these, one STEC isolate originated from salad and one from a cake sample.
Shiga toxin nomenclature.
The nomenclature proposed by Scheutz et al. (39) is used here for the designation of Shiga toxins. The more recently detected toxin variants stx1d (9), stx2-NV206 (1), and stx2g (26) are designated as described previously (3). We use the designation Stx2d for the mucus-activatable Stx2 type, as recently suggested (39, 43). Two types of mucus-activatable genes, called stx2d1 and stx2d2, have been described previously (42). stx2d1 was associated with the toxin B subunit type stx2v-ha and stx2d2 with the stx2v-hb B subunit (42). Further investigations have shown that the association of the mucus-activatable stx2d type with B-subunit variants is not absolute and that other variants, except stx2v-ha and stx2v-hb, can be associated with the mucus-activatable stx2d type (1, 17). Therefore, we use the term stx2d in this work to designate all types of mucus-activatable stx2 genes. The term Stx2d was used previously for the description of another Stx2 variant (VT2d or VT2d-Ount) (35) that was later renamed Stx2-O118 (39).
Microbiological investigation.
All 219 strains were identified as E. coli by biochemical reactions performed as described by Ewing (13). The strains were investigated for production of Stx with a Vero cell cytotoxicity assay and with a commercially obtainable enzyme immunoassay (Ridascreen-EIA; R-Biopharm AG, Darmstadt, Germany) (3). Serotyping of O (lipopolysaccharide) and H (flagellar) antigens was performed with O-specific and H-specific rabbit antisera prepared at the BfR according to standard methods (32). Antigens O1 to O181 and H1 to H56 were investigated, and nonmotile strains were analyzed for their flagellar (fliC) genotypes by PCR and restriction fragment length polymorphism (RFLP) of HhaI-digested fliC PCR products (4).
PCR detection of STEC virulence genes.
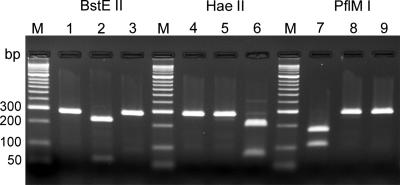
All strains were tested by PCR for the presence of the eae gene, which encodes intimin, and the EHEC plasmid-encoded hemolysin (hlyA) gene (4). Subtyping of eae genes was performed as described previously (4). Identification of stx genes and subtyping of stx1 and stx2 variant genes were performed by PCR as described previously (3) (Table 1). A new PCR/RFLP protocol based on nucleotide differences in the B subunits of the stx1, stx1c, and stx1d genes was used for distinguishing between genes of the Stx1 family. Common primers (Stx1com-F and Stx1com-R) for amplification of the B subunits of the different stx1 genes (Table 1) were derived from published DNA sequences (GenBank accession numbers are in parentheses) for stx1 (M16625), stx1-O48 (Z36899), stx1-CB168 (Z36900), stx1c (AJ413986 and Z36901), and stx1d (AY170851 and AY986980). These primers were shown to amplify all known types of stx1 and stx1 variant genes, yielding a 282-bp (stx1, stx1-O48, stx1-CB168, and stx1d) or a 283-bp (stx1c) PCR fragment. The PCR fragments were digested separately with BstEII, HaeII, and PflMI, yielding RFLP patterns characteristic of stx1, stx1c, and stx1d, respectively (Fig. 1; Table 2). As a control, the stx1c and stx1d genes were specifically amplified by PCR with their genotype-specific primers as listed in Table 1.
TABLE 1.
Detection of stx genes by PCR
| Target gene(s) | Primer | Nucleotide sequence (5′-3′) | PCR conditions | Size of PCR product (bp) | Source or reference |
|---|---|---|---|---|---|
| stx1, stx1-CB168, stx1-O48, stx1c, stx1d | Stx1com-F | (C/T)AGTTGAGGGGGGTAAAATG | 94.0°C, 30 s; 51.3°C, 60 s; 72.0°C, 40 s | 282-283 | This work |
| Stx1com-R | CG(A/G)AAAATAAC(C/T)TCGCTGAATC | ||||
| stx1c | Lin-up | GAACGAAATAATTTATATGT | 94.0°C, 60 s; 48.1°C, 90 s; 72.0°C, 90 s | 555 | 24 |
| 1OX3 | CTCATTAGGTACAATTCT | ||||
| stx1d | VT1AvarF | CTTTTCAGTTAATGCGATTGCT | 94.0°C, 60 s; 62.0°C (5 cycles, 60 s each); 58.0°C (5 cycles, 60 s each); 54.0°C (20 cycles, 60 s each); 72°C, 60 s | 192 | 9 |
| VT1AvarR | AACCCCATGATATCGACTGC | ||||
| stx2, stx2c, stx2-O118, stx2-Ox3:H21, stx2-OX392, stx2e, stx2g, stx2-NV206, stx2v-ha, stx2v-hb, stx2d | LP43 | ATCCTATTCCCGGGAGTTTACG | 94.0°C, 30 s; 57.0°C, 60 s; 72.0°C, 60 s | 584 | 3 |
| LP44 | GCGTCATCGTATACACAGGAGC | ||||
| stx2, stx2c, stx2-OX392, stx2g, stx2-NV206, stx2v-ha, stx2v-hb, stx2d | GK5 | ATGAAGAAGATGTTTATG | 94.0°C, 30 s; 52.0°C, 60 s; 72.0°C, 40 s | 269 | 3 |
| GK6 | TCAGTCATTATTAAACTG | ||||
| stx2-O118, stx2-Ox3:H21 | VTAM1 | AGGGCCCACTCTTTAAATACATCC | 94.0°C, 30 s; 52.0°C, 30 s; 72.0°C, 30 s | 246 | 3 |
| VTAM2 | CGTCATTCCTGTTAACTGTGCG | ||||
| stx2e | SLT-IIBv1 | ATGAAGAAGATGTTTATAGCG | 94.0°C, 30 s; 62.0°C, 60 s; 72.0°C, 30 s | 264 | 3 |
| SLT-IIBv2 | GTTAAACTTCACCTGGGCAAAG | ||||
| stx2f | 128-1 | AGATTGGGCGTCATTCACTGGTTG | 94.0°C, 30 s; 57.0°C, 60 s; 72.0°C, 60 s | 428 | 40 |
| 128-2 | TACTTTAATGGCCGCCCTGTCTCC | ||||
| stx2g | 209F | GTTATATTTCTGTGGATATC | 94.0°C, 30 s; 46.0°C, 60 s; 72.0°C, 60 s | 573 | 26 |
| 781R | GAATAACCGCTACAGTA |
FIG. 1.
PCR/RFLP typing of genes of the Stx1 family. PCR amplification of Stx1 family genes with primers Stx1com-F and Stx1com-R results in a 282-bp (stx1, stx1-O48, stx1-CB168, and stx1d) or a 283-bp (stx1c) fragment. The PCR fragments were digested separately with BstEII (lanes 1 to 3), HaeII (lanes 4 to 6), or PflMI (lanes 7 to 9), yielding RFLP patterns characteristic of stx1, stx1c, and stx1d, respectively (Table 2). Lanes M, molecular size standards; lanes 1, 4, and 7, C600(H19) stx1 (GenBank accession no. M16625); lanes 2, 5, and 8, DG131/3 stx1c (GenBank accession no. Z36901); lanes 3, 6, and 9, MHI813 stx1d (GenBank accession no. AY17085).
TABLE 2.
PCR/RFLP typing of Stx1 family genes with BstEII, HaeII, and PflMI
| Restriction enzyme | Size(s) (bp) of restriction fragment(s)a
|
||
|---|---|---|---|
| stx1b | stx1cc | stx1dd | |
| BstEII | 282e | 224, 59 | 282e |
| HaeII | 282e | 283e | 202, 80 |
| PflMI | 174, 108 | 283e | 282e |
Fragments were obtained by enzymatic digestion of PCR products obtained with primers Stx1com-F and Stx1com-R.
Results for stx1 (GenBank accession no. M16625), stx1-O48 (GenBank accession no. Z36899), and stx1-CB168 (GenBank accession no. Z36900).
Results for stx1c (GenBank accession no. AJ413986 and Z36901).
Results for stx1d (GenBank accession no. AY170851 and AY986980).
Undigested.
Primers LP43 and LP44 were used for the detection of all known genetic variants of the Stx2 family except stx2f. stx2f genes were searched separately by PCR with specific primers 128-1 and 128-2 (Table 1). All strains found positive by use of primers LP43 and LP44 were examined additionally in PCRs with primers GK5 and GK6 (stx2 and stx2d), VTAM1 and VTAM2 (stx2-O118), SLT-IIBv1 and SLT-IIBv2 (stx2e), and 209F and 781R (stx2g) (Table 1).
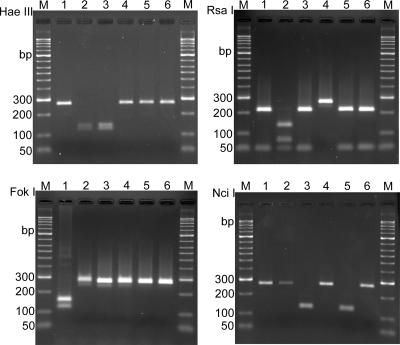
PCR/RFLP subtyping of stx2 and stx2d genes according to nucleotide sequence differences in the B subunits was performed with amplified products obtained with PCR primers GK5 and GK6, as described by Gobius et al. (17). The 269-bp PCR products were digested separately with HaeIII, RsaI, FokI, and NciI, yielding patterns specific for the B subunits of stx2, stx2v-ha, stx2v-hb, stx2g, and stx2-NV206 genes (Table 3; Fig. 2).
TABLE 3.
PCR/RFLP typing of Stx2 family genes with HaeIII, RsaI, FokI, and NciI
| Restriction enzyme | Size(s) (bp) of restriction fragment(s)a
|
|||||
|---|---|---|---|---|---|---|
| stx2b | stx2v-hac | stx2v-hbd | stx2ge | stx2-NV206f | stx2-EC1586g | |
| HaeIII | 269h | 142, 127 | 142, 127 | 269h | 269h | 269h |
| RsaI | 219, 50 | 139, 80, 50 | 219, 50 | 269h | 219, 50 | 219, 50 |
| FokI | 154, 115 | 269h | 269h | 269h | 269h | 269h |
| NciI | 269h | 269h | 140, 129 | 269h | 140, 129 | 269h |
Fragments were obtained by enzymatic digestion of PCR products obtained with primers GK5 and GK6.
GenBank accession no. AJ605767 and AF879828.1.
GenBank accession no. AF479829.2, AF500190, and AF500191.
Undigested.
FIG. 2.
PCR/RFLP typing of genes of the Stx2 family. PCR amplification of Stx2 family genes was performed with primers GK5 and GK6, resulting in a 269-bp fragment. The PCR fragment was digested separately with HaeIII, RsaI, FokI, and NciI, yielding RFLP patterns characteristic of stx2, stx2v-ha, stx2v-hb, stx2g, stx2-NV206, and stx2-EC1586 (Table 3). Lanes M, molecular size standards; lanes 1, C600(W34) stx2 (GenBank accession no. X07865); lanes 2, CB2851 stx2v-ha (GenBank accession no. AJ605767); lanes 3, CB7753 stx2v-hb (GenBank accession no. AF479829); lanes 4, 12422 stx2g (GenBank accession no. AY286000); lanes 5, NV206 stx2-NV206 (GenBank accession no. AF329817); lanes 6, RL481/06 stx2-EC1586 (GenBank accession no. AM498375).
The presence of mucus-activatable stx2d genes in STEC strains from food samples was investigated in all strains typed as stx2, stx2v-ha, stx2v-hb, or stx2-NV206. It was reported previously that the stx2d variant is found among STEC strains carrying these types of stx2 genes (3, 17, 21, 30). The activatable stx2d type differs from nonactivatable stx2 by two amino acid substitutions in the N-terminal region of the toxin A subunit. This results in the absence of a PstI site in the stx2d A-subunit gene, which can be taken as an indicator for the presence of an stx2d genotype (5, 17, 21, 30). For the detection of stx2d toxin types, PCR products of all stx2, stx2v-ha, stx2v-hb, and stx2-NV206 strains were generated with primers SLT-II-vc and CKS2, yielding an 890-bp product. PCR products generated from nonactivatable stx2 are cleaved with PstI into 504- and 386-bp fragments, whereas the stx2d gene remains uncleaved (17, 21).
Nucleotide sequencing.
To obtain the complete coding sequence of the B subunit of the stx2d gene present in strain RL481/06, common primers Stx2B-f (5′ ATGGGAAAGTAATACAGCAGC-3′) and Stx2B-r (5′-CACTTGTTACCCACATACCAC-3′) were designed from the published gene sequences of stx2 and stx2d genes. Stx2B-f and Stx2B-r are positioned in highly conserved DNA stretches upstream and downstream of the toxin B subunits of stx2 and stx2d strains. PCR products were purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany), used for sequencing by applying dye terminator chemistry (PE Applied Biosystems, Darmstadt, Germany), and separated on an automated DNA sequencer (ABI PRISM 3100 genetic analyzer; Applied Biosystems, Foster City, CA). The sequences were analyzed using Lasergene software (DNASTAR, Madison, WI) and Mac Vector software (Oxford Molecular Group, Campell, CA).
Nucleotide sequence accession number.
The nucleotide sequence of the complete coding regions of the stx2 B subunit of STEC strain RL481/06 was submitted to the EMBL database under the accession number AM498375.
RESULTS
Production of Stx and presence of stx genes in STEC strains isolated from food.
All 219 E. coli strains were shown to carry one or more stx genes, as tested by PCR (Table 4). Production of Stx was tested by a Vero cell cytotoxicity assay and an Stx enzyme-linked immunosorbent assay (ELISA). Cytotoxic activity on Vero cells was detected in 216 (98.6%) strains, and a positive Stx ELISA was found with 207 (94.5%) strains. stx1 or stx1 variant genes were detected in 88 (40.2%) and stx2 genes and/or variants in 177 (80.8%) of the 219 STEC strains. Among the 88 strains of the Stx1 family, 61 (69.3%) carried an stx1 gene, 24 (27.3%) an stx1c gene, and 3 (3.4%) an stx1d gene. Differentiation between stx1, stx1c, and stx1d genes was performed by PCR/RFLP analysis as described in Materials and Methods (Table 1; Fig. 1). The results from PCR/RFLP typing were in agreement with those obtained with stx1c and stx1d genotype-specific PCRs (Table 1). By PCR/RFLP typing, we could show that only one type of stx1 gene was present in each of the strains of the Stx1 family (Table 4).
TABLE 4.
Frequencies and combinations of stx genotypes in STEC strains from food
| stx gene(s)a | No. (%) of strains |
|---|---|
| stx1 | 22 (10.0) |
| stx1c | 16 (7.3) |
| stx1d | 3 (1.4) |
| stx1 + stx2/stx2d | 37 (16.9) |
| stx1 + stx2-O118 | 2 (0.9) |
| stx1c + stx2-O118 | 8 (3.7) |
| stx2/stx2d | 52 (23.7) |
| stx2/stx2d + stx2-O118 | 3 (1.4) |
| stx2/stx2d + stx2g | 3 (1.4) |
| stx2-O118 | 25 (11.4) |
| stx2e | 42 (19.2) |
| stx2g | 6 (2.7) |
stx2/stx2d indicates that the strains carry the stx2 and/or the stx2d gene. Further association of stx2 and stx2d genes with different stx types is presented in Table 5.
The presence of any stx2 gene except stx2f was indicated for 177 STEC strains by a positive PCR result with primers LP43 and LP44 (Table 1) (3). Among the members of the Stx2 family, types stx2 and stx2d are associated with highly virulent STEC strains (5, 21, 30). We were therefore interested to determine whether stx2 and/or stx2d strains are present in STEC from food. For detection of these strains, we investigated all 177 strains that had reacted with primers LP43 and LP44 by use of a second PCR with primers GK5 and GK6. GK5 and GK6 are specific for the B subunits of the stx2 and stx2d genes (17). A positive PCR result was obtained with 101 (57.1%) of the 177 Stx2 family strains. It was described previously that multiple copies of the stx2 and stx2d genes can coexist in the same STEC strain (1, 5, 21, 43). In order to examine this possibility, we digested the 269-bp PCR products obtained with primers GK5 and GK6 with HaeIII, RsaI, FokI, and NciI. The resulting RFLP patterns were attributed to the different subtypes of stx2 and stx2d genes, such as stx2v-ha, stx2v-hb, and stx2-NV206, as previously described (1, 17) (Table 3; Fig. 2). The PCR/RFLP analysis revealed the presence of single and multiple copies of stx2, stx2v-ha, stx2v-hb, and stx2-NV206 genes in 94 STEC strains from food (Table 5). In addition, two other Stx2 type genes were identified by the PCR/RFLP analysis (Table 3; Fig. 2). One RFLP pattern characteristic of the recently described stx2g variant was found in nine STEC strains. The other RFLP pattern (stx2-EC1586) was found in only one STEC strain (RL481/06), from raw milk (Table 3; Fig. 2). Because it did not correspond to patterns of previously described stx2 B variants, we determined the nucleotide sequence of the stx2 B subunit of RL481/06 (GenBank accession no. AM498375). It was found to be homologous to that of a recently described stx2d variant gene of an STEC O174:H8 strain from livestock sources in Australia (GenBank accession no. AF500188) (17).
TABLE 5.
Association of Stx B-subunit types with stx2 and stx2d genes and serotypes of STEC strains from food
| stx2 and/or stx2d B-subunit genotype (no. of strains) | No. of strains with stx2 and/or stx2d
|
||
|---|---|---|---|
| stx2 | stx2d | stx2 and stx2d | |
| stx2v-ha (14) | 8a | 6b | 0 |
| stx2 + stx2v-ha (3) | 0 | 0 | 3c |
| stx2v-hb (17) | 1d | 16e | 0 |
| stx2 + stx2v-hb (12) | 1f | 2g | 9h |
| stx2 + stx2g (3) | 3i | 0 | 0 |
| stx2-NV206 (4) | 0 | 4j | 0 |
| stx2 + stx2-NV206 (1) | 0 | 0 | 1k |
| stx2 (40) | 21l | 16m | 3n |
| stx2-EC1586 (1) | 0 | 1o | 0 |
O174:H21 (n = 4), O157:[H7] (n = 1), O178:H19 (n = 1), O8:H21 (n = 1), and ONT:HNT (n = 1).
O113:H21 (n = 2), O22:H8 (n = 1), O46:H38 (n = 1), and ONT:H8/48 (n = 2).
O91:H21 (n = 2) and ONT:H21 (n = 1).
O174:H8 (n = 1).
O113:H21 (n = 1), O120:H11 (n = 1), O153:H4 (n = 1), O163:[H27] (n = 1), O174:H2 (n = 2), O22:H8 (n = 3), O22:H16 (n = 1), O91:H21 (n = 2), O91:H42 (n = 1), and ONT:H19/46 (n = 3).
O178:H19 (n = 1).
O104:H21 (n = 1) and ONT:H7 (n = 1).
O91:H21 (n = 4), O113:H21 (n = 1), O153:H1 (n = 1), O8:H19 (n = 1), and O82:H8 (n = 2).
O109:H16 (n = 2) and ONT:NM (n = 1).
O6:H10 (n = 1), O4:H4 (n = 1), O113:H4 (n = 1), and ONT:H29 (n = 1).
ONT:H19 (n = 1).
O113:H21 (n = 1), O172:H25 (n = 1), O174:H2 (n = 1), O178:H19 (n = 2), O179:H8 (n = 4), O2:H27 (n = 1), O2:H42 (n = 1), O46:H38 (n = 1), O8:H19 (n = 1), ONT:H11/18/19/21/28 (n = 7), and O146:H28 (n = 1).
O22:H8 (n = 5), O88:H25 (n = 2), O113:H21 (n = 1), O153:H25 (n = 1), O6:H34 (n = 1), O74:H8 (n = 1), O76:H4 (n = 1), ONT:NM (n = 1), and ONT:H18/19/48 (n = 3).
O113:H21 (n = 3).
ONT:H4 (n = 1).
Finally, in order to distinguish between stx2 and stx2d we amplified the stx2 genes present in all 95 strains carrying stx2, stx2v-ha, stx2v-hb, stx2-NV206, and stx2-EC1586 genes with primers SLT-II-vc and CKS2. The 890-bp PCR product was digested with PstI, and stx2d genes were discriminated from nonactivatable stx2 by lack of a PstI site, as described previously (5, 17, 21). stx2 was present in 50 (52.6%) and stx2d in 61 (64.2%) of the strains (Table 5). Interestingly, 16 (16.8%) of the 95 STEC strains carried both an stx2 and an stx2d gene.
Other genetic variants of the Stx2 family that were detected in STEC strains from food were stx2e (42 strains), stx2-O118 (38 strains), and stx2g (nine strains) (Table 4). In contrast, none of the 219 STEC strains carried an stx2f gene.
Association between serotype, stx type, and other virulence markers of STEC.
The 219 STEC strains from food belonged to 45 E. coli O serogroups and to 31 H types. Fifty-four (24.7%) of the STEC strains showed nontypeable (ONT) O antigens different from O1 to O181, and 35 (16.0%) were nonmotile. However, only 10 serogroups (O8, O21, O22, O91, O100, O113, O146, O174, O178, and O179) accounted for 94 (42.9%) of all STEC isolates (Table 6). The association with the H type was even more pronounced, as only four flagellar types (H8, H9, H19, and H21) accounted for 97 (44.3%) of all STEC strains from food. Other STEC virulence markers were present, such as production of the EHEC hemolysin gene (hlyA) in 99 (45.2%) and the eae gene in 11 (5.0%) of the STEC strains. Only four (1.8%) STEC strains from food belonged to the group of typical EHEC strains. These were two O26:[H11] (stx1, eae-beta, and hlyA), one O103:H2 (stx1, eae-epsilon, and hlyA), and one O157:[H7] (stx1, stx2, eae-gamma, and hlyA) strain, all of which originated from meat.
TABLE 6.
Origins and properties of STEC strains from food belonging to the most important serotypes
| Serotype(s) (no. of strains) | Stx2 family gene(s) (no. of strains) | Other virulence gene(s) (no. of strains) | Sourcesa (no. of strains) |
|---|---|---|---|
| O8:H19 (3), O8:H21 (1) | stx2 (2), stx2 + stx2d (1), stx2e (1) | hlyA (3) | B (1), B/P (1), P (1), W (1) |
| O8:H4 (3), O8:H9 (11) | stx2e (14) | None | B (1), B/P (6), M (1), P (6) |
| O21:H21 (9) | stx2-O118 (9) | hlyA (2) | B (3), M (1), W (5) |
| O22:[H8] (9) | stx2d (9) | stx1 (8), hlyA (9) | B (4), B/P (2), M (1), P (1), W (1) |
| O91:[H14] (6) | stx2-O118 (2) | stx1 (6), hlyA (2) | B (1), B/P (2), L (1), P (2) |
| O91:[H21] (8) | stx2 + stx2d (6), stx2(2) | stx1 (6), hlyA (7) | B (1), B/P (1), M (2), P (2), W (2) |
| O100:[H30] (8) | stx2e | None | B (2), B/P (3), M (1), P (2) |
| O113:H21 (9) | stx2 (1), stx2d (4), stx2 + stx2d(4) | stx1 (1), hlyA (7) | B (3), B/P (1), P (3), W (2) |
| O146:H21 (4), O146:H28 (5) | stx2 + stx2-O118 (1), stx2-O118 (8) | stx1c (2), hlyA (3) | B (1), W (8) |
| O174:H2 (3) | stx2 (1), stx2d (2) | stx1 (3), hlyA (3) | B (2), M (1) |
| O174:H21 (5), O174:[H8] (2) | stx2 (5), stx2-O118 (2) | stx1c (2), hlyA (3) | B (4), W (3) |
| O178:H19 (4) | stx2 (4) | stx1 (2), hlyA (3) | B (3), L (1) |
| O179:H8 (4) | stx2 (4) | hlyA (4) | B (2), W (2) |
B, beef; W, wildlife meat; L, lamb; P, pork; B/P, mixed beef/pork meat; M, milk.
On the other hand, 95 (44.0%) of the STEC strains from food were shown to carry stx2 and/or stx2d genes (Table 5). stx2 and/or stx2d genes were found to be closely associated with atypical EHEC serotypes O8:H19, O22:[H8], O91:H21, O113:H21, O174:H2, O174:H21, O178:H19, and O179:H8 (Table 6).
The stx2e gene was present in other E. coli serotypes (O2, O8:H4/H9, O36:H19, O59:H21, O100:[H30], O141:H4, and ONT), and none of the 42 (19.2%) STEC isolates from this study carried other virulence markers except stx2e (Table 4). The stx2-O118 variant was detected in 37 (16.9%) of the STEC isolates from food. It was associated with certain serotypes (O21:H21, O22:H16, O91:H14, O128:H2, O146:H21/H28, and ONT) of strains, with toxin types stx2 or stx2d (three strains) and stx1 or stx1c (10 strains), and with the hlyA gene (14 strains) (Table 4). The stx2g variant was present in six STEC strains belonging to rare serotypes (O179:H31, O74:H28, O15:H16, O36:H14, and O59:H21) and occurred in combination with stx2 in three strains (Table 5).
DISCUSSION
Control of food samples for possible contamination with STEC is performed routinely by governmental laboratories situated in different parts of Germany. Most commonly, these laboratories employ commercially obtainable Stx-specific ELISA kits and colony immunoblotting techniques for detection and subsequent isolation of STEC from food (3, 23, 44). This procedure allows detection of STEC strains independent of their serotypes, which is important to identify virtually all STEC types present in a sample (2). Correspondingly, we obtained a wide spectrum of different serotypes of STEC from food samples for analysis. One interesting finding was that typical EHEC O26, O103, and O157 strains represented only 1.8% of STEC isolates from food. In contrast, EHEC O26:H11 and O157:H7 strains were reported as being isolated most frequently from human patients infected with STEC in Germany and other countries (4, 5, 15). In a similar way, intimin (eae gene)-positive strains accounted for only 11 (5.0%) STEC strains from food. Intimin-positive STEC strains were described as most frequently isolated from 1- to 3-year-old pediatric patients suffering from diarrhea and HUS (4, 47). Our data indicate that in Germany food is less important as a source of infection with typical EHEC strains. This finding corroborates a recently published risk analysis for STEC infections in Germany stating that direct transmission from food plays a limited role for infections in children under 3 years of age (47).
On the other hand, about half of the STEC strains from this study were positive for stx2 and/or stx2d. Production of these toxins is an indicator for severe clinical outcome in the infected patients (5, 6, 15, 21). In our study, stx2 and stx2d were common in STEC strains of serotypes O22:H8, O91:H21, O113:H21, O174:H2/H21, and O178:H19 and most of these strains originated from meat (Table 6). STEC strains belonging to these serotypes were associated with cattle as an animal reservoir (http://www.microbionet.com.au/vtectable.htm) and were isolated from patients of all age groups with diarrhea in Germany (4, 5, 47). Some types, such as STEC O91:H21, O113:H21, and O174:H21, were identified as agents of HUS in patients from different countries (7, 20, 33, 36).
In 2005, STEC was detected in 6.7% of 535 investigated meat samples and in 1.9% of 2,681 raw-milk samples in Germany (18). Consumption of meat was identified as a major risk factor of patients older than 10 years for getting infected with STEC (47). More than 80% of STEC strains from our study originated from meat and 87 (48.3%) of 180 STEC-positive meat samples were beef or contained beef together with lamb or pork meat. Seventy-two (82.8%) of the STEC strains isolated from pure beef or beef-containing products carried the stx2, stx2d, or stx2g gene. Beef and its products may therefore be regarded as most critical in regard to contamination with potentially human-virulent STEC strains.
Twenty-six (14.4%) of the STEC-positive samples from this study contained only pork meat. The stx2e gene was present in 15 (57.7%) of the 26 STEC strains isolated from pork but in only 3 (5.9%) of 51 samples containing only beef. Stx2e, also designated the porcine edema disease toxin (11), is associated mainly with STEC from pigs and is rarely found in other species, including humans (4, 15, 41). The linkage of the stx2e gene with pork meat could indicate that most of the products are contaminated directly by fecal material from pigs after slaughter. The serotypes of stx2e strains from our study were similar to those described to occur in pigs from a study performed in the United States (14). The pathogenicity of Stx2e-producing STEC strains for humans is regarded as low, but some types of Stx2e-producing strains were isolated from more severe cases of diarrhea in patients (4, 15, 41).
In this work, we developed a rapid PCR/RFLP method used for differentiation between members of the Stx1 family in regard to nucleotide sequence differences in the B subunits of stx1, stx1c, and stx1d (Fig. 1; Table 2). This method was found useful for identification of stx1 variants and provides the advantage that strains carrying multiple types of stx1 become detectable. In contrast to findings for genes of the Stx2 family, we did not detect more than one stx1 gene in any of the 88 investigated strains. This might indicate that the bacteriophages encoding different stx1 types do not coexist frequently in the same strain. Similar findings were reported for human and animal STEC strains investigated for stx1 genes (8, 25).
It has been described previously that the stx1c gene is associated with sheep and goats as the animal reservoir (8, 24), and 8 of 24 samples containing stx1c originated from lamb meat products, sheep milk, or goat cheese. The finding that 11 of the samples containing stx1c STEC strains were isolated from deer meat points to wild animals as a possible reservoir for these STEC types.
PCR/RFLP typing of the B subunits of the stx2 and stx2d genes was found useful for investigation of both the presence of genetic variants and that of multiple types of stx2 and stx2d in STEC (17). Apart from identification of stx2 and stx2d B-subunit variants, this method allowed the detection of the stx2g toxin type. Stx2g was recently described as a new, rarely occurring stx2 variant in bovine STEC that is not activatable by intestinal mucus (26). Like stx1d, stx2g genes carrying STEC have not yet been reported to occur in humans and their role as agents of disease is not clear. In any case, the finding that stx1d and stx2g STEC strains are present in food samples makes it likely that humans can be exposed to these STEC types along the food chain. In contrast, stx2f genes carrying STEC appear to be uncommon in meat and milk samples.
In this work, we have shown that the combination of serotyping and stx genotyping is very suitable for identification and for assignment of food-borne STEC strains to groups of lower and higher levels of potential virulence for humans. The finding that stx2 and/or stx2d STEC strains of known human-pathogenic serotypes are present in about half of the isolates indicates that raw-meat and -milk products present a risk for consumers of all age groups. Further improvement of molecular detection methods will contribute to the rapid identification of highly virulent STEC strains independent of their serotype. These methods will be very useful in the investigation of food-borne outbreaks and sporadic cases caused by new or unusual types of STEC strains.
Acknowledgments
We thank all colleagues from the Federal Food Inspection Laboratories in Germany for providing information on the origins of the STEC isolates investigated in this study.
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Bertin, Y., K. Boukhors, N. Pradel, V. Livrelli, and C. Martin. 2001. stx2 subtyping of Shiga toxin-producing Escherichia coli isolated from cattle in France: detection of a new Stx2 subtype and correlation with additional virulence factors. J. Clin. Microbiol. 39:3060-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettelheim, K. A., and L. Beutin. 2003. Rapid laboratory identification and characterization of verocytotoxigenic (Shiga toxin-producing) Escherichia coli (VTEC/STEC). J. Appl. Microbiol. 95:205-217. [DOI] [PubMed] [Google Scholar]
- 3.Beutin, L., H. Steinruck, G. Krause, K. Steege, S. Haby, G. Hultsch, and B. Appel. 2007. Comparative evaluation of the Ridascreen verotoxin enzyme immunoassay for detection of Shiga-toxin producing strains of Escherichia coli (STEC) from food and other sources. J. Appl. Microbiol. 102:630-639. [DOI] [PubMed] [Google Scholar]
- 4.Beutin, L., G. Krause, S. Zimmermann, S. Kaulfuss, and K. Gleier. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielaszewska, M., A. W. Friedrich, T. Aldick, R. Schurk-Bulgrin, and H. Karch. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160-1167. [DOI] [PubMed] [Google Scholar]
- 6.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet, R., B. Souweine, G. Gauthier, C. Rich, V. Livrelli, J. Sirot, B. Joly, and C. Forestier. 1998. Non-O157:H7 Stx2-producing Escherichia coli strains associated with sporadic cases of hemolytic-uremic syndrome in adults. J. Clin. Microbiol. 36:1777-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brett, K. N., V. Ramachandran, M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2003. stx1c is the most common Shiga toxin 1 subtype among Shiga toxin-producing Escherichia coli isolates from sheep but not among isolates from cattle. J. Clin. Microbiol. 41:926-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burk, C., R. Dietrich, G. Acar, M. Moravek, M. Bulte, and E. Martlbauer. 2003. Identification and characterization of a new variant of Shiga toxin 1 in Escherichia coli ONT:H19 of bovine origin. J. Clin. Microbiol. 41:2106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caprioli, A., S. Morabito, H. Brugereb, and E. Oswald. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289-311. [DOI] [PubMed] [Google Scholar]
- 11.Degrandis, S., H. Law, J. Brunton, C. Gyles, and C. A. Lingwood. 1989. Globotetraosylceramide is recognized by the pig edema disease toxin. J. Biol. Chem. 264:12520-12525. [PubMed] [Google Scholar]
- 12.Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Investig. 107:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewing, W. H. 1986. Differentiation of Enterobacteriaceae by biochemical reactions, p. 47-72. In W. H. Ewing (ed.), Edwards and Ewing's identification of Enterobacteriaceae. Elsevier Science Publishing Co., New York, NY.
- 14.Fratamico, P. M., L. K. Bagi, E. J. Bush, and B. T. Solow. 2004. Prevalence and characterization of Shiga toxin-producing Escherichia coli in swine feces recovered in the National Animal Health Monitoring System's Swine 2000 study. Appl. Environ. Microbiol. 70:7173-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich, A. W., J. Borell, M. Bielaszewska, A. Fruth, H. Tschape, and H. Karch. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41:2448-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gobius, K. S., G. M. Higgs, and P. M. Desmarchelier. 2003. Presence of activatable Shiga toxin genotype (stx2d) in Shiga toxigenic Escherichia coli from livestock sources. J. Clin. Microbiol. 41:3777-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartung, M. 2007. Zoonoses agents 2005 at the official Food Control in Germany. Fleischwirtschaft 2:98-106. (In German.) [Google Scholar]
- 19.Hussein, H. S., and T. Sakuma. 2005. Prevalence of Shiga toxin-producing Escherichia coli in dairy cattle and their products. J. Dairy Sci. 88:450-465. [DOI] [PubMed] [Google Scholar]
- 20.Ito, H., A. Terai, H. Kurazono, Y. Takeda, and M. Nishibuchi. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91-H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47-60. [DOI] [PubMed] [Google Scholar]
- 21.Jelacic, J. K., T. Damrow, G. S. Chen, S. Jelacic, M. Bielaszewska, M. Ciol, H. M. Carvalho, A. R. Melton-Celsa, A. D. O'Brien, and P. I. Tarr. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719-729. [DOI] [PubMed] [Google Scholar]
- 22.Karch, H., M. Bielaszewska, M. Bitzan, and H. Schmidt. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229-243. [DOI] [PubMed] [Google Scholar]
- 23.Klie, H., M. Timm, H. Richter, P. Gallien, K. W. Perlberg, and H. Steinruck. 1997. Detection and occurrence of verotoxin-forming and/or shigatoxin producing Escherichia coli (VTEC and/or STEC) in milk. Berl. Muench. Tieraerztl. Wochenschr. 110:337-341. (In German.) [PubMed] [Google Scholar]
- 24.Koch, C., S. Hertwig, R. Lurz, B. Appel, and L. Beutin. 2001. Isolation of a lysogenic bacteriophage carrying the stx1OX3 gene, which is closely associated with Shiga toxin-producing Escherichia coli strains from sheep and humans. J. Clin. Microbiol. 39:3992-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuczius, T., M. Bielaszewska, A. W. Friedrich, and W. Zhang. 2004. A rapid method for the discrimination of genes encoding classical Shiga toxin (Stx) 1 and its variants, Stx1c and Stx1d, in Escherichia coli. Mol. Nutr. Food Res. 48:515-521. [DOI] [PubMed] [Google Scholar]
- 26.Leung, P. H., J. S. Peiris, W. W. Ng, R. M. Robins-Browne, K. A. Bettelheim, and W. C. Yam. 2003. A newly discovered verotoxin variant, VT2g, produced by bovine verocytotoxigenic Escherichia coli. Appl. Environ. Microbiol. 69:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loukiadis, E., M. Kerouredan, L. Beutin, E. Oswald, and H. Brugere. 2006. Characterization of Shiga toxin gene (stx)-positive and intimin gene (eae)-positive Escherichia coli isolates from wastewater of slaughterhouses in France. Appl. Environ. Microbiol. 72:3245-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maule, A. 2000. Survival of verocytotoxigenic Escherichia coli O157 in soil, water and on surfaces. J. Appl. Microbiol. 88:71S-78S. [DOI] [PubMed] [Google Scholar]
- 29.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melton-Celsa, A. R., S. C. Darnell, and A. D. O'Brien. 1996. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect. Immun. 64:1569-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orskov, F., and I. Orskov. 1984. Serotyping of Escherichia coli, p. 43-112. In T. Bergan (ed.), Methods in microbiology, vol. 14. Academic Press, London, United Kingdom. [Google Scholar]
- 33.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pradel, N., K. Boukhors, Y. Bertin, C. Forestier, C. Martin, and V. Livrelli. 2001. Heterogeneity of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients, cattle, and food samples in central France. Appl. Environ. Microbiol. 67:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rangel, J. M., P. H. Sparling, C. Crowe, P. M. Griffin, and D. L. Swerdlow. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. Mcgee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 39.Scheutz, F., L. Beutin, D. Pierard, and H. Smith. 2001. Nomenclature of verotoxins, p. 447-452. In G. Duffy, P. Garvey, and D. A. McDowell (ed.), Verocytotoxigenic E. coli. Food and Nutrition Press, Trumbull, CT.
- 40.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonntag, A. K., M. Bielaszewska, A. Mellmann, N. Dierksen, P. Schierack, L. H. Wieler, M. A. Schmidt, and H. Karch. 2005. Shiga toxin 2e-producing Escherichia coli isolates from humans and pigs differ in their virulence profiles and interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 71:8855-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonntag, A. K., E. Zenner, H. Karch, and M. Bielaszewska. 2005. Pigeons as a possible reservoir of Shiga toxin 2f-producing Escherichia coli pathogenic to humans. Berl. Muench. Tieraerztl. Wochenschr. 118:464-470. [PubMed] [Google Scholar]
- 43.Teel, L. D., A. R. Melton-Celsa, C. K. Schmitt, and A. D. O'Brien. 2002. One of two copies of the gene for the activatable Shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect. Immun. 70:4282-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timm, M., H. Klie, H. Richter, P. Gallien, K. W. Perlberg, S. Lehmann, and D. Protz. 1999. Detection and prevalence of verotoxin-producing Escherichia coli (VTEC) in raw sausage. Berl. Muench. Tieraerztl. Wochenschr. 112:385-389. (In German.) [PubMed] [Google Scholar]
- 45.Urdahl, A. M., L. Beutin, E. Skjerve, S. Zimmermann, and Y. Wasteson. 2003. Animal host associated differences in Shiga toxin-producing Escherichia coli isolated from sheep and cattle on the same farm. J. Appl. Microbiol. 95:92-101. [DOI] [PubMed] [Google Scholar]
- 46.Verweyen, H. M., H. Karch, M. Brandis, and L. B. Zimmerhackl. 2000. Enterohemorrhagic Escherichia coli infections: following transmission routes. Pediatr. Nephrol. 14:73-83. [DOI] [PubMed] [Google Scholar]
- 47.Werber, D., S. C. Behnke, A. Fruth, R. Merle, S. Menzler, S. Glaser, L. Kreienbrock, R. Prager, H. Tschape, P. Roggentin, J. Bockemuhl, and A. Ammon. 2007. Shiga toxin-producing Escherichia coli infection in Germany: different risk factors for different age groups. Am. J. Epidemiol. 165:425-434. [DOI] [PubMed] [Google Scholar]