Abstract
Mycobacterium ulcerans is a slow-growing environmental bacterium that causes a severe skin disease known as Buruli ulcer. PCR has become a reliable and rapid method for the diagnosis of M. ulcerans infection in humans and has been used for the detection of M. ulcerans in the environment. This paper describes the development of a TaqMan assay targeting IS2404 multiplexed with an internal positive control to monitor inhibition with a detection limit of less than 1 genome equivalent of DNA. The assay improves the turnaround time for diagnosis and replaces conventional gel-based PCR as the routine method for laboratory confirmation of M. ulcerans infection in Victoria, Australia. Following analysis of 415 clinical specimens, the new test demonstrated 100% sensitivity and specificity compared with culture. Another multiplex TaqMan assay targeting IS2606 and the ketoreductase-B domain of the M. ulcerans mycolactone polyketide synthase genes was designed to augment the specificity of the IS2404 PCR for the analysis of a variety of environmental samples. Assaying for these three targets enabled the detection of M. ulcerans DNA in soil, sediment, and mosquito extracts collected from an area of endemicity for Buruli ulcer in Victoria with a high degree of confidence. Final confirmation was obtained by the detection and sequencing of variable-number tandem repeat (VNTR) locus 9, which matched the VNTR locus 9 sequence obtained from the clinical isolates in this region. This suite of new methods is enabling rapid progress in the understanding of the ecology of this important human pathogen.
Buruli ulcer (BU) is a skin disease caused by infection with Mycobacterium ulcerans, a slow-growing environmental mycobacterium (11). The disease has been reported in over 30 countries worldwide (9), predominantly in riverine areas with tropical and subtropical climates but also in temperate climates, such as southern Australia (10, 30). Infection with M. ulcerans can lead to extensive destruction of skin and soft tissue with the formation of large ulcers. The characteristic necrosis and ulceration is induced by an unusual diffusible cytotoxic macrocyclic polyketide called mycolactone, which is produced by M. ulcerans. Mycolactone is the product of three large multienzyme complexes called polyketide synthases that are encoded by the genes mlsA1 (51 kb), mlsA2 (7 kb), and mlsB (42 kb). These genes are located on the M. ulcerans virulence plasmid known as pMUM001 (25, 29).
Although not generally a fatal condition, BU lesions can become extensive and heal by scarring. When diagnosis and treatment are delayed, sufferers are frequently left with long-term physical and cosmetic disabilities. In areas of endemicity, the combination of a suspicious skin lesion and a smear positive for acid-fast bacilli is highly suggestive of BU and can be definitively diagnosed by culture. However, due to the very slow growth of the organism, culture confirmation may take 8 to 12 weeks and treatment needs to be initiated much sooner than this to ensure an optimal outcome for the patient. The use of PCR for diagnosis of BU has been a major step forward, particularly in Australia where we have access to molecular diagnostics and where BU is increasingly common. The most commonly used target sequence for PCR is IS2404, a multicopy insertion sequence that encodes a 328-amino-acid transposase (26). Provided that diagnostic PCR is performed in a laboratory with high standards to avoid false positives, it is a reliable and rapid method for laboratory confirmation of disease caused by M. ulcerans (15).
Despite strong epidemiological evidence linking the source of M. ulcerans to swamps and slow-flowing water (16), the precise mode of transmission has yet to be elucidated. Understanding the ecology of M. ulcerans has been severely hampered by the extreme difficulty of culturing the organism directly from the environment (13, 16, 22, 24). This difficulty may be due partly to contamination by less fastidious mycobacteria that outgrow M. ulcerans. IS2404 PCR performed on DNA extracted from environmental samples has proved a major advance since its first use in the mid-1990s (22). The bacterium has now been detected in water and detritus (22, 24), aquatic insects (17) and plants (13), snails (12), and small fish (5). However, while IS2404 PCR is highly specific and sensitive for testing diagnostic specimens from humans, its application to the analysis of environmental samples is less straightforward due to PCR inhibitors and the existence of other environmental mycobacteria that may carry IS2404 (14, 18-20, 32). Approaches for increasing confidence in the interpretation of PCR-positive tests for environmental samples and for overcoming the issues surrounding primer specificity are the use of an internal probe, such as in the TaqMan assay, to confirm the identity of a PCR product and to use two or more DNA targets.
In the Australian state of Victoria, there has been a significant recent increase in the incidence of BU (also known locally as Bairnsdale ulcer) (8). With increasing numbers of clinical specimens, we sought to develop a real-time PCR method based on IS2404 to improve the turnaround time for diagnosis of M. ulcerans infection. We also wanted to develop additional real-time PCR assays targeting different regions in the M. ulcerans genome that could be used as confirmatory assays for the analysis of environmental samples. This paper describes the development of two multiplex real-time PCR assays targeting three distinct repeated sequences in the M. ulcerans genome: IS2404, IS2606 (26), and a sequence encoding the ketoreductase B domain (KR), present in 15 copies within the mycolactone polyketide synthase genes mlsA1, mlsA2, and mlsB (25). These assays have facilitated the rapid diagnosis of M. ulcerans disease in clinical specimens and the analysis of environmental samples in less time and with greater confidence than conventional PCR.
MATERIALS AND METHODS
Mycobacterial strains.
Mycobacterial isolates used in this study are listed in Table 1 (also see Table S1 in the supplemental material).
TABLE 1.
Specificity of IS2404, IS2606, and KR real-time PCR assays
| Mycobacterial species (strain) | Origin | Sourceb | IS2404 | IS2606 | KR |
|---|---|---|---|---|---|
| M. ulcerans (ATCC 19423) | Victoria, Australia | VIDRL | +c | + | + |
| M. ulcerans (63/04) | Victoria, Australia | VIDRL | + | + | + |
| M. ulcerans (40/02) | Western Australia | VIDRL | + | + | + |
| M. ulcerans (75/04) | Northern Territory, Australia | + | + | + | |
| M. ulcerans (20213/91) | Queensland, Australia | VIDRL | + | + | + |
| M. ulcerans (38/02) | Queensland, Australia | VIDRL | + | + | + |
| M. ulcerans (71/05) | Papua New Guinea | VIDRL | + | + | + |
| M. ulcerans (5145) | Zaire | ITM | + | + | + |
| M. ulcerans (98912) | People's Republic of China | ITM | + | + | + |
| M. ulcerans (842) | Suriname | ITM | + | + | + |
| M. ulcerans (9118) | Mexico | ITM | + | + | + |
| M. ulcerans (186510) | Malaysia | VIDRL | + | + | + |
| M. liflandii (128FXT) | California | UTK | + | + | + |
| M. pseudoshottsii (L15) | Chesapeake Bay, United States | VIMS | + | + | + |
| M. marinum (DL045)a | Mediterranean Sea, Greece | NCM | + | + | + |
| M. marinum (CC240299)a | Fresh water, Israel | NCM | + | NDd | + |
| M. marinum (DL240490)a | Red Sea, Israel | NCM | + | + | + |
| M. tuberculosis (H37Rv) | Sequence strain | VIDRL | ND | ND | ND |
| M. paratuberculosis (field strain) | VIDRL | ND | ND | ND | |
| M. marinum (NCTC 2275) | VIDRL | ND | ND | ND | |
| M. marinum (ITM 00-1026) | France | IP | ND | ND | ND |
| M. kansasii (NCTC 10268) | VIDRL | ND | ND | ND | |
| M. xenopi (NCTC 10042) | VIDRL | ND | ND | ND | |
| M. szulgai (NCTC 10831) | VIDRL | ND | ND | ND | |
| M. simiae (ATCC 25275) | VIDRL | ND | ND | ND | |
| M. marinum (NCTC 2275) | VIDRL | ND | ND | ND | |
| M. malmoense (NCTC 11298) | VIDRL | ND | ND | ND | |
| M. asiaticum (21674) | VIDRL | ND | ND | ND | |
| M. avium (ATCC 25291) | VIDRL | ND | ND | ND | |
| M. intracellulare (ATCC 13950) | VIDRL | ND | ND | ND | |
| M. lentiflavum (A) | Patient isolate | VIDRL | ND | ND | ND |
| M. lentiflavum (B) | Patient isolate | VIDRL | ND | ND | ND |
| M. lentiflavum (C) | Patient isolate | VIDRL | ND | ND | ND |
These isolates were originally designated M. marinum because of their growth characteristics, but they have since been shown to produce mycolactone and are now described as “other mycolactone-producing mycobacteria” (18, 32).
IP, Institut Pasteur, Paris, France; ITM, Institute for Tropical Medicine, Antwerp, Belgium; NCM, National Centre for Mariculture, Eilat, Israel; UTK, Department of Microbiology, University of Tennessee, Knoxville, TN; VIDRL, Victorian Infectious Diseases Reference Laboratory, Victoria, Australia; VIMS, Virginia Institute of Marine Science, Gloucester Point, VA.
+, presence of.
ND, not detected.
Clinical specimens.
A total of 415 primary clinical specimens received at the Victorian Mycobacterium Reference Laboratory from July 2003 to June 2005 were included in the study. These specimens consisted of 244 swabs (dry or in bacterial transport medium), 159 fresh tissue biopsies, 11 formalin-fixed, paraffin-embedded tissue sections, and 1 bone sample. Swab and fresh tissue specimens were split for DNA extraction and culture following maceration and/or homogenization in a bottle containing glass beads and phosphate-buffered saline. Specimens were cultured in BACTEC 12B bottles and on Brown and Buckle medium (11) and incubated at 31°C for up to 12 weeks.
Environmental samples.
Mosquitoes and soil samples were collected from various locations in an area of Victoria where there is currently an outbreak of M. ulcerans infection. Mosquitoes were trapped using overnight mosquito traps baited with carbon dioxide and light and stored at −70°C. Both wet and dry surface soil samples from drains and soak pits were collected in sterile containers and stored for no more than 5 days at 4°C until DNA extraction was performed.
DNA preparation.
Extraction of DNA from mycobacterial cultures was performed as previously described (6). DNA was extracted directly from swabs and homogenized fresh tissue biopsies by using the Roche AMPLICOR respiratory specimen preparation kit (Roche Diagnostics Co., Indianapolis, IN), followed by a cleanup to remove potential PCR inhibitors by using the QIAamp DNA Mini kit (QIAGEN Inc., Valencia, CA). DNA was extracted from formalin-fixed, paraffin-embedded tissue sections following a 24-h proteinase K digestion of dewaxed samples, using the QIAamp DNA Mini kit. DNA from mosquitoes was extracted using the FastDNA kit with the FastPrep instrument (Qbiogene, Inc., Carlsbad, CA), according to the manufacturer's instructions for insects, with the addition of an extra ceramic bead to the lysing matrix. DNA from 100- to 200-mg aliquots of soil and sediment samples was extracted by using the FastDNA SPIN kit for soil with the FastPrep instrument (Qbiogene) according to the manufacturer's instructions. DNA extracts were stored at −20°C.
PCR amplification of a region of IS2404 containing the TaqMan real-time PCR target.
Amplification of a 515-bp product to be used as a positive control in the IS2404 TaqMan assay for clinical specimens was performed using the primers MU1-new (5′-GAT CAA GCG TTC ACG AGT GA-3′) and MU2 (5′-GGC AGT TAC TTC ACT GCA CA-3′). The 50-μl PCR contained 5 μl 10× PCR buffer with 15 mM MgCl2 (QIAGEN), primers MU1 and MU2 at a final concentration of 1 μM, deoxynucleoside triphosphates (200 μM each) and 2.5 U Taq polymerase (QIAGEN), and 1 ng of M. ulcerans genomic DNA. Amplification was performed in an Eppendorf gradient thermocycler using the following cycling conditions: 1 cycle of 94°C for 4 min, 35 cycles of 94°C for 40 s, 60°C for 40 s, and 72°C for 40 s, and a final extension of 72°C for 5 min. The primers and PCR protocols are those described by the WHO for the molecular diagnosis of M. ulcerans infection (31) and varies from the original protocol described by Ross et al. (23). PCR products were visualized following electrophoresis through a 2% agarose gel, and subsequent purification was performed using the High Pure PCR product purification kit (Roche), according to the manufacturer's instructions.
DNA sequence analysis.
Sequence analysis of purified PCR products was performed using the BigDye Terminator, version 3.1, cycle sequencing kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Reactions were analyzed on an Applied Biosystems 3730S genetic analyzer (Applied Biosystems). Sequence data were edited using Bionumerics, version 4.0 (Applied Maths BVBA, Ghent, Belgium), and database searches of GenBank were performed using the BLASTN algorithm.
TaqMan real-time PCR.
Primers and TaqMan MGB probes (Applied Biosystems) (Table 2) were selected from regions of the sequences for IS2404, IS2606, and KR present on the plasmid pMUM001 (GenBank accession no. BX649209) by using the Primer Express, version 2.0, software program (Applied Biosystems). Probes IS2404TP and KRTP (Table 2) were labeled with the fluorescent dye 6-carboxyfluorescein (FAM) at the 5′ end and a nonfluorescent quencher at the 3′ end (Applied Biosystems). Probe IS2606TP (Table 2) was labeled with the fluorescent dye VIC at the 5′ end and a nonfluorescent quencher at the 3′ end (Applied Biosystems). IS2404 real-time PCR mixtures contained 1 μl of template DNA, 0.9-μM concentrations of each primer, a 0.25 μM concentration of the probe, ABsolute QPCR ROX (500 nM) mix (ABgene), and TaqMan exogenous internal positive control (IPC) reagents (Applied Biosystems) in a total volume of 25 μl. IS2606 and KR assays were performed as a multiplex assay (without IPC). Amplification and detection were performed with the ABI Prism 7000 sequence detection system (Applied Biosystems) using the following program: 1 cycle of 50°C for 2 min, 1 cycle of 95°C for 15 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. DNA extracts were tested in at least duplicate, and negative controls were included in each assay.
TABLE 2.
Primers and probes designed for real-time PCR assays targeting IS2404, IS2606, and KR
| Primer or probea | Sequence (5′-3′) | Nucleotide positionsb | Amplicon size (bp) | Putative gene function (reference) | No. of copies of amplicon per plasmid/chromosomec |
|---|---|---|---|---|---|
| IS2404 TF | AAAGCACCACGCAGCATCT | 27746-27762 | 59 | Transposase (21) | 4/201d |
| IS2404 TR | AGCGACCCCAGTGGATTG | 27787-27804 | |||
| IS2404 TP | 6 FAM-CGTCCAACGCGATC-MGBNFQ | 27768-27781 | |||
| IS2606 TF | CCGTCACAGACCAGGAAGAAG | 28912-28932 | 58 | Transposase (21) | 8/82 |
| IS2606 TR | TGCTGACGGAGTTGAAAAACC | 28947-28969 | |||
| IS2606 TP | VIC-TGTCGGCCACGCCG-MGBNFQ | 28933-28946 | |||
| KRTF | TCACGGCCTGCGATATCA | 3178-3195 | 65 | KR-B domain (20) | 15/0 |
| KRTR | TTGTGTGGGCACTGAATTGAC | 3222-3242 | |||
| KRTP | 6 FAM-ACCCCGAAGCACTG-MGBNFQ | 3199-3212 |
TF, forward primer; TR, reverse primer; TP, probe.
Numbering based on the first copy of the amplicon in pMUM001 (GenBank accession no. BX649209).
Determined by BLAST analysis of the amplicon sequence in WASABI, an in-house, web-interfaced MySQL database for genome analysis.
A total of 171 were identical copies, with an additional 30 copies identified with 1-nucleotide substitutions within the primer sites which would be unlikely to prevent amplification.
Nested PCR protocol for the amplification of variable-number tandem repeat (VNTR) typing in M. ulcerans (MUVNTR) locus 9 in environmental samples.
Previous research has shown that the concentration of M. ulcerans DNA in extracts from environmental samples can be low (24), so a nested protocol was designed to increase the sensitivity of the PCR for VNTR loci which are present in single copies per genome. MUVNTR locus 9 was chosen in the first instance, as the size and nucleotide sequence of the amplicon were specific for M. ulcerans isolates from Victoria and Africa and were different from those of other Australian strains and other mycolactone-producing bacteria. The first-round PCR used primers MUVNTR9NF (5′-ACTGCCCAGACATGGCGA-3′) and MUVNTR9NR (5′-ACGCGAGGTGGAACAAAGC-3′), designed to flank the published VNTR locus 9 primers (2). Amplification was performed in an Eppendorf gradient thermocycler using the following cycling conditions: 1 cycle of 94°C for 2 min, 35 cycles of 94°C for 15 s, 59°C for 30 s, and 72°C for 30 s, and a final elongation of 72°C for 5 min. First-round PCR products were used as templates for a second-round PCR, performed as described by Ablordey et al. (2). PCR products were visualized on a 2% agarose gel, and sizes were estimated by comparing fragment sizes with a 100-bp DNA ladder (Promega). Products of the expected size were purified and sequenced as described above.
Statistical analyses.
Statistical analyses were performed using STATA 8.0 (STATA Corporation, College Station, TX). Median differences in cycle threshold (ΔCT) values (IS2606-IS2404) were compared using the Mann-Whitney nonparametric test.
RESULTS
Development of real-time PCR assays targeting IS2404, IS2606, and KR.
Three TaqMan real-time PCR assays were designed using the Primer Express software package (Applied Biosystems) to detect different regions in the M. ulcerans genome, the insertion sequences IS2404 and IS2606 and the KR domain of the mls genes. These targets were chosen because they were reported to be present in multiple copies in the M. ulcerans genome and are absent in the closely related species Mycobacterium marinum (7, 26). Primers and probes were selected from the IS2404, IS2606, and KR sequences present on the M. ulcerans virulence plasmid pMUM001 (GenBank accession no. BX649209). There are two distinct sequence types among the KR domains on the plasmid, KR-A (3 copies) and KR-B (15 copies). The primers and probe for the KR real-time assay were selected from the KR-B sequence. Table 2 presents the nucleotide positions of the primers and probes designed, the corresponding amplicon sizes, the putative function of the target, and the number of copies of each amplicon on the plasmid and chromosome. Studies of pMUM001 copy number have shown that it is present in one to two copies per cell (28). Thus, the theoretical copy numbers for the IS2404, IS2606, and KR-B target sequences in the genome of M. ulcerans strain Agy99 are 205 to 209, 90 to 98, and 15 to 30 copies per cell, respectively (Table 2).
Sensitivity testing of the IS2404, IS2606, and KR real-time PCR assays.
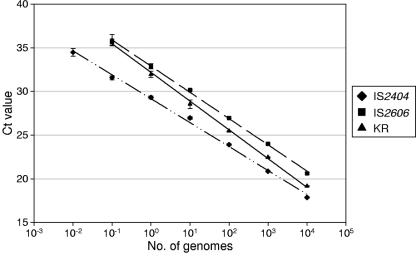
The sensitivities of the IS2404/IPC and the IS2606/KR multiplex assays were tested by performing real-time PCR on dilutions of purified M. ulcerans genomic DNA from strain Agy99 (Table 1). During validation, the IS2606 and KR assays were evaluated with and without multiplexing. The results indicated that multiplexing the two assays did not reduce the sensitivity (data not shown). The standard curves obtained with 10-fold serially diluted genomic DNA preparations were linear over six orders of magnitude for the IS2404 assay and over five orders of magnitude for both the IS2606 and the KR assays (Fig. 1). Based on the complete DNA sequence, the predicted mass of a single copy of the M. ulcerans genome is 5.8 fg (27). Given the actual number of copies of the target in the M. ulcerans genome (Table 2), the IS2404 assay reliably detected 0.01 genomes, equivalent to two copies of the target. As expected, the IS2606 and KR assays were less sensitive due to the presence of fewer target copies per genome, with a reliable detection limit of 0.1 genomes (equivalent to 9 copies of IS2606 and 1.5 to 3 copies of KR-B). It was noted that the ΔCT between the IS2606 and IS2404 assays [ΔCT (IS2606-IS2404)] ranged from 2.77 (for 104 genomes) to 4.1 (for 0.1 genomes), which was slightly higher than expected based on the 2.3-fold difference in target copy numbers (Table 2). The ΔCT between the KR and IS2404 assays increased from 1.3 (for 104 genomes) to 4.3 (for 0.1 genomes) but was generally less than that for the ΔCT (IS2606-IS2404), further indicating that the IS2606 assay is less sensitive than the KR assay. To compare the sensitivity of the IS2404 real-time PCR with the conventional gel-based PCR targeting IS2404 (31), 10-fold dilutions of purified M. ulcerans genomic DNA were tested using both methods. The results indicated that the IS2404 TaqMan real-time PCR is 100 to 1,000 times more sensitive than the conventional single-round gel-based PCR (data not shown).
FIG. 1.
Standard curve generated using a logarithmic scale by the analysis of known amounts of genomic M. ulcerans DNA with the IS2404, IS2606, and KR TaqMan real-time PCR assays. Each 10-fold dilution was performed in quadruplicate, and the means of these replicates were used as data points. One standard deviation on either side of the mean is shown. The regression lines calculated for the data points for the IS2404, IS2606, and KR assays were as follows: y = −1.1892Ln(x) + 29.156, y = −1.3043Ln(x) + 32.886, and y = −1.428Ln(x) + 32.174, respectively. For each assay, the coefficient of correlation was greater than 0.99.
Specificity testing of IS2404, IS2606, and KR real-time PCR assays.
To test the specificity of the assays, we conducted BLASTN searches using the IS2404, IS2606, and KR TaqMan amplicon sequences (Table 2) and performed the IS2404/IPC and IS2606/KR multiplex real-time PCRs on purified genomic DNA from (i) a diverse collection of M. ulcerans isolates, (ii) five other recently described mycolactone-producing mycobacteria (MPM), and (iii) 24 non-MPM (Table 1), including the isolate (ITM 00-1026) previously considered to be a “missing link” between M. ulcerans and M. marinum because it was PCR positive for IS2404 (3). Three isolates of Mycobacterium lentiflavum were included, as it had previously been reported that IS2606 had been detected in a strain of this species (26).
BLASTN searches performed using the target sequences failed to identify significant homology with any other sequences in the NCBI databases. PCR amplification of all three targets was achieved for M. ulcerans isolates from all geographic regions tested, demonstrating that the assays are able to detect M. ulcerans strains from different parts of the world where M. ulcerans disease occurs (Table 1). All three targets were also detected in the other MPM tested, with the exception of the strain designated M. marinum CC240299, for which IS2606 was not detected (Table 1). None of the three targets were detected in any of the 24 non-MPM, including M. lentiflavum (Table 1), demonstrating the specificity of the assays for MPM. It was noteworthy that the “missing link” strain, M. marinum ITM 00-1026 (3), was not detected by the real-time assay targeting IS2404, despite the fact that a PCR product was amplified using the IS2404-specific primers MU1-new and MU2 (31). A BLASTN search of the GenBank database using the sequence amplified from strain ITM 00-1026 (GenBank accession no. EF164897) containing the 59-bp target region revealed only 86% identity with the equivalent region of IS2404 (GenBank accession no. BX649209), with eight nucleotide differences in the 59-bp region targeted in the TaqMan assay.
The ΔCT value for the detection of IS2404 and IS2606 distinguishes between M. ulcerans subsp. ulcerans and other MPM.
The term M. ulcerans subsp. ulcerans has been proposed to describe the M. ulcerans genotypes which cause disease outbreaks in humans, are phylogenetically clustered on the basis of multilocus sequence types (MLST), and have high copy numbers of IS2606. These MLST types have been isolated from Africa (ST17), Southeast Asia and northern Australia (ST18), and southeastern Australia (ST20) (32). Other members of the MPM were shown to have fewer (or no) copies of IS2606 per genome. To determine whether the ΔCT (IS2606-IS2404) could distinguish between isolates with multiple and few copies of IS2606, regardless of the DNA extraction procedure, DNA extracts from isolates, clinical specimens, and environmental samples spiked with M. ulcerans isolate 63/04 were appropriately diluted to give CT IS2404 values of 20 to 25 and assayed for IS2606. ΔCT (IS2606-IS2404) values were then determined (see Table S1 in the supplemental material).
For the M. ulcerans subsp. ulcerans isolates and the DNA preparations from Victorian patient specimens and spiked environmental samples, the median ΔCT value was 2.37 (95% confidence interval, 2.17 to 2.79), indicating a similar ratio of IS2404-to-IS2606 copy number, which has been estimated to be 2.3:1 for the sequenced strain Agy99 (27). However, M. ulcerans strains from China (ST19) and Suriname (ST16), Queensland isolate 20213/91, and the other MPM (ST9, -10, -11, -12, and -14) had consistently higher ΔCT values (median, 7.60; 95% confidence interval, 6.94 to 8.07), indicating fewer copies of IS2606 relative to IS2404. This difference in ΔCT values between the subcluster of M. ulcerans isolates with MLST types ST17, ST18, and ST20 and isolates with the other MLST types was statistically significant (P < 0.001) and represents a valuable means of distinguishing between those M. ulcerans strains causing disease in endotherms (including humans) and other closely related mycobacteria that also harbor IS2404 and IS2606.
Application of the IS2404 TaqMan real-time assay for the detection of M. ulcerans DNA in clinical specimens.
The IS2404 TaqMan assay replaced gel-based PCR as the principal method for laboratory diagnosis of suspected cases of M. ulcerans infection in Victoria in 2003. To evaluate the sensitivity and specificity of the test in a clinical context, we compared real-time PCR results with culture results for primary specimens tested over a 2-year period from July 2003 to June 2005. During that period, we tested a total of 415 primary specimens consisting of 244 swabs (dry or in transport medium), 159 fresh tissue biopsies, 11 paraffin-embedded, fixed-tissue sections and 1 bone specimen. Of these specimens, 410 were from humans (Homo sapiens), 4 were from ringtail possums (Pseudocheirus peregrinus), and 1 specimen was from a koala (Phascolarctos cinereus). Forty-three specimens (10.4%) were PCR positive (22 fresh tissue biopsies, 18 swabs, and 3 paraffin sections). The CT values ranged from 16.01 to 35.52 and averaged 25.83. Of the 40 PCR-positive specimens that were suitable for culture (excluding paraffin-embedded, fixed-tissue sections), 35 were culture positive, indicating that real-time PCR was 12.5% more sensitive than culture. All specimens that were PCR negative were culture negative. More recently, we have been able to detect M. ulcerans DNA in fine-needle aspirates from preulcerative lesions by using the IS2404 real-time PCR assay.
Application of the IS2404, IS2606, and KR real-time assays to the analysis of environmental samples.
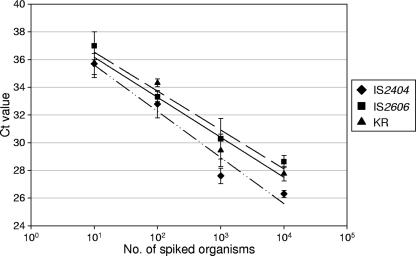
To determine the ability of the assays to detect M. ulcerans DNA in environmental samples, we first performed the IS2404/IPC and IS2606/KR multiplex PCR assays on DNA extracts from mosquito and soil samples that had been spiked with known numbers of M. ulcerans. This served to test both the inhibitory effects of mosquitoes/soil on the real-time PCR assays and the efficiency of the DNA extraction method (FastDNA kit). The results showed that all three targets were detected in DNA extracted from the sample spiked with approximately 10 organisms, with mean CT values of 35.6 for IS2404, 37.0 for IS260, and 36.8 for KR (Fig. 2). A single organism per sample with a CT value of 38 to 40 was detected intermittently by using the IS2404 assay; however, the sensitivities of IS2606 and KR assays were insufficient to detect less than 10 organisms. No inhibition was observed (based on the CT values obtained for the IPC [data not shown]). Similar results were obtained with spiked soil samples extracted using the FastDNA SPIN kit for soil, except that for certain soil samples, PCR inhibitors were present in the DNA extracts, necessitating dilution of the extracts up to 10-fold prior to inclusion in the assay.
FIG. 2.
Standard curve generated using a logarithmic scale by the analysis of DNA extracted from pools of 15 Aedes camptorhynchus mosquitoes spiked with known numbers of M. ulcerans organisms with the IS2404, IS2606, and KR TaqMan real-time PCR assays. Each 10-fold dilution was performed in quadruplicate, and the means of these replicates were used as data points. One standard deviation on either side of the mean is shown.
To test whether the assays could be used to detect M. ulcerans in association with mosquitoes caught in an area where cases of infection were occurring, DNA was extracted from 42 pools of insects trapped in six different locations on the same night. Prior to extraction, mosquitoes were sorted according to species and sex. Pools contained between 1 and 23 mosquitoes. Results indicated that two DNA extracts, from pools of 22 and 23 female Aedes camptorhynchus mosquitoes caught in the same trap, were positive for IS2404, with CT values of 32.4 and 33.0, respectively. These extracts also tested positive for IS2606 and KR, with ΔCT (IS2606-IS2404) values of 2.7 and 2.8, respectively, suggesting that M. ulcerans DNA was being detected and not DNA from other MPM that we have shown to have higher ΔCT values (see Table S1 in the supplemental material). All extracts for which IS2404 was not detected were also negative for IS2606 and KR. DNA extracts from soil and an associated earthworm (class Oligochaeta) collected from a nearby drain were also positive for all three targets, with ΔCT (IS2606-IS2404) values of 3.3 and 2.5, respectively, again suggesting that M. ulcerans DNA was present.
Further evidence that environmental samples contain the outbreak strain of M. ulcerans.
To gain further evidence that the strain being detected in environmental samples by our real-time assays is the Victorian M. ulcerans outbreak strain, we developed a nested PCR targeting MUVNTR locus 9. The nested PCR was at least 10-fold more sensitive than the conventional single-round PCR. This locus was chosen, as the size and nucleotide sequence of the amplicon are specific for M. ulcerans isolates from Victoria and Africa and were different from those of other Australian strains and other MPM. The amplification of a PCR product of the expected size was achieved for the DNA extracts from a mosquito pool, soil from a soak pit, and the earthworm. Sequencing of the PCR products confirmed that they were identical to the sequence determined for locus 9 from Victorian M. ulcerans strains which contain repeats A9 and C9 (1).
DISCUSSION
The number of cases of M. ulcerans disease in Australia continues to rise, with 66 new human cases reported in 2006 compared with 41 in 2005 and 26 in 2004 (4). With increasing numbers of clinical specimens for diagnosis as well as the need to determine the mode of transmission and the natural reservoir of the organism, rapid, sensitive, and specific molecular tests are needed. We have developed two multiplex, TaqMan, real-time PCR assays targeting three independent regions in the M. ulcerans genome for the detection of M. ulcerans DNA in clinical and environmental samples.
A TaqMan assay targeting IS2404 has been described previously (21); however, the sequence used for the design of this assay (GenBank accession no. AF003002) has been shown subsequently to differ from the IS2404 sequence derived from pMUM001 (GenBank accession no. BX649209) and the complete M. ulcerans genome (27). It is now known that primer F1 described by Rondini et al. (21) has a single-nucleotide mismatch, and the probe sequence (P1) has a deletion relative to the true sequence of IS2404. Consequently, the assay described in the present study is more sensitive (with a detection limit of 0.01 genome copies compared with 0.2 genome copies) and robust, particularly when applied to DNA extracts derived from clinical and environmental samples, which may contain potential PCR inhibitors. The sensitivity of the assay targeting the KR-B domain was consistent with the expected number of copies of this target, although the IS2606 assay was slightly less sensitive, with a detection limit of 9 copies rather than 1.5 to 3 copies. Reasons for this were not explored as the assay was robust over a range of sample types and always provided consistent results.
The application of the IS2404 real-time PCR to 415 clinical samples demonstrated that it is a rapid, reliable, sensitive, and specific assay for the diagnosis of M. ulcerans infection using a wide range of clinical specimens, including swabs, biopsies (fresh as well as formalin-fixed, paraffin-sectioned tissue), and fine-needle aspirates. Comparison with culture revealed only five discrepant results. For these five PCR-positive, culture-negative specimens, all lesions were clinically consistent with M. ulcerans infection and the patients had epidemiological links with known regions of endemicity. Possible reasons for the failure of culture could include (i) small numbers of organisms in the specimen (the CT values ranged from 23.2 to 33.3, equivalent to approximately 500 and 0.5 genomes per well, respectively), (ii) delay in receiving the specimen for processing, or (iii) prior antibiotic treatment. As well as being 100 to 1,000 times more sensitive than the conventional gel-based IS2404 PCR, the simpler closed-tube format, which avoids post-PCR handling of samples containing high numbers of amplicons, may make it easier to perform and provide quality assurance in more remote locations than gel-based PCR.
In addition to its important role in the diagnosis of M. ulcerans infection in humans and other animals, the IS2404 real-time PCR has a broad range of applications, including the analysis of environmental samples. Since environmental samples often contain complex mixtures of DNA that may generate false positives, we developed the multiplex TaqMan assay targeting IS2606 and the KR-B domain to complement the specificity of the IS2404 PCR. The ability to quantify the amount of M. ulcerans DNA in the sample using real-time PCR enabled the detection of differences in the relative numbers of copies of IS2404, IS2606, and KR in any given sample. The ΔCT values between the IS2404 and IS2606 assays allowed for the distinction between M. ulcerans subsp. ulcerans isolates with MLST types ST17, ST18, and ST20 and other closely related mycobacteria within the M. ulcerans/M. marinum complex that also harbor IS2404 and IS2606. The results obtained in this study regarding IS2606 copy numbers are consistent with the recent findings of Yip et al. (32).
Using these assays, M. ulcerans DNA was detected in mosquitoes and soil, without the need to use sequence capture, which has been relied on previously (24). The presence of all three targets with the expected ΔCT (IS2606-IS2404), together with the demonstration that the size and sequence of VNTR locus 9 were the same as that amplified from the local outbreak strain, confirms that M. ulcerans DNA consistent with that of the same strain was present in these samples. However, the presence of viable bacteria has yet to be confirmed by culture.
The ability to reliably detect M. ulcerans DNA in mosquito samples was an important first step in our study investigating the hypothesis that mosquitoes play a role in the transmission of M. ulcerans infection, which is the subject of another report (P. D. R. Johnson et al., submitted for publication).
The development of this real-time IS2404 PCR assay for the diagnosis of BU has improved laboratory efficiency and reduced the risk of contamination compared with gel-based methods. Furthermore, the development of a suite of assays targeting multiple regions in the M. ulcerans genome has been a major advance for the screening of environmental samples, which can now be analyzed with much greater confidence and with a reduced risk of false positives due to contamination. This major new tool will assist research into the identification of the natural reservoir, the route of infection, and the mechanism of transmission of this increasingly important environmental pathogen.
Supplementary Material
Acknowledgments
This project was supported by a Victorian State Government Department of Human Services public health research grant (2004-5). DHS Victoria was not involved in study design, study conduct, or manuscript preparation.
We thank Michaela Riddell and the Epidemiology Unit at VIDRL for performing statistical analyses.
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ablordey, A., M. Hilty, P. Stragier, J. Swings, and F. Portaels. 2005. Comparative nucleotide sequence analysis of polymorphic variable-number tandem-repeat loci in Mycobacterium ulcerans. J. Clin. Microbiol. 43:5281-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ablordey, A., J. Swings, C. Hubans, K. Chemlal, C. Locht, F. Portaels, and P. Supply. 2005. Multilocus variable-number tandem repeat typing of Mycobacterium ulcerans. J. Clin. Microbiol. 43:1546-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chemlal, K., G. Huys, F. Laval, V. Vincent, C. Savage, C. Gutierrez, M. Laneelle, J. Swings, W. Meyers, M. Daffe, and F. Portaels. 2002. Characterization of an unusual mycobacterium: a possible missing link between Mycobacterium marinum and Mycobacterium ulcerans. J. Clin. Microbiol. 40:2370-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Department of Human Services. 30 April 2007, accession date. Notifications of infectious diseases. Department of Human Services, Victoria, Australia. http://www.health.vic.gov.au/ideas/downloads/daily_reports/rptVictorianSummary.pdf.
- 5.Eddyani, M., D. Ofori-Adjei, G. Teugels, D. De Weirdt, D. Boakye, W. Meyers, and F. Portaels. 2004. Potential role for fish in transmission of Mycobacterium ulcerans disease (Buruli ulcer): an environmental study. Appl. Environ. Microbiol. 70:5679-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson, K., R. Edwards, D. Leslie, and J. Hayman. 1995. Molecular method for typing Mycobacterium ulcerans. J. Clin. Microbiol. 33:2250-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkin, G., T. Stinear, P. Johnson, and J. Davies. 2003. Subtractive hybridization reveals a type I polyketide synthase locus specific to Mycobacterium ulcerans. J. Bacteriol. 185:6870-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, P., J. Hayman, T. Quek, J. Fyfe, G. Jenkin, J. Buntine, E. Athan, M. Birrell, J. Graham, and C. Lavender. 2007. Consensus recommendations for the diagnosis, treatment and control of Mycobacterium ulcerans infection (Bairnsdale or Buruli ulcer) in Victoria, Australia. Med. J. Aust. 186:64-68. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, P., T. Stinear, P. Small, G. Pluschke, R. Merritt, F. Portaels, K. Huygen, J. Hayman, and K. Asiedu. 2005. Buruli ulcer (M. ulcerans infection): new insights, new hope for disease control. PLoS Med. 2:282-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, P. D., M. G. Veitch, D. E. Leslie, P. E. Flood, and J. A. Hayman. 1996. The emergence of Mycobacterium ulcerans infection near Melbourne. Med. J. Aust. 164:76-78. [DOI] [PubMed] [Google Scholar]
- 11.MacCallum, P., J. Tolhurst, G. Buckle, and H. Sissons. 1948. A new mycobacterial infection in man. J. Pathol. Bacteriol. 60:93-122. [PubMed] [Google Scholar]
- 12.Marsollier, L., T. Severin, J. Aubry, R. Merritt, J. Saint Andre, P. Legras, A. Manceau, A. Chauty, B. Carbonnelle, and S. Cole. 2004. Aquatic snails, passive hosts of Mycobacterium ulcerans. Appl. Environ Microbiol. 70:6296-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsollier, L., T. Stinear, J. Aubry, J. Saint Andre, R. Robert, P. Legras, A. Manceau, C. Audrain, S. Bourdon, H. Kouakou, and B. Carbonnelle. 2004. Aquatic plants stimulate the growth of and biofilm formation by Mycobacterium ulcerans in axenic culture and harbor these bacteria in the environment. Appl. Environ. Microbiol. 70:1097-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mve-Obiang, A., R. E. Lee, E. S. Umstot, K. A. Trott, T. C. Grammer, J. M. Parker, B. S. Ranger, R. Grainger, E. A. Mahrous, and P. L. C. Small. 2005. A newly discovered mycobacterial pathogen isolated from laboratory colonies of Xenopus species with lethal infections produces a novel form of mycolactone, the Mycobacterium ulcerans macrolide toxin. Infect. Immun. 73:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips, R., C. Horsfield, S. Kuijper, A. Lartey, I. Tetteh, S. Etuaful, B. Nyamekye, P. Awuah, K. M. Nyarko, F. Osei-Sarpong, S. Lucas, A. H. Kolk, and M. Wansbrough-Jones. 2005. Sensitivity of PCR targeting the IS2404 insertion sequence of Mycobacterium ulcerans in an assay using punch biopsy specimens for diagnosis of Buruli ulcer. J. Clin. Microbiol. 43:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portaels, F. 1995. Epidemiology of mycobacterial diseases. Clin. Dermatol. 13:207-222. [DOI] [PubMed] [Google Scholar]
- 17.Portaels, F., P. Elsen, A. Guimaraes-Peres, P. Fonteyne, and W. Meyers. 1999. Insects in the transmission of Mycobacterium ulcerans infection. Lancet 353:986. [DOI] [PubMed] [Google Scholar]
- 18.Ranger, B. S., E. A. Mahrous, L. Mosi, S. Adusumilli, R. E. Lee, A. Colorni, M. Rhodes, and P. L. Small. 2006. Globally distributed mycobacterial fish pathogens produce a novel plasmid-encoded toxic macrolide, mycolactone F. Infect. Immun. 74:6037-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes, M., H. Kator, S. Kotob, P. van Berkum, I. Kaattari, W. Vogelbein, F. Quinn, M. Floyd, W. Butler, and C. Ottinger. 2003. Mycobacterium shottsii sp. nov., a slowly growing species isolated from Chesapeake Bay striped bass (Morone saxatilis). Int. J. Syst. Evol. Microbiol. 53:421-424. [DOI] [PubMed] [Google Scholar]
- 20.Rhodes, M., H. Kator, A. McNabb, C. Deshayes, J. Reyrat, B. Brown-Elliott, R. J. Wallace, K. Trott, J. Parker, B. Lifland, G. Osterhout, I. Kaattari, K. Reece, W. Vogelbein, and C. Ottinger. 2005. Mycobacterium pseudoshottsii sp. nov., a slowly growing chromogenic species isolated from Chesapeake Bay striped bass (Morone saxatilis). Int. J. Syst. Evol. Microbiol. 55:1139-1147. [DOI] [PubMed] [Google Scholar]
- 21.Rondini, S., E. Mensah-Quainoo, H. Troll, T. Bodmer, and G. Pluschke. 2003. Development and application of real-time PCR assay for quantification of Mycobacterium ulcerans DNA. J. Clin. Microbiol. 41:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross, B., P. Johnson, F. Oppedisano, L. Marino, A. Sievers, T. Stinear, J. Hayman, M. Veitch, and R. Robins-Browne. 1997. Detection of Mycobacterium ulcerans in environmental samples during an outbreak of ulcerative disease. Appl. Environ. Microbiol. 63:4135-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross, B., L. Marino, F. Oppedisano, R. Edwards, R. Robins-Browne, and P. Johnson. 1997. Development of a PCR assay for rapid diagnosis of Mycobacterium ulcerans infection. J. Clin. Microbiol. 35:1696-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stinear, T., J. Davies, G. Jenkin, J. Hayman, F. Oppedisano, and P. Johnson. 2000. Identification of Mycobacterium ulcerans in the environment from regions in Southeast Australia in which it is endemic with sequence capture-PCR. Appl. Environ. Microbiol. 66:3206-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stinear, T., A. Mve-Obiang, P. Small, W. Frigui, M. Pryor, R. Brosch, G. Jenkin, P. Johnson, J. Davies, R. Lee, S. Adusumilli, T. Garnier, S. Haydock, P. Leadlay, and S. Cole. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. USA 101:1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stinear, T., B. Ross, J. Davies, L. Marino, R. Robins-Browne, F. Oppedisano, A. Sievers, and P. Johnson. 1999. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 37:1018-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stinear, T., T. Seemann, S. Pidot, W. Frigui, G. Reysset, T. Garnier, G. Meurice, D. Simon, C. Bouchier, L. Ma, M. Tichit, J. Porter, J. Ryan, P. Johnson, J. Davies, G. Jenkin, P. Small, L. Jones, T. F., F. Laval, M. Daffé, J. Parkhill, and S. Cole. 2007. Reductive evolution and niche-adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stinear, T. P., M. J. Pryor, J. L. Porter, and S. T. Cole. 2005. Functional analysis and annotation of the virulence plasmid pMUM001 from Mycobacterium ulcerans. Microbiology 151:683-692. [DOI] [PubMed] [Google Scholar]
- 29.van der Werf, T., T. Stinear, Y. Stienstra, W. van der Graaf, and P. Small. 2003. Mycolactones and Mycobacterium ulcerans disease. Lancet 362:1062-1064. [DOI] [PubMed] [Google Scholar]
- 30.Veitch, M., P. Johnson, P. Flood, D. Leslie, A. Street, and J. Hayman. 1997. A large localized outbreak of Mycobacterium ulcerans infection on a temperate southern Australian island. Epidemiol. Infect. 119:313-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization. 6 November 2006, accession date. Diagnosis of Mycobacterium ulcerans disease (Buruli ulcer). World Health Organization, Geneva, Switzerland. http://whqlibdoc.who.int/hq/2001/WHO_CDS_CPE_GBUI_2001.4.pdf.
- 32.Yip, M., J. Porter, J. Fyfe, C. Lavender, F. Portaels, M. Rhodesm, H. Kator, A. Colorni, G. Jenkin, and T. Stinear. 2007. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J. Bacteriol. 189:2021-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.