Abstract
This investigation addresses the following question: what are the important factors for maintenance of a high catabolic capacity under various starvation conditions? Saccharomyces cerevisiae was cultured in aerobic batch cultures, and during the diauxic shift cells were transferred and subjected to 24 h of starvation. The following conditions were used: carbon starvation, nitrogen starvation in the presence of glucose or ethanol, and both carbon starvation and nitrogen starvation. During the starvation period changes in biomass composition (including protein, carbohydrate, lipid, and nucleic acid contents), metabolic activity, sugar transport kinetics, and the levels of selected enzymes were recorded. Subsequent to the starvation period the remaining catabolic capacity was measured by addition of 50 mM glucose. The results showed that the glucose transport capacity is a key factor for maintenance of high metabolic capacity in many, but not all, cases. The results for cells starved of carbon, carbon and nitrogen, or nitrogen in the presence of glucose all indicated that the metabolic capacity was indeed controlled by the glucose transport ability, perhaps with some influence of hexokinase, phosphofructokinase, aldolase, and enolase levels. However, it was also demonstrated that there was no such correlation when nitrogen starvation occurred in the presence of ethanol instead of glucose.
Microorganisms are often exposed to large and sometimes rapid variations in the quality and availability of nutrients (13). This is true in many natural habitats, but large-scale industrial fermentations can also offer similar challenges due to, e.g., imperfect mixing and/or propagation procedures with periods of nongrowth or starvation. Hence, knowledge concerning the physiological responses of cells when energy and/or nutrients are severely limited or depleted is not only of fundamental interest but can be of crucial importance for applied biotechnological processes (39).
The yeast Saccharomyces cerevisiae is a key player in many biotechnological processes, not the least because of its ability to produce and tolerate high levels of ethanol, even under harsh industrial conditions (49). In addition, it is a common host for production of recombinant proteins (34). Finally, S. cerevisiae is frequently used as a model for higher organisms due to its eukaryotic nature (26). In spite of its huge importance for mankind there is still a considerable gap in our knowledge concerning the role and importance of various metabolic adaptations that occur during starvation in S. cerevisiae (14).
For instance, the ability to maintain the high capacity of the catabolic machinery during starvation is a key characteristic in many applications of S. cerevisiae. Irrespective of whether S. cerevisiae is used for fuel (ethanol) production or for manufacturing heterologously expressed products, a high production rate in combination with a high yield is crucial for process economy. However, a high product yield requires no or limited biomass formation, and this usually affects the production rate in a negative way. The mechanism(s) employed for maintaining a high catabolic capacity during conditions of starvation is far from fully understood. It is known that the history of the cells (that is, the condition of the cells when they are subjected to starvation) is important. Stationary-phase cells are in general much more stress and starvation tolerant than exponentially growing cells (45, 53). Similarly, cells originating from the diauxic shift from respirofermentative growth on glucose to respiratory growth on ethanol in batch cultures have superior starvation tolerance compared to cells growing logarithmically on glucose (29, 30). Another important factor for adjustment of catabolic capacity is whether cells are challenged with carbon or nitrogen starvation, but the response is quite complex. Cells from the diauxic shift are much more severely affected by nitrogen starvation than by carbon starvation (31), while aerobic exponentially growing cells respond similarly irrespective of the starvation conditions (37). Cells growing exponentially under anaerobic conditions, on the other hand, lose almost all their catabolic capacity and even viability if they are subjected to carbon starvation (45, 46). In some instances, the reduced catabolic capacity can be attributed to specific events, such as the deterioration of sugar transporters during nitrogen starvation in the presence of a fermentable carbon source (6, 20). Furthermore, the loss of viability and catabolic capacity accompanying carbon starvation in anaerobic cells was due to an insufficient supply of storage carbohydrates and thus a shortage of energy during the adaptation period (44). However, in many conditions it is not known what actually causes the diminished catabolic capacity frequently observed during starvation. Apart from the glucose transport capacity and the level of storage carbohydrates, the concentration of allosteric effectors such as adenine nucleotides and the level of glycolytic enzymes have been put forward as factors that may be important for regulating or controlling glycolytic flux capacity during starvation as well as growth (22, 23, 44, 45, 47, 48). In summary, no single factor can explain the behavior of S. cerevisiae. Instead, the importance of the suggested candidates varies substantially depending on the conditions.
The present investigation was undertaken in order to (i) carefully describe the biomass composition during different types of starvation conditions, (ii) determine what causes the declining metabolic capacity following starvation, and (iii) address the question why different investigations during starvation have resulted in such dramatically different conclusions concerning the importance of sugar uptake capacity. To this end, S. cerevisiae was cultivated in aerobic batch cultures, harvested in the diauxic shift, and subjected to various types of starvation, such as carbon starvation, nitrogen starvation in the presence of a fermentable carbon source (i.e., glucose) or a nonfermentable carbon source (i.e., ethanol), and both carbon starvation and nitrogen starvation. The physiological consequences of such treatments in terms of biomass accumulation and composition, metabolic activity, changes in sugar transport kinetics, and changes in the levels of glycolytic and other key enzymes during the 24-h starvation period were assessed. Subsequent to the starvation period the remaining catabolic capacity was measured by challenging the cells with 50 mM glucose. The resulting sugar consumption rates were compared with the sugar uptake capacity, measured as the zero trans-influx rate (41).
MATERIALS AND METHODS
Strain, media, and culture conditions.
The diploid laboratory strain X-2180aα from the Yeast Genetic Stock Center (Berkeley, CA) was used. The medium contained 1.7 g liter−1 yeast nitrogen base without amino acids and ammonium sulfate, 100 mM glucose as a carbon source, and 5 g liter−1 ammonium sulfate as a nitrogen source in 0.1 M potassium phthalate buffer (pH 5.0). The cells were grown in semiaerobic Erlenmeyer flasks with a liquid volume that was less than 50% of the total volume at 30°C in a rotary shaker. The cultures were started by adding cells from a 24-h inoculum culture, and the cells were grown overnight until 1 h after glucose depletion into the diauxic shift (checked with Diabur-Test 5000 test strips [Roche]). Subsequently, cells were harvested by centrifugation (4°C, 7 min, 5,000 × g), transferred to starvation medium at the original density using a liquid volume that was less than 25% of the total volume, and starved for 24 h ± 0.5 h at 30°C in a rotary shaker.
The starvation media lacked the carbon source, the nitrogen source, or both the carbon and nitrogen sources of the medium. The medium for nitrogen starvation experiments contained either 177 mM glucose, which was sufficient to prevent depletion of glucose during the starvation period (each culture was checked with Diabur-Test 5000 glucose test strips [Roche]), or 345 mM ethanol as the carbon source. The initial sugar concentration for nitrogen starvation was determined in separate experiments to avoid glucose depletion during 24 h of starvation. Equal amounts of cells were transferred to media with different levels of glucose (120, 160, and 290 mM). After 24 h it was found that equal amounts of glucose (106 to 112 mM) had been consumed, irrespective of the initial level. Thus, to avoid depletion of glucose, the initial concentration selected was 177 mM.
Determination of dry weight, ash content, viability, and intracellular volume.
Dry weight was determined as described previously (2), and ash content was determined as recommended by Gurakan et al. (15).
The numbers of viable cells were determined by determining the numbers of CFU on YPD plates (20 g liter−1 peptone, 10 g liter−1 yeast extract, 20 g liter−1 glucose, 20 g liter−1 agar). A 10-fold dilution series (five to seven dilutions) was prepared, and 100-μl portions of the appropriate dilutions were spread on plates. The plates were incubated for 2 days at 30°C before the numbers of colonies were counted. The numbers of cells obtained from CFU determinations were compared with the total numbers counted with a Coulter Counter, and it was found that the percentages of viable cells based on the total numbers were similar for both diauxic shift cells and all types of starved cells with an average viability value of 89%.
In principle, the intracellular volume was determined as described by Larsson et al. (21). Inulin was used as the tracer excluded by the cell wall, and sorbitol was used as the tracer excluded by the cell membrane. The incubation buffer contained morpholineethanesulfonic acid (MES) (10 mM, pH 4.5) and glucose (80 mM). Radioactive tracers (Amersham) were added to buffer as follows: 1 μl ml−1 of 3H2O (185 MBq ml−1) and 2.5 μl ml−1 of inulin [14C]carboxylic acid (592 MBq mmol−1; dissolved to obtain 7.40 MBq ml−1) or 2.5 μl ml−1 of d-[U-14C]sorbitol (7.40 MBq ml−1). Cells from 40 ml of a culture were resuspended in the incubation buffer.
Sampling for determination of nitrogenous compounds, carbohydrates, total fatty acids, and elemental composition of cells.
Depending on the type of analysis, either samples were taken immediately from the cultures or cells were harvested by centrifugation, washed once with cold MilliQ water, and resuspended in synthetic S6 freshwater containing (per liter H2O) 84 mg NaHCO3, 12 mg NaNO3, 11.1 mg CaCl2, 6.0 mg MgSO4, 1.74 mg K2HPO4, and 0.49 mg Na2SiO3. Samples were then taken from the cell suspension. Freeze-dried samples were prepared by centrifugation of 10 to 40 ml (depending on the type of cells) at 4°C for 5 min at 2,500 × g. Cell pellets were frozen in liquid nitrogen, stored at −20°C, and freeze-dried for at least 24 h.
Determination of nitrogen-containing compounds.
Total protein content was measured using a modified biuret method (50), scaled down to suit samples in which cells from 1 ml of culture broth were resuspended in 1 ml of 1 M NaOH and stored at −20°C. A 900-μl aliquot of the cell suspension was boiled for 5 min and cooled in an ice bath, and 300 μl of CuSO4·5H2O (39 g liter−1) was added. The mixed sample was centrifuged (16,000 × g, 5 min), and the absorbance at 555 nm of the supernatant was measured. Bovine serum albumin was used as the standard. In some cases the method of Lowry et al. (24) was used for determination of the protein content. Samples of whole cells were obtained as they were for the biuret method, and samples of crude extracts were mixed 1:1 with 2 M NaOH, yielding 1 M solutions. The thawed samples were boiled for 15 min, cooled for some minutes, and centrifuged, and the supernatant was used in the protein assay. Bovine serum albumin was used as the standard, and measurements were obtained with laboratory robots (COBAS Fara [Roche] or Bio-Mek 2000 laboratory automation workstation [Beckman] equipped with a Fluostar spectrophotometric detector for microtiter plates [BMG Labtechnologies]), with working volumes adapted to the specific robot. The assay used to obtain protein data is indicated below. The biuret assay gave higher values for identical samples, and the conversion factors (grams of protein determined by the biuret method/grams of protein determined by the method of Lowry et al.) for the cell types are as follows: diauxic shift, 1.47; C starved, 1.42; N and C starved, 1.47; N starved with glucose,1.74; and N starved with ethanol, 1.51.
The total cellular RNA content was determined as described previously (4), with some modifications. Cells from 1-ml samples (harvested by centrifugation) were frozen in liquid nitrogen and stored at −20°C. The cells were washed three times with 600 μl of ice-cold 0.7 M HClO4 and resuspended in 300 μl of 0.3 M KOH. During incubation at 37°C for 90 min the RNA was selectively hydrolyzed, after which the samples were cooled in an ice bath and 100 μl of 3 M HClO4 was added. The total volume was adjusted to 1 ml with 0.5 M HClO4, and the supernatant was obtained by centrifugation (16,000 × g, 5 min, 4°C). The RNA was quantified by measurement of absorbance at 260 nm after appropriate dilution with 0.5 M HClO4 (ɛ = 10,800 M−1 cm−1; molecular weight of average nucleotide, 340 g mol−1).
Measurement of free amino acids was generally done as described by Holmes et al. (17), with slight modifications. The cell pellets from 10-ml samples obtained after centrifugation were resuspended in 1 ml of 600 mM ice-cold perchloric acid, mixed rigorously for 2 min, and centrifuged for 5 min. Each supernatant was neutralized with 2 M KCO3 (200 to 220 μl), and the crystals obtained were removed by centrifugation (2 min, 16,000 × g). The samples were frozen in liquid nitrogen and stored at −20°C. Thawed samples were analyzed by staining the amino nitrogen with ninhydrin (12), using glycine as the standard.
Determination of carbohydrates.
Samples for determination of glycogen and trehalose contents (1 ml) were centrifuged, and each cell pellet was stored at −20°C. Extraction (under alkaline conditions with Na2CO3 at 95°C; neutralized with acetic acid/acetate, resulting in 0.2 M Na acetate [pH 5.2]) and hydrolysis were done as described previously (33) using enzyme concentrations of 0.13 to 0.20 U ml−1 of trehalase (pig kidney; Sigma) and 1.6 to 2.4 U ml−1 of amyloglucosidase (Aspergillus niger; Roche). The released glucose was determined using an enzymatic kit assay (Roche) in microtiter plates with a Fluostar spectrophotometric plate reader (BMG Labtechnologies).
For measurement of total carbohydrates, 0.5-ml samples were withdrawn, centrifuged, washed twice with 9 g liter−1 ice-cold NaCl, resuspended in 1 ml of distilled water, and stored at −20°C. The samples were measured after appropriate dilutions with water by the phenol method (16). In some experiments samples were taken from the extracted glycogen-trehalose samples and after appropriate dilution with water were analyzed by the phenol method. These different ways of sampling produced identical results. The measurements were also corrected for the RNA and DNA contents, using average residue weights of 340 and 309 g mol−1, respectively (4, 16).
Determination of fatty acids.
Using 22 to 25 mg of freeze-dried samples, the total fatty acids were determined by extraction with KOH-ethanol as described previously (3) and converting the liberated fatty acids to methyl esters as follows. After evaporation at 45°C under a flow of nitrogen gas, the samples were dissolved in 1 ml toluene, and 1 ml of an acetylchloride-methanol solution (10% acetylchloride in dry methanol) was added. The samples were incubated at 70°C for 2 h, and then 200 μl of MilliQ water was added to separate the toluene and methanol phases. The samples were extracted twice with 2 ml of petroleum ether, and the two extracts were pooled and evaporated at 50°C under a nitrogen atmosphere. The dry samples were dissolved in 300 μl of isooctane, and 400 μg of 17:0 fatty acid and 400 μg of 21:0 fatty acid were used as internal standards. The samples were analyzed by gas chromatography (HP 5890; Hewlett Packard, Avondale, PA) using a DB-wax column (length, 30 m; inside diameter, 0.25 mm; film thickness, 0.25 μm; J&W Scientific, Folsom, CA) and He as the carrier gas. The sample volume was 2 μl, and the split was set to 1:10. The injection temperature was 225°C, and the following temperature gradient was used: 160°C at the start, increasing at a rate of 5°C/min up to 250°C, and then 250°C for 8 min. The fatty acid methyl esters were detected using a flame ionization detector at 300°C.
Determination of elemental composition
Elemental analysis was done using freeze-dried samples with a CHN analyzer and was performed at MikroKemi AB (Uppsala, Sweden).
Determination of sugar uptake kinetics.
Cells were harvested by centrifugation, concentrated five- or sixfold in S6 water, and kept on ice until measurement. The initial glucose uptake rate was measured over 5 s as described previously by Walsh et al. (51). The concentrations of glucose and fructose used were 0.05 to 50 mM dissolved in S6 water, while the mannose concentrations ranged from 0.05 to 100 mM. When fructose uptake and mannose uptake were determined, the quenching buffer contained either 500 mM unlabeled fructose or mannose instead of glucose. The kinetic parameters were determined by nonlinear regression, assuming a single Michaelis-Menten component. The uptake data were checked to verify the existence of only one uptake component.
The uptake assay has been shown to be affected by a lack of ATP (52), indicated by a decreasing rate during the 5-s measurement period. Since depletion of ATP has been reported to be important for activity of starved cells (44, 45), the linearity of the uptake assay was checked for the cells used in this study. Thus, the zero trans-influx rates were determined during a 3- to 12-s time course. The 5-s assay was verified to give constant rates for all cell types for at least 7 s, as described previously (52).
Preparation of crude extracts for determination of enzyme activities.
Crude cell extracts were prepared in KH2PO4 (or Tris-HCl as indicated below) buffer (pH 7.5), as described previously (1), from 50-ml portions of samples taken directly from the cultures. The crude extracts were used immediately for analysis of enzyme activity at 30°C either with a COBAS Fara Autoanalyser (Roche), with a Fluostar spectrophotometric plate reader (BMG Labtechnologies) equipped with an internal reagent pump for addition of the starting substrate, or manually with a Shimadzu spectrophotometer (UV-240 with the option program/interface OPI-1). The extracts used for determination of glycerol phosphate dehydrogenase and glycerol phosphate phosphatase activities were desalted on a PD-10 column (Amersham Pharmacia Biotech) prior to measurement.
Determination of enzyme activities.
The enzymatic activities were measured after appropriate dilution of the extracts with 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (pH 7.0), and the reactions were started by adding one substrate. Enzyme activity (expressed in U [μmol min−1]) was determined from the difference in the slope of NAD(P)H absorbance (340 nm; ɛ = 6.3 mM−1 cm−1) before and after addition of the substrate. The activities were measured in 50 mM PIPES buffer (pH 7.0) with 100 mM KCl and 5 mM MgSO4, using assays partly based on the assays described by Teusink et al. (42), unless stated otherwise. Enzymes were obtained from Roche Diagnostics or Sigma-Aldrich. The contents of the reaction mixtures were as follows. The hexokinase reaction mixture contained 1 mM ATP, 0.4 mM NADP, and 0.9 U ml−1 glucose-6-phosphate dehydrogenase with 10 mM glucose as the substrate; the reaction mixture for the fructose-specific hexokinase activity also contained 3.5 U ml−1 phosphoglucose isomerase with 100 mM fructose as the substrate. The phosphoglucose isomerase reaction mixture contained 0.4 mM NADP and 0.9 U ml−1 glucose-6-phosphate dehydrogenase with 20 mM fructose-6-phosphate as the substrate. The phosphofructokinase reaction mixture contained 20 mM KH2PO4, 0.01 mM fructose-2,6-bisphosphate, 0.15 mM NADH, 3 mM fructose-6-phosphate, 0.85 U ml−1 glycerol-3-phosphate dehydrogenase, 0.45 U ml−1 aldolase, and 2.5 U ml−1 triose phosphate isomerase with 2 mM ATP. The aldolase reaction mixture contained 0.15 mM NADH, 0.85 U ml−1 glycerol-3-phosphate dehydrogenase, and 10 U ml−1 triose phosphate isomerase with 30 mM fructose-1,6-bisphosphate. The triose phosphate isomerase reaction mixture contained 0.9 mM Na2EDTA, 2 mM NAD, 10 mM disodium hydrogen arsenate, and 1.5 U ml−1 glyceraldehyde-3-phosphate dehydrogenase with 10 mM Li-dihydroxyacetone phosphate (based on the description of Nickbarg and Knowles [28]). The glyceraldehyde-3-phosphate dehydrogenase reaction mixture contained 2 mM NAD, 10 mM KH2PO4, 0.9 mM Na2EDTA, and 0.2 mM dithiothreitol (DTT) with 1 mM glyceraldehyde-3-phosphate. The phosphoglycerate kinase reaction mixture contained 0.15 mM NADH, 1 mM ATP, 0.9 mM Na2EDTA, and 1.5 U ml−1 glyceraldehyde-3-phosphate dehydrogenase with 5 mM 3-phosphoglycerate (measured backwards). The phosphoglycerate mutase reaction mixture contained 0.15 mM NADH, 1 mM 2,3-bisphosphoglycerate, 1 mM ADP, 1.9 U ml−1 lactate dehydrogenase, 1 U ml−1 pyruvate kinase, and 0.4 U ml−1 enolase with 5 mM 3-phosphoglycerate (extraction in Tris-HCl). The enolase reaction mixture contained 0.15 mM NADH, 1 mM ADP, 1.9 U ml−1 lactate dehydrogenase, and 1 U ml−1 pyruvate kinase with 1 mM 2-phosphoglycerate (extraction in Tris-HCl). The pyruvate kinase reaction mixture contained 0.15 mM NADH, 2 mM phosphoenolpyruvate, 1 mM fructose-1,6-bisphosphate, and 1.9 U ml−1 lactate dehydrogenase with 25 mM ADP. The pyruvate decarboxylase reaction mixture contained 0.15 mM NADH, 0.33 mM thiamine pyrophosphate, 3.3 mM DTT, and 22 U ml−1 alcohol dehydrogenase with 25 mM pyruvate.
The alcohol dehydrogenase reaction mixture contained 2 mM NAD, 1 mM oxidized glutathione, and 10 mM semicarbazide with 200 mM ethanol. For possible determination of only the isoenzyme Adh2p, 200 mM 2-butanol was used as the substrate (25, 54). The kinetics of alcohol dehydrogenase at alcohol concentrations between 2 and 620 mM was determined by nonlinear regression to possibly distinguish between Adh1p (Km, 45 mM) and Adh2p (Km, 0.7 mM) (42). Neither of these methods showed any activity of Adh2p.
The reaction mixture for acetaldehyde dehydrogenase (NAD or NADP dependent, based on the description of Bruinenberg et al. [5]) contained 0.4 mM NAD or NADP, 15 mM pyrazole, and 0.2 mM DTT with 0.4 mM acetaldehyde.
The reaction mixture for glycerol-3-phosphate dehydrogenase (extraction in Tris-HCl) contained 1 mM Na2EDTA, 1 mM DTT, and 0.15 mM NADH with 5 mM Li-dihydroxyacetone phosphate as the substrate. The reaction mixture did not contain any MgSO4 or KCl.
The activity of glycerol-3-phosphate phosphatase (extraction in Tris-HCl) was determined manually at 30°C with crude extracts in the general buffer (including MgSO4 and KCl) containing 20 mM l-glycerol-3-phosphate. The reaction in aliquots of the reaction mixture (320 μl) was stopped with 80 μl of trichloroacetic acid (250 g liter−1) at appropriate time intervals after extract addition (up to 2.5 min). Subsequent determination of released phosphate was performed as described by Dryer et al. (9), giving the enzymatic activity directly from the rate of phosphate release.
The glucose-6-phosphate dehydrogenase reaction mixture contained 0.4 mM NADP with 5 mM glucose-6-phosphate (based on the descriptions of Kuby and Noltmann [19] and Postma et al. [35]). The phosphoenolpyruvate carboxykinase reaction mixture contained 50 mM NaHCO3 2 mM MnCl2, 2 mM reduced glutathione, 2.5 mM ADP, 0.15 mM NADH, and 3 U ml−1 malate dehydrogenase with 2.5 mM sodium phosphoenolpyruvate (based on the description of de Jong-Gubbels et al. [7] but with the same buffer containing KCl and MgSO4 that was used for the glycolytic enzymes).
Determination of glucose consumption and product formation rates.
Cells were harvested by centrifugation, washed once in cold distilled water, and kept on ice until measurement. Each sample was slightly concentrated in S6 water (prewarmed to 30°C) to allow for sugar addition. In some experiments the cells were already resuspended in the appropriate volume of S6 water and kept on ice until measurement. Each cell suspension was incubated at 30°C for 5 to 10 min and stirred with a magnetic stirrer. The cells were challenged with sugar to obtain a concentration of cells that was the same as the concentration in the original culture sample and a sugar concentration of 50 mM (glucose, fructose, or mannose). Samples were withdrawn at about 2-min intervals for 30 to 35 min for determination of extracellular compounds (using supernatant after centrifugation for 1 min at 16,000 × g) and protein concentration. The cell pellet after centrifugation was resuspended in a volume of 1 M NaOH equal to the volume of the original culture sample for protein determination.
Sugars were measured by endpoint determination of NADPH formation in HEPES buffer (69 mM, pH 7.6) with 1.4 mM NADP, 1.7 mM ATP, and 7 mM MgSO4, using the combined action of 0.05 U ml−1 glucose-6-phosphate dehydrogenase and 1.6 U ml−1 hexokinase for glucose determination. To determine the fructose content, 1.9 U ml−1 phosphoglucose isomerase was also added, and to determine the mannose content, 1.9 U ml−1 phosphoglucose isomerase and 0.5 U ml−1 phosphomannose isomerase were added. In some experiments glucose was measured by a commercial enzymatic kit assay (Roche). Ethanol, glycerol, and acetate concentrations were measured with enzymatic kit assays (Roche). Measurements were obtained by using laboratory robots (COBAS Fara [Roche] and Bio-Mek 2000 laboratory automation workstation [Beckman]) equipped with a Fluostar spectrophotometric detector for microtiter plates (BMG Labtechnologies) or manually in microtiter plates by determining absorbance with the Floustar detector. The working volumes were adapted to the specific robot.
The protein concentrations were averaged and used to normalize the extracellular concentration data. The consumption and production rates were estimated by determining the slope calculated by linear regression using all data from independent cultures. The standard deviations were also obtained in the regression analysis. During the whole time period rates were constant unless indicated otherwise.
Determination of respiration rates.
Cells were harvested by centrifugation, concentrated 10-fold in S6 water, and kept on ice until measurement. Oxygen consumption was measured with an oxygen electrode in a stirred thermostatically controlled closable chamber at 30°C. S6 water was added to the chamber and incubated until the baseline (100% dissolved O2) was stable. Cells were incubated at 30°C for 5 to 10 min. An appropriate amount of cells (maximum, 1/10 of the total volume) was added to the chamber to obtain suitable rates, and the chamber was closed. When the rate was stable enough to obtain the endogenous respiration rate, a small amount of glucose (from a 2.5 M stock solution) was added to obtain 50 mM glucose in the chamber in order to obtain the initial respiration rate with glucose. The zero baseline was obtained by either adding some crystals of NaS2O3 or flushing pure nitrogen gas into the full working volume of liquid in the measuring chamber.
RESULTS
Cells were cultivated in aerobic batch cultures, harvested in the diauxic shift between respirofermentative growth on glucose and respiratory growth on ethanol, and subjected to various forms of starvation for 24 h. The starvation conditions included carbon starvation, nitrogen starvation in the presence of glucose, nitrogen starvation in the presence of ethanol, and both carbon starvation and nitrogen starvation. Cells subjected to carbon starvation or both carbon starvation and nitrogen starvation did not produce any ethanol or glycerol during the starvation period, and there was no consumption of the nitrogen source, NH4+ (when available). Nitrogen-starved cells that had access to a carbon source, on the other hand, showed substantial metabolic activity. In the presence of glucose a sugar consumption rate of 147 μmol g protein−1 min−1 was recorded together with ethanol, glycerol, and acetic acid production rates of 107, 13, and 3 μmol g protein−1 min−1, respectively. Nitrogen starvation using ethanol as a carbon source resulted in an ethanol consumption rate of 77 μmol g protein−1 min−1 and no formation of glycerol.
On average, the number of viable cells in the transition phase, at the time of harvest before starvation, was 1.4 × 1011 cells g protein−1, and there was no increase in this number or the dry weight during carbon starvation. Nitrogen starvation, on the other hand, resulted in increases of 5.2 × 1010 cells g protein−1 (total number, 1.9 × 1011 cells g protein−1) and 4.0 × 1010 cells g protein−1 (total number, 1.8 × 1011 cells g protein−1) after 24 h in the presence of glucose and in the presence of ethanol, respectively, while the dry weight increased by 3.3 and 1.4 g liter−1, respectively. No budding cells were detected after the starvation period irrespective of the conditions.
Changes in biomass composition during different types of starvation.
Nitrogen-starved cells, especially in the presence of glucose, accumulated carbohydrates as well as lipids, although the increase in the latter compounds was statistically significant only at a P value of <0.1 (Table 1). Consequently, as the total amount of protein remained constant during the starvation treatment, there was a reduction in the protein content of the biomass. The increase in carbohydrates could be attributed to accumulation of glycogen and trehalose, while the amount of the remaining carbohydrates, presumably consisting of the structural carbohydrates glucan and mannan, remained fairly constant during the 24-h starvation period (Table 1). The lipid composition was also affected by starvation, and there was an increase in the proportion of unsaturated fatty acids. This was most obvious during nitrogen starvation, but carbon-starved cells and cells starved of both carbon and nitrogen responded in a similar albeit less severe way (Table 1). Nitrogen starvation also provoked a substantial reduction in the content of RNA. However, when nitrogen starvation was combined with carbon starvation, the level of this nucleic acid was close to the levels found following carbon starvation. In fact, the biomass compositions were indistinguishable for carbon-starved cells and cells starved of both carbon and nitrogen; a two-tailed Student t test failed to show any significant difference even at a P value of <0.2. The changes in cellular content induced by starvation resulted in different elemental compositions, as shown by the C molar formulas of cells from different conditions (Table 1). The most significant result was the reduction in the nitrogen content in comparison to the carbon content following nitrogen starvation. A low level of intracellular nitrogenous compounds during nitrogen starvation, especially in the presence of glucose, was also manifested by a very low level of free amino acids in the cells. The dry weight of a single cell before starvation was 18 pg, and the volume was 3.6 ml g protein−1 or 22 fl cell−1. There was in most cases a general trend towards an increasing volume as a consequence of starvation. However, the only significant difference detected (P < 0.01) was found for cells starved of nitrogen in the presence of glucose, where the volume increased to 5.7 ml g protein−1.
TABLE 1.
Composition of cells from the diauxic shift and after subsequent starvation (24 h; carbon, carbon and nitrogen, nitrogen with glucose, nitrogen with ethanol) determined with at least triplicate cultures using duplicate samplesa
| Type of cells | Protein concn (mg [g dry wt]−1)b | Carbohydrate concn (mg [g dry wt]−1)
|
Fatty acid content (lipids)
|
RNA concn (mg [g dry wt]−1)d | Amt of ash (mg [g dry wt]−1) | Total (g [g dry wt]−1)g | Free amino acid concn (mg N [g dry wt]−1) | Elemental compositione | C-mol wt (g dry wt [C-mol]−1)h | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Glycogen | Trehalose | Glucan/mannanc | Total (mg [g dry wt]−1) | 14:0 (mol%) | 16:0 (mol%) | 16:1 (mol%) | 18:0 (mol%) | 18:1 (mol%) | Saturated/unsaturated (g [g dry wt]−1) | ||||||||
| Diauxic shift | 472 | 365 | 31 | 28 | 306 | 49 | 2.03 | 22.3 | 40.7 | 7.2 | 27.8 | 0.452 | 51 | 68.5 | 1.010 | 1.14 | CH1.74O0.57N0.15 | 26.72 |
| C starved | 489 | 385 | 3.2*** | 40 | 342 | 40 | 1.18* | 17.0*** | 42.3 | 6.9 | 32.6*** | 0.330*** | 48 | 65.8d | 1.033 | 0.15* | CH1.72O0.53N0.13 | 25.86 |
| N and C starvedd | 456 | 349 | 3.3*** | 32 | 313 | 34 | 0.93*** | 15.9*** | 40.8 | 7.5 | 34.9*** | 0.317*** | 43 | 68.8 | 0.956 | NDf | ND | ND |
| N starved with glucose | 249*** | 589*** | 115*** | 143*** | 330 | 101 | 1.38 | 15.9*** | 47.0*** | 4.9*** | 30.7 | 0.281*** | 22*** | 72.0d | 1.036 | 0.01* | CH1.81O0.60N0.05 | 25.72 |
| N starved with ethanol | 354*** | 453* | 41 | 94*** | 319 | 76 | 0.77*** | 15.1*** | 44.1 | 5.6 | 34.3*** | 0.271*** | 29*** | 86.7***d | 1.002 | 0.18 | CH1.79O0.59N0.10 | 26.93 |
| SD (%) | 7.2 | 6.1 | 17.0 | 14.6 | 9.8 | 24.0 | 16.8 | 4.2 | 3.3 | 8.4 | 3.4 | 5.1 | 11.5 | 3.3 | 39.5 | |||
The mean standard deviation was estimated using data for all cell types. The effects of the various starvation conditions were statistically evaluated by using a two-tailed Student t test and comparison with nonstarved cells. Significant differences are indicated as follows: three asterisks, P < 0.01; one asterisk, P < 0.05. The degrees of freedom ranged from 14 to 28.
Determined by the modified biuret method.
Total carbohydrates other than glycogen and trehalose.
Determined using duplicate cultures.
Determined using a single culture.
ND, not determined.
Including DNA content, assumed to be 11 mg (g protein)−1 (38) for all cells.
1 C-mol is the amount of the organic compound containing 1 mol, i.e., 12 g, of carbon. C-mol wt is the weight of 1 C-mol of the organic compound.
Changes in hexose uptake kinetics during different types of starvation.
There was a decrease in the hexose uptake capacity and a decrease in affinity as a result of all types of starvation; i.e., there was a decrease in Vmax and an increase in Km during starvation (Table 2). Glucose and fructose yielded similar Vmax values, but the Km was substantially higher for fructose. The most severe reduction in hexose uptake capacity was obtained for cells starved of nitrogen in the presence of glucose (Table 2). On the other hand, if nitrogen starvation was performed in the presence of ethanol, which is a nonfermentable carbon source, a large part of the uptake capacity was preserved. In fact, the Vmax value recorded after nitrogen starvation using ethanol was higher than the corresponding values during carbon starvation and during dual carbon and nitrogen starvation (Table 2).
TABLE 2.
Kinetic parameters of hexose uptake in cells from the diauxic shift and after subsequent starvation (carbon, both carbon and nitrogen, nitrogen with glucose, nitrogen with ethanol)a
| Type of cells |
Vmax (μmol g protein−1 min−1)
|
Km (mM)
|
||
|---|---|---|---|---|
| Glucose | Fructose | Glucose | Fructose | |
| Diauxic shiftb | 502 ± 12 | 455 ± 6 | 2.1 ± 0.2 | 9.2 ± 0.4 |
| C starved | 244 ± 6*** | 198 ± 4*** | 4.7 ± 0.5*** | 13.5 ± 0.7*** |
| N and C starved | 172 ± 4*** | 185 ± 5*** | 4.8 ± 0.5*** | 14.0 ± 1.0*** |
| N starved with glucose | 63 ± 4*** | 82 ± 6*** | 5.0 ± 1.1** | 14.6 ± 2.4* |
| N starved with ethanol | 354 ± 12*** | NDc | 4.3 ± 0.5*** | ND |
The data were determined using at least duplicate cultures and two series of rate measurements. The values are means ± standard deviations of the parameters obtained in the regression. Protein contents were determined using the method of Lowry et al. (24). The effects of the various starvation conditions were statistically evaluated by using a two-tailed Student t test and comparison with nonstarved cells. Significant differences are indicated as follows: three asterisks, P < 0.01; two asterisks, P < 0.02; one asterisk, P < 0.05. The degrees of freedom were always more than 70.
For mannose Vmax was determined to be 552 ± 10 μmol g protein−1 min−1 and Km was determined to be 29 ± 1 mM.
ND, not determined.
Changes in activities of selected enzymes during different types of starvation.
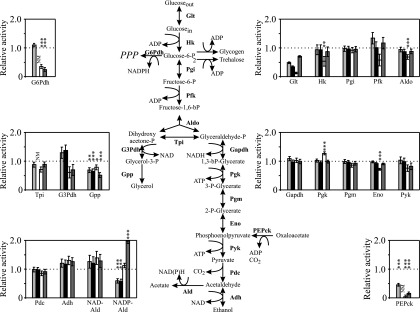
The activities of various enzymes were measured before and after starvation. The values obtained for nonstarved cells are presented in Table A1 in the Appendix, while Fig. 1 shows the activity levels for starved cells in comparison to those for nonstarved cells. Glycolytic enzymes, such as hexokinase, phosphoglucose isomerase, phosphofructokinase, aldolase, triose phosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, phosphoglycerate mutase, enolase, and pyruvate kinase, as well as the ethanol branch enzymes, pyruvate decarboxylase and alcohol dehydrogenase, seemed in most cases to be largely unaffected by the different starvation conditions; i.e., similar activities were recorded before and after starvation irrespective of the conditions (Fig. 1). There was, however, an indication of somewhat lower activities of the enzymes hexokinase, phosphofructokinase, aldolase, and enolase as a consequence of nitrogen starvation in the presence of glucose. An activity that seemed to be critically dependent on starvation and starvation conditions was the activity of acetaldehyde dehydrogenase, which has several NAD- and NADP-coupled isoforms (27). Activities connected to NAD seemed to be resistant, while NADP-coupled activities showed great variation depending on the type of starvation (Fig. 1). Carbon starvation, alone or with nitrogen starvation, induced a reduction, while nitrogen starvation alone, especially in the presence of ethanol, resulted in increased acetaldehyde dehydrogenase activity. Glycerol-3-phosphate dehydrogenase, one of the enzymes in the glycerol branch, showed decreasing activity during nitrogen starvation but preserved or even slightly elevated activity during carbon starvation or dual carbon and nitrogen starvation. The other enzyme of the pathway, glycerol-3-phosphate phosphatase, responded by lower activity with all types of starvation (Fig. 1). Entry into the pentose phosphate pathway is controlled by the enzyme glucose-6-phosphate dehydrogenase, whose activity was severely restricted during nitrogen starvation. Also, the gluconeogenic enzyme phosphoenolpyruvate carboxykinase displayed a drastic decrease in activity during nitrogen starvation, but in contrast to glucose-6-phosphate dehydrogenase, carbon starvation also resulted in a reduction in activity, although not to the same extent (Fig. 1).
TABLE A1.
Enzyme activities in diauxic shift cellsa
| Metabolic pathway | Enzyme | Activity (U g protein−1) (mean ± SD) |
|---|---|---|
| Glycolysis | Hexokinaseb | 1,580 ± 200 |
| Phosphoglucose isomerase | 1,960 ± 110 | |
| Phosphofructokinase | 250 ± 40 | |
| Aldolase | 420 ± 30 | |
| Triose phosphate isomerase | 3,570 ± 440 | |
| Glyceraldehyde phosphate dehydrogenase | 4,750 ± 230 | |
| Phosphoglycerate kinase | 7,840 ± 390 | |
| Phosphoglycerate mutase | 1,070 ± 700 | |
| Enolase | 1,840 ± 120 | |
| Pyruvate kinase | 3,060 ± 250 | |
| Ethanol and acetate branch | Pyruvate decarboxylase | 1,680 ± 120 |
| Alcohol dehydrogenase | 2,650 ± 380 | |
| NAD-aldehyde dehydrogenase | 19 ± 2 | |
| NADP-aldehyde dehydrogenase | 55 ± 4 | |
| Glycerol branch | Glycerol-3-phosphate dehydrogenase | 184 ± 31 |
| Glycerol-3-phosphate phosphatase | 106 ± 18 | |
| Others | Glucose-6-phosphate dehydrogenase | 115 ± 7 |
| Phosphoenolpyruvate carboxykinase | 118 ± 9 |
Means were determined using two to four independently grown cultures, and the mean standard deviations were estimated using also the data for starved cells shown in Fig. 1. Protein contents were determined using the method of Lowry et al. (24).
The ratio of the activity with fructose as the substrate to the activity with glucose as the substrate was 1.62.
FIG. 1.
Activities of selected enzymes. The influence of starvation on the activities is expressed as ratios of activity after 24 h of starvation to activity in nonstarved cells. Light gray columns, carbon starvation; black columns, dual carbon and nitrogen starvation; open columns, nitrogen starvation in the presence of glucose; dark gray columns, nitrogen starvation in the presence of ethanol. The data are averages for two to four independently grown cultures, and the mean standard deviations, estimated using data for all cell types, are indicated by error bars. Abbreviations: G6Pdh, glucose-6-phosphate dehydrogenase; Glt, glucose transport; Hk, hexokinase; Pgi, phosphoglucose isomerase; Pfk, phosphofructokinase; Aldo, aldolase; Tpi, triose phosphate isomerase; G3Pdh, glycerol-3-phosphate dehydrogenase; Gpp, glycerol-3-phosphate phosphatase; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; Pgk, phosphoglycerate kinase; Pgm, phosphoglycerate mutase; Eno, enolase; Pyk, pyruvate kinase; Pdc, pyruvate decarboxylase; Adh, alcohol dehydrogenase; NAD-Ald, NAD-acetaldehyde dehydrogenase; NADP-Ald, NADP-acetaldehyde dehydrogenase; PEPck, phosphoenolpyruvate carboxykinase; NM, not measured. The effects of the various starvation conditions were statistically evaluated by using a two-tailed Student t test and comparison with nonstarved cells. Significant differences at P values of <0.01 (three asterisks), <0.02 (two asterisks), and <0.05 (one asterisk) are indicated. The degrees of freedom ranged from 4 to 22.
Changes in catabolic capacity of S. cerevisiae during different types of starvation.
The catabolic capacity after 24 h of starvation was assessed by transferring the cells to S6 water (see Materials and Methods) and adding 50 mM (final concentration) glucose. Subsequently, the initial respiratory rates, glucose consumption rates, and rates of formation of ethanol, glycerol, and acetate were measured. To a large extent, the initial respiratory capacity was preserved during the starvation period for all different starvation conditions (Table 3). The lowest values were recorded for cells starved of nitrogen in the presence of glucose and after dual carbon and nitrogen starvation, but in these cases more than 50% of the original activity was maintained. It should also be mentioned that carbon-starved cells, as well as dual carbon- and nitrogen-starved cells, had very low endogenous respiratory activity (i.e., respiratory activity without external addition of glucose). The explanation for this is probably that these cells had very low levels of internal sources that could be mobilized rapidly, such as storage carbohydrates (Table 1). However, as soon as glucose was added, these cells immediately exhibited respiratory activity that was more or less the same as that of their nonstarved counterparts (Table 3). Ethanol production or fermentative capacities were much higher than respiration capacities both before and after starvation (Table 3). There was, however, a severe reduction in the ethanol-forming capacity when glucose was used in conjunction with nitrogen starvation. Although there was a reduction in fermentative capacity during the other starvation conditions tested, it was not at all to the same extent. This was also reflected in the level of glucose consumption, whose values were lowest when nitrogen starvation occurred in the presence of glucose (Table 3). Similarly, this condition also resulted in the most severe reduction in glycerol-forming activity. A different picture emerged when the ability to form acetate was examined. For some reason, nitrogen starvation together with glucose induced an acetic acid production capacity that was more than twice the capacity before starvation (Table 3).
TABLE 3.
Steady-state glucose consumption rates, rates of production of ethanol, glycerol, and acetate, and initial oxygen consumption rates after addition of 50 mM glucose to cells harvested in the diauxic shift and cells starved for 24 h for carbon, carbon and nitrogen, nitrogen with glucose, or nitrogen with ethanola
| Type of cells | Rate (μmol g protein−1 min−1)
|
||||
|---|---|---|---|---|---|
| Glucose consumption | Ethanol productionb | Glycerol production | Acetic acid production | Oxygen consumptionc | |
| Diauxic shift | 294 ± 25d | 388 ± 6 | 32 ± 3 | 4.0 ± 0.6 | 74 |
| C starved | 250 ± 22d | 267 ± 5***e | 16 ± 2*** | 0***f | 62 |
| N and C starved | 185 ± 22*** | 211 ± 6*** | NDg | ND | 43* |
| N starved with glucose | 81 ± 18***d | 99 ± 2*** | 5.8 ± 0.3*** | 8.5 ± 0.4*** | 41*** |
| N starved with ethanol | 183 ± 28*** | 289 ± 5*** | 34 ± 2 | 0.7 ± 0.3*** | 70 |
Each rate was calculated using data from duplicate independent cultures in one linear regression of the slope, and the values are means ± standard deviations from the regression. Protein contents were determined using the method of Lowry et al. (24). The effects of the various starvation conditions were statistically evaluated by using a two-tailed Student t test and comparison with nonstarved cells. Significant differences are indicated as follows: three asterisks, P < 0.01; one asterisk, P < 0.05. The degrees of freedom ranged from 27 to 114.
For C-starved cells and N- and C-starved cells the first points at 0 to 5 min were excluded because of nonlinearity.
The mean standard deviation for the oxygen consumption rate was 15% and was estimated using data for all cell types.
Determined using data from four cultures.
Determined using data from three cultures.
Before the steady-state level was reached with neither consumption nor production, there was accumulation and then consumption of acetate during the first 20 min.
ND, not determined.
Role of hexose transport capacities in overall glucose consumption rates after different types of starvation.
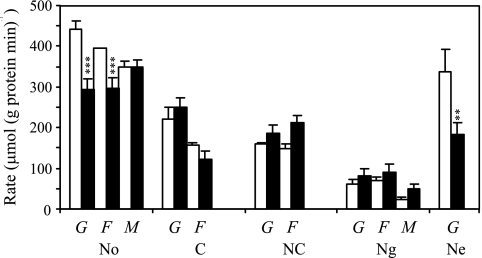
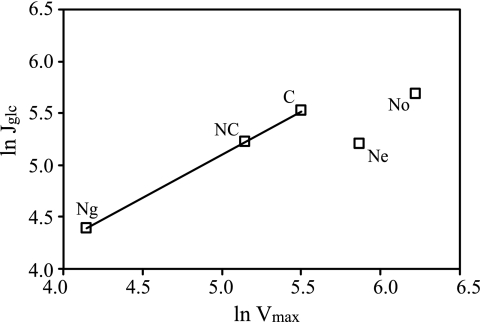
A comparison was made between the sugar consumption rate and the sugar uptake capacity (measured as the zero trans-influx rate) of starved and nonstarved cells subjected to an extracellular sugar concentration of 50 mM (Fig. 2). In most cases, the sugar consumption rate was very close to the uptake capacity irrespective of what sugar was tested (i.e., glucose, fructose, or mannose) or from what type of conditions the cells originated. A notable exception was cells that were not starved or were starved for nitrogen in the presence of ethanol, which showed a glucose uptake capacity much higher than the recorded consumption rates (Fig. 2). This suggests that the uptake capacity is not the limiting factor for glucose consumption and does not exert metabolic flux control in these cells. A plot of the natural logarithm of the fluxes versus the natural logarithm of the maximal uptake rates is a way in metabolic control analysis to determine the control coefficient of a certain enzymatic step in a metabolic network. The control coefficient indicates how large an influence the enzyme has on the overall flux (Fig. 3). The high value (0.83) of the slope in our case indicates a high degree of control (a slope of 1 means full control) for sugar uptake on glycolytic flux when diauxic shift cells have been starved for carbon, both carbon and nitrogen, or nitrogen in the presence of glucose.
FIG. 2.
Comparison between zero trans-influx rates (open columns) and consumption rates (black columns) at an extracellular sugar concentration of 50 mM in cells from the diauxic shift (No) and after subsequent starvation (C, carbon starvation; NC, dual nitrogen and carbon starvation; Ng, nitrogen starvation with glucose; Ne, nitrogen starvation with ethanol). The sugars used were glucose (G), fructose (F), and mannose (M). Glucose consumption rates were obtained from Table 3, the zero trans-influx rates were measured by determining [14C]glucose uptake, and the protein content was measured by the method of Lowry et al. (24). The standard deviations are indicated by error bars. The differences between sugar consumption rates and uptake capacities were statistically evaluated by using a two-tailed Student t test. Significantly lower consumption rates compared to the uptake capacity are indicated by three asterisks (P < 0.01) and two asterisks (P < 0.02). The degrees of freedom were always more than 30.
FIG. 3.
Plot of influx of glucose (Jglc) versus the maximal initial uptake rate (Vmax) for the diauxic shift (No) and for starved cells (C, carbon starvation; NC, dual nitrogen and carbon starvation; Ng, nitrogen starvation with glucose; Ne, nitrogen starvation with ethanol). The linear relationship for carbon-starved cells, nitrogen- and carbon-starved cells, and nitrogen-starved cells in the presence of glucose is indicated, with a slope of 0.83 (R2 = 0.999).
DISCUSSION
The ability to maintain a metabolic capacity when a nutrient and/or energy source was abruptly removed from cells was critically dependent on the starvation conditions. Nitrogen starvation was much more detrimental in this respect than carbon starvation. There have been conflicting results concerning the importance of the glucose uptake capacity for the overall glucose consumption rate obtained after starvation. In the classical work of Lagunas et al. (6, 20) it was shown that the glucose transport capacity deteriorated when cells of S. cerevisiae were subjected to nitrogen starvation in the presence of a fermentable carbon source, such as glucose. Hence, under such conditions the (remaining) glucose uptake capacity determines the overall glucose consumption rate. The view that glucose transport capacity to a large extent determines the glycolytic flux in yeast is not limited to starved cells but is also frequently expressed concerning actively growing cells (8, 32, 37). However, others have questioned this view and suggested or shown that other factors, such as glycolytic enzyme activities, are important as well (10, 18). The truth of the matter is probably that the importance of glucose transport characteristics for overall glucose consumption rates varies with the growth conditions as well as the phase of growth. A glucose-limited chemostat culture offers very different conditions than unlimited, glucose-excess growth at the maximum growth rate in a batch culture. Similarly, in a batch culture using glucose as the carbon source there is a progressive change in conditions from exponential respirofermentative growth on glucose to the diauxic shift and subsequently exponential growth on ethanol.
There has been a similar discrepancy in opinion concerning the importance of the glucose transport capacity for starved cells. While some reports have stressed the role of glucose transport for glycolytic flux control, at least following nitrogen starvation (36, 37), other reports have considered this factor more or less unimportant (31, 44). It is now clear that that this also depends very much on the conditions encountered during the starvation period. The glucose consumption capacity was almost entirely governed by the uptake capacity of cells subjected to carbon starvation, carbon and nitrogen starvation, and nitrogen starvation in the presence of glucose (Fig. 2 and 3). On the other hand, cells starved of nitrogen in the presence of ethanol or nonstarved cells did not show any correlation between consumption rate and glucose uptake capacity. It should be noted that this is in agreement with the earlier observations reported by Nilsson et al. (31) and by Rossell et al. (37), since the former used ethanol and the latter used glucose during nitrogen starvation experiments.
Not only glucose transport capacities but also the levels of glycolytic and/or ethanol-forming enzymes have been shown to be important for the rate of glycolysis, at least under certain conditions (11, 18, 48). However, most studies have failed to show any strict correlation between levels of enzymes and flux through the glycolytic pathway (22, 31, 40, 43, 47). Similarly, most attempts to increase the ethanol production rate in S. cerevisiae by overexpression of a single or several glycolytic enzymes have been unsuccessful. Also, the present study failed to show any clear-cut correlation between enzyme levels and glycolytic capacity (Fig. 1), although two of the enzymes regarded as key components for glycolytic flux control, hexokinase and phosphofructokinase, did indeed show their lowest expression during nitrogen starvation in the presence of glucose, conditions which also provoked the largest reduction in glycolytic capacity (Table 3).
Another factor that is potentially important for preservation of viability and metabolic capacities during starvation is the level of endogenous carbon and energy sources, such as glycogen and trehalose. Access to storage carbohydrates as a source of energy during transfer to carbon and energy starvation conditions was proven to be of crucial importance for successful adaptation of anaerobic cells (44, 45). Under aerobic conditions, on the other hand, it does not seem that the levels of storage carbohydrates are critical for performance during starvation (31). Probably, this is due to the fact that many other cellular constituents (e.g., proteins and lipids) may serve as energy sources under aerobic (but not anaerobic) conditions. In agreement with this, no positive correlation could be detected between storage carbohydrate content and the ability to withstand starvation in the present investigation performed aerobically (Tables 1 and 3). However, starvation in the presence of a potential energy source (e.g., glucose or ethanol) did indeed provoke massive accumulation of not only storage carbohydrates but also fatty acids. These compounds, especially fatty acids, are very energy rich and could fulfill maintenance requirements for a long time. Hence, this provides an advantage to the cells during conditions of growth arrest and might lead to prolonged survival. Access to an external energy source during growth arrest seems to be the key determinant for the cellular response to starvation. This was reflected in the fact that additional nitrogen starvation in combination with carbon starvation resulted in almost no significant change in the cellular response compared to the response observed with carbon starvation. There was, however, a significant difference compared to cells starved of nitrogen in the presence of a carbon and energy source.
To summarize, the metabolic capacity of cells aerobically starved of carbon, carbon and nitrogen, or nitrogen in the presence of glucose is controlled by the glucose transport capacity, perhaps with some influence of hexokinase and/or phosphofructokinase and/or aldolase and/or enolase activities. It is still not known what governs the metabolic capacity during nitrogen starvation in the presence of ethanol.
Acknowledgments
The Swedish Energy Administration (Energimyndigheten) is acknowledged for financial support. The support to L.G. by the Chalmers Bioscience initiative is also acknowledged.
The hospitality of and valuable help provided by Karel van Dam (Amsterdam, The Netherlands) in introducing E.A. to the world of “zero trans-influx rate” measurements are very much appreciated.
APPENDIX
The activities of various enzymes for nonstarved cells are shown in Table A1.
Footnotes
Published ahead of print on 1 June 2007.
REFERENCES
- 1.Albers, E., V. Laize, A. Blomberg, S. Hohmann, and L. Gustafsson. 2003. Ser3p (Yer081wp) and Ser33p (Yil074cp) are phosphoglycerate dehydrogenases in Saccharomyces cerevisiae. J. Biol. Chem. 278:10264-10272. [DOI] [PubMed] [Google Scholar]
- 2.Albers, E., C. Larsson, G. Lidén, C. Niklasson, and L. Gustafsson. 1996. Influence of the nitrogen source on Saccharomyces cerevisiae anaerobic growth and product formation. Appl. Environ. Microbiol. 62:3187-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andlid, T., C. Larsson, C. Liljenberg, I. Marison, and L. Gustafsson. 1995. Enthalpy content as a function of lipid accumulation in Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 42:818-825. [Google Scholar]
- 4.Benthin, S., J. Nielsen, and J. Villadsen. 1991. A simple and reliable method for the determination of cellular RNA content. Biotechnol. Tech. 5:39-42. [Google Scholar]
- 5.Bruinenberg, P. M., J. P. van Dijken, and W. A. Scheffers. 1983. An enzymic analysis of NADPH production and consumption in Candida utilis. J. Gen. Microbiol. 129:965-971. [DOI] [PubMed] [Google Scholar]
- 6.Busturia, A., and R. Lagunas. 1986. Catabolite inactivation of the glucose transport system in Saccharomyces cerevisiae. J. Gen. Microbiol. 132:379-385. [DOI] [PubMed] [Google Scholar]
- 7.de Jong-Gubbels, P., P. Vanrolleghem, S. Heijnen, J. P. van Dijken, and J. T. Pronk. 1995. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast 11:407-418. [DOI] [PubMed] [Google Scholar]
- 8.Diderich, J. A., B. Teusink, J. Valkier, J. Anjos, I. Spencer-Martins, K. van Dam, and M. C. Walsh. 1999. Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast Saccharomyces bayanus. Microbiology 145:3447-3454. [DOI] [PubMed] [Google Scholar]
- 9.Dryer, R. L., A. R. Tammes, and J. I. Routh. 1957. The determination of phosphorus and phosphatase with N-phenyl-p-phenylenediamine. J. Biol. Chem. 225:177-183. [PubMed] [Google Scholar]
- 10.Elbing, K., C. Larsson, R. M. Bill, E. Albers, J. L. Snoep, E. Boles, S. Hohmann, and L. Gustafsson. 2004. Role of hexose transport in control of glycolytic flux in Saccharomyces cerevisiae. Appl. Environ Microbiol. 70:5323-5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbing, K., A. Ståhlberg, S. Hohmann, and L. Gustafsson. 2004. Transcriptional responses to glucose at different glycolytic rates in Saccharomyces cerevisiae. Eur. J. Biochem. 271:4855-4864. [DOI] [PubMed] [Google Scholar]
- 12.European Brewery Convention. 1987. Free amino nitrogen, p. E141-E142. In Analytica-EBC, 4th ed. European Brewery Convention, Zurich, Switzerland.
- 13.Gagiano, M., F. F. Bauer, and I. S. Pretorius. 2002. The sensing of nutritional status and the relationship to filamentous growth in Saccharomyces cerevisiae. FEMS Yeast Res. 2:433-470. [DOI] [PubMed] [Google Scholar]
- 14.Gray, J. V., G. A. Petsko, G. C. Johnston, D. Ringe, R. A. Singer, and M. Werner-Washburne. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurakan, T., I. W. Marison, U. von Stockar, L. Gustafsson, and E. Gnaiger. 1990. Proposals for a standardized sample handling procedure for the determination of elemental composition and enthalpy of combustion of biological material. Thermochim. Acta 172:251-266. [Google Scholar]
- 16.Herbert, D., P. J. Phipps, and R. E. Strange. 1971. Chemical analysis of microbial cells. Methods Microbiol. 5B:209-344. [Google Scholar]
- 17.Holmes, A. R., G. S. McNaughton, R. D. More, and M. G. Shepherd. 1991. Ammonium assimilation by Candida albicans and other yeasts: a 13N isotope study. Can. J. Microbiol. 37:226-232. [DOI] [PubMed] [Google Scholar]
- 18.Jansen, M. L., J. A. Diderich, M. Mashego, A. Hassane, J. H. de Winde, P. Daran-Lapujade, and J. T. Pronk. 2005. Prolonged selection in aerobic, glucose-limited chemostat cultures of Saccharomyces cerevisiae causes a partial loss of glycolytic capacity. Microbiology 151:1657-1669. [DOI] [PubMed] [Google Scholar]
- 19.Kuby, S. A., and E. A. Noltmann. 1966. Glucose 6-phosphate dehydrogenase (crystalline) from brewers’ yeast. Methods Enzymol. 9:116-125. [Google Scholar]
- 20.Lagunas, R., C. Dominguez, A. Busturia, and M. J. Saez. 1982. Mechanism of appearance of the Pasteur effect in Saccharomyces cerevisiae: inactivation of sugar transport systems. J. Bacteriol. 152:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsson, C., C. Morales, L. Gustafsson, and L. Adler. 1990. Osmoregulation of the salt-tolerant yeast Debaryomyces hansenii grown in a chemostat at different salinities. J. Bacteriol. 172:1769-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson, C., A. Nilsson, A. Blomberg, and L. Gustafsson. 1997. Glycolytic flux is conditionally correlated with ATP concentration in Saccharomyces cerevisiae: a chemostat study under carbon- or nitrogen-limiting conditions. J. Bacteriol. 179:7243-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson, C., I.-L. Påhlman, and L. Gustafsson. 2000. The importance of ATP as a regulator of glycolytic flux in Saccharomyces cerevisiae. Yeast 16:797-809. [DOI] [PubMed] [Google Scholar]
- 24.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 25.Lutstorf, U., and R. Megnet. 1968. Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae. I. Physiological control of ADH-2 and properties of ADH-2 and ADH-4. Arch. Biochem. Biophys. 126:933-944. [DOI] [PubMed] [Google Scholar]
- 26.Mustacchi, R., S. Hohmann, and J. Nielsen. 2006. Yeast systems biology to unravel the network of life. Yeast 23:227-238. [DOI] [PubMed] [Google Scholar]
- 27.Navarro-Avino, J. P., R. Prasad, V. J. Miralles, R. M. Benito, and R. Serrano. 1999. A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast 15:829-842. [DOI] [PubMed] [Google Scholar]
- 28.Nickbarg, E. B., and J. R. Knowles. 1988. Triosephosphate isomerase: energetics of the reaction catalyzed by the yeast enzyme expressed in Escherichia coli. Biochemistry 27:5939-5947. [DOI] [PubMed] [Google Scholar]
- 29.Nilsson, A., C. Larsson, and L. Gustafsson. 1995. Catabolic capacity of Saccharomyces cerevisiae in relation to the physiological state and maintenance requirement. Thermochim. Acta 250:233-245. [Google Scholar]
- 30.Nilsson, A., J. Norbeck, R. Oelz, A. Blomberg, and L. Gustafsson. 2001. Fermentative capacity after cold storage of bakers’ yeast is dependent on the initial physiological state but not correlated to the levels of glycolytic enzymes. Int. J. Food Microbiol. 71:111-124. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson, A., I.-L. Påhlman, P.-Å. Jovall, A. Blomberg, C. Larsson, and L. Gustafsson. 2001. The catabolic capacity of Saccharomyces cerevisiae is preserved to a higher extent during carbon compared to nitrogen starvation. Yeast 18:1371-1381. [DOI] [PubMed] [Google Scholar]
- 32.Oehlen, L. J. W. M., M. E. Scholte, W. de Konig, and K. van Dam. 1994. Decrease in glycolytic flux in Saccharomyces cerevisiae cdc35-1 cells at restrictive temperature correlates with a decrease in glucose transport. Microbiology 140:1891-1898. [DOI] [PubMed] [Google Scholar]
- 33.Parrou, J. L., and J. Francois. 1997. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 248:186-188. [DOI] [PubMed] [Google Scholar]
- 34.Porro, D., M. Sauer, P. Branduardi, and D. Mattanovich. 2005. Recombinant protein production in yeasts. Mol. Biotechnol. 31:245-259. [DOI] [PubMed] [Google Scholar]
- 35.Postma, E., C. Verduyn, W. A. Scheffers, and J. P. Van Dijken. 1989. Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 55:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossell, S., C. C. van der Weijden, A. Kruckeberg, B. M. Bakker, and H. V. Westerhoff. 2002. Loss of fermentative capacity in baker's yeast can partly be explained by reduced glucose uptake capacity. Mol. Biol. Rep. 29:255-257. [DOI] [PubMed] [Google Scholar]
- 37.Rossell, S., C. C. van der Weijden, A. L. Kruckeberg, B. M. Bakker, and H. V. Westerhoff. 2005. Hierarchical and metabolic regulation of glucose influx in starved Saccharomyces cerevisiae. FEMS Yeast Res. 5:611-619. [DOI] [PubMed] [Google Scholar]
- 38.Schulze, U. 1995. Anaerobic physiology of Saccharomyces cerevisiae. Ph.D. thesis. Technical University of Denmark, Lyngby.
- 39.Schweder, T., and M. Hecker. 2004. Monitoring of stress responses. Adv. Biochem. Eng. Biotechnol. 89:47-71. [DOI] [PubMed] [Google Scholar]
- 40.Sierkstra, L. N., J. M. A. Verbakel, and C. T. Verrips. 1992. Analysis of transcription and translation of glycolytic enzymes in glucose-limited continuous cultures of Saccharomyces cerevisiae. J. Gen. Microbiol. 138:2559-2566. [DOI] [PubMed] [Google Scholar]
- 41.Teusink, B., J. A. Diderich, H. V. Westerhoff, K. van Dam, and M. C. Walsh. 1998. Intracellular glucose concentration in derepressed yeast cells consuming glucose is high enough to reduce the glucose transport rate by 50%. J. Bacteriol. 180:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teusink, B., J. Passarge, C. A. Reijenga, E. Esgalhado, C. C. van der Weijden, M. Schepper, M. C. Walsh, B. M. Bakker, K. van Dam, H. V. Westerhoff, and J. L. Snoep. 2000. Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur. J. Biochem. 267:5313-5329. [DOI] [PubMed] [Google Scholar]
- 43.Thomsson, E. 2005. Glycolytic flux regulation in Saccharomyces cerevisiae during anaerobic growth and starvation. Ph.D. thesis. Chalmers University of Technology, Göteburg, Sweden.
- 44.Thomsson, E., L. Gustafsson, and C. Larsson. 2005. Starvation response of Saccharomyces cerevisiae grown in anaerobic nitrogen- or carbon-limited chemostat cultures. Appl. Environ. Microbiol. 71:3007-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomsson, E., C. Larsson, E. Albers, A. Nilsson, C. J. Franzen, and L. Gustafsson. 2003. Carbon starvation can induce energy deprivation and loss of fermentative capacity in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:3251-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomsson, E., M. Svensson, and C. Larsson. 2005. Rapamycin pre-treatment preserves viability, ATP level, and catabolic capacity during carbon starvation of Saccharomyces cerevisiae. Yeast 22:615-623. [DOI] [PubMed] [Google Scholar]
- 47.van Hoek, P., J. P. van Dijken, and J. T. Pronk. 2000. Regulation of fermentative capacity and levels of glycolytic enzymes in chemostat cultures of Saccharomyces cerevisiae. Enzyme Microb. Technol 26:724-736. [DOI] [PubMed] [Google Scholar]
- 48.van Hoek, W. P. M., J. P. van Dijken, and J. T. Pronk. 1998. Effect of specific growth rate on fermentative capacity of bakers’ yeast. Appl. Environ. Microbiol. 64:4226-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Maris, A. J., D. A. Abbott, E. Bellissimi, J. van den Brink, M. Kuyper, M. A. Luttik, H. W. Wisselink, W. A. Scheffers, J. P. van Dijken, and J. T. Pronk. 2006. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Leeuwenhoek 90:391-418. [DOI] [PubMed] [Google Scholar]
- 50.Verduyn, C. 1991. Physiology of yeasts in relation to biomass yields. Antonie Leeuwenhoek 60:325-353. [DOI] [PubMed] [Google Scholar]
- 51.Walsh, M. C., H. P. Smits, M. Scholte, and K. van Dam. 1994. Affinity of glucose transport in Saccharomyces cerevisiae is modulated during growth on glucose. J. Bacteriol. 176:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walsh, M. C., H. P. Smits, and K. van Dam. 1994. Respiratory inhibitors affect incorporation of glucose into Saccharomyces cerevisiae cells, but not the activity of glucose transport. Yeast 10:1553-1558. [DOI] [PubMed] [Google Scholar]
- 53.Werner-Washburne, M., E. Braun, G. Johnston, and R. Singer. 1993. Stationary phase in Saccharomyces cerevisiae. Microbiol. Rev. 57:384-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiesenfeld, M., L. Schimpfessel, and R. Crokaert. 1975. Multiple forms of mitochondrial alcohol dehydrogenase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 405:500-512. [DOI] [PubMed] [Google Scholar]