Abstract
Burkholderia cepacia MBA4 is a bacterium that can utilize 2-haloacids as carbon and energy sources for growth. It has been proposed that dehalogenase-associated permease mediates the uptake of haloacid. In this paper, we report the first cloning and characterization of such a haloacid permease. The structural gene, designated deh4p, was found 353 bases downstream of the dehalogenase gene deh4a. Quantitative analysis of the expression of deh4p showed that it was induced by monochloroacetate (MCA), to a level similar to the MCA-induced level of deh4a. The nucleotide sequence of deh4p was determined, and an open reading frame of 1,656 bp encoding a putative peptide of 552 amino acids was identified. Deh4p has a putative molecular weight of 59,414 and an isoelectric point of 9.88. Deh4p has the signatures of sugar transport proteins and integral membrane proteins of the major facilitator superfamily. Uptake of [14C]MCA into the cell was Deh4p dependent. Deh4p has apparent Kms of 5.5 and 8.9 μM and Vmaxs of 9.1 and 23.1 nmol mg−1 min−1 for acetate and MCA, respectively. A mutant with a transposon-inactivated haloacid operon failed to grow on MCA even when deh4a was provided in trans.
Burkholderia spp. are gram-negative bacteria with versatile capability. They have been isolated in various niches including soil, water, plants, the rhizosphere, and animals including humans. Burkholderia cepacia strains have been shown to be plant and/or animal pathogens, and yet they have also been found to possess antibacterial and antifungal activity. Moreover, they have also been shown to be involved in the bioremediation of many types of environmental pollutants (2, 34, 44). Burkholderia cepacia MBA4 was isolated from soil by an enrichment method using monobromoacetic acid (MBA) as the sole carbon and energy source. B. cepacia MBA4 is also able to grow on monochloroacetate (MCA), 2-monochloropropionate (2MCPA), and 2-monobromopropionate (47). This bacterium produces a haloacid-induced dehalogenase (renamed Deh4a) in batch culture conditions. 2-Haloacid dehalogenases or halidohydrolases are hydrolytic enzymes that cleave the halogen-carbon bond(s) in halogenated aliphatic acids, yielding hydroxy- or oxoalkanoic acids from mono- or disubstituted substrate, respectively (12). Deh4a has been purified and characterized (47). Its structural gene (renamed deh4a) has also been isolated and characterized and encodes 231 residues. Deh4a exists as a dimeric protein of 45 kDa, composed of two identical subunits of 23 kDa each (27, 45). The regions involved in the dimerization and the activity functions of the enzyme have also been elucidated (30, 46).
Deh4a is mainly found in the cytoplasm of B. cepacia MBA4, and no signature of any signal sequence has been identified. In order for the cell to utilize haloacid as a growth substrate, the availability of a transporter protein specialized in carrying the substrate into the cell would be necessary. Previous study on the toxicity of 2-haloacids to Pseudomonas putida suggested the presence of a dehalogenase-associated permease. Cells expressing inducible haloacid dehalogenase were more sensitive to haloacid than mutant cells having a decreased level of expression (53). While the presence of a haloacid permease has been suggested for a long time, the isolation and characterization of this haloacid transporter have never been achieved. Dehalogenases have also been isolated in other microorganisms (17, 22), and other enzymes that acted on haloalkanes or haloalcohols have also been identified (4, 7, 43). Nonetheless, no transport system for these halogenated compounds has been identified and characterized.
It is well known that permeases have overlapping specificity. Permease LldP of the E. coli lactate operon and transporter GlcA of the glycolate operon are both involved in the uptake of 2-hydroxymonocarboxylic acids (28, 29). An acetyl-coenzyme A synthetase gene (acs)-contranscribed gene, yjcG (or actP), which was initially classified as encoding a member of the sodium-solute symporter family, has been shown to be an acetate permease (10). MctP of Rhizobium leguminosarum is able to transport alanine and other monocarboxylates (18). These studies revealed that the transport of monocarboxylic acids can be managed by specific transporters of versatile capability. It is therefore possible that the transport of haloacids is also mediated by a specific permease.
In prokaryotes, genes coding for products that perform related functions were usually grouped together and controlled under the same regulatory sequence. This operon type organization provides a convenient means of detecting genes involved in a metabolic pathway. By means of chromosome walking, we aimed to identify the permease gene involved in the uptake of haloacids. Here we report the first cloning and characterization of a dehalogenase-associated permease in B. cepacia MBA4. The corresponding gene, deh4p, is located 353 bases downstream of deh4a and encodes a transmembrane protein of 552 residues. This protein is a member of the major facilitator superfamily (MFS) and is responsible for the uptake of haloacetate. Our results showed that deh4a and deh4p form an operon that mediates the utilization of haloacetate in B. cepacia MBA4.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. cepacia MBA4 is a soil isolate that can utilize 2-haloacids as the carbon and energy source for growth (47). Escherichia coli TOP10 (Invitrogen) was used as the host for genetic manipulation. B. cepacia MBA4 was grown at 30°C in Luria broth without NaCl (1% tryptone, 0.5% yeast extract) or in defined medium with an appropriate carbon source (0.5 g carbon liter−1) as described previously (47). E. coli TOP10 was grown at 30°C or 37°C in Luria broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl) with antibiotics.
RNA isolation and analytical RT-PCR.
Total RNA was isolated from monobromoacetate-grown B. cepacia MBA4 cells with the High Pure RNA isolation kit (Boehringer-Mannheim) according to the supplier's protocol. Oligonucleotides deh4aC (5′-CCGCT GCTAG CGAAG AACGT TACA-3′) and deh4pN (5′-CGCAC GTCCT TCAAT AGTCG CCAT-3′) were used to amplify the intergenic region between deh4a and deh4p. Primers deh4a5 (5′-ATTAA GGTCA TGGTG GATTC ACTCC GC-3′) and deh4p3 (5′-AAGCT TGTCA GAACT GGAGA CGATT AGTCC-3′) were used to amplify the region between the 5′ end of deh4a and the 3′ end of deh4p. Reverse transcriptase PCR (RT-PCR) was performed with SuperScript one-step RT-PCR system with Platinum Taq (Invitrogen). The reaction was carried out in 1× reaction mixture (0.2 mM of each of the deoxynucleoside triphosphates and 1.2 mM MgSO4), 1.6 ng total RNA, 0.5 μl RT-Taq mixture, and 0.1 μM of each of the forward and reverse primers in a final volume of 25 μl. A similar reaction with Taq DNA polymerase (Invitrogen) instead of RT-Taq mixture was also set up. Reverse transcription and predenaturation were performed at 50°C for 30 min and 94°C for 2 min, respectively. The conditions for 35 cycles of PCR were as follows: 94°C for 2 min, 60°C for 30 s, and 72°C for 35 s. The PCR products were analyzed on a 1% agarose gel.
Quantitative real-time RT-PCR.
The reverse transcription was performed with Maloney murine leukemia virus RT (Promega) in 1× reaction mixture with 1 μg total RNA, 20 pmol reverse primer, 1 μl of RNasin (Promega), and water to 20 μl. The reaction mixture was incubated at 42°C for 1 h. The quantitative PCR was set up with 4 μl of reaction mixture from a LightCycler FastStart DNA Master Plus SYBR green I kit (Roche), 2 μl of RT product, 1 μl each of forward and reverse primers (10 μM), and water to a final volume of 20 μl. The quantitative PCR was carried out in a LightCycler (Roche) at 94°C (10 s), 63°C (5 s), and 72°C (11 s) for 45 cycles. The initial amount of the specific transcript was quantified mathematically by a relative method (33). The fit point method in the LightCycler software 3.3 (Roche) was used, and the crossing point (CP) was determined at a fluorescence value of 0.5. The CP is the point where the fluorescence rises above the background fluorescence. The amounts of deh4a and deh4p were estimated with reference to the 16S rRNA level in each sample. The relative expression ratio is expressed as follows: ratio = (Etarget)ΔCPtarget(control − sample)/(Eref)ΔCPref(control − sample). ΔCPtarget and ΔCPref are the CP differences of the relative fluorescence levels of target (deh4a or deh4p) and reference (16S rRNA), respectively. Control and sample are preparations from cells grown in pyruvate or MCA. E stands for the efficiency of amplification, which is determined separately for each primer pair used in this study. The efficiency was calculated by the equation E = 10(−1/slope) (35). The slopes were obtained from real-time PCR runs by using serial dilutions of the DNA template. The cycle numbers of CP were plotted against the input in log concentration. The E value obtained for primers 16SF (5′-GGTAA TACGT AGGGT GCAAG CGTT-3′) and 16SR (5′-CACCA ATGCA GTTCC CAGGT TAAG-3′) was 1.965. The E value for primers deh4aF (5′-GGCTG ATGAG TGCGT ACAAG GA-3′) and deh4aR (5′-CTCAT CGTTA CCGTT CGAAA GGA-3′) was 1.752, and the E value for primers deh4pF (5′-CTGCA GATGG CGACT ATTGA AGGAC GTGCG-3′) and deh4pR (5′-ATATC GCAAG TGAAC CAGCG AGAT-3′) was 1.741. Mean CPs were obtained from triplicates of three experiments.
Transport assays.
B. cepacia MBA4 cells were cultured in minimal medium with pyruvate, acetate, or MCA to late logarithmic phase with an optical density at 600 nm (OD600) of 0.5 to 0.7. Cells were harvested by centrifugation and washed twice with phosphate-buffered saline (PBS; Fluka) and resuspended in PBS to an OD600 of 0.5. [2-14C]MCA (Sigma-Aldrich) or [2-14C]acetate (Moravek) was diluted to 1 mM in PBS. The diluted radiolabeled substrate (100 μl) was mixed with washed cells (400 μl), and 100-μl samples were taken immediately and at 1 min, 2 min, 4 min, and 6 min. The samples were filtered through 0.45-μm-pore-size nitrocellulose filter disks (Millipore) and washed with 2 ml PBS under continuous suction (QiaVac 24 Plus vacuum manifold; QIAGEN). Filter disks with trapped cells were put into vials with 2 ml scintillation fluid (Ready-Solv EP; Beckman), and the amounts of carbon 14 were measured by a scintillation counter (LS6500; Beckman). Total protein in each sample was measured with protein assay reagent (Bio-Rad). All uptake assays were performed at room temperature.
For the competition study diluted radiolabeled substrate was mixed with 10× competing substrate (10 mM in PBS) at a 1:1 ratio before mixture with prepared cells. Percent inhibition was calculated as [1 − (uptake rate with competing substrate/uptake rate without competing substrate)] × 100%. The kinetic constants of MCA and acetate uptake by B. cepacia MBA4 were determined using various [2-14C]MCA or [2-14C]acetate concentrations in a standard uptake assay.
Transposon mutagenesis.
The haloacid operon was inactivated by insertion of the Tn5 transposon. Tn5 carrying plasmid pOT182 (26) was delivered into B. cepacia MBA4 via electroporation (8). Briefly, late-logarithmic growing cells (100 ml) were harvested by centrifugation at 1,600 × g for 15 min at 4°C, washed twice with 20 ml of ice-cold electroporation buffer (1 mM MgCl2, 1 mM HEPES, pH 7.0), resuspended in 2 ml buffer, and stored on ice until use. Plasmid DNA was gently mixed with 100 μl cell, and the mixture was incubated at 4°C for at least 5 min before electroporation. The cell-DNA mixture was transferred to a prechilled (4°C) sterile electroporation cuvette with a 0.2-cm electrode gap. A Gene Pulser with a pulse controller unit (Bio-Rad) was used for the electroporation. The settings were 12.5 kV cm−1, 25 μF, and 200 Ω. After the discharge through the cells, 900 μl of SOC medium (tryptone, 2%; yeast extract, 0.5%; NaCl, 0.06%; KCl, 0.02%; MgCl2·6H2O, 0.2%; MgSO4, 0.12%; glucose, 0.18%; all percentages are wt/vol) was immediately added and the mixture was mixed gently. A volume (100 μl) of this transformation mixture was plated on an LB− agar plate containing the appropriate antibiotics. The plate was incubated at 30°C for 2 to 3 days. Tetracycline-resistant transformants were screened for their ability to grow on MCA. Clones that failed to grow on MCA were further analyzed for the site of integration (26).
Construction of Deh4a- and Deh4a-Deh4p-producing plasmids.
DNA fragments containing deh4a or deh4a plus deh4p were amplified from genomic DNA by PCR using primers PstI-4a-F (5′-CTGCA GTATC GATCG AGCCT CGTGA CCGAC C-3′) and KpnI-4a-R (5′-GGTAC CGCGT TGTCT CCTGA TCTAA TCGTA TAAGT C-3′) or primers PstI-4a-F and KpnI-4ap-R (5′-GGTACC GACGC GCTTG CTGTC CGGCT C-3′), respectively. The DNA fragments were cloned into PstI- and KpnI-cut pUCP28T (41) to form plasmids p28T4A (containing deh4a only) and p28T4AP (containing deh4a and deh4p). These plasmids were transformed into mutant 15D02 by electroporation, and transformants were selected on the basis of their resistance to trimethoprim and tetracycline.
Nucleotide and protein sequence accession numbers.
The nucleotide sequence of the deh4p open reading frame (ORF) has been deposited with GenBank under accession number AF439266, and the associated protein sequence has been deposited under UniProt accession number Q7X4L6.
RESULTS
Identification of haloacid permease activity in B. cepacia MBA4.
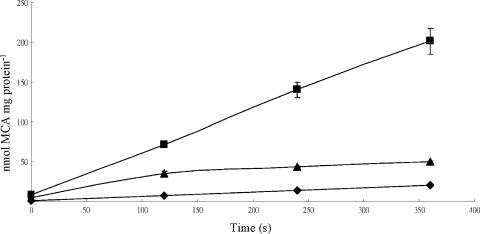
Expression of the haloacid-metabolizing enzyme, dehalogenase, in B. cepacia MBA4 is regulated. The gene deh4a (previously hdlIVa) is induced only when haloacid is the sole carbon source (47). If an associated permease is present in B. cepacia MBA4, the expression of this permease is most likely also regulated. B. cepacia MBA4 cells were therefore grown in defined medium with pyruvate, acetate, or MCA as the carbon and energy source, and [2-14C]MCA transport was examined. Figure 1 shows that cells grown on pyruvate transport MCA into the cell at a rate of 3.9 nmol mg protein−1 min−1. Cells grown on acetate have an initial transport rate of 17.5 nmol mg protein−1 min−1, but this rate was reduced to a rate similar to that of pyruvate-grown cells after 2 min. MCA-grown cells, on the other hand, have an uptake rate of 35.5 nmol mg protein−1 min−1, and this rate was maintained throughout the assay period of 6 min. This suggested that MCA induced a transporter protein or proteins that facilitated the specific uptake of MCA.
FIG. 1.
Uptake of monochloroacetate by B. cepacia MBA4. Uptake of 200 μM [2-14C]MCA was assayed by a filtration method for cells grown in minimal medium containing pyruvate (⧫), acetate (▴), or MCA (▪). Data shown are the means of three independent experiments with error bars.
Property of the haloacid permease activity.
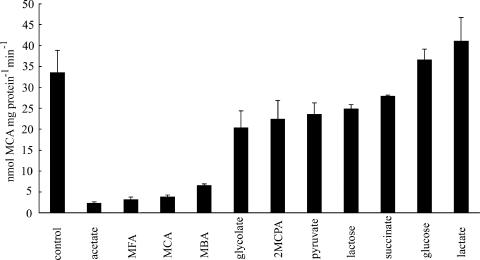
In order to characterize the substrate specificity of the induced permease, MCA uptake assays were carried out in the presence of a 10-fold excess of a number of solutes. Competing substrates selected were 2-haloacids (monofluoroacetate [MFA], MCA, MBA, and 2MCPA), monocarboxylates (acetate, glycolate, pyruvate, and lactate), dicarboxylate (succinate), monosaccharide (glucose), and disaccharide (lactose). Figure 2 shows the inhibition of [2-14C]MCA uptake by MCA-grown cells in the presence of competing substrate. The results showed that only acetate and derivatives (MFA, MCA, and MBA) are strong inhibitors of MCA uptake (between 80 and 93% inhibition). Other solutes tested did not inhibit MCA uptake (glucose and lactate) or inhibited MCA uptake weakly (2MCPA, glycolate, pyruvate, succinate, and lactose), between 17 and 39%. The inhibition by acetates seems to be dependent on molecular size, with the smaller compounds showing more inhibitory effect (acetate > MFA > MCA > MBA). These results revealed that the MCA-induced permease is a specific transporter for acetates.
FIG. 2.
Inhibition of MCA uptake by other solutes. Uptake of 200 μM [2-14C]MCA by MCA-grown MBA4 was assayed by a filtration method. Competing solutes were added to a final concentration of 2 mM. Data shown are the means of two independent experiments with error bars.
Since the MCA-induced permease activity was able to transport both MCA and acetate, it is necessary to characterize its properties in more detail. MCA-grown cells were used, and different concentrations (3 to 100 μM) of [2-14C]acetate or [2-14C]MCA were used to determine the Km and Vmax of the permease activity. The Kms for acetate and MCA were 5.5 and 8.9 μM, respectively, and the Vmaxs were 9.1 and 23.1 nmol mg−1 min−1, respectively.
Identification of a putative transporter gene downstream of deh4a of B. cepacia MBA4.
The discovery of MCA-induced haloacetate permease activity prompted us to look for the permease gene. Since genes of related functions are usually clustered together in bacteria as an operon, we strived to look for a related gene(s) involved in the degradation of 2-haloacids. The deh4a gene (GenBank accession no. X66249) was isolated on a 1.6-kb EcoRI fragment, and E. coli cells harboring this fragment were able to produce Deh4a (45). This suggested that a promoter present on this fragment probably drove the expression of Deh4a in E. coli. Computational analysis of the sequences upstream of deh4a by the Neural Network Promoter Prediction program (36) has revealed the presence of a putative bacterial promoter. The transcription initiation site has recently been identified by primer extension to start at an A at −26 upstream of the initiation codon of deh4a (J. S. H. Tsang and L. Sam, unpublished data). Oligonucleotides were synthesized and used to unravel the sequence downstream of the deh4a gene. An ORF of 1,659 bp, designated deh4p for permease, was identified 353 bp downstream of the deh4a gene. No terminator sequence was identified between deh4a and this ORF. On the other hand, sequences that exhibit structural properties of intrinsic transcriptional terminators (11, 40) were identified downstream of this putative gene.
A postulated Shine-Dalgarno region (GGAGA) was found at the right proximity close to the ATG of the gene at position 307. Analysis of the putative amino acid sequence of Deh4p revealed that it is probably a member of the MFS transporters (31). It has the signature of family 1: sugar transporter family proteins. The signature [LIVMF]-X-G-[LIVMFA]-X(2)-G-X(8)-[LIFY]-X(2)-[EQ]-X(6)-[RK] is found between residues 130 and 155. The putative gene product Deh4p has an apparent molecular weight of 59,414 and an isoelectric point of 9.88. The nucleotide sequence of deh4p exhibits up to 74% similarity with putative MFS transporter genes of many Proteobacteria. Analysis of the predicted amino acid sequence with SOUSI (16) shows that Deh4p is a putative integral membrane protein containing 12 transmembrane spanners. Moreover, residues 81 to 90, GRLGD MIGRK, are also consistent with the consensus region connecting helices 2 and 3 of a typical MFS member (15).
deh4p is the second member of a haloacid operon.
The identification of a putative transporter protein gene downstream of deh4a prompted us to consider the possibility of an operon containing deh4a and deh4p, responsible for the utilization of haloacids in B. cepacia MBA4. Quantitative RT-PCR analysis was used to check the abundance of the transcripts in different growth conditions. The results showed that both genes were concomitantly expressed in the presence of MCA and turned off in its absence (growth in pyruvate). Moreover, the transcript amounts in the induced condition for both genes were roughly the same (260-fold for deh4a and 280-fold for deh4p). This suggested that deh4a and deh4p were probably expressed in series in a haloacid operon.
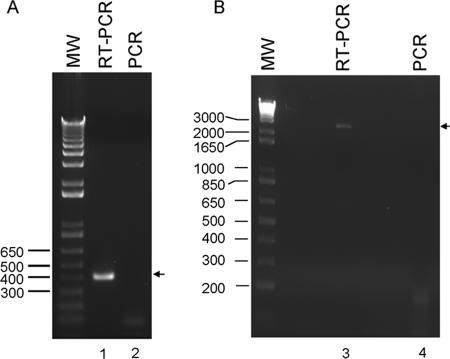
In order to confirm that a single transcript was made for both genes, RT-PCR was performed. A pair of specific primers was used to anneal to the 3′ end of deh4a and 5′ end of deh4p. An expected RT-PCR product of about 400 bp was amplified (Fig. 3A, lane 1), while the negative control (no reverse transcription) showed no observable band (Fig. 3A, lane 2). Oligonucleotide primers were also designed to amplify transcripts from the 5′ end of deh4a to the 3′ end of deh4p. The results showed that an RT-PCR product of around 2.7 kb was detected (Fig. 3B, lane 3). The size of this RT-PCR product matches the sum of the deh4a and the deh4p genes and some 5′ and 3′ untranslated regions. Similarly, no PCR product was detected in the RNA preparation when reverse transcription was omitted (Fig. 3B, lane 4). These results suggested that the RT-PCR products were not the consequence of contaminated genomic DNA and that deh4a and deh4p were expressed as a single transcript.
FIG. 3.
Agarose gel electrophoresis of RT-PCR products. Total RNAs were isolated from MCA-grown MBA4, and primers deh4pN (for lane 1) and deh4p3 (for lane 3) were used for first-strand synthesis. Primers deh4aC and deh4a5, respectively, were used for the PCR. PCR of the corresponding RNA samples without reverse transcription (lanes 2 and 4) was performed with similar primer pairs.
Complementation of mutant defective in the haloacid operon.
To show that Deh4p is essential for the cell to transport and for the uptake of haloacids, it is necessary to confirm its function by complementation of a Deh4p-negative mutant. While the generation of gene-specific deletion or disruption has not been successful for B. cepacia MBA4, we used transposon mutagenesis (42) to generate a mutant with a defective deh4p. Transposon Tn5 was introduced into B. cepacia MBA4 by electroporation, and transformants were screened for their inability to grow on MCA. A transformant, 15D02, able to grow on pyruvate but not on MCA was isolated. This mutant was characterized by cloning the DNA fragment containing the transposon and sequences flanking the insertion site. The transposon was found to insert at nucleotide 300 of the deh4a gene, resulting in a truncated nonfunctional protein. This truncated protein is unlikely to be functional because D181 of Deh4a is essential for dehalogenase activity (30). Since deh4p is the second gene of the haloacid operon, it is likely that its expression was also disrupted due to a polar effect.
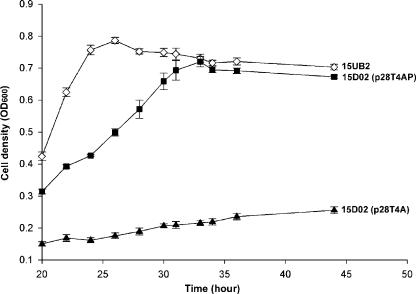
The failure of the mutant to grow on MCA could be explained by the lack of a functional dehalogenase rather than the requirement for a coexpressed permease. Strain 15D02 was transformed with plasmid p28T4A, which contains a wild-type deh4a. Figure 4 shows that this cell failed to grow on MCA. This suggested that the provision of a functional dehalogenase in trans was not sufficient to complement the haloacid utilization phenotype. This suggested that the expression of deh4p was also inhibited by the insertion of the transposon. Conversely, a cell transformed with plasmid p28T4AP, containing both deh4a and deh4p, was able to utilize MCA as the carbon and energy source. This indicated the importance of Deh4p in MCA utilization and that deh4a and deh4p are organized as an operon. Tn5 insertional mutants with wild-type deh activities have higher MCA growth rates than 15D02(p28T4AP), and a typical growth curve for one of these strains, 15UB2, carrying a pUCP28T-derived plasmid is shown in Fig. 4.
FIG. 4.
Growth of mutant 15D02 carrying various constructs. Mutant 15D02, containing plasmid p28T4A (▴) or p28T4AP (▪), was grown at 30°C in a defined medium containing MCA, tetracycline, and trimethoprim. A randomly selected Tn5-generated mutant, 15UB2 (⋄), with wild-type dehalogenase activity was used as the dehalogenase-positive control. Data shown are the means of at least three independent experiments with the error bars indicated.
DISCUSSION
In this study, we have demonstrated that the uptake of MCA for B. cepacia MBA4 was induced when MCA was the sole carbon source. This property was in line with the expression of the deh4p gene located immediately downstream of the dehalogenase IVa gene, deh4a. The expression of deh4a is inducible and is essential for MBA4 to degrade MCA. Structural analysis of a dehalogenase-producing Pseudomonas putida strain (strain PP3) has also identified a putative heavy-metal-associated cation efflux transporter gene adjacent to a DehI dehalogenase gene (52). This transporter, however, is truncated, and its haloacid uptake function has not been demonstrated. A similar genomic structure has also been found in Agrobacterium sp. strain NHG3, where putative transporter gene dehP (GenBank/EBI Data Bank accession number CAD32751) was found 66 bp upstream of d-specific and l-specific 2-chloropropionic acid dehalogenases (13). The function of this putative transporter, however, has not been investigated. The current study is the first to report the presence of a haloacid operon that mediated the uptake and degradation of haloacid in bacteria. Haloacetate or 2-haloacid dehalogenase genes have been identified in a wide variety of bacteria (14). Comparative analysis of the genomes (24) of these bacteria showed that, while a putative dehalogenase gene can be found in many Proteobacteria such as Burkholderia cenocepacia (http://img.jgi.doe.gov/; Integrated Microbial Genome [IMG] gene object accession no. 636436945, 636439065, 636442040, and 636446170), Burkholderia mallei (IMG accession no. 5771450 and 5789940), Agrobacterium tumefaciens (IMG accession no. 922710, 955800, 957310, 957340, and 972890), Ralstonia spp. (IMG accession no. 623760310, 623760340, 623782300, 3498150, and 3509420), and Rhizobium spp. (IMG accession no. 635185875, 635188675, 635188740, 635193460, 635211910, 635211950, 636299245, 636315300, and 636319835), none of these genes were found next to a gene encoding an MFS transporter.
Comparison of the protein sequence of Deh4p with the translated genomic sequences of various Proteobacteria has identified many putative proteins of the MFS. These putative MFS proteins, found in Burkholderia sp. strain 383 (NCBI accession number ABB09576), Burkholderia xenovorans LB400 (NCBI accession number ABE32266), Burkholderia pseudomallei (NCBI accession number YP_332381), B. mallei (NCBI accession number AAU48731), B. cepacia (NCBI accession number EA043951), Burkholderia vietnamiensis (NCBI accession number EAM26890), and Burkholderia dolosa (NCBI accession number ZP_00987315), exhibit around 80% identities with Deh4p. Inspection of the locations of the corresponding genes failed to discover any putative dehalogenase gene in juxtaposed positions. It is likely that the haloacid operon, containing dehalogenase and permease genes, is not common in Proteobacteria. It is also possible that the haloacid operon was evolved fairly recently and that those related transporters have a different natural substrate.
Haloacetic acids are disinfection by-products (6, 20) and have been shown to possess toxic and mutagenic properties (21, 38). It is thus reasonable to identify haloacetate dehalogenase genes in various bacteria because the acquisition of the gene helps the bacterium to detoxify and survive (50). To further eliminate the unwelcome chemical, the presence of an efflux transporter that pumps the compound out of the cell would be favorable. On the other hand the acquisition of a transporter that enhanced the uptake of haloacid will damage and impair the growth of the organism. The evolution of a haloacid operon containing a dehalogenase gene and a permease gene suggests that B. cepacia MBA4 has adapted to an environment that allows the cell not just to tolerate the presence of haloacids but to utilize these mutagenic compounds as a growth substrate. The regulated expression of these genes by haloacetate further confirms their role in substrate utilization and metabolism. The genome of MBA4 has the cryptic dehalogenase gene chd1, which was not normally expressed (48). This dehalogenase has a leader sequence that targeted the protein to the periplasm but the affinity for monobromoacetate of this protein is only 1/10 that of Deh4a.
The suggestion that there is an operon containing deh4a and deh4p is supported by (i) the concomitant induced expression of these genes to similar levels; (ii) detection of reverse transcription-PCR products spanning the two genes from mRNA and not from DNA; (iii) the presence of a terminator sequence at the end of deh4p and not at the end of deh4a; and (iv) the polar effect of Tn5 insertion in deh4a on deh4p expression, as shown by the lack of growth of strain 15D02(p28T4A). The possession of a haloacid operon containing deh4a, encoding a haloacid dehalogenase, and deh4p, encoding a haloacetate transporter, helps the cell to grow on haloacetates. The regulated expression of deh4a and deh4p in response to haloacids implied that they are involved in the metabolism of the halogenated compound. Figure 4 shows that strain 15D02(p28T4AP) was able to grow on MCA, but its growth was not as efficient as other Tn5 insertion mutants with the wild-type dehalogenase phenotype (e.g., strain 15UB2). This can be explained by the fact that the regulatory proteins required for the expression of the haloacid operon bound to the deh promoters on the chromosome and on the plasmids. The binding of the regulatory factor to the promoter located on the chromosome will produce only a truncated transcript and nonfunctional peptide. This may have quenched the activities of the promoters on the plasmids, and full-length transcripts and functional proteins were not produced to their utmost levels.
In order to confirm that the MCA-induced haloacid permease activity was due to the expression of Deh4p, we have cloned the structural gene of Deh4p and expressed it in E. coli cells. The deh4p gene was expressed in E. coli TOP10 using a ribosomal s12 promoter. E. coli cells harboring deh4p (on plasmid p28tS12-4p) have a higher (threefold) MCA uptake rate than cells harboring the vector (p28tS12-E) only (54). One would expect a more dramatic difference in uptake rate by using stronger promoters. We in fact used an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible trc promoter initially, but the growth of the cell was severely hindered even under a repressed condition. Moreover, in the presence of IPTG E. coli cells produced transporter proteins such as LacY (3) that transported nonspecific substrates (23, 39) including monochloroacetate (54).
Comparative analysis of the primary sequence of Deh4p with the transport classification database (TCDB; http://www.tcdb.org/) has placed it in MFS 2.A.1. Uptake inhibition studies for MCA-grown MBA4 cells suggested that the induced permease has narrow substrate specificity for short-chain aliphatic monocarboxylates. This is similar to the acetate permease ActP of E. coli (10). The apparent Km for acetate for ActP is 5.4 μM, which is comparable to that for Deh4p. However, the Vmax for ActP is 19.6 nmol mg−1 min−1, which is much higher than that for Deh4p. Preliminary results suggest the presence of 12 transmembrane helices, as predicted by the majority of topology programs (5, 9, 16, 19, 25, 32, 37, 49, 51, 55). Most of these predictions suggested that the N terminus is located in the cytoplasm. By tagging PhoA-LacZ dual reporters (1) at various positions of Deh4p and expressing them in E. coli we have confirmed that the N terminus is indeed located in the cytoplasm (Y. M. Tse and J. S. H. Tsang, unpublished data). The topology of Deh4p, however, still awaits further analysis.
Acknowledgments
We thank H. Schweizer for plasmid pUCP28T, A. Weightman for plasmid pOT182, W. K. Yip for the use of the scintillation counter, and J. Brabyn for reading the manuscript.
Y.-W.F. and W.Y.K.C. thank the University of Hong Kong for studentships. This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region, China (project no. HKU7536/06 M).
Footnotes
Published ahead of print on 1 June 2007.
REFERENCES
- 1.Alexeyev, M. F., and H. H. Winkler. 1999. Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J. Mol. Biol. 285:1503-1513. [DOI] [PubMed] [Google Scholar]
- 2.Chiarini, L., A. Bevivino, C. Dalmastri, S. Tabacchioni, and P. Visca. 2006. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 14:277-286. [DOI] [PubMed] [Google Scholar]
- 3.Chuang, S. E., D. L. Daniels, and F. R. Blattner. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Copley, S. D. 1998. Microbial dehalogenases: enzymes recruited to convert xenobiotic substrates. Curr. Opin. Chem. Biol. 2:613-617. [DOI] [PubMed] [Google Scholar]
- 5.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 6.Dalvi, A. G. I., R. Al-Rasheed, and M. A. Javeed. 2000. Haloacetic acids (HAAs) formation in desalination processes from disinfectants. Desalination 129:261-271. [Google Scholar]
- 7.de Jong, R. M., and B. W. Dijkstra. 2003. Structure and mechanism of bacterial dehalogenases: different ways to cleave a carbon-halogen bond. Curr. Opin. Struct. Biol. 13:722-730. [DOI] [PubMed] [Google Scholar]
- 8.Dennis, J. J., and P. A. Sokol. 1995. Electrotransformation of Pseudomonas. Methods Mol. Biol. 47:125-133. [DOI] [PubMed] [Google Scholar]
- 9.Gardy, J. L., C. Spencer, K. Wang, M. Ester, G. E. Tusnady, I. Simon, S. Hua, K. deFays, C. Lambert, K. Nakai, and F. S. Brinkman. 2003. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 31:3613-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gimenez, R., M. F. Nunez, J. Badia, J. Aguilar, and L. Baldoma. 2003. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J. Bacteriol. 185:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gusarov, I., and E. Nudler. 1999. The mechanism of intrinsic transcription termination. Mol. Cell 3:495-504. [DOI] [PubMed] [Google Scholar]
- 12.Hardman, D. J. 1991. Biotransformation of halogenated compounds. Crit. Rev. Biotechnol. 11:1-40. [DOI] [PubMed] [Google Scholar]
- 13.Higgins, T. P., S. J. Hope, A. J. Effendi, S. Dawson, and B. N. Dancer. 2005. Biochemical and molecular characterisation of the 2,3-dichloro-1-propanol dehalogenase and stereospecific haloalkanoic dehalogenases from a versatile Agrobacterium sp. Biodegradation 16:485-492. [DOI] [PubMed] [Google Scholar]
- 14.Hill, K. E., J. R. Marchesi, and A. J. Weightman. 1999. Investigation of two evolutionarily unrelated halocarboxylic acid dehalogenase gene families. J. Bacteriol. 181:2535-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirai, T., J. A. Heymann, P. C. Maloney, and S. Subramaniam. 2003. Structural model for 12-helix transporters belonging to the major facilitator superfamily. J. Bacteriol. 185:1712-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 17.Hope, S. J., and J. H. Slater. 1995. Cryptic dehalogenase and chloroamidase genes in Pseudomonas putida and the influence of environmental conditions on their expression. Arch. Microbiol. 163:57-64. [DOI] [PubMed] [Google Scholar]
- 18.Hosie, A. H., D. Allaway, and P. S. Poole. 2002. A monocarboxylate permease of Rhizobium leguminosarum is the first member of a new subfamily of transporters. J. Bacteriol. 184:5436-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 20.Kanokkantapong, V., T. F. Marhaba, S. Wattanachira, B. Panyapinyophol, and P. Pavasant. 2006. Interaction between organic species in the formation of haloacetic acids following disinfection. J. Environ. Sci. Health Part A 41:1233-1248. [DOI] [PubMed] [Google Scholar]
- 21.Kargalioglu, Y., B. J. McMillan, R. A. Minear, and M. J. Plewa. 2002. Analysis of the cytotoxicity and mutagenicity of drinking water disinfection by-products in Salmonella typhimurium. Teratogen. Carcinogen. Mutagen. 22:113-128. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki, H., T. Toyama, T. Maeda, H. Nishino, and K. Tonomura. 1994. Cloning and sequence analysis of a plasmid-encoded 2-haloacid dehalogenase gene from Pseudomonas putida no. 109. Biosci. Biotechnol. Biochem. 58:160-163. [DOI] [PubMed] [Google Scholar]
- 23.Kreimer, D. I., H. Malak, J. R. Lakowicz, S. Trakhanov, E. Villar, and V. L. Shnyrov. 2000. Thermodynamics and dynamics of histidine-binding protein, the water-soluble receptor of histidine permease. Implications for the transport of high and low affinity ligands. Eur. J. Biochem. 267:4242-4252. [DOI] [PubMed] [Google Scholar]
- 24.Markowitz, V. M., F. Korzeniewski, K. Palaniappan, E. Szeto, G. Werner, A. Padki, X. Zhao, I. Dubchak, P. Hugenholtz, I. Anderson, A. Lykidis, K. Mavromatis, N. Ivanova, and N. C. Kyrpides. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34:D344-D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 26.Merriman, T. R., and I. L. Lamont. 1993. Construction and use of a self-cloning promoter probe vector for gram-negative bacteria. Gene 126:17-23. [DOI] [PubMed] [Google Scholar]
- 27.Murdiyatmo, U., W. Asmara, J. S. H. Tsang, A. J. Baines, A. T. Bull, and D. J. Hardman. 1992. Molecular biology of the 2-haloacid halidohydrolase IVa from Pseudomonas cepacia MBA4. Biochem. J. 284:87-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunez, M. F., O. Kwon, T. H. Wilson, J. Aguilar, L. Baldoma, and E. C. Lin. 2002. Transport of L-lactate, D-lactate, and glycolate by the LldP and GlcA membrane carriers of Escherichia coli. Biochem. Biophys. Res. Commun. 290:824-829. [DOI] [PubMed] [Google Scholar]
- 29.Nunez, M. F., M. T. Pellicer, J. Badia, J. Aguilar, and L. Baldoma. 2001. The gene yghK linked to the glc operon of Escherichia coli encodes a permease for glycolate that is structurally and functionally similar to L-lactate permease. Microbiology 147:1069-1077. [DOI] [PubMed] [Google Scholar]
- 30.Pang, B. C. M., and J. S. H. Tsang. 2001. Mutagenic analysis of the conserved residues in dehalogenase IVa of Burkholderia cepacia MBA4. FEMS Microbiol. Lett. 204:135-140. [DOI] [PubMed] [Google Scholar]
- 31.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persson, B., and P. Argos. 1996. Topology prediction of membrane proteins. Protein Sci. 5:363-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen, R. 2001. Quantification on the LightCycler, p. 21-34. In S. Meuer, C. Wittwer, and K. Nakagawara (ed.), Rapid cycle real-time PCR, methods and applications. Springer, Heidelberg, Germany.
- 36.Reese, M. G. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26:51-56. [DOI] [PubMed] [Google Scholar]
- 37.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5:1704-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakai, A., H. Shimizu, K. Kono, and E. Furuya. 2005. Monochloroacetic acid inhibits liver gluconeogenesis by inactivating glyceraldehyde-3-phosphate dehydrogenase. Chem. Res. Toxicol. 18:277-282. [DOI] [PubMed] [Google Scholar]
- 39.Sandermann, H., Jr. 1977. Beta-D-galactoside transport in Escherichia coli: substrate recognition. Eur. J. Biochem. 80:507-515. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz, A., A. R. Rahmouni, and M. Boudvillain. 2003. The functional anatomy of an intrinsic transcription terminator. EMBO J. 22:3385-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer, H. P., T. Klassen, and T. Hoang. 1996. Improved methods for gene analysis and expression in Pseudomonas spp., p. 229-237. In T. Nakazawa, K. Fukukawa, D. Hass, and S. Silver (ed.), Molecular biology of pseudomonads. ASM Press, Washington, DC.
- 42.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol. Gen. Genet. 196:413-420. [DOI] [PubMed] [Google Scholar]
- 43.Slater, J. H., A. T. Bull, and D. J. Hardman. 1997. Microbial dehalogenation of halogenated alkanoic acids, alcohols and alkanes. Adv. Microb. Physiol. 38:133-176. [DOI] [PubMed] [Google Scholar]
- 44.Tsang, J. S. H. 2004. Molecular biology of the Burkholderia cepacia complex. Adv. Appl. Microbiol. 54:71-91. [DOI] [PubMed] [Google Scholar]
- 45.Tsang, J. S. H., D. J. Hardman, and A. T. Bull. 1988. Cloning and expression of 2-haloalkanoic acid dehalogenase of Pseudomonas cepacia MBA4 in Escherichia coli and in Pseudomonas putida, p. 231-239. In S. T. Chang, K.-Y. Chan, and N. Y. S. Woo (ed.), Recent advances in biotechnology and applied biology. Chinese University Press, Hong Kong.
- 46.Tsang, J. S. H., and B. C. M. Pang. 2000. Identification of the dimerization domain of dehalogenase IVa of Burkholderia cepacia MBA4. Appl. Environ. Microbiol. 66:3180-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsang, J. S. H., P. J. Sallis, A. T. Bull, and D. J. Hardman. 1988. A monobromoacetate dehalogenase from Pseudomonas cepacia MBA4. Arch. Microbiol. 150:441-446. [Google Scholar]
- 48.Tsang, J. S. H., and L. Sam. 1999. Cloning and characterization of a cryptic haloacid dehalogenase from Burkholderia cepacia MBA4. J. Bacteriol. 181:6003-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 50.Van der Ploeg, J. R., J. Kingma, E. J. De Vries, J. G. Van der Ven, and D. B. Janssen. 1996. Adaptation of Pseudomonas sp. GJ1 to 2-bromoethanol caused by overexpression of an NAD-dependent aldehyde dehydrogenase with low affinity for halogenated aldehydes. Arch. Microbiol. 165:258-264. [DOI] [PubMed] [Google Scholar]
- 51.Viklund, H., and A. Elofsson. 2004. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 13:1908-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weightman, A. J., A. W. Topping, K. E. Hill, L. L. Lee, K. Sakai, J. H. Slater, and A. W. Thomas. 2002. Transposition of DEH, a broad-host-range transposon flanked by ISPpu12, in Pseudomonas putida is associated with genomic rearrangements and dehalogenase gene silencing. J. Bacteriol. 184:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weightman, A. J., A. L. Weightman, and J. H. Slater. 1985. Toxic effects of chlorinated and brominated alkanoic acids on Pseudomonas putida PP3: selection at high frequencies of mutations in genes encoding dehalogenases. Appl. Environ. Microbiol. 49:1494-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, M., and J. S. H. Tsang. 2006. Use of ribosomal promoters from Burkholderia cenocepacia and Burkholderia cepacia for improved expression of transporter protein in Escherichia coli. Protein Expr. Purif. 49:219-227. [DOI] [PubMed] [Google Scholar]
- 55.Zhai, Y., and M. H. Saier, Jr. 2001. A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J. Mol. Microbiol. Biotechnol. 3:501-502. [PubMed] [Google Scholar]