Abstract
An increasing number of studies utilize confocal laser scanning microscopy (CLSM) for in situ visualization of biofilms and rely on the use of image analysis programs to extract quantitative descriptors of architecture. Recently, designed programs have begun incorporating procedures to automatically determine threshold values for three-dimensional CLSM image stacks. We have found that the automated threshold calculation is biased when a stack contains images lacking pixels of biological significance. Consequently, we have created the novel program Auto PHLIP-ML to resolve this bias by iteratively excluding extraneous images based on their area coverage of biomass. A procedure was developed to identify the optimal percent area coverage value used for extraneous image removal (PACVEIR). The optimal PACVEIR was defined to occur when the standard deviation of mean thickness, determined from replicate image stacks, was at a maximum, because it more accurately reflected inherent structural variation. Ten monoculture biofilms of either Ralstonia eutropha JMP228n::gfp or Acinetobacter sp. strain BD413 were tested to verify the routine. All biofilms exhibited an optimal PACVEIR between 0 and 1%. Prior to the exclusion of extraneous images, JMP228n::gfp appeared to develop more homogeneous biofilms than BD413. However, after the removal of extraneous images, JMP228n::gfp biofilms were found to form more heterogeneous biofilms. Similarly, JMP228n::gfp biofilms grown on glass surfaces vis-à-vis polyethylene membranes produced significantly different architectures after extraneous images had been removed but not when such images were included in threshold calculations. This study shows that the failure to remove extraneous images skewed a seemingly objective analysis of biofilm architecture and significantly altered statistically derived conclusions.
The importance of biofilms as the primary habitat for bacteria and other microorganisms in natural and engineered environments is widely recognized (9, 16, 44, 46). A quintessential goal of biofilm analysis is to provide insights into the relationship between a biofilm's physical architecture and biological function. Such analyses can be used to either promote or inhibit biofilm processes. For example, thwarting biofilm growth is essential to prevent biofouling of surfaces (12, 13, 32) and biofilm-induced infections in humans (2, 17). Conversely, promoting biofilms is a proven method to achieve effective wastewater treatment (33, 52) and, when used as biocontrol agents, a means to aid in the growth of plants (1, 11). To further our knowledge of the relationship between biofilm structure and function, it is necessary to standardize reliable and efficient methodology for biofilm analysis.
Direct visualization by confocal laser scanning microscopy (CLSM) is arguably one of the most important tools to study the architecture of biofilms. CLSM allows for nondestructive, in situ, three-dimensional investigation of biofilms in their naturally hydrated state, detecting fluorescence from specific constituents such as cells or extracellular polymeric substances (28). CLSM captures the biofilm's physical structure through a series of digital images that occupy the same x-y plane but vary along the z- axis, often called a “z-stack” or an “image stack.” Typically, multiple z-stacks are acquired either systematically (22, 27) or randomly (21, 52) from the same biofilm to obtain a statistically significant representation of inherent structural variability (26, 45). Confocal microscopes equipped with a motorized stage allow for automated acquisition by utilizing user-specified (27) or proprietary macros (25). Image stacks are analyzed by using image analysis software packages such as COMSTAT (21), ISA3D (5), DAIME (10), or PHLIP (34) that calculate biofilm architectural metrics including biovolume, mean thickness, roughness, percent area coverage, porosity, area-to-volume ratio, spatial spreading, and fractal dimension.
Although biofilm image analysis has not reached the point of being completely automated, new methods of automation and improvements to the current stages of automation are continually being made (10, 34, 47). One key function that has improved the automation of biofilm analysis is the implementation of automated image thresholding for three-dimensional CLSM z-stacks (49). Previously thought to be unsuitable for automation (21, 27), the automated thresholding of z-stacks is now seen to be a benefit by eliminating the user subjectivity of manual thresholding (6, 34), where values are set to account for what the user believes to be the best representation of the biofilm. Employing automated methods makes biofilm image analysis results more reproducible (51) and allows for the full automation of biofilm quantification (50).
In addition, because automated acquisition makes it easy to produce a great number of biofilm image stacks, automated thresholding is rapidly becoming an essential tool for biofilm studies using CLSM. For instance, a representative area of 1 × 105 (26) or 2 × 105 (45) μm2 would be sufficient to account for the inherent variability of cell coverage in a Pseudomonas fluorescens or Sphingomonas sp. strain L138 biofilm, respectively, while a representative area of 106 μm2 might be needed for a more variable parameter (45). At ×63 magnification (146.25-by-146.25-μm image area), an area of 106 μm2 would require 47 image stacks. In a time course experiment with five sampling events and four experimental biofilms, an astounding 940 image stacks would be collected.
Although automated acquisition eases user involvement when attaining z-stacks, images that lack architectural information are acquired by default and complicate downstream automated image processing. These extraneous images occur when acquisition extends beyond the biofilm-substratum and bulk-medium interface boundaries or when every pixel value of an image does not exceed the calculated threshold value. It follows that extraneous images need to be removed prior to analysis because they bias the automated calculation of the threshold. However, the manual identification of unsuitable images is tedious and subjective and leads to bottlenecks in automated image processing.
The objectives of the present study were to develop an automated procedure to remove extraneous images and eliminate automated thresholding-induced bias. We describe a newly developed program called Auto PHLIP-ML and a routine to calculate an unbiased Otsu threshold (39) by selecting the optimal percent area coverage value used for extraneous image removal (PACVEIR). Ten monoculture biofilms were investigated to verify the robustness of the routine. Image stacks were acquired by using CLSM and then processed, with or without extraneous image removal by Auto PHLIP-ML, using the MatLab-based image analysis toolbox PHLIP (34).
MATERIALS AND METHODS
Bacterial strains and biofilm growth.
Six biofilms of R. eutropha JMP228n::gfp and four biofilms of Acinetobacter sp. strain BD413 (24) were grown in stainless steel, multilane flow cells under continuous flow conditions. R. eutropha JMP228n was chromosomally labeled with gfp by random insertion with the Tn5 transposon containing kanamycin resistance genes and genes for green fluorescent protein (GFP) production (H. McCaslin, unpublished data) and displayed homogeneous expression of gfp throughout the biofilm. Flow cells were sealed by glass coverslips with silicone glue. A single flow lane had dimensions of either 4 mm by 4 mm by 40 mm (height by width by length) for four BD413 biofilms (Ac1, Ac2, Ac3, and Ac4) and four JMP228n::gfp biofilms (Jmp1, Jmp2, Jmp3, and Jmp4) or 4 mm by 150 mm by 40 mm for two JMP228n::gfp biofilms (Ral1 and Ral2). Each continuous flow system was comprised of a growth medium reservoir, peristaltic pump, flow cell, and waste reservoir interconnected by either Tygon or Teflon tubing. The entire system (except peristaltic pump) was autoclaved prior to experimentation. Prior to flow cell inoculation, all cultures were washed three times in 1× phosphate-buffered saline after having been grown overnight in LB (BD413) or LB including 20 μg of tetracycline/ml (JMP228n::gfp) at 30°C. Cells were then enumerated by using a hemacytometer. Equal volumes of cells were inoculated into flow cell lanes by using a hypodermic needle and syringe and allowed to settle for 2 h before beginning medium flow.
The two JMP228n::gfp biofilms Ral1 and Ral2 grew from an inoculation of 5 × 107 cells with a medium flow of 3.75 ml h−1 (Re = 0.0068). Biofilms were grown at a constant temperature of 30°C for 4 days in LB medium containing 20 μg of tetracycline/ml and then for two more days in MMO medium (7) containing 0.78 mM 3-chloroaniline (3-CA). Ral1 was grown on a glass coverslip, while Ral2 was grown on a polyethylene membrane commonly used for laser microdissection and pressure catapulting (36). CLSM images were scanned after 6 days of biofilm growth.
The four JMP228n::gfp biofilms Jmp1, Jmp2, Jmp3, and Jmp4 grew from an inoculation of 2.5 × 105 cells with a medium flow rate of 3.75 ml h−1 (Re = 0.13). Biofilms were grown at a constant temperature of 30°C for 5 days in LB medium containing 20 μg of tetracycline/ml. Afterwards the medium was switched for 2 days to MMO containing an organic carbon source mixture adapted from Bathe et al. (3) with potassium acetate (0.03 M), sodium citrate (0.01 M), sodium gluconate (0.01 M), and d(+)-glucose (0.01 M); these substrate concentrations together correspond to a chemical oxygen demand value equal to that of 0.78 mM 3-CA. Biofilms Jmp1 and Jmp2 continued to receive MMO and the organic carbon source mixture; biofilms Jmp3 and Jmp4 were switched to MMO containing 0.78 mM 3-CA as the sole carbon source. CLSM images were scanned after 10 days of biofilm growth.
The four BD413 biofilms Ac1, Ac2, Ac3, and Ac4 were supplied with LB medium at room temperature for 5 days at a flow rate of 2.5 ml h−1 (Re = 0.089) prior to being scanned by CLSM. To visualize the biofilms, the inlet medium for each lane was switched to LB containing 5 μM Syto9 (Molecular Probes, Eugene, OR), a nucleic acid stain. After each lane had received 1.5 ml of LB plus Syto9, the inlet was switched back to the original LB medium and allowed to rinse for 1 h prior to visualization.
Automated CLSM image acquisition.
Multichannel flow cells were mounted on a motorized stage of a Zeiss 510 META confocal laser scanning microscope (Carl Zeiss, Jena, Germany) and visualized with a 63×/1.2 NA (C-Apochromat) water immersion objective lens. All biofilms were scanned with a 488-nm argon laser set at 25% intensity with a 505-nm long-pass filter to visualize Syto9 (BD413) or GFP expression (JMP228n::gfp). Multiple CLSM image stacks were acquired from each biofilm to obtain a representative sample of architectural variation (26). Automatic acquisition was accomplished by using the MultiTime series macro supplemental to Zeiss's CLSM interface software. The MultiTime series macro was programmed to capture multiple locations within the biofilm, which allowed for automated CLSM scanning.
A total of 24 scan locations were acquired for BD413 biofilms; 28 scan locations were acquired for biofilms Ral1 and Ral2; and 18 scan locations were acquired for the biofilms Jmp1, Jmp2, Jmp3, and Jmp4. Scan locations were separated 2 mm in the x direction and 0.5 mm in the y direction, making an x-y grid of either 8 by 3 (BD413 biofilms), 7 by 4 (Ral1 and Ral2), or 6 by 3 (Jmp1, Jmp2, Jmp3, and Jmp4). Scanning locations were limited to the central 20-mm region of the flow lane to exclude entrance and exiting flow effects on biofilm architecture occurring 10 mm from the inlet and outlet, respectively. Scanning was also limited to 1.5 mm from the channel walls to exclude excessive biofilm accumulation. z-stacks scanned from a single biofilm contained the same number of images. The number of images was set to capture the thickest part of the biofilm. Images collected for the four BD413 biofilms and the four JMP228n::gfp biofilms Jmp1, Jmp2, Jmp3, and Jmp4 had a pixel resolution of 0.285 μm/pixel; the two JMP228n::gfp biofilms Ral1 and Ral2 had a pixel resolution of 0.190 μm/pixel. The z-step for images in a z-stack was 1 μm. In accordance with optimal settings described by Sekar et al. (42), images were acquired utilizing a ×1 digital magnification, a pinhole setting of 1 airy unit, and a scan average of 2; the detector gain (500 to 550 arbitrary units) and amplifier offset (0 to 0.05 arbitrary units) were set to obtain adequately contrasted grayscale images based on the brightest region of the biofilm that was scanned. Image stacks that did not capture the entire biofilm thickness due to errors in automated acquisition were removed prior to image analysis. Two image stacks were removed from the biofilm Ac4, and seven image stacks were removed from the biofilm Ral2. Inadequate image stacks were identified both automatically by Auto PHLIP-ML and manually by visual inspection.
Image analysis.
Automated image analysis was performed by utilizing PHLIP (34), a MatLab-based image analysis toolbox freely available from the PHLIP website (http://www.itqb.unl.pt:1141/∼webpages/phlip/). PHLIP automatically calculates the Otsu threshold (39) and the architectural parameters biovolume, mean thickness, percent area coverage, roughness, spatial spreading, two-dimensional fractal dimension, and surface area/volume ratio for each z-stack. The architectural parameters biovolume and mean thickness were utilized for biofilm analysis. For each biofilm, the average and standard deviation of biovolume and mean thickness were calculated using the results from replicate z-stacks. The terms “average mean thickness” and “average biovolume” refer to the average of the mean thickness or biovolume values.
The percent relative standard deviation of the average biovolume (B-%RSD) and the mean thickness (MT-%RSD) were used to measure the effect of extraneous images on the quantification of biovolume and mean thickness, respectively. MT-%RSD was calculated as follows:
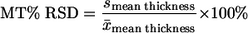 |
(1) |
where x̄ is the average mean thickness from replicate z-stacks and s is the sample standard deviation. B-%RSD was calculated by replacing mean thickness with biovolume. %RSD is a percentage measurement of the dimensionless coefficient of variation and allows for the comparison of variation between significantly different mean values. If the %RSD decreased, then the exclusion of extraneous images was considered to reduce the variation in values between replicate z-stacks, whereas an increase meant that extraneous image removal enhanced the variation between z-stacks. Little or no change in %RSD indicated that the removal of extraneous images did not affect the variability of replicate z-stacks.
MT-%RSD was also used to measure biofilm homogeneity. A biofilm with a low MT-%RSD was considered to be more homogeneous than a biofilm with a higher MT-%RSD value. It should be noted that MT-%RSD as defined here effectively calculates the roughness coefficient that is representative of the biofilm for the sampled region. In contrast, PHLIP uses the same method (equation 1) to calculate the roughness coefficient for a single z-stack where x̄ is the average of the pixel height distribution and s is the standard deviation of that distribution (34). Measuring the thickness variability of a biofilm or an individual z-stack with MT-%RSD is comparable to the roughness coefficient method described by Murga et al. (35) and used by COMSTAT (21).
CLSM image stacks were exported from their native LSM format to either TIFF or BMP lossless formats using a Zeiss batch converter. Exported images were imported utilizing PHLIP-ML (PHLIP markup language), PHLIP's proposed format to standardize the handling of CLSM images and data. A PHLIP-ML file harbors the information about z-stacks required for automated processing. Required information includes the directory location and names of images, pixel dimension, the distance between images in the z direction (z-step), and the number of detection channels that were used. An explanation of the structure of a PHLIP-ML file is given elsewhere (34). Analysis results calculated by PHLIP can also be saved in PHLIP-ML files and later reloaded into PHLIP for additional or alternative processing. PHLIP-ML files were either written automatically using Auto PHLIP-ML (Fig. 1) or written manually.
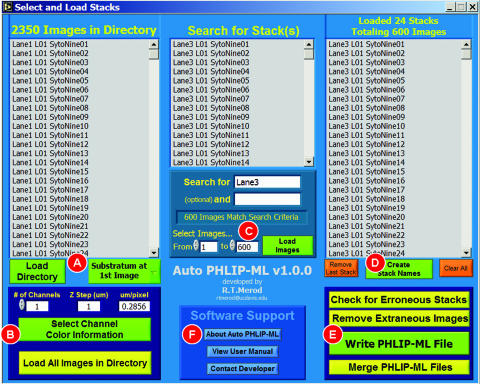
FIG. 1.
Graphical user interface (GUI) of Auto PHLIP-ML. The GUI is divided into three columns for processing (A and B, C, and D and E) and a user help area (F). In the first column, users load image stacks stored within the same directory, select the location of the substratum (A), and input the image resolution and number of detection channels shared among the z-stacks (B). Users can either load all of the z-stacks in the directory (B) or specify individual z-stacks (C) requiring analysis. In the third column, names are given to the z-stacks to undergo processing (D) either automatically derived from image file names or by loading a TXT file containing the desired names created by the user. If the image format of the images is in either a BMP or an uncompressed TIFF format the user can check for erroneous stacks and remove extraneous images (E). A PHLIP-ML file can be created (E) with or without image removal and for any image format (e.g., JPG, PNG, etc.). Separate PHLIP-ML files can be combined into a single PHLIP-ML file (E) to expedite PHLIP image analysis. Information regarding Auto PHLIP-ML such as version update notes, the user manual, and a means of contacting the developer via e-mail can be accessed in the software support section (F).
Statistical analysis.
For each biofilm a two-tailed Student t test (P < 0.05) was used to determine whether the removal of extraneous images had a significant effect on the quantification of the architectural metrics biovolume and mean thickness. A two-tailed Student t test (P < 0.05) was also used to investigate whether extraneous images affected the comparison of biofilms grown on different surface materials (Ral1 and Ral2) as well as replicate biofilms (Jmp1 and Jmp2; Jmp3 and Jmp4). A one-way analysis of variance (ANOVA) with Tukey's test (P < 0.05) was used to determine the significant differences between pair-wise comparisons of the replicate biofilms Ac1, Ac2, Ac3, and Ac4 before and after the removal of extraneous images. The Student t test was performed by using Microsoft Office Excel 2003 (Microsoft Corp.). One-way ANOVA was performed with SigmaStat v2.0 (SPSS, Inc.).
Extraneous image removal using Auto PHLIP-ML.
The program Auto PHLIP-ML, downloadable from the SourceForge website (http://sourceforge.net/projects/auto-phlip-ml/), was developed with LabVIEW 7.1 (National Instruments). However, the installation of LabVIEW is not required to run Auto PHLIP-ML. Auto PHLIP-ML provides users with an automated means to remove extraneous images from biofilm image stacks. Multiple z-stacks can be loaded into Auto PHLIP-ML by using the graphical user interface (Fig. 1).
The iterative process of identifying and removing extraneous images from biofilm z-stacks based on percent area coverage begins by determining the Otsu threshold for the z-stack (Table 1). The threshold is calculated by concatenating all of the images in the stack and then performing the Otsu algorithm (34). The resulting threshold is used for image segmentation, a process that distinguishes the fluorescent signal from void space. Pixels with values equal to or above the threshold are considered to be a fluorescent signal, and pixels with values less than the threshold are void space. After image segmentation, the percent area coverage (the number of pixels representing fluorescent signal divided by the total number of pixels) is determined for the segmented images. Two separate percent area coverage values are used to identify and preclude extraneous images. One PACVEIR identifies the substratum of the biofilm, and the other identifies the bulk-medium interface. Image removal starts at the base of the biofilm and continues through the image stack to the bulk-medium interface. Basal images with a percent area coverage value less than the substratum PACVEIR are removed. When an image's percent area coverage value equals or exceeds the substratum PACVEIR, that image is set as the new substratum and the criterion for image removal switches to the bulk-medium interface PACVEIR. Once an image is found to be lower than the interface PACVEIR, it and subsequent images are removed from the image stack (Table 1, original stack). A new Otsu threshold is then calculated for the trimmed z-stack to determine new percent area coverage values for the remaining images (Table 1, first iteration). As before, images that do not meet the specified PACVEIR are removed. The iterative process continues until the first and last images meet the selected substratum and interface PACVEIR criteria (Table 1, second iteration). Individual images from the middle of the stack are not removed.
TABLE 1.
Example of PACVIER method utilizing 1% for the substratum and 0.1% for the bulk-medium interfacea
| z-stack image no.c | Biomass area coverage (%)b
|
||
|---|---|---|---|
| Original stack (threshold = 43) | First iteration (threshold = 52) | Second iteration (threshold = 54) | |
| 1 (Substratum) | 3.18 | 2.60 | 1.52 |
| 2 | 15.87 | 12.29 | 11.64 |
| 3 | 40.14 | 32.67 | 31.17 |
| 4 | 61.16 | 50.83 | 48.68 |
| 5 | 67.72 | 56.05 | 53.55 |
| 6 | 65.61 | 52.57 | 49.89 |
| 7 | 59.65 | 45.84 | 43.06 |
| 8 | 51.86 | 37.63 | 34.92 |
| 9 | 42.95 | 29.27 | 26.81 |
| 10 | 34.06 | 21.53 | 19.36 |
| 11 | 24.89 | 14.36 | 12.67 |
| 12 | 17.49 | 9.15 | 7.88 |
| 13 | 11.24 | 5.37 | 4.55 |
| 14 | 6.71 | 2.77 | 2.27 |
| 15 | 3.78 | 1.43 | 1.17 |
| 16 | 1.98 | 0.67 | 0.53 |
| 17 | 0.92 | 0.26 | 0.20 |
| 18 | 0.40 | 0.09 | |
| 19 | 0.17 | 0.03 | |
| 20 | 0.06 | ||
| 21 | 0.03 | ||
| 22 | 0.01 | ||
| 23 | 0.00 | ||
| 24 | 0.00 | ||
A threshold is calculated for the original z-stack by concatenating all of the images and performing the Otsu algorithm. Image removal begins at the substratum; images with a cellular area coverage value of <1% (or a value set by the user) are removed. Once an image with a cellular area coverage value of ≥1% is identified, it is set as the new substratum and the value of comparison switches to 0.1% (or a value set by the user) to identify the bulk-medium interface. Once an image with a cellular area coverage value of <0.1% is found, that image and all remaining images are removed from the stack (boldface values). A new Otsu threshold is calculated for the remaining images, and image removal is repeated. The process continues until the substratum image and bulk-medium interface image have a cellular area coverage value of greater than or equal to 1 and 0.1%, respectively.
To calculate the cellular percent area coverage, each image is segmented using the calculated Otsu threshold. The number of pixels representing fluorescent signals is then divided by the total number of pixels in the image and multiplied by 100%.
The z-step between images was 1 μm.
For all biofilms in the present study, a PACVEIR between 0 and 1% was used to identify the bulk-medium interface. The PACVEIR used to find the substratum was held constant at 1%. The image information for trimmed z-stacks was saved in a PHLIP-ML file automatically written by Auto PHLIP-ML (Fig. 1). Auto PHLIP-ML also saves the Otsu threshold for each z-stack in the PHLIP-ML file. This prevents PHLIP from unnecessarily performing the Otsu algorithm a second time. Ten batch PHLIP-ML files were created, one for each biofilm. Information of trimmed z-stacks resulting from a single bulk-medium interface PACVEIR comprised one sample in a batch file.
Artificially adding extraneous images.
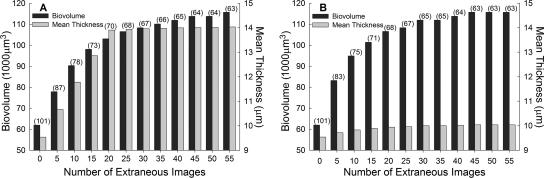
Extraneous images were artificially added to a random z-stack to investigate the extent that they impact the calculation of threshold and the architectural metrics biovolume and mean thickness. The z-stack originally contained images 1 through 25, with image 1 being the substratum. After visual inspection, images 1 through 19 were found to contain fluorescent pixels. After an interface PACVEIR of 0% had been applied to the image stack, images 1 through 13 remained. Hence, images 14 through 19 were considered to be extraneous because they did not contain biofilm-representing pixels after image segmentation. In addition to the six extraneous images 14 through 19, 49 images of image 20, which had a mean pixel value of zero, were added as images 20 through 68. The threshold, biovolume, and mean thickness were calculated for the image stack containing no extraneous images (images 1 through 13) and up to 55 extraneous images (Fig. 2A). To exclude the effects of the extraneous images that were visually considered to contain biofilm information, images 14 through 19 were excluded from analysis, and 55 images of image 20 were added as images 14 through 68. As before, the threshold, biovolume and mean thickness were calculated for the image stack containing no extraneous images and up to 55 extraneous images (images 1 through 68; Fig. 2B).
FIG. 2.
Effect of extraneous images on Otsu threshold (numbers in parentheses), biovolume, and mean thickness calculations. Of 25 images in the z-stack, images 1 through 19 were visually identified to contain fluorescent pixels. However, after a PACVEIR of 0% was applied images 1 through 13 remained and were defined to contain zero extraneous images. Images 14 through 19 were therefore considered to be extraneous due to the lack of architecture information after image segmentation. (A) Up to 49 supplementary extraneous images with a mean pixel value of zero were added in addition to images 14 through 19. (B) To exclude the effects of images 14 through 19, up to 55 extraneous images with a mean pixel value of zero were added in addition to images 1 through 13. The effects of images 14 through 19 had a more dramatic effect on mean thickness (A) than when they were excluded (B).
RESULTS AND DISCUSSION
Effect of extraneous images on image analysis.
The architecture descriptors biovolume and mean thickness were used to investigate the effect that extraneous images have on automated thresholding. Because of their fundamental representation of biofilm architecture, biovolume and mean thickness are widely accepted parameters to obtain biologically relevant information from biofilm images (19, 43, 48). As seen with PHLIP, the presence of extraneous images dramatically biased the Otsu threshold, consequently affecting the quantification of biovolume and mean thickness (Fig. 2). Mean thickness was more affected when the extraneous images 14 through 19 were present, images that contained fluorescent pixels but lacked cellular coverage after threshold segmentation (Fig. 2A). Applying an algorithm for intensity correction (29, 47) to z-stacks prior to image analysis would be expected to minimize the presence of such images. However, even when the extraneous images 14 through 19 were excluded, the Otsu threshold and biovolume were still affected when extraneous images with a mean pixel value of zero were present (Fig. 2B). In both scenarios, remarkably different results were obtained with only a difference of five extraneous images. Hence, obtaining reproducible architecture quantification requires the removal of extraneous images, whether the images result from overshooting the biofilm boundaries during CLSM acquisition or as a result of image segmentation.
Resolving automated thresholding-induced bias.
Removing extraneous images from CLSM stacks was paramount to eliminate automated thresholding bias. Auto PHLIP-ML was developed to calculate an unbiased Otsu threshold for each z-stack by automatically removing extraneous images based on their percent area coverage of biomass. The PACVEIR was held constant for the biofilm substratum because the sudden appearance of cells was not ambiguous to identify. A relatively low value of 1% was used to capture the first appearance of cells at the substratum due to the glass coverslip not being perfectly level. In cases where the coverslip is aligned or an alignment algorithm has been applied to the z-stacks, a greater PACVEIR could be used. In contrast, the bulk-medium interface was ambiguous to identify due to the gradual disappearance of cells. Therefore, a range of PACVEIRs were tested between 0 and 1%. Because the calculated threshold directly depends on the chosen PACVEIR, a method of selection was established to determine the optimal bulk-medium interface PACVEIR.
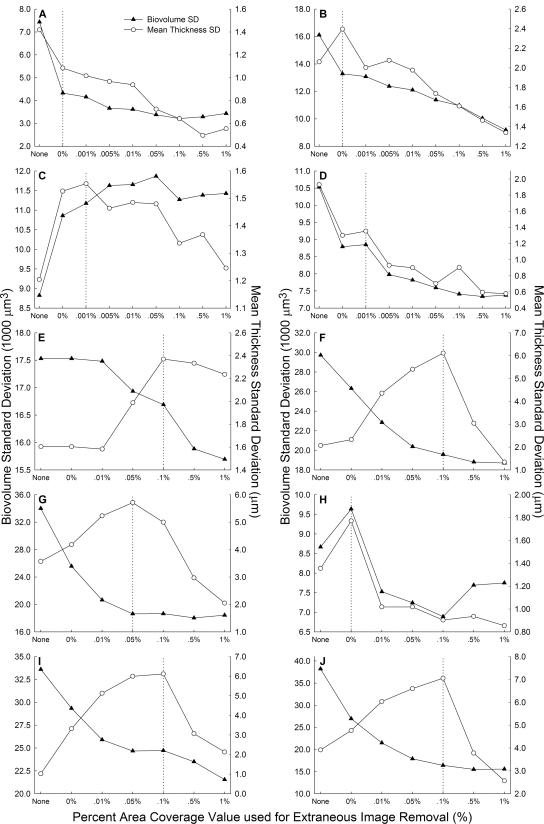
The basis for selecting the most favorable PACVEIR was to optimize the representation of biofilm architecture by digital images. Ideal representation was determined to take place when the standard deviation of the average mean thickness was at its maximum value between bulk-medium interface PACVEIRs of 0 and 1% (Fig. 3). This result may at first appear counterintuitive because it is generally desirable to minimize the standard deviation of relevant architectural descriptors when attempting to cultivate replicate biofilms (15, 20, 23). However, our study suggests that selecting the bulk-medium interface PACVEIR corresponding to the maximal standard deviation of biofilm thickness ensures that the inherent variability of biofilm architecture is being preserved. Another benefit of utilizing the optimal PACVEIR is that Otsu threshold values are set objectively and become reproducible between different investigators.
FIG. 3.
Effects of various interface PACVEIRs on the standard deviation of biovolume and mean thickness of BD413 biofilms Ac1 to Ac4 (A to D, respectively), JMP228n::gfp biofilms Ral1 and Ral2 (E and F, respectively), and JMP228n::gfp biofilms Jmp1 to Jmp4 (G to J, respectively). The percent area coverage criterion for the substratum was held constant at 1%. The vertical dashed line indicates the maximum standard deviation of mean thickness and the corresponding optimal PACVEIR. Utilizing the identified percent area coverage value as the criterion for image removal maintains the inherent structural variation of the biofilm and sets a threshold value that is both unbiased and reproducible. BD413 biofilms (A to D) had a characteristic PACVEIR that was nearly 0%, while JMP228n::gfp biofilms (E to J) had an average interface PACVEIR near 0.1%. The x axes are not to scale.
A characteristic value for the optimal PACVEIR was observed for the two bacterial species. The JMP228n::gfp biofilms on average had a value near 0.1% (Fig. 3E to J), and BD413 biofilms had an optimal value very close to 0% (Fig. 3A to D). This suggests that different types of biofilms will have a characteristic PACVEIR that can be used to optimally remove extraneous images. The biofilm Jmp2 (Fig. 3H) was the only biofilm to dramatically deviate from this trend. Although deviations are to be expected, their occurrence cannot yet be predicted and necessitates the testing of a full range of PACVEIRs.
Architecture quantification based on the optimal PACVEIR versus no removal.
Given the significant number of references to COMSTAT (21), the majority of reports investigating biofilm architecture utilize manual thresholding (30, 31, 38, 41), which is neither unbiased nor reproducible. However, heedless implementation of automated thresholding can also lead to a biased threshold that, depending on the number of extraneous images present, may not be reproducible either. Utilizing a skewed threshold leads to irreproducible and biased calculations of biofilm architecture metrics. Because only recently developed image analysis programs (5, 10, 34) provide a means to automatically determine a threshold value, the extensive use of automated methods has not yet become prevalent in the literature. Investigators utilizing such programs, including Auto PHLIP-ML, are advised to utilize automated techniques with circumspection, especially when coupled with automated CLSM image acquisition. For instance, it is possible that the bulk-medium PACVIER might find a false bulk-medium interface for a biofilm that is discontinuous in the z-direction (however, this is unlikely for PACVEIR values of ≤0.1%). In this case, utilizing a PACVEIR of 0% may be better than using the identified “optimal” PACVEIR. Alternatively, the acquisition of an additional constituent, such as EPS, may be more appropriate to represent the boundaries of the biofilm.
In the present study, the quantification of biovolume and/or mean thickness for 9 of the 10 biofilms was significantly affected (P < 0.05) by extraneous images. In contrast to JMP228n::gfp biofilms (Table 2), BD413 biofilms maintained consistent average values of B-%RSD and MT-%RSD that did not differ by more than 0.5% (Table 3). This suggests that BD413 formed relatively homogeneous biofilms, which coincides with the results of Perumbakkam et al. (40), who observed a low variation of mean thickness of BD413 when monitored in the central region of a flow cell. By definition, homogeneous biofilms are presumed to maintain a relatively constant thickness. Therefore, extraneous images would not contribute to a variation in statistical results because each image stack would contain approximately the same number of extraneous images and be a common factor among replicate z-stacks. As hypothesized, the one-way ANOVA results on the replicate BD413 biofilms Ac1, Ac2, Ac3, and Ac4 did not change after precluding extraneous images (data not shown).
TABLE 2.
Effect of extraneous images on R. eutropha JMP228n::gfp biofilms
| Biofilm and parameterf | No image removal
|
Extraneous image removala
|
||
|---|---|---|---|---|
| Avgb ± SD | %RSDc | Avgb ± SD | %RSDc | |
| Jmp1d | ||||
| Threshold | 110 ± 8 | 126 ± 16 | ||
| Biovolume (103 μm3)f | 171.0 ± 34.0 | 19.9 | 138.9 ± 18.6 | 13.4 |
| Mean thickness (μm)f | 32.1 ± 3.6 | 11.2 | 23.9 ± 5.7 | 23.8 |
| Jmp2d | ||||
| Threshold | 76 ± 3 | 87 ± 4 | ||
| Biovolume (103 μm3)f | 179.6 ± 8.7 | 4.8 | 143.4 ± 9.6 | 6.7 |
| Mean thickness (μm)f | 25.9 ± 1.4 | 5.4 | 23.7 ± 1.8 | 7.6 |
| Jmp3d | ||||
| Threshold | 59 ± 6 | 68 ± 11 | ||
| Biovolume (103 μm3)f | 145.3 ± 33.6 | 23.1 | 111.3 ± 24.7 | 22.2 |
| Mean thickness (μm)f | 36.0 ± 1.0 | 2.8 | 23.6 ± 6.1 | 25.8 |
| Jmp4d | ||||
| Threshold | 63 ± 7 | 72 ± 14 | ||
| Biovolume (103 μm3)f | 126.8 ± 38.2 | 30.1 | 96.5 ± 16.4 | 17.0 |
| Mean thickness (μm)f | 29.0 ± 3.9 | 13.4 | 22.0 ± 7.1 | 32.3 |
| Ral1e | ||||
| Threshold | 100 ± 9 | 100 ± 9 | ||
| Biovolume (103 μm3) | 62.4 ± 17.5 | 28.0 | 61.6 ± 16.7 | 27.1 |
| Mean thickness (μm) | 30.3 ± 1.6 | 5.3 | 29.2 ± 2.4 | 8.2 |
| Ral2e | ||||
| Threshold | 56 ± 20 | 60 ± 22 | ||
| Biovolume (103 μm3) | 56.3 ± 29.7 | 52.8 | 47.6 ± 19.6 | 41.2 |
| Mean thickness (μm)f | 31.1 ± 2.1 | 6.8 | 25.2 ± 6.1 | 24.2 |
| Avg B-%RSDg | 26.5 | 21.3 | ||
| Avg MT-%RSDf,h | 7.5 | 20.3 | ||
Extraneous image removal values based on the optimal PACVEIR determined by maximum mean thickness standard deviation.
Averaged from replicate z-stacks.
Percent relative standard deviation (SD/average·100%).
Illustrated in Fig. 3G to J.
Illustrated in Fig. 3E and F.
Significant difference (P < 0.05) between average values after extraneous image removal.
The average percent relative standard deviation of biovolume.
The average percent relative standard deviation of mean thickness.
TABLE 3.
Effect of extraneous images on Acinetobacter sp. strain BD413 biofilms
| Biofilm and parameter | No image removal
|
Extraneous image removala
|
||
|---|---|---|---|---|
| Avgb ± SD | %RSDc | Avgb ± SD | %RSDc | |
| Ac1d | ||||
| Threshold | 27 ± 5 | 32 ± 8 | ||
| Biovolume (103 μm3)e | 68.3 ± 7.4 | 10.8 | 55.3 ± 4.3 | 7.8 |
| Mean thickness (μm)e | 11.5 ± 1.4 | 12.2 | 10.6 ± 1.1 | 10.4 |
| Ac2d | ||||
| Threshold | 49 ± 12 | 55 ± 13 | ||
| Biovolume (103 μm3)e | 72.9 ± 16.1 | 22.1 | 62.5 ± 13.3 | 21.3 |
| Mean thickness (μm) | 16.4 ± 2.1 | 12.8 | 15.8 ± 2.4 | 15.2 |
| Ac3d | ||||
| Threshold | 74 ± 13 | 87 ± 17 | ||
| Biovolume (103 μm3)e | 90.7 ± 8.8 | 9.7 | 73.4 ± 11.2 | 15.3 |
| Mean thickness (μm)e | 13.0 ± 1.2 | 9.2 | 11.9 ± 1.6 | 13.4 |
| Ac4d | ||||
| Threshold | 23 ± 4 | 26 ± 5 | ||
| Biovolume (103 μm3)e | 55.1 ± 10.5 | 19.1 | 46.7 ± 8.9 | 19.1 |
| Mean thickness (μm)e | 12.1 ± 1.9 | 15.7 | 10.7 ± 1.4 | 13.1 |
| Avg B-%RSDf | 15.4 | 15.9 | ||
| Avg MT-%RSDg | 12.5 | 13.0 | ||
Extraneous image removal values based on the optimal PACVEIR determined by maximum mean thickness standard deviation.
Averaged from replicate z-stacks.
Percent relative standard deviation (SD/average × 100%).
Illustrated in Fig. 3A to D.
Significant difference (P < 0.05) between average values after extraneous image removal.
The average relative standard deviation of biovolume.
The average relative standard deviation of mean thickness.
Conversely, in addition to the significant differences in architecture quantification, the statistical comparison of heterogeneous biofilms would also be expected to be significantly affected due to a variable number of extraneous images in each z-stack. Compared to BD413, the average B-%RSD for the six JMP228n::gfp biofilms showed a greater decrease of 5.2% (26.5 to 21.3%) and a significant increase (P < 0.05) of 12.8% (7.5 to 20.3%) for the average MT-%RSD (Table 2). Although the architecture of JMP228n::gfp biofilms has not previously been described, this observation suggests that biofilms of this strain are heterogeneous. As predicted for heterogeneous biofilms, the statistical results comparing JMP228n::gfp biofilms were altered due to the presence of extraneous images (Table 4). Prior to precluding extraneous images, t test analyses detected significant differences (P < 0.05) between the average mean thicknesses of the replicate biofilms Jmp1 and Jmp2 and the replicate biofilms Jmp3 and Jmp4. Upon removal, the difference in the average mean thickness was negligible, which would be expected (and preferred) for replicate biofilms. In addition, once extraneous images were removed from the biofilms Ral1 and Ral2, the effects on biovolume and mean thickness from a glass coverslip versus a polyethylene membrane as an attachment medium became statistically significant (P < 0.05; Table 4). These results are of notable relevance when mean thickness is utilized as the measure of experimental reproducibility as by Heydorn et al. (20) or used to develop conclusions about differences in architecture, which is popularly done (4, 8, 19, 35, 43).
TABLE 4.
Statistical comparison of R. eutropha JMP228n::gfp biofilms
| Biofilm(s) and parameter | Statistically significant differences between biofilmsa
|
|
|---|---|---|
| No image removal | Extraneous image removalb | |
| Jmp1 and Jmp2c | ||
| Avg biovolume | - | - |
| Avg mean thickness | + | - |
| Jmp3 and Jmp4c | ||
| Avg biovolume | - | - |
| Avg mean thickness | + | - |
| Ral1 and Ral2d | ||
| Avg biovolume | - | + |
| Avg mean thickness | - | + |
Student t test with P < 0.05; “+” represents a significant difference, and “−” signifies no difference.
Extraneous image removal values based on the optimal PACVEIR determined by maximum mean thickness standard deviation.
Replicate biofilms grown under identical conditions.
Ral1 was grown on a glass coverslip, and Ral2 was grown on a polyethylene membrane.
For instance, when the effect of extraneous images was ignored, JMP228n::gfp biofilms were found to form more homogeneous biofilms than BD413 biofilms based on their average MT-%RSD: 7.5 and 12.5%, respectively (Tables 2 and 3). However, once extraneous images were removed using their optimal PACVEIR, the architectural differences between the two strains changed. JMP228n::gfp biofilms were then found to form more heterogeneous biofilms than BD413: 20.3 versus 13.0%, respectively. Such alterations of results emphasize the necessity to preclude extraneous images when automated thresholding is used.
Auto PHLIP-ML streamlines automated CLSM image processing.
The image analysis software packages COMSTAT, ISA-3D and PHLIP have previously been compared (34). Although one program is not clearly better than the other in terms of utility and user friendliness, they have limitations in common. The first bottleneck that occurs in automated image analysis is the method of importing acquired CLSM images for analysis. Unfortunately, ISA-3D's method of importing confocal images was not available for comparison. Although COMSTAT and PHLIP directly support Leica TCS4D formatted CLSM stacks (PHLIP additionally supports Leica TCS-NT), other confocal microscope image formats such as Zeiss's proprietary format (LSM) must be converted to a supported image format such as TIFF or BMP. In order to import converted images into PHLIP or COMSTAT an auxiliary file must be created containing essential information about the images (e.g., name of images, pixel size, and z-step). This file is tedious and time-consuming to create manually, especially for extremely large data sets. Auto PHLIP-ML was also designed to overcome this bottleneck of automated image analysis by automatically creating the PHLIP-ML file that is required to import converted images into PHLIP.
In comparison, a PHLIP-ML file containing 96 z-stacks (four independent biofilms each with 24 replicate z-stacks) that was written manually took the user an average of 2 h to create, while the same file created by Auto PHLIP-ML took the user less than 5 min. In a more complex scenario, such as the method of removing extraneous images from the same 96 z-stacks, a manually written PHLIP-ML file took the user an average of 8 h to perform one iteration utilizing a single PACVEIR. With Auto PHLIP-ML the user was able to identify the optimal PACVEIR in less than 3 h.
The PHLIP-ML format was chosen because of its expandability and potential to become a universal medium to standardize the format of CLSM data (34). Utilizing PHLIP as the program for image processing allows for automated analysis of multiple z-stacks and, due to its extendable design and open source license, provides the user with the potential to expand input and output functionality. To our knowledge, PHLIP is currently the only image analysis package besides DAIME (10) that supports automated analysis of multichannel CLSM stacks, which is important for ever more elaborate biofilm experiments requiring the detection of multiple fluorescent markers and subsequent colocalization analysis (14, 18, 37). Taking advantage of PHLIP's functionality, Auto PHLIP-ML also supports multichannel CLSM stacks when removing extraneous images. The user can select which detection channel best represents the boundaries of the biofilm. Another benefit of extraneous image removal by Auto PHLIP-ML is the automated identification of the biofilm's substratum, a necessity to quantify mean thickness and implement algorithms such as connected volume filtration (21).
In conclusion, removal of bias from automated image analysis is crucial for correctly correlating and interpreting biofilm structure and function relationships. Auto PHLIP-ML augments PHLIP's novel approach to automated thresholding of three-dimensional image stacks by determining an Otsu threshold unbiased by the number of extraneous images present. As a result, the quantification of biofilm architecture metrics becomes more reproducible. Further development of Auto PHLIP-ML will provide integration with other automated image analysis programs that use alternative algorithms to automatically calculate the threshold. A single intensity threshold, as utilized in the present study, may not be suitable in all cases for segmentation of three-dimensional image stacks when considering sources of error such as photobleaching during image recording, various expression levels of reporter genes such as gfp, or irregular immunofluorescence fluorophore brightness. More elaborate segmentation techniques that make use of edge information, the spatial coherence of segmented voxels, and local intensity thresholds in optical sections and subvolumes are already being applied during image analysis of computer tomography data; in the future they may be refined to analyze microscopic biofilm images.
Acknowledgments
We thank Gregory R. Stanczak for advice with LabView concerning the programming of Auto PHLIP-ML, Joao Xavier for recommendations on Auto PHLIP-ML's user interface, and Belinda McSwain for suggestions pertaining to the method of selecting the optimal percent area coverage value used for extraneous image removal.
R.T.M. was partially funded by an industry supported fellowship under the Training Program in Biomolecular Technology at the University of California, Davis, and from the NEAT-IGERT program sponsored by the National Science Foundation (IGERT grant DGE-9972741). J.E.W. was a fellow of the NIH Training Grant in Biomolecular Technology supported by grant number T32-GM08799 from the National Institute of General Medical Sciences (NIGMS), National Institutes of Health (NIH). H.M. was a fellow of the Ecotoxicology Lead Campus Program and the Toxic Substances Research and Teaching Program, and the project described was also supported by grant number 5 P42 ES004699 from the National Institute of Environmental Health Sciences (NIEHS), NIH.
The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS, NIEHS, or NIH.
Footnotes
Published ahead of print on 1 June, 2007.
REFERENCES
- 1.Bais, H. P., R. Fall, and J. M. Vivanco. 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban, N., P. Stoodley, C. A. Fux, S. Wilson, J. W. Costerton, and G. Dell'Acqua. 2005. Prevention of staphylococcal biofilm-associated infections by the quorum sensing inhibitor RIP. Clin. Orthop. Relat. Res. 437:48-54. [DOI] [PubMed] [Google Scholar]
- 3.Bathe, S., T. V. K. Mohan, S. Wuertz, and M. Hausner. 2004. Bioaugmentation of a sequencing batch biofilm reactor by horizontal gene transfer. Water Sci. Technol. 49:337-344. [PubMed] [Google Scholar]
- 4.Battin, T. J., L. A. Kaplan, J. D. Newbold, X. H. Cheng, and C. Hansen. 2003. Effects of current velocity on the nascent architecture of stream microbial biofilms. Appl. Environ. Microbiol. 69:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyenal, H., C. Donovan, Z. Lewandowski, and G. Harkin. 2004. Three-dimensional biofilm structure quantification. J. Microbiol. Methods 59:395-413. [DOI] [PubMed] [Google Scholar]
- 6.Beyenal, H., Z. Lewandowski, and G. Harkin. 2004. Quantifying biofilm structure: facts and fiction. Biofouling 20:1-23. [DOI] [PubMed] [Google Scholar]
- 7.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2001. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl. Environ. Microbiol. 67:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, B. B., J. A. J. Haagensen, A. Heydorn, and S. Molin. 2002. Metabolic commensalism and competition in a two-species microbial consortium. Appl. Environ. Microbiol. 68:2495-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton, J. W. 1995. Overview of microbial biofilms. J. Ind. Microbiol. 15:137-140. [DOI] [PubMed] [Google Scholar]
- 10.Daims, H., S. Lucker, and M. Wagner. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200-213. [DOI] [PubMed] [Google Scholar]
- 11.Emmert, E. A., and J. Handelsman. 1999. Biocontrol of plant disease: a (gram-)positive perspective. FEMS Microbiol. Lett. 171:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Flemming, H. C. 1995. Biofouling and biocorrosion: effects of undesired biofilms. Chem-Ing-Tech 67:1425-1430. [Google Scholar]
- 13.Fletcher, M. 1994. Bacterial biofilms and biofouling. Curr. Opin. Biotechnol. 5:302-306. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, E. S., A. Khlebnikov, S. E. Cowan, and J. D. Keasling. 2001. Analysis of biofilm structure and gene expression using fluorescence dual labeling. Biotechnol. Prog. 17:1180-1182. [DOI] [PubMed] [Google Scholar]
- 15.GrayMerod, R., L. Hendrickx, L. N. Mueller, J. B. Xavier, and S. Wuertz. 2005. Effect of nucleic acid stain Syto9 on nascent biofilm architecture of Acinetobacter sp. BD413. Water Sci. Technol. 52:195-202. [Google Scholar]
- 16.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 17.Hall-Stoodley, L., and P. Stoodley. 2005. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13:7-10. [DOI] [PubMed] [Google Scholar]
- 18.Hendrickx, L., M. Hausner, and S. Wuertz. 2003. Natural genetic transformation in monoculture Acinetobacter sp. strain BD413 biofilms. Appl. Environ. Microbiol. 69:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heydorn, A., B. Ersboll, J. Kato, M. Hentzer, M. R. Parsek, T. Tolker-Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heydorn, A., B. K. Ersboll, M. Hentzer, M. R. Parsek, M. Givskov, and S. Molin. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146:2409-2415. [DOI] [PubMed] [Google Scholar]
- 21.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 22.Hu, Z. Q., G. Hidalgo, P. L. Houston, A. G. Hay, M. L. Shuler, H. D. Abruna, W. C. Ghiorse, and L. W. Lion. 2005. Determination of spatial distributions of zinc and active biomass in microbial biofilms by two-photon laser scanning microscopy. Appl. Environ. Microbiol. 71:4014-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson, G., H. Beyenal, W. M. Rees, and Z. Lewandowski. 2001. Growing reproducible biofilms with respect to structure and viable cell counts. J. Microbiol. Methods 47:1-10. [DOI] [PubMed] [Google Scholar]
- 24.Juni, E., and A. Janik. 1969. Transformation of Acinetobacter calcoaceticus (Bacterium anitatum). J. Bacteriol. 98:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Junker, L. M., J. E. Peters, and A. G. Hay. 2006. Global analysis of candidate genes important for fitness in a competitive biofilm using DNA-array-based transposon mapping. Microbiology 152:2233-2245. [DOI] [PubMed] [Google Scholar]
- 26.Korber, D. R., J. R. Lawrence, M. J. Hendry, and D. E. Caldwell. 1992. Programs for determining statistically representative areas of microbial biofilms. Binary 4:204-210. [Google Scholar]
- 27.Kuehn, M., M. Hausner, H. J. Bungartz, M. Wagner, P. A. Wilderer, and S. Wuertz. 1998. Automated confocal laser scanning microscopy and semiautomated image processing for analysis of biofilms. Appl. Environ. Microbiol. 64:4115-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence, J. R., D. R. Korber, B. D. Hoyle, J. W. Costerton, and D. E. Caldwell. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, S. C., and P. Bajcsy. 2006. Intensity correction of fluorescent confocal laser scanning microscope images by mean-weight filtering. J. Microsc. 221:122-136. [DOI] [PubMed] [Google Scholar]
- 30.Lydmark, P., M. Lind, F. Sorensson, and M. Hermansson. 2006. Vertical distribution of nitrifying populations in bacterial biofilms from a full-scale nitrifying trickling filter. Environ. Microbiol. 8:2036-2049. [DOI] [PubMed] [Google Scholar]
- 31.Mai-Prochnow, A., J. S. Webb, B. C. Ferrari, and S. Kjelleberg. 2006. Ecological advantages of autolysis during the development and dispersal of Pseudoalteromonas tunicata biofilms. Appl. Environ. Microbiol. 72:5414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall, K. 1990. Biofilms, biofouling, and biocorrosion. Trends Biotechnol. 8:337-338. [Google Scholar]
- 33.Martin-Cereceda, M., B. Perez-Uz, S. Serrano, and A. Guinea. 2002. An integrated approach to analyze biofilms of a full scale wastewater treatment plant. Water Sci. Technol. 46:199-206. [PubMed] [Google Scholar]
- 34.Mueller, L. N., J. F. de Brouwer, J. S. Almeida, L. J. Stal, and J. B. Xavier. 2006. Analysis of a marine phototrophic biofilm by confocal laser scanning microscopy using the new image quantification software PHLIP. BMC Ecol. 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murga, R., P. S. Stewart, and D. Daly. 1995. Quantitative-analysis of biofilm thickness variability. Biotechnol. Bioeng. 45:503-510. [DOI] [PubMed] [Google Scholar]
- 36.Nagasawa, Y., M. Takenaka, Y. Matsuoka, E. Imai, and M. Hori. 2000. Quantitation of mRNA expression in glomeruli using laser-manipulated microdissection and laser pressure catapulting. Kidney Int. 57:717-723. [DOI] [PubMed] [Google Scholar]
- 37.Nancharaiah, Y. V., V. P. Venuopalan, S. Wuertz, P. A. Wilderer, and M. Hausner. 2005. Compatibility of the green fluorescent protein and a general nucleic acid stain for quantitative description of a Pseudomonas putida biofilm. J. Microbiol. Methods 60:179-187. [DOI] [PubMed] [Google Scholar]
- 38.O'Connell, H. A., G. S. Kottkamp, J. L. Eppelbaum, B. A. Stubblefield, S. E. Gilbert, and E. S. Gilbert. 2006. Influences of biofilm structure and antibiotic resistance mechanisms on indirect pathogenicity in a model polymicrobial biofilm. Appl. Environ. Microbiol. 72:5013-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otsu, N. 1979. A threshold selection method from gray level histograms. IEEE Trans. Syst. Man Cybern. 9:62-66. [Google Scholar]
- 40.Perumbakkam, S., T. F. Hess, and R. L. Crawford. 2006. A bioremediation approach using natural transformation in pure-culture and mixed-population biofilms. Biodegradation 17:545-557. [DOI] [PubMed] [Google Scholar]
- 41.Searcy, K. E., A. I. Packman, E. R. Atwill, and T. Harter. 2006. Capture and retention of Cryptosporidium parvum oocysts by Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 72:6242-6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sekar, R., T. Griebe, and H. C. Flemming. 2002. Influence of image acquisition parameters on quantitative measurements of biofilms using confocal laser scanning microscopy. Biofouling 18:47-56. [Google Scholar]
- 43.Staudt, C., H. Horn, D. C. Hempel, and T. R. Neu. 2004. Volumetric measurements of bacterial cells and extracellular polymeric substance glycoconjugates in biofilms. Biotechnol. Bioeng. 88:585-592. [DOI] [PubMed] [Google Scholar]
- 44.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 45.Venugopalan, V. P., A. Kuehn, M. Hausner, D. Springael, P. A. Wilderer, and S. Wuertz. 2005. Architecture of a nascent Sphingomonas sp. biofilm under varied hydrodynamic conditions. Appl. Environ. Microbiol. 71:2677-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, H. X., and L. Ji. 2005. Fully automated intensity compensation for confocal microscopic images. J. Microsc. 220:9-19. [DOI] [PubMed] [Google Scholar]
- 48.Xavier, J. B., C. Picioreanu, and M. C. van Loosdrecht. 2004. Assessment of three-dimensional biofilm models through direct comparison with confocal microscopy imaging. Water Sci. Technol. 49:177-185. [PubMed] [Google Scholar]
- 49.Xavier, J. B., A. Schnell, S. Wuertz, R. Palmer, D. C. White, and J. S. Almeida. 2001. Objective threshold selection procedure (OTS) for segmentation of scanning laser confocal microscope images. J. Microbiol. Methods 47:169-180. [DOI] [PubMed] [Google Scholar]
- 50.Xavier, J. B., D. C. White, and J. S. Almeida. 2003. Automated biofilm morphology quantification from confocal laser scanning microscopy imaging. Water Sci. Technol. 47:31-37. [PubMed] [Google Scholar]
- 51.Yang, X., H. Beyenal, G. Harkin, and Z. Lewandowski. 2001. Evaluation of biofilm image thresholding methods. Water Res. 35:1149-1158. [DOI] [PubMed] [Google Scholar]
- 52.Yun, M. A., K. M. Yeon, J. S. Park, C. H. Lee, J. Chun, and D. J. Lim. 2006. Characterization of biofilm structure and its effect on membrane permeability in MBR for dye wastewater treatment. Water Res. 40:45-52. [DOI] [PubMed] [Google Scholar]