Abstract
Biofilm formation is controlled by an array of coupled physical, chemical, and biotic processes. Despite the ecological relevance of microbial biofilms, their community formation and succession remain poorly understood. We investigated the effect of flow velocity, as the major physical force in stream ecosystems, on biofilm community succession (as continuous shifts in community composition) in microcosms under laminar, intermediate, and turbulent flow. Flow clearly shaped the development of biofilm architecture and community composition, as revealed by microscopic investigation, denaturing gradient gel electrophoresis (DGGE) analysis, and sequencing. While biofilm growth patterns were undirected under laminar flow, they were clearly directed into ridges and conspicuous streamers under turbulent flow. A total of 51 biofilm DGGE bands were detected; the average number ranged from 13 to 16. Successional trajectories diverged from an initial community that was common in all flow treatments and increasingly converged as biofilms matured. We suggest that this developmental pattern was primarily driven by algae, which, as “ecosystem engineers,” modulate their microenvironment to create similar architectures and flow conditions in all treatments and thereby reduce the physical effect of flow on biofilms. Our results thus suggest a shift from a predominantly physical control to coupled biophysical controls on bacterial community succession in stream biofilms.
Biofilms are matrix-enclosed, attached microbial communities that can develop highly differentiated architectures (i.e., physical structure), including mushroom-like structures, ripples and ridges, or filamentous streamers floating in the bulk liquid (for examples, see references 14, 17, and 43). Intensive work with single- or multispecies bacterial biofilms unraveled a wide array of factors, ranging from quorum sensing, competition, and predation to the hydrodynamics of the bulk liquid, that collectively drive structural differentiation (2, 29, 33). This research has traditionally focused on structural development, described as a sequence of events from attachment to the formation and differentiation of bacterial microcolonies as expressed by distinct changes in physical traits (e.g., thickness, roughness, and porosity). Similarly, the succession of algae in phototrophic biofilms has traditionally received much attention (40). However, few have investigated the combined succession of algae and bacteria, both in their architecture and community composition, in environmental biofilms (26).
While succession is a prominent research avenue in classical ecology, we have no clear understanding of how biofilm communities assemble and which factors drive their succession. Building evidence (19, 26, 27) suggests that initial community formation is stochastic and largely driven by the recruitment of species from the bulk liquid. As microbial cells become more abundant with biofilm growth, competition for resources may become increasingly important and less-competitive microorganisms are out-competed. Diversity would decrease during this developmental phase, with a few competitors becoming predominant. Finally, as biofilms mature, the community may become more diverse through niche diversification and internal recycling of resources. Lyautey and colleagues (26) also identified a suite of possible environmental variables (e.g., temperature and light) that may shape biofilm succession in a river. Finally, differential aggregation behavior of bacteria may also lead to higher diversity in biofilms growing under low flow than in biofilms growing under high flow (35). Despite these various pieces of evidence, we still lack a mechanistic understanding of how physical and biotic controls may collectively affect biofilm formation and succession.
Recently, Battin and colleagues (8) suggested viewing biofilms as microbial landscapes to explicitly link the hydrodynamics of the bulk liquid, the biofilm topography (i.e., the landscape), and the immigration rate of propagule cells. Their framework thus recognizes the function of coupled physical processes (e.g., hydrodynamics and topography) and demographic biological processes that are largely stochastic in nature (e.g., death and reproduction) in the assembly of biofilm communities.
The goal of this study was to test the effect of flow velocity, as the primary physical force in streams, on biofilm community succession. While chemistry (e.g., nutrients and dissolved organic carbon) is rather invariant at smaller scales in streams, as a result of continuous mixing, flow velocity is highly patchy and controls multiple ecological processes (1). In analogy to the relationship between flow velocity and deposition of suspended particles (7), we hypothesized that increased flow velocity would increase the flux of microorganisms from the bulk liquid to biofilms, thereby generating higher richness in these communities. Alternatively, flow-mediated changes in biofilm architecture could also affect community composition. To isolate flow as the governing physical determinant, we grew biofilms from raw stream water in specially designed microcosms (37) under laminar, intermediate, and turbulent flow regimes, yet exposed to the same source community and water chemistry. Our study shows clear successional trajectories of biofilm community composition, as revealed by denaturing gradient gel electrophoresis (DGGE) analysis of the bacterial 16S rRNA gene. It further suggests a shift from a predominantly physical control to coupled biological and physical controls on community composition during biofilm succession.
MATERIALS AND METHODS
Experimental setup.
Biofilms were grown in microcosms from raw stream water as described by Singer et al. (37). Briefly, the microcosms consisted of flumes made of acrylic glass (length, 1.3 m; width, 2 cm; height, 2 cm), each paved with 100 sterile unglazed ceramic coupons (1 by 2 cm) that served as the substratum for biofilm growth. The water flow was adjusted to yield a constant flow environment with specific velocity, depth, and bulk-flow Reynold's number. Three hydrodynamic treatments with laminar, intermediate, and turbulent flows were achieved by adjusting the slope and flow rate in each flume individually (Table 1), and the flow patterns were visualized by rhodamine injection (37). The flow velocities achieved are representative for midgradient streams (6, 38). We assembled sets of 12 replicate flumes for each flow treatment. Water recirculated within and among treatments to ensure complete mixing of the inoculum and water chemistry. Controlled light (mean photon flux density of 31.6 μmol m−2 s−1 during a 12-h light period per day) and temperature (15°C) further ensured constant environmental conditions. Water recirculating in the flumes was exchanged for new stream water every third day; 50% of the water was exchanged twice within one hour, resulting in a total exchange of 75%. The stream water (Piesting, Austria) was filtered (60 μm) to remove major particles and instars of macrofauna. The stream water nutrient concentration averages ± standard deviations were 21 ± 5 μg NH4-N liter−1 and 779 ± 138 μg NO3-N liter−1; the soluble reactive phosphorus was usually below the detection limit (<50 μg liter−1). The conductivity was 435 ± 6 μS cm−1.
TABLE 1.
Reynold's numbers and velocities of flow treatments
| Flow treatment | Reynold's no. | Velocity (m s−1) |
|---|---|---|
| Laminar | 257 | 0.065 |
| Transitional | 642 | 0.162 |
| Turbulent | 1541 | 0.390 |
Sampling.
We monitored bulk biomass, architecture, and community composition (bacteria and algae) during a 12-week period. Coupons were collected for analysis of bacterial abundance, chlorophyll a concentrations, and bacterial community composition on 10 occasions, and samples were collected for analysis of algal community composition on 5 occasions. For bacterial abundance and algal community composition, three coupons were sampled from the 12 flumes in a randomized way, yielding triplicate samples on each sampling date. One coupon per flume and sampling date was collected for analysis of chlorophyll a concentrations, resulting in 12 replicates. For DGGE analysis, coupons were collected from 12 replicate flumes, and four of each (from flumes 1 to 4, 5 to 8, and 9 to 12) were pooled to yield triplicate samples. The stream water was sampled every week to get a DGGE fingerprint of the source community; microbial cells were harvested from the stream water through filtration (0.2-μm GSWP filter; Millipore Inc.) and frozen pending further processing. This resulted in more than 110 samples for DGGE analysis.
Microcosms were tested for reproducibility at several levels of the experimental setup in previous work; no longitudinal gradients could be detected in the individual flumes, and reproducibility within treatments proved to be excellent (37). Coupons were collected from downstream to upstream to avoid changes in hydrodynamics.
Chlorophyll a concentrations and bacterial and cyanobacterial abundance.
Chlorophyll a was extracted with p.a.-grade acetone (12 h, 4°C) in the dark. Samples were vortexed, and the supernatant filtered (GF/F; Whatman) and assayed fluorometrically (EX435/EM675), using spinach (Sigma) as a standard. Coupons with biofilm were incubated with 0.025 mmol/liter tetrasodium pyrophosphate solution, shaken for 1 h, and subsequently sonicated (180 s, 40 W output; Branson) to detach and disaggregate cells (44). An aliquot of the suspension was stained with SYBR green I (Invitrogen) and filtered onto a 0.2-μm black filter (GTBP; Millipore Corp., Bedford, MA). Bacteria and cyanobacteria were enumerated in 30 randomly selected fields to account for 300 to 500 cells by using epifluorescence microscopy (Nikon E800).
Microscopy of biofilms.
Confocal laser scanning microscopy was performed with an inverse LSM 510 (Zeiss, Jena, Germany). Biofilms were carefully transferred into chambered coverglass units (Nalge Nunc Intl., IL) and stained “in situ” with SYTO 13 (Invitrogen). Series of 22 to 59 optical sections were recorded by using a water immersion lens objective (C-Apochromat 40×/1.2 W Korr.). Two-dimensional projections of the image stacks were used to estimate the percent coverage using ImageJ (http://rsb.info.nih.gov/ij/). Color-coded depth profiles of the three-dimensional biofilm structure were reconstructed using the manufacturer's software. Dark-field and epifluorescence microscopy (Nikon E800) were used to further examine structural details of the biofilms.
Identification and quantification of algae.
Algae were removed from the coupons by sonication and preserved in Lugol's solution. At least 400 cells per sample were counted with an inverted microscope. At least 30 cells of each taxon were measured, and their biovolume determined using standard geometric formulae (18).
PCR and DGGE.
DNA from biofilms and the source community was extracted and purified with the UltraClean soil DNA isolation kit from MoBio (Carlsbad, CA). Filters were cut in pieces, using ethanol-flamed scissors and tweezers, to enhance the extraction efficiency. Noncolonized ceramic coupons and unused filters served as negative controls. DNA concentrations were determined fluorometrically, using a fluorescent DNA quantitation kit (Bio-Rad Laboratories Inc., CA). The primers used for PCR of the 16S rRNA gene were 341F, with a GC clamp (underlined) on its 5′ end (5′-CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC CCC TAC GGG AGG CAG CAG-3′), and 907R (5′-CCG TCA ATT CCT TTG AGT TT-3′) (30). Each 50-μl PCR mixture contained both primers at 1 μmol/liter (Thermo Electron GmbH, Germany), each deoxynucleoside triphosphate at 0.25 mmol/liter, MgCl at 1.75 mmol/liter, 50 μg bovine serum albumin, 1.25 U of Taq polymerase, and the recommended PCR buffer (all from MBI Fermentas). Samples were amplified using the following protocol: an initial denaturation step of 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 55°C for 40 s, and extension at 72°C for 1 min. Cycling was completed by a final extension at 72°C for 15 min. The integrity of the PCR products was checked on 1% agarose gels (MBI Fermentas).
DGGE was performed as described by Muyzer et al. (30), using a DCode universal mutation detection system (Bio-Rad). PCR products (400 ng to 600 ng per lane) were applied to 8% polyacrylamide gels (acrylamide-bisacrylamide, 37.5:1) with gradients ranging from 30% to 70% (where 100% denaturant contains 7 mol/liter urea and 40% deionized formamide) and stacking gels on top (0% denaturant). The DGGE gels were run at a constant voltage of 100 V at 60°C for 16 h. The gels were poststained with SYBR green I (Invitrogen), and the bands visualized with a UV Transilluminator. DGGE images were edited and analyzed using ImageJ, and the results transferred into a band presence-absence matrix. DGGE bands are subsequently referred to as operational taxonomic units (OTUs).
Phylogenetic analysis of selected DGGE bands.
Eight different prominent bands were excised from DGGE gels for phylogenetic analysis and confirmation of correct alignment. The gel slices were overlaid with 45 μl ultrapure water, and DNA was extracted overnight at 4°C. One microliter of a 1/50 dilution was reamplified using the PCR mixture described above, except that it contained MgCl at 1.5 mmol/liter and no bovine serum albumin. The cycling protocol consisted of an initial denaturation step at 94°C for 3 min, followed by 12 touchdown cycles of denaturation at 94°C for 40 s, annealing at 62°C for 40 s decreasing every other cycle by 1°C, and extension at 72°C for 1 min. Another 10 cycles of 94°C for 40 s, 56°C for 40 s, 72°C for 1 min, and a final extension step at 72°C for 15 min were performed. The purity and correct position of the PCR products of excised bands were confirmed by DGGE as described above. Reamplified DNA was purified with the QIAquick (QIAGEN) PCR purification kit according to the manufacturer's recommendations. Amplicons were added to the sequencing reactions using the 907R primer and the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems), and sequences were obtained on an ABI 3130 XL genetic analyzer (Applied Biosystems).
Sequences were checked for chimeric artifacts using the Pintail program (5). Further analysis included comparison to the sequence libraries of GenBank (9) and Ribosomal Database Project II (13) (Table 2).
TABLE 2.
Analysis of sequences obtained for selected OTUs
| DGGE band | GenBank accession no. | Closest relative (and encoded structure)a (GenBank accession no.) | Similarity (%) | Closest cultured relativea,b (GenBank accession no.) | Similarity (%) | Phylogenetic group or encoded structure |
|---|---|---|---|---|---|---|
| OTU 1 | EF396239 | Uncultured Betaproteobacterium Gitt-GS-139 (AJ582191) | 99.0 | Hydrogenophaga atypica (AJ585992) | 98.4 | Betaproteobacteria |
| OTU 2 | EF396241 | Uncultured bacterium RB041 (AB240293) | 99.2 | Herbaspirillum seropedicae (Y10146) | 94.4 | Betaproteobacteria |
| OTU 3 | EF396240 | Uncultured Betaproteobacterium KB17 (AB074944) | 100.0 | Hydrogenophaga atypica (AJ585992) | 98.0 | Betaproteobacteria |
| OTU 4 | EF396242 | Uncultured bacterium EV818SWSAP42 (DQ337076) | 99.8 | Azoarcus evansii (X77679) | 94.3 | Betaproteobacteria |
| OTU 5 | EF396243 | Zygnema circumcarinatum chloroplast (AY958086) | 89.4 | Chloroplast | ||
| OTU 6 | EF451826 | Uncultured bacterium SXAU023 (AY863079) | 91.2 | Lewinella cohaerens (AF039292) | 89.3 | Bacteroidetes |
| OTU 7 | EF451827 | Uncultured bacterium S011D (AM158337) | 96.2 | Lysobacter antibioticus (AB019582) | 92.1 | Gammaproteobacteria |
| OTU 8 | EF188846 | Uncultured bacterium A2Sp-18 (AJ965830) | 99.2 | “Candidatus Odyssella thessalonicensis” (AF069496) | 92.2 | Alphaproteobacteria |
Estimated by comparison to the Ribosomal Database Project II database (February 2007).
Species type strains.
Statistical analysis.
Bacterial abundance and chlorophyll a concentrations were z-standardized within each sampling date to remove temporal variability, as described by Singer et al. (37). Kruskal-Wallis analysis of variance (ANOVA) and the Tamhane test were used to calculate the effect of flow velocity on biofilm biomass. We computed a similarity matrix including all DGGE band presence/absence data according to the Soerensen index, 2S/(A1 + A2), where S is the number of OTUs shared between communities 1 and 2 and A1 and A2 are the total numbers of OTUs in the respective communities. This similarity matrix was subjected to multidimensional scaling (MDS) analysis to visualize the community composition dynamics. Similarity values between samples of adjacent dates or between flow treatments were further used to assess differences between biofilms and the source community. The mean similarity indices and mean numbers of OTUs were tested for significant differences using the Kruskal-Wallis ANOVA and the Tamhane test. The software package SPSS 12.0 for Windows was used for all statistical analyses.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study were submitted to GenBank and are available under the accession numbers given in Table 2.
RESULTS
Biomass development.
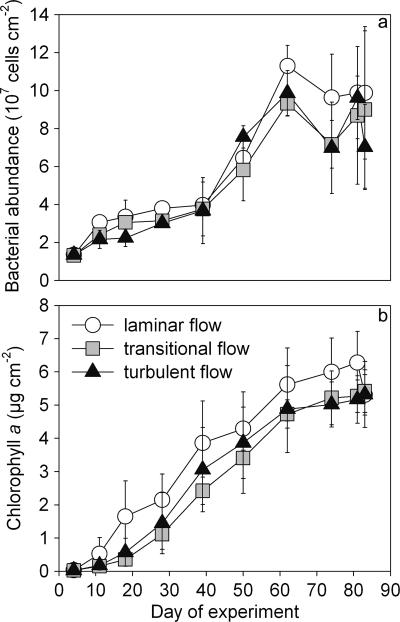
The mean bacterial abundance ranged from 1.32 × 107 to 1.13 × 108 cells cm−2, and reached stationary growth at between 50 and 60 days (Fig. 1a). The concentration of chlorophyll a increased from 0.03 to 6.28 μg cm−2, and growth leveled off after 70 days (Fig. 1b). Both bacterial abundance and concentrations of chlorophyll a were significantly higher with the laminar flow treatment than with the transitional and turbulent treatments (P < 0.001). The transitional and turbulent flow treatments exhibited no significant differences; in fact, the concentration of chlorophyll a in the turbulent flow treatment was often higher than in the transitional flow treatment. Cyanobacterial abundance ranged from 1.75 × 105 to 9.17 × 106 cells cm−2 and did not exhibit any clear pattern related to flow treatment (data not shown).
FIG. 1.
Biofilm growth, monitored by bacterial abundance (a) and chlorophyll a concentrations (b) over a time period of 83 days. Symbols and error bars represent means ± standard deviations of results for replicate samples.
Development of biofilm architecture.
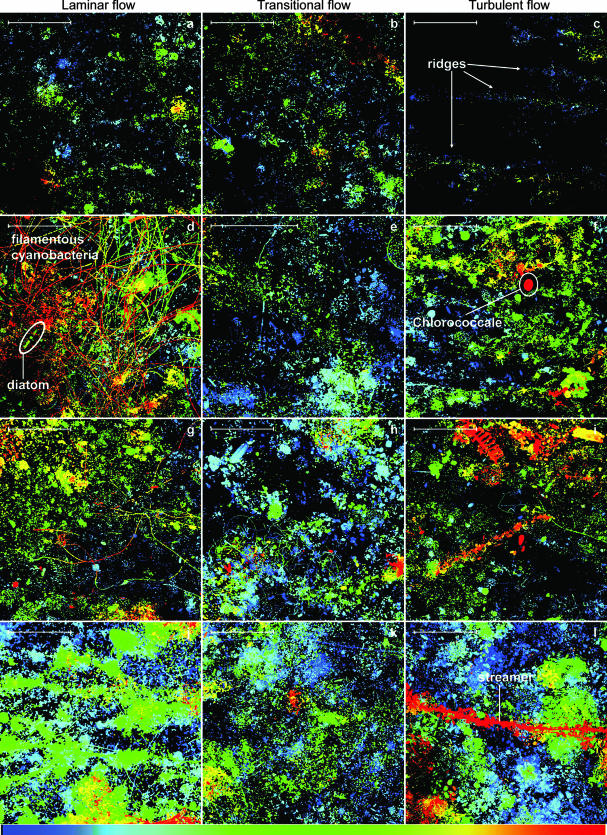
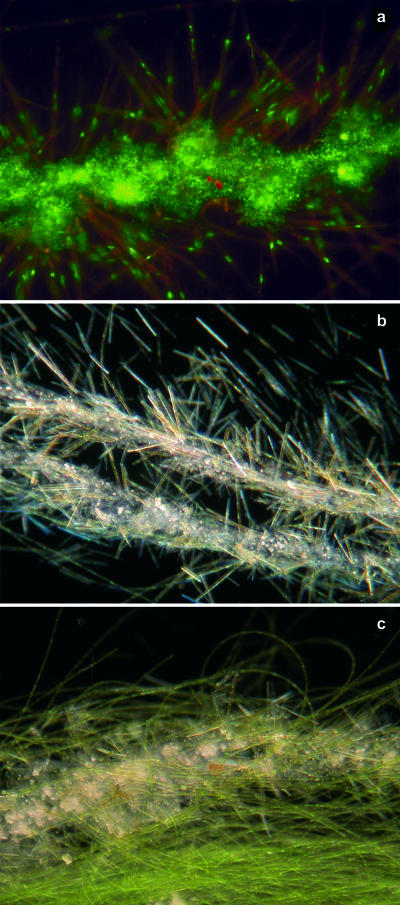
During initial (<6 days) growth, biofilm biomass was distributed rather randomly in the laminar and transitional treatments, but followed directional ridge-like patterns in the turbulent flow treatment (Fig. 2a to c). Diatoms occurred in all treatments during that initial phase, whereas filamentous bacteria and cyanobacteria increasingly characterized the biofilm landscape in the laminar flow treatment (Fig. 2d and g). The mean coverage was 10% after 6 days in all treatments and increased to about 50% around day 20; the highest coverage (66%) occurred in the laminar treatment. Around day 20, the first streamers, initially consisting of cyanobacteria and attached bacteria, emerged in the turbulent flow treatment (Fig. 2l). Between day 30 and day 40, macroscopically visible streamers developed in the turbulent and transitional flow treatments. Bacteria embedded in copious amounts of extracellular polymeric substances formed backbones densely colonized by diatoms (Fig. 3a and b), conspicuous structures that were never observed under laminar flow conditions. These “garlands” were then successively replaced by filamentous green algae (Fig. 3c), and biofilms from all three flow treatments became increasingly similar in structure.
FIG. 2.
Nascent biofilm growth visualized by confocal laser scanning microscopy at day 6 (a to c), at day 11 (d to f), at day 16 (g to i), and at day 21 (j to l) of the experiment. Images are depth coded, with blue representing the base layer and red the canopy of the biofilms. The scale bars at the upper left corners represent 100 μm.
FIG. 3.
Microstructures of streamers formed under transitional and turbulent flows. Garland streamer formed from bacteria and diatoms at day 47, stained with SYTO 13, using epifluorescence microscopy (a) and as seen by dark-field microscopy (b). (c) Garland streamers are successively overgrown by filamentous green algae (day 75).
Algal community composition.
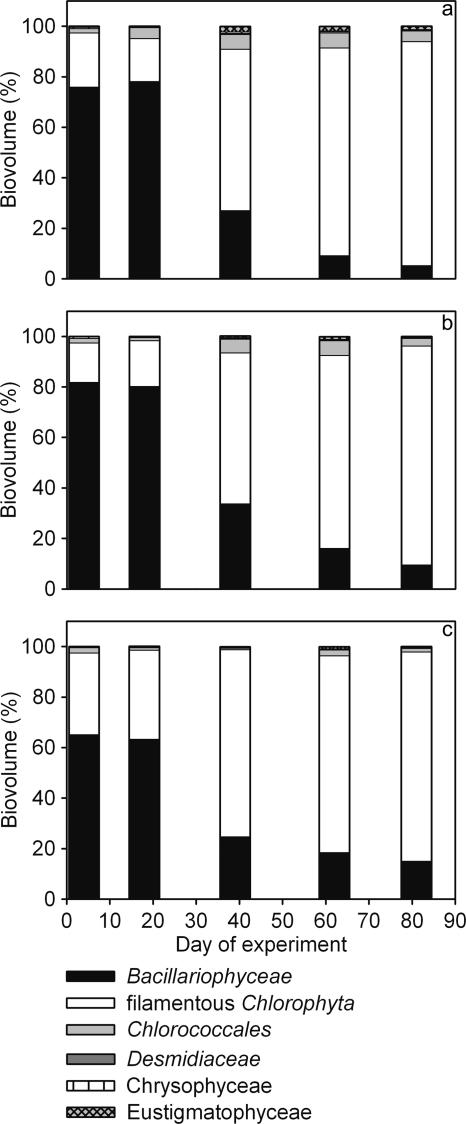
Congruent with results from confocal scanning microscopy, algal biovolumes clearly showed abundant diatom cells, accounting for most of the algal biomass in early biofilms in all flow treatments (Fig. 4). This changed dramatically between day 20 and day 40, when filamentous chlorophytes became the dominant algal group. Garland-like streamers still developed for several weeks, though in decreased numbers. Coccal chlorophytes generally exhibited higher biomass in the laminar and transitional flow treatments than in the turbulent flow treatment.
FIG. 4.
Percentages of algal groups in total algal biomass in laminar (a), transitional (b), and turbulent (c) flow treatments.
Shifts in bacterial community composition.
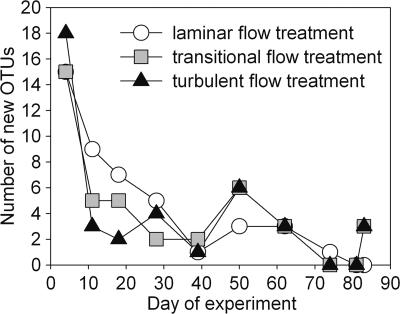
DGGE yielded reproducible results within flow treatments and revealed substantially more OTUs in the biofilms than in the stream water. In total, we identified 51 biofilm OTUs, of which only 19 also occurred in stream water samples. The number of OTUs, averaged over the experiment, was significantly (P < 0.001) lower in the stream water (8.4 ± 1.1) than in the biofilms (laminar, 15.5 ± 2.7; transitional, 12.6 ± 1.6; turbulent, 14.2 ± 1.7). Absolute richness in the biofilms did not vary significantly among treatments or over time (data not shown). However, there was a clear decreasing trend of OTUs appearing for the first time for a given sampling date during early development. (Fig. 5). This initial decline of new OTUs was more pronounced in the transitional and turbulent flow treatments, meaning that more OTUs invaded the biofilm grown under laminar flow during the first 30 days.
FIG. 5.
Numbers of new OTUs appearing in the biofilms, as inferred from the appearance of new DGGE bands in at least one of the triplicate samples.
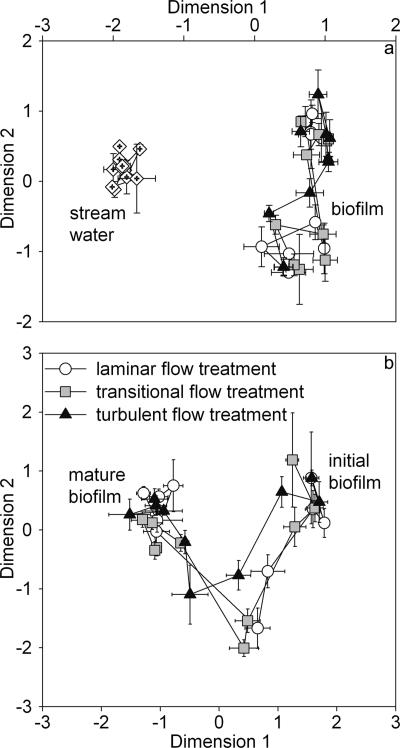
MDS analysis of these OTUs identified distinct bacterial communities in the stream water and the biofilms (Fig. 6a). Furthermore, similarity analysis (Soerensen index) of samples from consecutive dates indicated that the community composition over time was significantly (P < 0.001) less variable (0.73 ± 0.09) in the stream water than in the biofilms (laminar, 0.60 ± 0.11; transitional, 0.63 ± 0.16; turbulent, 0.64 ± 0.06).
FIG. 6.
MDS analysis of DGGE gels. (a) Mapping of stream water and biofilm communities. Kruskal's standardized stress S is 0.170. (b) Development of bacterial community compositions among the flow treatments. Kruskal's standardized stress S is 0.210.
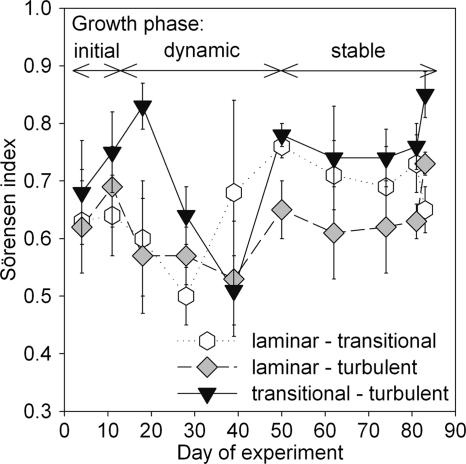
We computed the Soerensen index between community compositions of the three treatments to compare the successional trajectories (Fig. 7). This comparison revealed three major phases (<10 days, 11 to 50 days, and 51 to 90 days), corresponding to initial biofilm formation, both late lag phase and early log phase of microbial growth, and mature biofilms (Fig. 1a). After an initial phase of high similarity between treatments, communities were highly dynamic between day 10 and day 50, and started varying with little variation around a mean of high similarity during mature growth (>50 days). Remarkably, during this late stage, the highest similarity was encountered between the transitional and turbulent flow treatments and the lowest similarity between the laminar and turbulent flow treatments.
FIG. 7.
Mean similarities (Soerensen index) between biofilm bacterial communities among flow treatments, showing an early, dynamic growth phase and a more-stable later phase. The Soerensen indices compare triplicate samples from a given flow treatment with each respective triplicate sample from another flow treatment, resulting in nine similarity values per flow treatment comparison and sampling date. Given are the means ± standard deviations of these nine values.
MDS analysis of the Soerensen similarity matrix showed clear shifts in community composition (Fig. 6b). The analysis closely mapped all three flow treatments during early biofilm growth and revealed a clear divergence of the biofilm in the turbulent flow treatment between day 10 and day 50 and convergence during mature growth.
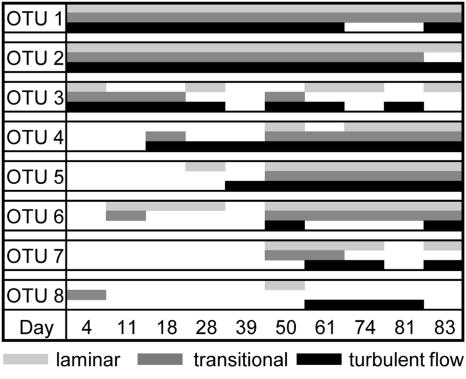
In order to identify the dominant bacteria at the typical stages of biofilm development, the sequences of prominent DGGE bands were compared with those in the Ribosomal Database Project II database. This analysis revealed sequences affiliated with Alpha-, Beta-, and Gammaproteobacteria and Bacteroidetes—all of them more closely related to environmentally derived sequences than to cultivated organisms (Table 2). One band referred to an unidentified chloroplast. Remarkably, OTUs of which the closest cultured relatives include strains from Hydrogenophaga and Herbaspirillum occurred during the entire biofilm development (Fig. 8), whereas OTUs affiliated with Azoarcus, Lewinella, and Lysobacter characterized biofilms during mature growth (>50 days), though with some differences among flow treatments. An alphaproteobacterium putatively affiliated with a bacterial endosymbiont living in amoebae occurred sporadically in all flow treatments.
FIG. 8.
Scheme of the appearance of sequenced OTUs during biofilm development. Shaded blocks indicate the presence of the respective organism in the biofilms as revealed by DGGE. OTUs occurring in at least two of the triplicate samples were considered.
DISCUSSION
Our findings indicate biophysical controls on the community succession in stream biofilms. These observations complement studies showing short-term and long-term succession of biofilms in rivers (26), lakes (19), drinking water distribution systems (27), and fixed-film reactors (28). Our study documents the effects of flow velocity on the successional trajectories of biofilm communities. It further suggests that biophysical coupling (i.e., algal growth modulating the physical microenvironment) attenuates the initial effect of flow as biofilms develop into mature communities. This is of relevance in streams, where flow is the primary physical template shaping biofilm structure-function coupling (6) and drives numerous ecological and ecosystem processes (1) in general. Our study cannot, however, demonstrate whether the observed changes are attributable to directional succession, where successive replacement of species (eventually with multiple equilibrium states) leads to a common point in community composition.
Development of biofilm architecture.
The flow treatments had noticeable effects on the spatial distribution of biomass and its dynamics. Biomass distribution was directed (i.e., nonrandom) in biofilms grown in the turbulent flow treatment, where ridges developed during initial growth (<10 days), as reported in previous studies (6, 31). Filamentous building blocks characterized biofilms in the laminar flow; initial filamentous bacteria and cyanobacteria were successively replaced by filamentous green algae. These structures formed increasingly dense mats, trapping particles from the bulk liquid and thereby achieving high coverage and biomass.
The development of streamers is well known from multispecies laboratory biofilms, where they develop up to a few hundred μm in length (41). The streamers we observed in the turbulent flow grew up to 50 mm in length and constituted a striking interaction between bacteria and diatoms and a great adaptation to the water flow. The bacterial backbone (including extracellular polymeric substances) is likely viscoelastic (42), offers ample opportunities for diatoms to colonize, and ensures continuous solute replenishment in a turbulent environment. The biofilm thus enlarges its biomass and bioreactive surface without getting eroded.
As growth progressed, biofilms in the turbulent flow treatments successively shifted from diatom- to filamentous green alga-dominated communities, followed by the reduction of the conspicuous garland-like streamers. This shift caused the overall biofilm architecture to converge among flow treatments.
Microbial community succession.
The comparison between stream water and biofilm community dynamics suggests that the observed succession in biofilms was not affected by variations in the source community. Though biofilms must have developed from microorganisms present in the stream water, many of these were likely not abundant enough to be detected by DGGE analysis. In fact, DGGE is unlikely to detect OTUs accounting for less than 1% of the total community (30). Bacterial populations potentially able to invade biofilms may indeed be present in the stream water, but the dominance of other populations makes them unlikely to be detected. Similar differences between communities in the biofilms and bulk liquid were reported previously (34, 35).
Strikingly, flow clearly shaped the succession of biofilm community composition: successional trajectories diverged from an initial common community to increasingly converge towards mature biofilms. This points towards a shift from predominantly stochastic to deterministic processes, as was recently predicted for microbial landscapes (8). Initial colonization is random, as it greatly depends on the immigration from a source community (i.e., stream water) (15) and yields almost identical communities in all flow treatments. As microorganisms in the nascent biofilms develop closer ties with their physical microenvironment, communities start to diverge—a phenomenon that is well known from plant succession, for instance (45). Given that the experimental design of our experiments clearly isolates the flow effect, our results obviously suggest that the flow drives the diverging successional trajectories during this early growth.
Divergence was most pronounced when the garland-like streamers dominated the biofilm canopy in the turbulent flow treatment. As growth progressed, community compositions converged among flow treatments to finally reach a common community composition. This occurred concurrently with the disappearance of the garland-like streamers and the establishment of filamentous algae in all flow treatments. This would agree with general succession theory predicting that, given initial landscape and environmental heterogeneity, convergence requires biologically based processes with low stochasticity (45).
We suggest the prominent shift in the algal community as such a biologically based process that triggers shifts in the microbial community. Algae, as the larger building blocks, are most responsive to the flow environment and, at the same time, can easily change the overall architecture of biofilms and thereby their hydrodynamic microenvironment and local solute supply (10, 40). We therefore propose biofilm algae as ecosystem engineers (20) that physically modify the fluid dynamics and create similar flow microenvironments in all flow treatments during late growth. Such physical modulation by organisms often entails changes in the community composition of a given system (20). As algae grow and coalesce to form larger entities, they form less heterogeneous biofilms and finally reduce the physical effect of flow on bacteria. In this way, filamentous green algae form a structural template on which bacterial communities develop. Underlying mechanisms may include even distribution of space and resources (e.g., algal exudates) among flow treatments. Our results thus indicate a shift from predominantly physical to coupled biophysical controls on bacterial diversity in stream biofilms and support our initial hypothesis.
Biofilms from the laminar flow treatment had higher appearances of new OTUs during early growth. As the absolute richness did not vary significantly among treatments, this points to higher turnover of OTUs in laminar than in turbulent flow biofilms. We suspect that this dynamic in community composition relates to the architectural dynamics of biofilms. After an initial phase, an extended boundary layer typically allows biofilms to grow thicker in the laminar flow, where they may develop stronger internal material cycling and establish chemical gradients (6, 7, 23, 32). This may result in more pronounced niche diversification in biofilms growing under laminar flow, and thus, in higher opportunities for propagules to successfully establish. Such a scenario would refute our initial hypothesis that the immigration rate, highest in the turbulent flow, would affect succession. Immigration rate is, in fact, a function of propagule abundance, which is the same in all treatments, and of advective delivery, which increases with flow velocity.
Phylogenetic shifts.
The phylogenetic shifts we observed reasonably support the notion that biofilm architecture influences community composition. Generally, phylogenetic analysis of selected DGGE bands identified bacteria associated with groups typically found in freshwater biofilms (Alpha-, Beta-, and Gammaproteobacteria and Bacteroidetes). Four of eight sequences belonged to the Betaproteobacteria, which have been found to be the most abundant bacterial group in freshwater ecosystems (11, 16, 24). Araya and colleagues (4) proposed that Betaproteobacteria may attach more easily to surfaces during initial biofilm formation than other groups of bacteria and, thus, dominate biofilm succession.
During the dynamic early growth phase, community compositions exhibited rapid changes, the assemblages of adjacent sampling dates having only a few OTUs in common, as shown by the MDS analysis (Fig. 6b). It was thus difficult to identify OTUs “typical” for this early growth phase. However, some OTUs which colonized biofilms during early growth obviously persisted throughout the entire experiment. More OTUs established “permanently” during the less-dynamic growth phase dominated by filamentous green algae. Three prominent DGGE bands present over the entire experiment represented Hydrogenophaga and Herbaspirillum species. These genera are known for their diverse metabolic phenotypes and ability to use a wide range of low-molecular-weight organic matter (21, 36). On the other hand, OTUs prominent during the stable growth phase were affiliated with bacterial groups that may perform rather specialized functions in a microbial community. Lysobacter, for instance, is known to lyse and degrade bacteria, cyanobacteria, and eukaryotic algae (25). Members of the Bacteroidetes group were found to degrade refractive and high-molecular-weight organic matter in biofilms and freshwater aggregates (22, 36), while members of the genus Azoarcus can use a wide range of organic acids, aromatic compounds, and amino acids (3, 39, 46). Though it is certainly difficult to infer the physiological properties of bacteria from phylogenetic relationships, this pattern might reflect a change of organic matter availability and metabolic capabilities within biofilms. In fact, nascent biofilms may predominantly rely on external resources of organic matter (from the bulk liquid), whereas mature biofilms may be increasingly supplied from internal sources (7, 23). Together with an increasingly complex architecture, this might have led to a higher number of potential niches for heterotrophic and predatory bacteria.
Massol-Deya and colleagues (28) observed similar convergence in fixed-film reactors treating aromatic hydrocarbons in groundwater, where biofilms developed from different chemical (carbon source) starting conditions to stable mature communities. Along with our findings, this suggests that successional trajectories as described from general ecology may be more common in microbial ecology than hitherto admitted.
Implications for stream ecosystems.
The streambed is an inherently heterogeneous landscape where flow velocity is patchily distributed, mainly as a result of changes in local topography and slope. Our experimental results suggest that this structural heterogeneity may translate into diverse biofilms with differing successional trajectories. Divergence among flow treatments was most pronounced during early exponential growth (days 20 to 40). Strikingly, this brackets the interflood period, with an average biofilm turnover of 30 days, in our study streams (except during snowmelt) (38). Successional divergence between major flood events would thus maximize biofilm diversity within a given stream reach. This may have further consequences for ecosystem functioning, given the relationship between biodiversity and ecosystem functioning in general (12) and the role of microbial biofilms in stream ecosystems (7).
Acknowledgments
We thank two anonymous reviewers for their comments on an earlier draft and Kilian Stoecker for confocal laser scanning microscopy support.
This research was supported by the Austrian Science Fund (FWF grant P16935-B03) to T.J.B.
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Allan, J. D. 1995. Stream ecology: structure and function of running waters. Chapman & Hall, London, United Kingdom.
- 2.An, D., T. Danhorn, C. Fuqua, and M. R. Parsek. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. USA 103:3828-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders, H.-J., A. Kaetzke, P. Kämpfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 45:327-333. [DOI] [PubMed] [Google Scholar]
- 4.Araya, R., K. Tani, T. Takagi, N. Yamaguchi, and M. Nasu. 2003. Bacterial activity and community composition in stream water and biofilm from an urban river determined by fluorescent in situ hybridization and DGGE analysis. FEMS Microbiol. Ecol. 43:111-119. [DOI] [PubMed] [Google Scholar]
- 5.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least one in twenty 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battin, T. J., L. A. Kaplan, J. D. Newbold, X. Cheng, and C. Hansen. 2003. Effects of current velocity on the nascent architecture of stream microbial biofilms. Appl. Environ. Microbiol. 69:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battin, T. J., L. A. Kaplan, J. D. Newbold, and C. M. E. Hansen. 2003. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 426:439-442. [DOI] [PubMed] [Google Scholar]
- 8.Battin, T. J., W. T. Sloan, S. Kjelleberg, H. Daims, I. M. Head, T. P. Curtis, and L. Eberl. 2007. Microbial landscapes: new paths to biofilm research. Nat. Rev. Microbiol. 5:7-12. [DOI] [PubMed] [Google Scholar]
- 9.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2004. GenBank: update. Nucleic Acids Res. 32:D23-D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borchardt, M. A. 1996. Nutrients, p. 184-227. In R. J. Stevenson, M. L. Bothwell, and R. L. Lowe (ed.), Algal ecology: freshwater benthic ecosystems. Academic Press, Inc., San Diego, CA.
- 11.Brümmer, I. H. M., A. Felske, and I. Wagner-Döbler. 2003. Diversity and seasonal variability of β-proteobacteria in biofilms of polluted rivers: analysis by temperature gradient gel electrophoresis and cloning. Appl. Environ. Microbiol. 69:4463-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardinale, B. J., D. S. Srivastava, J. E. Duffy, J. P. Wright, A. L. Downing, M. Sankaran, and C. Jouseau. 2006. Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature 443:989-992. [DOI] [PubMed] [Google Scholar]
- 13.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 15.Curtis, T. P., I. M. Head, M. Lunn, S. Woodcock, P. D. Schloss, and W. T. Sloan. 2006. What is the extent of prokaryotic diversity? Philos. Trans. R. Soc. Lond. B 361:2023-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 18.Hillebrand, H., C.-D. Dürselen, D. Kirschtel, U. Pollingher, and T. Zohary. 1999. Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 35:403-424. [Google Scholar]
- 19.Jackson, C. R., P. F. Churchill, and E. E. Roden. 2001. Successional changes in bacterial assemblage structure during epilithic biofilm development. Ecology 82:555-566. [Google Scholar]
- 20.Jones, C. G., J. H. Lawton, and M. Shackak. 1994. Organisms as ecosystem engineers. Oikos 69:373-386. [Google Scholar]
- 21.Kämpfer, P., R. Schulze, U. Jäckel, K. A. Malik, R. Amann, and S. Spring. 2005. Hydrogenophaga defluvii sp. nov. and Hydrogenophaga atypica sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 55:341-344. [DOI] [PubMed] [Google Scholar]
- 22.Kindaichi, T., T. Ito, and S. Okabe. 2004. Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl. Environ. Microbiol. 70:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kühl, M., R. N. Glud, H. Ploug, and N. B. Ramsing. 1996. Microenvironmental control of photosynthesis and photosynthesis-coupled respiration in an epilithic cyanobacterial biofilm. J. Phycol. 32:799-812. [Google Scholar]
- 24.Loy, A., W. Beisker, and H. Meier. 2005. Diversity of bacteria growing in natural mineral water after bottling. Appl. Environ. Microbiol. 71:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lueders, T., R. Kindler, A. Miltner, M. W. Friedrich, and M. Kaestner. 2006. Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl. Environ. Microbiol. 72:5342-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyautey, E., C. R. Jackson, J. Cayrou, J.-L. Rols, and F. Garabetian. 2005. Bacterial community succession in natural river biofilm assemblages. Microb. Ecol. 50:589-601. [DOI] [PubMed] [Google Scholar]
- 27.Martiny, A. C., H.-J. Albrechtsen, E. Arvin, and S. Molin. 2005. Identification of bacteria in biofilm and bulk water samples from a nonchlorinated model drinking water distribution system: detection of a large nitrite-oxidizing population associated with Nitrospira spp. Appl. Environ. Microbiol. 71:8611-8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massol-Deya, A., R. Weller, L. Rios-Hernandez, J.-Z. Zhou, R. F. Hickey, and J. M. Tiedje. 1997. Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl. Environ. Microbiol. 63:270-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matz, C., D. McDougald, A. M. Moreno, P. Y. Yung, F. H. Yildiz, and S. Kjelleberg. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 102:16819-16824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muyzer, G., E. C. DeWaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neu, T. R., and J. R. Lawrence. 1997. Development and structure of microbial biofilms in river water studied by confocal laser scanning microscopy. FEMS Microbiol. Ecol. 24:11-25. [Google Scholar]
- 32.Picioreanu, C., M. C. M. van Loosdrecht, and J. J. Heijnen. 2000. A theoretical study on the effect of surface roughness on mass transport and transformation in biofilms. Biotechnol. Bioeng. 68:355-369. [DOI] [PubMed] [Google Scholar]
- 33.Purevdorj, B., J. W. Costerton, and P. Stoodley. 2002. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 68:4457-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rickard, A. H., A. J. McBain, R. G. Ledder, P. S. Handley, and P. Gilbert. 2003. Coaggregation between freshwater bacteria within biofilm and planktonic communities. FEMS Microbiol. Lett. 220:133-140. [DOI] [PubMed] [Google Scholar]
- 35.Rickard, A. H., A. J. McBain, A. T. Stead, and P. Gilbert. 2004. Shear rate moderates community diversity in freshwater biofilms. Appl. Environ. Microbiol. 70:7426-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweitzer, B., I. Huber, R. Amann, W. Ludwig, and M. Simon. 2001. α- and β-Proteobacteria control the consumption and release of amino acids on lake snow aggregates. Appl. Environ. Microbiol. 67:632-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singer, G., K. Besemer, I. Hödl, A.-K. Chlup, G. Hochedlinger, P. Stadler, and T. J. Battin. 2006. Microcosm design and evaluation to study stream microbial biofilms. Limnol. Oceanogr. Methods 4:436-447. [Google Scholar]
- 38.Singer, G., M. Panzenböck, G. Weigelhofer, C. Marchesani, J. Waringer, W. Wanek, and T. J. Battin. 2005. Flow history explains temporal and spatial variation of carbon fractionation in stream periphyton. Limnol. Oceanogr. 50:706-712. [Google Scholar]
- 39.Song, B., M. M. Häggblom, J. Zhou, J. M. Tiedje, and N. J. Palleroni. 1999. Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of Azoarcus toluvorans sp. nov. and Azoarcus toluclasticus sp. nov. Int. J. Syst. Bacteriol. 49:1129-1140. [DOI] [PubMed] [Google Scholar]
- 40.Stevenson, R. J. 1996. The stimulation and drag of current, p. 321-340. In R. J. Stevenson, M. L. Bothwell, and R. L. Lowe (ed.), Algal ecology: freshwater benthic ecosystems. Academic Press, Inc., San Diego, CA.
- 41.Stoodley, P., Z. Lewandowski, J. D. Boyle, and H. M. Lappin-Scott. 1998. Oscillation characteristics of biofilm streamers in turbulent flowing water as related to drag and pressure drop. Biotechnol. Bioeng. 57:536-544. [DOI] [PubMed] [Google Scholar]
- 42.Stoodley, P., Z. Lewandowski, J. D. Boyle, and H. M. Lappin-Scott. 1999. Structural deformation of bacterial biofilms caused by short-term fluctuations in fluid shear: an in situ investigation of biofilm rheology. Biotechnol. Bioeng. 65:83-92. [PubMed] [Google Scholar]
- 43.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 44.Velji, M. I., and L. J. Albright. 1986. Microscopic enumeration of attached marine bacteria of seawater, marine sediment, fecal matter, and kelp blade samples following pyrophosphate and ultrasound treatments. Can. J. Microbiol. 32:121-126. [Google Scholar]
- 45.Walker, L. R., and R. del Moral. 2003. Primary succession and ecosystem rehabilitation. Cambridge University Press, Cambridge, United Kingdom.
- 46.Zhou, J., M. R. Fries, J. C. Chee-Sanford, and J. M. Tiedje. 1995. Phylogenetic analysis of a new group of denitrifyers capable of anaerobic growth on toluene and description of Azoarcus tolulyticus sp. nov. Int. J. Syst. Bacteriol. 45:500-506. [DOI] [PubMed] [Google Scholar]