Abstract
A successful vaccine vector for human immunodeficiency virus type 1 (HIV-1) should induce anti-HIV-1 T-cell immune responses at mucosal sites. We have constructed recombinant Mycobacterium bovis bacillus Calmette-Guérin (rBCG) expressing an HIV-1 group M consensus envelope (Env) either as a surface, intracellular, or secreted protein as an immunogen. rBCG containing HIV-1 env plasmids engineered for secretion induced optimal Env-specific T-cell gamma interferon enzyme-linked immunospot responses in murine spleen, female reproductive tract, and lungs. While rBCG-induced T-cell responses to HIV-1 envelope in spleen were lower than those induced by adenovirus prime/recombinant vaccinia virus (rAd-rVV) boost, rBCG induced comparable responses to rAd-rVV immunization in the female reproductive tract and lungs. T-cell responses induced by rBCG were primarily CD4+, although rBCG alone did not induce anti-HIV-1 antibody. However, rBCG could prime for a protein boost by HIV-1 envelope protein. Thus, rBCG can serve as a vector for induction of anti-HIV-1 consensus Env cellular responses at mucosal sites.
A major roadblock in human immunodeficiency virus type 1 (HIV-1) vaccine development is the lack of a vaccine vector that addresses HIV-1 diversity and induces durable protective immune responses at mucosal and systemic sites. Protection at mucosal surfaces is critical for prevention of AIDS, since the majority of transmission is via this route, and the loss of memory T cells occurs in gut compartments within the first weeks of infection (7).
To address the issue of HIV-1 genetic diversity, an artificial consensus ancestral envelope has been developed, termed the group M CON6 envelope (6). The CON6 envelope was derived from 1,999 HIV-1 sequences from the Los Alamos HIV-1 sequence database (6). CON6 envelope bound monoclonal antibodies (MAbs) that recognize linear, conformational, and glycan-dependent antibody epitopes in HIV-1 Env glycoproteins. The CON6 Env mediates infection of CD4/CCR5-expressing cells and induces a breadth of T-cell responses similar to that induced by wild-type subtype A, B, or C envelope immunogens (6, 28).
Mycobacterium bovis bacillus Calmette-Guérin (BCG) is widely used as a tuberculosis (TB) vaccine with a low incidence of serious complications (2). Other potential advantages of recombinant BCG (rBCG) as a vaccine include cost-effective vaccine production, safety, the ability to express large insert genes, and adjuvant activity (2, 26).
In this paper, we demonstrate the expression of full-length HIV-1 Env in rBCG as a secreted protein capable of inducing anti-HIV-1 mucosal T-cell responses. Moreover, we show that rBCG expressing HIV-1 Env could boost an rBCG prime for induction of anti-HIV-1 CD4+ T-cell responses and that rBCG can prime for an HIV-1 Env protein boost for induction of anti-Env antibody.
MATERIALS AND METHODS
Generation of rBCG and expression of HIV-1 envelopes.
DNA manipulation and cloning steps were performed in Escherichia coli DH5α (Invitrogen, CA) (21). BCG-Pasteur (BCG-P) and BCG-Danish (BCG-D) strains were used for vector development and for expression of HIV-1 envelopes. rBCG strains generated are listed in Table 1. rBCG was grown in Middlebrook 7H9 broth (Difco, Sparks, MD) containing 10% albumin-dextrose saline, 0.5% glycerol, 0.05% Tween 80 (8). For transformation into mycobacteria, cells were prepared in 10% glycerol and plasmids were introduced into the BCG-P parent strain or the BCG-D parent strain using the Gene Pulser (Bio-Rad, CA) set at 2.5 kV, 25 μF, with the pulse controller resistance set at 1,000 Ω (24, 27). Transformed BCG strains were selected on Middlebrook 7H10 (Difco, Sparks, MD) agar plates supplemented with 10% albumin-dextrose saline containing 30 μg/ml kanamycin. To monitor the expression of HIV-1 gp120 or gp140CF, single colonies of rBCG were grown in Middlebrook 7H9-albumin-dextrose saline-Tween broth with 30 μg/ml of kanamycin. After being rinsed with sterile phosphate-buffered saline (PBS), mycobacterial cells were extracted by modified extraction buffer with glass beads (106-μm glass beads; Sigma, St Louis, MO) (8) and cell lysates were separated from cell debris by centrifugation. rBCG lysate was fractionated on 4 to 20% sodium dodecyl sulfate-polyacrylamide gels and blotted onto nitrocellulose filters (Schleicher & Schuell, Germany). MAbs (T8 and 7B2) were used at 1 μg/ml followed by goat anti-mouse immunoglobulin G (IgG) or goat anti-human IgG conjugated with alkaline phosphatase (Sigma, St. Louis, MO). T8 is a mouse MAb that maps to the gp120 C1 region (gift from Pat Earl, NIH, Bethesda, MD). 7B2, a human MAb against the gp41 region on gp140, was the gift of James Robinson (Tulane Medical School, New Orleans, LA).
TABLE 1.
Recombinant BCG strains generated for expression of HIV-1 envelope
| Strain | Descriptiona |
|---|---|
| BCG-P | BCG-P parent |
| 222empPas | Empty vector pJH22 in BCG-P |
| 120surPas | CON6 gp120 for surface form in BCG-P |
| 120intPas | CON6 gp120 for intracellular form in BCG-P |
| 120secPas | CON6 gp120 for secreted form in BCG-P |
| 140surPas | CON6 gp140CF for surface form in BCG-P |
| 140intPas | CON6 gp140CF for intracellular form in BCG-P |
| 140secPas | CON6 gp140CF for secreted form in BCG-P |
| BCG-D | BCG-D parent |
| 222empDan | Empty vector pJH22 in BCG-D |
| 120surDan | CON6 gp120 for surface form in BCG-D |
| 120intDan | CON6 gp120 for intracellular form in BCG-D |
| 120secDan | CON6 gp120 for secreted form in BCG-D |
| 140surDan | CON6 gp140CF for surface form in BCG-D |
| 140intDan | CON6 gp140CF for intracellular form in BCG-D |
| 140secDan | CON6 gp140CF for secreted form in BCG-D |
All constructs have a kanamycin resistance gene for selection and were cloned into ApaI and HpaI restriction endonuclease sites in vectors. C, cleavage site deletion; F, fusion domain deletion. All gp140 constructs have the transmembrane and cytoplasmic domains deleted.
Mycobacterial vectors were constructed as previously described (30). In pJH152-gp120 and pJH152-gp140CF, the env gene is fused in frame to the Mycobacterium tuberculosis 19-kDa signal sequence and expressed for surface expression. In pJH153-gp120 and pJH153-gp140CF, the HIV-1 env gene is regulated by the α-antigen promoter, although this vector does not have a localization signal, and the insert is targeted for intracellular expression. In pJH154-gp120 and pJH154-gp140CF, the expression cassette consists of the HIV-1 env gene fused in frame to the M. tuberculosis α-antigen export signal and expressed under the control of the α-antigen promoter for secreted expression.
Immunizations.
Female BALB/c mice (Charles River Laboratory, Raleigh, NC) 6 to 8 weeks of age were used for immunogenicity studies of expressed HIV-1 envelopes. rBCG strains were resuspended in PBS containing 0.05% Tween 80. Mice were immunized intraperitoneally (i.p.) with one of three doses, 108 CFU, 107 CFU, or 106 CFU. Each mouse was immunized i.p. with 100 μl of mycobacterial mixture. For protein immunization, 50 μg of rCON6 gp140CF protein (6) was formulated with RiBi adjuvant (Sigma, MO) at a 1:1 (vol/vol) ratio and administered as 200 μl of a protein mixture by i.p. and subcutaneous routes. Mouse serum was collected 2 weeks after immunization. All animals were housed in the Duke Cancer Center Isolation Facility under AAALAC guidelines with animal use protocols approved by the Duke University Animal Use and Care Committee and the Duke University Institutional Biosafety Committee.
ELISA and antibody isotype analysis.
Serum antibody titers against HIV-1 CON6 gp140CF or M. tuberculosis whole-cell lysate (WCL) (Colorado State University, Fort Collins, CO) were determined in a standard enzyme-linked immunosorbent assay (ELISA). For isotype analysis of serum antibody, goat anti-mouse IgG (heavy chain specific), IgG1, IgG2a, IgG2b, IgG3, or IgM (Southern Biotechnology, AL) was used as a secondary reagent. Antibody endpoint titers were determined as the reciprocal of the highest dilution of the serum assayed against rHIV-1 CON6 protein or M. tuberculosis WCL, giving an optical density ratio of assay to control of ≥3.0. Geometric mean titers were determined based on the endpoint titer of each mouse serum. Statistical significance was assessed by comparing experimental and control groups using Student's t test.
IFN-γ ELISPOT assays.
Enzyme-linked immunospot (ELISPOT) assays were performed as described previously (20). A pool of 10 CON6 gp140 Env overlapping peptides (15-mers overlapping by 11 residues) previously shown to stimulate gamma interferon (IFN-γ) ELISPOT responses by restimulation of T cells from BALB/c mice immunized with CON6 Env immunogen was used for in vitro restimulation (6; E. Weaver, Z. Lu, H. X. Liao, B. Ma, M. S. Alam, R. M. Scearce, L. L. Sutherland, J. M. Decker, Z. Hartman, A. Amalfitano, B. T. Korber, B. H. Hahn, D. C. Montefiori, and B. Haynes, presented at the International AIDS Vaccine 2003 Meeting, New York, NY, 2003). Ninety-six-well flat-bottomed plates (Millipore, MA) were coated with anti-mouse IFN-γ antibody (Pharmingen, CA) before being washed with PBS and blocked. Freshly isolated spleen cells were mixed with either the pooled CON6 gp140 Env peptides or M. tuberculosis WCL and then incubated for 24 h at 37°C in 5% CO2. Following incubation the plates were washed, biotinylated anti-IFN-γ antibody (Pharmingen, CA) was added, and the plates were incubated overnight at 4°C. After the plates were washed with PBS, streptavidin-horseradish peroxidase (Pharmingen, CA) was added to each well and developed. To count spot-forming cells (SFC), spots in the ELISPOT plates were scanned into an ImmunoSpot Series I analyzer and quantified by using ImmunoSpot 2.1 software (CTL Analyzers, Cleveland, OH). Control cells included cells cultured in medium in the absence of peptide or M. tuberculosis WCL stimulation. For analysis of mucosal T-cell responses, we pooled the lungs and female reproductive tracts (FRT) of three to four mice. Statistical significance was assessed by comparing data from the control groups using Student's t test.
Isolation of mucosal lymphocytes.
Lymphocytes were isolated from mucosal tissues as previously described (20). The FRT included the ovaries, fallopian tubes, uterus, and vagina. All FRT and lung tissue were digested with RPMI 1640 with 5% fetal bovine serum, penicillin-streptomycin, HEPES, 2.5 mg of collagenase type A (Roche, CA)/ml, and 5 units of DNase I (Roche, CA)/ml. FRT and lung tissues were pooled from three to four mice/immunization group to provide sufficient cells to perform replicates of each IFN-γ ELISPOT experiment. Cells were purified through an LSM (MP Biomedicals, OH) cushion by centrifugation (20). The average yield of cells per mouse was 2.9 × 106 cells/FRT and 7.9 × 106 cells/lung. Purified FRT and lung cells contain approximately 32 to 37% CD3-positive T cells as determined by flow cytometry analysis (data not shown).
CD4 and CD8 depletion.
CD4+ and CD8+ lymphocytes were depleted from spleen samples using magnetically assisted cell sorting CD4 (L3T4) and CD8α (Ly-2) MicroBeads (Miltenyi, Auburn, CA) and following the protocol provided with the MicroBeads. Briefly, single-cell suspensions were incubated with MicroBeads for 15 min at 6°C. The cells were then washed with 5 ml of PBS, 0.5% fetal bovine serum, and 2 mM EDTA. The cell pellet was resuspended in 500 μl of wash buffer and placed onto a prewetted MS+ selection column (Miltenyi, Auburn, CA) in the separator. Following the separation the column was washed three times and the negatively selected cells were washed and pelleted before being counted and adjusted to the appropriate concentration for the ELISPOT assay.
RESULTS
Expression of HIV-1 CON6 gp120 and gp140CF in rBCG.
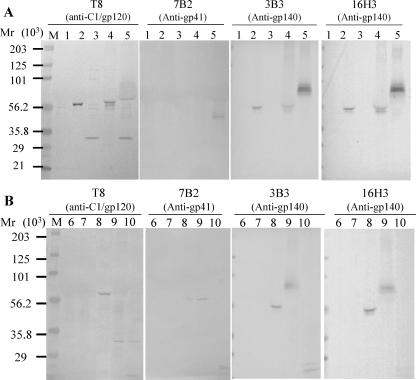
We used three different expression strategies for the expression of CON6 gp120 or gp140CF: surface, intracellular, and secreted. Figure 1 shows that the analysis of protein expression in rBCG could be detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis. Negative controls confirmed that untransformed BCG-D (lane 6) and rBCG strains (222empPas and 222empDan) transformed with an empty plasmid (lanes 1 and 7, respectively) produced no HIV-1 Env protein bands. Full-length expression of CON6 gp120 as the surface antigen, 120surPas (lane 2), was demonstrated in BCG-P, while the full length of the secreted CON6 gp120 antigen, 120secPas, was shown in both BCG-P (lane 4) and BCG-D (lane 8). Interestingly, a cleaved gp120 product was seen in BCG-P transformed with the intracellular expression plasmid pJH153-gp120 (lane 3), and it was reacted only with anti-C1 gp120-specific MAb T8 (Fig. 1A). The expression of CON6 gp140CF was also demonstrated in BCG-P (lane 5), and in BCG-D (lane 9) transformed by the surface expression vector pJH152-gp140CF. Both the intact and partially cleaved gp140 products were demonstrated in BCG-P transformed with the surface expression plasmid and immunoblotted with MAb T8 (anti-C1 gp120 region), gp41-specific MAb 7B2, anti-gp140 MAb 3B3, and anti-gp140 MAb 16H3 (lane 5). The intact and partially cleaved gp140 products were also seen in BCG-D transformed with the surface expression plasmid pJH152-gp140CF (lane 9) or the intracellular expression plasmid pJH153-gp140CF (lane 10). Analysis of different clones and batches of rBCG that expressed the cleaved CON6 Envs demonstrated consistent results (not shown). Thus, the cleaved CON6 Env produced by either strain of BCG is a property of this particular rBCG when transformed by the CON6 Env expression plasmid. The calculated nonglycosylated molecular mass based on amino acid sequences for CON6 gp120 was 53 kDa, and that for CON6 gp140CF was 71 kDa.
FIG. 1.
Expression of HIV-1 group M consensus gp120 and gp140 envelope proteins in BCG strains. (A) Expression of intact CON6 gp120 in rBCG-P transformed by the surface expression plasmid pJH152-gp120 (lane 2), the intracellular expression plasmid pJH153-gp120 (lane 3), and the secreted expression plasmid pJH154-gp120 (lane 4) in rBCG-P. The expression of CON6 gp140CF was also determined in rBCG-P transformed by the surface expression plasmid pJH152-gp140CF (lane 5). (B) The secreted expression of CON6 gp120 in pJH154-gp120 (lane 8), the surface expression of gp140CF in pJH152-gp140CF (lane 9), and the intracellular expression of gp140 in pJH153-gp140CF (lane 10) were demonstrated in rBCG-D. Both the intact and partially cleaved gp140 products were shown using gp120 MAb T8 (anti-C1 gp120 region), gp41-specific MAb 7B2, anti-gp140 MAb 3B3, and anti-gp140 MAb 16H3. Lane M is molecular weight standards in thousands (Bio-Rad, CA). Negative controls were untransformed BCG-D (lane 6) and rBCG strains transformed by empty plasmid pJH222 (lanes 1 and 7).
Determination of optimal rBCG vectors for HIV-1 Env immunogenicity.
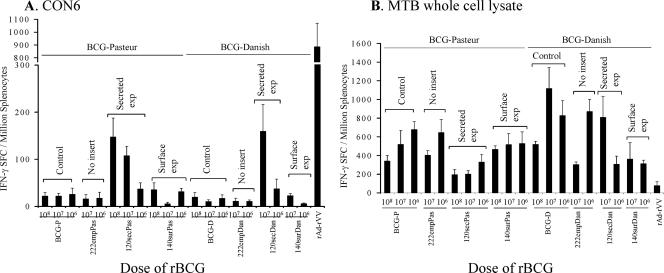
Next, animals were immunized with rBCG twice followed by a final boost with recombinant HIV-1 Env oligomer followed by an assay for T-cell responses. Lymphocytes were isolated from spleens and assayed for HIV-1 CON6 Env and BCG vector-specific T-cell responses with IFN-γ ELISPOT assays. All mice were immunized i.p. with either 108, 107, or 106 CFU of rBCG strains (Table 1). Control groups included mice immunized with untransformed BCG strains, BCG-P and BCG-D, or rBCG strains harboring an empty plasmid, 222empPas or 222empDan. HIV-1-specific IFN-γ SFC responses were between 100 and 150 spots/106 splenocytes assayed, and IFN-γ responses from constructs containing no plasmids expressing gp120 or gp140 but boosted with CON6 protein were between 25 and 30 spots/106 splenocytes. Mice immunized with 120secPas and 120secDan expressing CON6 gp120 as secreted antigen showed the optimal response in a dose-dependent manner (Fig. 2A). For example, mice primed and boosted with 108 CFU 120secPas produced 147 ± 40 SFC/106 cells, a significantly greater (P < 0.05) number of SFC than that for mice primed and boosted with lower doses of 107 CFU (100 ± 19 SFC) or 106 CFU (37 ± 13 SFC). In addition, 120secDan given at 107 CFU induced 159 ± 57 SFC/106 cells while 106 CFU induced 37 ± 20 SFC/106 cells (P < 0.01). In contrast, the responses with 140surPas or 140surDan (both designed for surface expression) were suboptimal (Fig. 2A). Thus, 108 CFU 140surPas induced 36 ± 14 SFC/106 cells, 107 CFU induced 6 ± 3 SFC/106 cells, and 106 CFU induced 31 ± 7 SFC/106 cells. Similarly, the 120intPas or 140intPas (each designed for intracellular expression) BCG strain did not give responses above background (not shown). However, it remains to be determined if the differences in immune responses were due to the levels of expression or due to the different strategies of insert expression by BCG.
FIG. 2.
Antigen-specific T-cell responses induced by different modes of insert expression by rBCG. HIV-1 envelope-specific (A) and BCG vector-specific (B) T-cell responses were assessed by using IFN-γ ELISPOT assays. Mice were immunized i.p. with various doses of rBCG strains or untransformed BCG strains such as BCG-P or BCG-D. The mean (± standard error of the mean) SFC per 106 cells derived from five mice per group are shown on the y axis. Mouse groups immunized with the indicated doses and designs of rBCG or control constructs are shown on the x axis. The mouse group indicated as CON6 was immunized with CON6 protein only. The mouse group indicated as rAd-rVV was primed with rAd-CON6 and boosted with rVV-CON6 as a positive control.
BCG vector-specific IFN-γ SFC responses were measured with WCL as a restimulating antigen (Fig. 2B). There was no difference between rBCG-P strains and rBCG-D strains with regard to induction of vector-specific T-cell responses. There was a definite trend of higher doses of BCG decreasing responses to the vector. For example, BCG-P induced vector-specific IFN-γ SFC at 108 CFU (334 ± 60 SFC/106 cells), at 107 CFU (519 ± 149 SFC), and at 106 CFU (677 ± 86 SFC). Thus, vector-specific T-cell responses from mice immunized with a low dose of BCG-P were significantly higher than those from mice immunized with high doses (108 CFU versus 107 CFU [0.02 < P < 0.05], 107 CFU versus 106 CFU [P = 0.06], and 108 CFU versus 106 CFU [P < 0.01], respectively).
Detection of HIV-1 envelope-specific IFN-γ SFC in spleen generated by rBCG-P and 120secPas prime/boost.
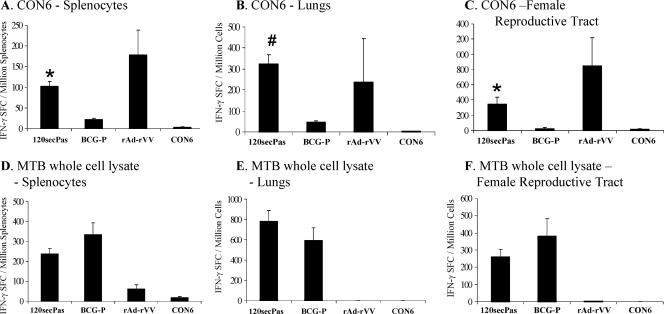
For determination of T-cell responses after prime boost with rBCG, lymphocytes were isolated from spleens, lungs, and FRT of all immunization groups of mice and assayed for HIV-1 CON6 Env and BCG vector-specific T-cell responses in IFN-γ ELISPOT assays. Mice were immunized with 120secPas expressing HIV-1 gp120 as a secreted antigen at 107 CFU in week 0 by i.p. immunization. Unimmunized mice served as negative controls, and recombinant adenovirus (rAd) prime-recombinant vaccinia virus (rVV) boost-immunized animals were positive controls. Splenocytes from animals immunized i.p. once with rBCG yielded little or no (<5 SFC/106 cells assayed) CON6 gp120 peptide-specific responses, comparable to those observed in mice immunized with 222empPas. When mice were boosted with 120secPas 8 weeks following the priming immunization, HIV-1 CON6 gp120-specific T-cell responses were easily detected by the IFN-γ SFC assay (Fig. 3A). Mice primed and boosted with 107 CFU 120secPas induced 102 ± 12 SFC/106 splenocytes, a significantly greater number of SFC than that induced by mice primed and boosted with BCG-P (22 ± 12 SFC) or mice immunized with CON6 protein alone (3 ± 1 SFC) (P < 0.0001), indicating that rBCG induced HIV-1-specific T-cell responses. Mice primed with rAd and boosted with rVV expressing CON6 were used as positive controls and induced 178 ± 60 IFN-γ SFC/106 splenocytes.
FIG. 3.
HIV-1 envelope-specific and vector-specific T-cell responses in spleen, lungs, and FRT of mice immunized with rBCG-P/120secPas. HIV-1-specific (A to C) and BCG vector-specific (D to F) T-cell responses in spleen (A and D), lungs (B and E), and FRT (C and F) were assessed by using IFN-γ ELISPOT assays. “CON6” indicates mice immunized with only CON6 protein. “rAd-rVV” indicates mice that were primed with rAd5 expressing CON6 Env and boosted with rVV expressing CON6 Env, used as positive controls. The mean (± standard error of the mean) SFC per 106 cells are shown on the y axis. The indicated immunization groups and doses are shown on the x axis. Assays were performed and analyzed on pooled lungs and FRT samples as described in Materials and Methods. Statistical significance was assessed by comparing rBCG-P/120secPas with control groups such as BCG-P or CON6 (*, P < 0.0001; #, P < 0.001).
rBCG elicited HIV-1 envelope-specific T-cell responses in lungs and the FRT.
In addition to antigen-specific responses to CON6 gp140 peptides in splenocytes, we determined the number of IFN-γ SFC/million lymphocytes induced in lungs and FRT of immunized mice. To obtain sufficient cells to perform antigen-specific IFN-γ ELISPOT assays, the lungs of three to four mice were removed and pooled for each group assayed and data from three groups of mice were analyzed. Mice immunized once with 120secPas produced no mucosal CON6 gp140 peptide-specific IFN-γ SFC. However, upon boosting with the same dose of 120secPas, CON6 gp120 peptide-specific responses were detected 2 weeks following the second immunization. In the lungs, mice immunized with 107 CFU 120secPas gave the highest frequency (323 ± 44 SFC) (Fig. 3B) of IFN-γ SFC, a significantly greater number than that given by mice immunized with BCG-P (46 ± 7 SFC) or with CON6 protein alone (2 ± 2 SFC) (P < 0.001 and P < 0.0001, respectively).
In addition to T-cell responses in the lungs, we found that 120secPas could also induce T-cell responses in the FRT (Fig. 3C). Just as with lung responses, FRT were pooled in order to obtain sufficient cells for IFN-γ SFC ELISPOT assays. After boosting with 120secPas, CON6 peptide-specific responses were easily detectable in the FRT (Fig. 3C). Lymphocytes from the FRT of mice primed and boosted with 107 CFU 120secPas produced 350 ± 88 IFN-γ SFC/106 cells isolated. Mice immunized with BCG-P or CON6 protein had low frequencies of IFN-γ SFC from cells isolated from the FRT (22 ± 19 and 15 ± 11 SFC, respectively).
Vector-specific T-cell responses.
Responses to M. tuberculosis WCL were also monitored in the spleen, lungs, and FRT. 120secPas given at 107 CFU induced 239 ± 27 SCF, BCG-P induced 336 ± 57 SFC, and CON6 protein induced 18 ± 4 SFC in splenocytes (Fig. 3D). Following a prime/boost with 120secPas, the induction of BCG vector-specific immunity in the lungs was 784 ± 106 SFC and that in FRT was 262 ± 42 SFC (Fig. 3E and 3F). BCG-P also induced BCG-specific T-cell responses in the lungs (595 ± 124 SFC) and in the FRT (382 ± 100 SFC) (Fig. 3E and 3F).
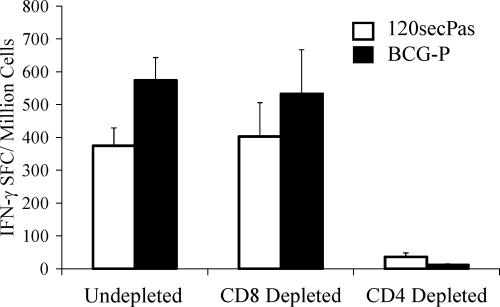
Next, to assess the responding anti-vector T-cell types induced by rBCG, we immunized mice with prime-boost 120secPas at 108 CFU. BCG-specific T-cell ELISPOT IFN-γ responses induced by rBCG-P/120secPas were primarily CD4+ dependent as depletion of CD4+ splenocytes eliminated ≥90% of the anti-vector ELISPOT responses (Fig. 4). Similarly, CON6-specific T-cell responses induced by 120secPas were also depleted by anti-CD4 MAb treatment (not shown).
FIG. 4.
M. tuberculosis WCL-specific IFN-γ production is dependent on CD4+ splenocytes. In order to access the T-cell population responsible for IFN-γ production following immunization with 120secPas and BCG-P, via the i.p. route, we removed the respective CD4 or CD8 population using magnetically labeled antibodies from the prepared splenocytes. CD4+ T-cell responses were predominant when mice were immunized with rBCG. The mean (± standard error of the mean) SFC per 106 cells derived from 15 mice per group are shown on the y axis.
rBCG effectively primed for a protein boost of anti-HIV-1 envelope antibody.
To further evaluate the immune responses after immunizations with rBCG expressing HIV-1 CON6 gp120 and gp140, female BALB/c mice were immunized with either rBCG-P strains or rBCG-D strains expressing the secreted and surface forms of CON6 gp120 or CON6 gp140 (Table 1). Mice were immunized with rBCG strains twice followed by a final boost with recombinant HIV-1 Env oligomer to test for antibody induction. Serum samples were collected 2 weeks after each immunization and were assayed by ELISA for antibody titers against recombinant CON6 gp140CF. We found no detectable antibody responses after two immunizations with the rBCG strain alone. Mice were then boosted with CON6 gp140 protein immunogen. Table 2 shows the summary of serum ELISA endpoint titers for the immunization groups after boosting with CON6 gp140 protein. We found that mice primed with rBCG expressing HIV-1 CON6 Env had significantly higher antibody titers than the control groups primed with either untransformed BCG strains (BCG-P or BCG-D) or rBCG strains harboring the empty plasmid (222empPas or 222empDan) (Table 2) or the group immunized once with CON6 protein (group CON6) (P < 0.02 and P < 0.001, respectively). Mice primed with untransformed BCG strains such as BCG-P or BCG-D had significantly higher anti-Env antibody titers than did animals injected with protein alone (P < 0.01). These latter results suggested that immunization with nontransformed BCG had an adjuvant effect on later immunizations with Env oligomers (2).
TABLE 2.
Antibody responses after rBCG CON6 prime and rHIV-1 CON6 gp140 protein boost in BALB/c micea
| Prime or boost immunogen | Expressed insert form | Geometric mean endpoint titer(s)b |
|---|---|---|
| BCG-P | Control | 6,925 |
| 222empPas | Control | 6,653 |
| 120secPas | Secreted | 28,900* |
| 140surPas | Surface | 20,564** |
| BCG-D | Control | 8,680 |
| 222empDan | Control | 9,313 |
| 120secDan | Secreted | 41,572** |
| 140surDan | Surface | 42,286** |
| CON6 | 959 |
The dose of rBCG was 107 or 106 CFU. The boost immunogen was CON6 gp140 with RiBi adjuvant.
Anti-HIV-1 gp140 geometric mean titer (n = 5 mice). *, statistically significant (P < 0.01) compared to the control groups that were immunized with BCG-P or 222empPas; **, statistically significant (0.01 < P < 0.02) compared to the control groups that were immunized with BCG-D or 222empDan.
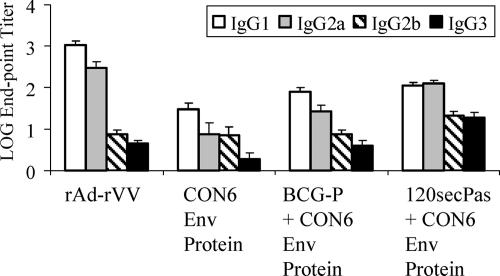
Next, HIV-1-specific IgG subclass responses were assayed by ELISA using sera from mice immunized with 120secPas at 107 CFU. Mice were primed and boosted i.p. with 120secPas strains followed by CON6 protein boosting. Control groups were immunized with rAd prime and rVV boost (rAd-rVV) or a single dose of CON6 gp140 protein. Immunization of mice with rAd-rVV or CON6 protein alone induced predominantly IgG1 HIV-1-specific antibodies with the IgG1/IgG2 ratio of 4.56 or 3.84, respectively (Fig. 5), which is characteristic of a Th2-like response. When sera from mice immunized with BCG-P and boosted with CON6 protein were analyzed, antibody responses to CON6 protein were again biased toward IgG1 with an IgG1/IgG2a ratio of 2.98. Interestingly, immunization with 120secPas generated mixed Th1-Th2 responses to HIV-1 immunogen, with an IgG1/IgG2a ratio of 0.89 (Fig. 5). In addition, analysis of IgG responses to the vector demonstrated typical Th1-like responses regardless of BCG immunogens, and the IgG1/IgG2a ratios of antibody in mice immunized with 120secPas and BCG-P were 0.58 and 0.28, respectively. Thus, priming immunization with CON6 Env in the form of rBCG (120secPas) shifted HIV-1-specific antibody responses from an IgG1/IgG2a ratio of greater than 1 to a ratio of less than 1.
FIG. 5.
HIV envelope-specific isotype induction by rBCG-P/120secPas. IgG subclasses including IgG1 (empty columns), IgG2a (gray columns), IgG2b (hatched columns), and IgG3 (solid columns) of HIV-1 envelope-specific serum antibodies induced by immunization with various immunization regimens (shown on the x axis) were determined by ELISA. The geometric mean log endpoint antibody titers (± standard errors of the means; n = 15) were plotted on the y axis.
Mechanism of limitation of rBCG induction of anti-HIV-1 antibody.
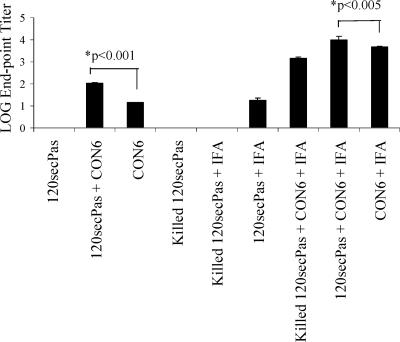
To determine the factors associated with lack of induction of anti-Env antibody by rBCG, we first determined if live rBCG was immunosuppressive for Env insert antibody responses. To test for vector-induced immunosuppression, live rBCG-P/120secPas was mixed with 50 μg of CON6 Env oligomer protein. Mice were immunized with this mixture and then compared to mice immunized with 50 μg CON6 Env oligomer protein alone or mice immunized with heat-killed 120secPas plus 50 μg CON6 Env. We found that rBCG did not suppress anti-Env antibody responses induced by coadministered rCON6 protein but rather that 120secPas had a significant adjuvant effect for anti-Env antibody induction (Fig. 6) (P < 0.001). Similarly, formulations of 120secPas in incomplete Freund's adjuvant (IFA) demonstrated that BCG-P expressing CON6 gp120 as the secreted antigen alone induced detectable levels of antibody responses in mice 4 weeks after a single immunization (Fig. 6). Thus, rBCG itself did not induce anti-Env antibody due to insufficient expression of HIV-1 Env.
FIG. 6.
Induction of antibody response with rBCG-P expressing HIV-1 gp120 in BALB/c mice with combinations of immunogen formulations. To evaluate the immunogenicity in mice of the rBCG-P expressing CON6 Env proteins, mice were immunized once with either killed or live rBCG-P/120secPas with or without IFA. The geometric mean endpoint titers (± standard errors of the means) for each group of mice (n = 5 per group) are shown. Abbreviations: 120secPas, rBCG-P expressing HIV-1 CON6 gp120 as the secreted protein; CON6, recombinant CON6 gp140CF protein.
DISCUSSION
In this paper, we have shown that rBCG can express full-length nonglycosylated HIV-1 Env and, when engineered for the secretion of plasmid-encoded protein, can induce mucosal HIV-1 IFN-γ T-cell responses. While unable to directly induce anti-HIV-1 Env B-cell responses alone, rBCG could effectively prime for an HIV-1 Env protein boost.
A key concern for use of mycobacterial vectors is that vector utility might be limited by preexisting immunity due to environmental mycobacterial colonization (1, 4). For example, prior exposure to environmental mycobacteria blocked the in vivo multiplication of BCG and thus potentially reduced its efficacy as a vaccine (1). In this regard, we found that rBCG can boost an rBCG prime for both insert and vector immune responses. We have found this to be the case for recombinant Mycobacterium smegmatis as well (30) but did not find this for BCG in a previous study using a BCG prime and boost and monitoring by assay for HIV-1 CD8+ T-cell tetramer responses (3). Since we are measuring only CD4+ T-cell responses in this study, our current hypothesis is that rBCG preexisting immunity limits CD8+ T-cell responses but not CD4+ T-cell responses. These data suggest that preexisting antimycobacterial immunity is relatively selective for inhibiting or killing CD8 T cells or their precursors versus CD4+ T cells. While the explanation for this is not known, one hypothesis is that preexisting antimycobacterial immunity modifies the cytokines that support CD8+ T-cell activation but does not interfere with cytokines that activate CD4+ T cells.
An important factor of mycobacterial vaccine vectors that influence immune responses to HIV envelope is antigen compartmentalization. The timing of the immune response and the pathway of presenting insert antigen to the immune system could be affected by different sites of localization of insert expression. It was of interest that the secreted form of insert was the optimal mode of expression of HIV-1 Env for induction of T-cell responses in both BCG-D and BCG-P. This optimal inducibility of rBCG with secreted inserts may relate to class II antigen presentation, degradation pathways of insert antigen, or other functions. Interestingly, BCG induces primarily CD4+ T-cell responses in immunized mice both to the insert and to the vector. While weak CD8+ T-cell responses are seen when the Env insert contains an immunodominant CD8 T-cell epitope (P18) (3), it appears that, overall, the majority of T-cell responses to both vector and insert are CD4+. This may relate to deficient levels of cross priming induced by BCG due to BCG not being able to access cytosolic compartments of infected host cells (17). BCG is considered an intraphagosomal bacterium; thus, antigens expressed by rBCG should be delivered to the phagolysosome. As a consequence, these antigens would be preferentially processed via the major histocompatibility complex (MHC) class II pathway to induce CD4+ T-cell responses (11). In contrast, antigens delivered in the cytosol are responsible for the preferential stimulation of CD8+ T cells via the MHC class I pathway (19). Therefore, intracellular bacteria such as attenuated Salmonella enterica serovar Typhimurium or BCG preferentially activate CD4+ T cells while Listeria monocytogenes preferentially stimulates CD8+ T cells, since MHC class I molecules transport antigens from the endoplasmic reticulum to the cell surface (5, 11). However, M. tuberculosis appears to be processed by a third scenario. M. tuberculosis presumably stays in the phagosome but induces apoptosis in infected macrophages, resulting in the formation of apoptotic vesicles that can be taken up by bystander dendritic cells (22, 29). Therefore, M. tuberculosis antigens not only are delivered to the MHC class I pathway but also stimulate a combination of CD4+ and CD8+ T cells (22). However, BCG may fail to induce this combination of T-cell responses because it induces only weak host infected-cell apoptosis and CD8+ T-cell induction (14). Weak induction of CD8+ T-cell responses could be one of the possible reasons for unsatisfactory protection by BCG against M. tuberculosis. Clearly, we have found optimal immunogenicity of both recombinant M. smegmatis (30) and rBCG (this report) when engineered for insert secretion.
In addition, rBCG strains that secrete a biologically active listeriolysin (Hly) fusion protein of L. monocytogenes could deliver antigens into the MHC class II and MHC class I pathway, thus stimulating both CD4+ and CD8+ T cells (10, 14). In this regard, it is important to develop an expression strategy that induces CD8+ T-cell responses to HIV-1 Env and to M. tuberculosis WCL. The ΔsecA2 mutant of M. tuberculosis, which has been shown to be defective in secreting superoxide dismutase, is being explored as a possible apoptosis-inducing strain (S. Porcelli and W. R. Jacobs, Jr., unpublished observations). Such a mutant might induce more cytotoxic T cells by antigen cross-priming. Therefore, the incorporation of the ΔsecA2 mutation into BCG strains might deliver antigens to the MHC class I cross-priming pathway.
That M. tuberculosis WCL responses as well as CON6-specific T-cell responses induced by rBCG were primarily CD4+ T-cell responses was surprising. Robust CD4+ T-cell responses are critical for protective immunity against M. tuberculosis; thus, the development of cytotoxic CD8+ effector cells in the lungs is substantially diminished in the absence of CD4+ T cells (23). CD4+ T cells also play an important role in the differentiation of CD8+ T cells into long-term functional memory cells (13, 18). Thus, the ability of the rBCG vector to elicit HIV-1 as well as vector-specific CD4+ T-cell responses could prove to be important for the induction of protective immunity for both M. tuberculosis and HIV-1.
Since the development of mycobacterial vectors has been hindered by the low level of heterologous gene expression, others have fused the V3 epitope of HIV-1 Env with mycobacterial antigen 85B and expressed the V3-85B fusion protein in BCG as an HIV immunogen (12, 15). Immunization of rhesus macaques with BCG expressing HIV-1 envelope V3 antigen was able to induce neutralizing antibody and to protect against homologous but not heterologous T-cell live-adapted simian-HIV-1 (25). However, we and others have recently shown the limitation of anti-V3 loop immunogens for practical use as an HIV-1 vaccine immunogen in vivo (9).
To address the issue of genetic diversity of HIV-1, we have begun to explore the immunogenicity of artificial consensus HIV-1 genes that induced a greater breadth of cross-clade T-cell responses than did a polyvalent subtype A, B, and C envelope immunogen (6; Weaver et al., AIDS Vaccine Meeting) in rBCG and recombinant M. smegmatis (30). The group M consensus CON6 Env induced cross-clade T-cell responses in mice (6). We have now produced a second-generation HIV-1 consensus Env, CONS, that has shortened variable loops and that has induced a greater breadth of neutralizing antibodies than CON6 Env has done to subtype A, B, and C primary isolates (16). rBCG vectors are currently being constructed with CONS gp140 inserts.
Even though rBCG induced detectable levels of antibody responses with IFA (Fig. 6), anti-HIV-1 antibody was not induced by rBCG alone, given that mycobacteria can be a component of complete Freund's adjuvant. It is interesting that IFA could augment rBCG expressing HIV-1 Env immunogenicity. However, it is important that rBCG could prime for an Env oligomer boost in mice and in guinea pigs. We also demonstrated that immunizations with rBCG (120secPas) prime/CON6 protein boost in guinea pigs were able to neutralize HIV-1 subtype SF162 (clade B) at low titers (J. Yu, B. Haynes, L. Liao, and S. Xia, unpublished data). Taken together, insufficient levels of HIV-1 CON6 envelope protein expression in rBCG were likely the cause of the lack of the induction of anti-Env antibody following prime/boost immunization by rBCG (30).
In conclusion, our data are proof of concept that rBCG vectors can induce mucosal cellular responses and prime for protein boosts of serum antibody. By optimizing rBCG ability to cross-prime for CD8+ T-cell responses and optimizing the vector for increased levels of insert expression, it should be possible to make rBCG a candidate to serve as a prime for heterologous vector boost in an HIV-1 candidate vaccine formulation.
Acknowledgments
We acknowledge the expert technical assistance of Mary Brock and Robert Parks.
This work was supported by NIAID, NIH PO1 grant A152816, and the Bill and Melinda Gates Foundation.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Brandt, L., J. F. Cunha, A. W. Olsen, B. Chilima, P. Hirsch, R. Appelberg, and P. Andersen. 2002. Failure of the Mycobacterium bovis BCG vaccine: some species of environmental mycobacteria block multiplication of BCG and induction of protective immunity to tuberculosis. Infect. Immun. 70672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burlein, J. E., C. K. Stover, S. Offutt, and M. S. Hanson. 1994. Expression of foreign genes in mycobacteria. American Society for Microbiology, Washington, DC.
- 3.Cayabyab, M. J., A. H. Hovav, T. Hsu, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, G. J. Fennelly, B. F. Haynes, W. R. Jacobs, Jr., and N. L. Letvin. 2006. Generation of CD8+ T-cell responses by a recombinant nonpathogenic Mycobacterium smegmatis vaccine vector expressing human immunodeficiency virus type 1 Env. J. Virol. 801645-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lisle, G. W., B. J. Wards, B. M. Buddle, and D. M. Collins. 2005. The efficacy of live tuberculosis vaccines after presensitization with Mycobacterium avium. Tuberculosis (Edinburgh) 8573-79. [DOI] [PubMed] [Google Scholar]
- 5.Finlay, B. B., and P. Cossart. 1997. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276718-725. [DOI] [PubMed] [Google Scholar]
- 6.Gao, F., E. A. Weaver, Z. Lu, Y. Li, H. X. Liao, B. Ma, S. M. Alam, R. M. Scearce, L. L. Sutherland, J. S. Yu, J. M. Decker, G. M. Shaw, D. C. Montefiori, B. T. Korber, B. H. Hahn, and B. F. Haynes. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group M consensus envelope glycoprotein. J. Virol. 791154-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossman, Z., M. Meier-Schellersheim, W. E. Paul, and L. J. Picker. 2006. Pathogenesis of HIV infection: what the virus spares is as important as what it destroys. Nat. Med. 12289-295. [DOI] [PubMed] [Google Scholar]
- 8.Hatfull, G. F., and W. R. Jacobs, Jr. 2000. Molecular genetics of mycobacteria. ASM Press, Washington, DC.
- 9.Haynes, B. F., B. Ma, D. C. Montefiori, T. Wrin, C. J. Petropoulos, L. L. Sutherland, R. M. Scearce, C. Denton, S. M. Xia, B. T. Korber, and H. X. Liao. 2006. Analysis of HIV-1 subtype B third variable region peptide motifs for induction of neutralizing antibodies against HIV-1 primary isolates. Virology 34544-55. [DOI] [PubMed] [Google Scholar]
- 10.Hess, J., D. Miko, A. Catic, V. Lehmensiek, D. G. Russell, and S. H. Kaufmann. 1998. Mycobacterium bovis Bacille Calmette-Guerin strains secreting listeriolysin of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 955299-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess, J., U. Schaible, B. Raupach, and S. H. Kaufmann. 2000. Exploiting the immune system: toward new vaccines against intracellular bacteria. Adv. Immunol. 751-88. [DOI] [PubMed] [Google Scholar]
- 12.Hiroi, T., H. Goto, K. Someya, M. Yanagita, M. Honda, N. Yamanaka, and H. Kiyono. 2001. HIV mucosal vaccine: nasal immunization with rBCG-V3J1 induces a long term V3J1 peptide-specific neutralizing immunity in Th1- and Th2-deficient conditions. J. Immunol. 1675862-5867. [DOI] [PubMed] [Google Scholar]
- 13.Kalams, S. A., and B. D. Walker. 1998. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J. Exp. Med. 1882199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufmann, S. H., and J. Hess. 1999. Impact of intracellular location of and antigen display by intracellular bacteria: implications for vaccine development. Immunol. Lett. 6581-84. [DOI] [PubMed] [Google Scholar]
- 15.Kawahara, M., A. Hashimoto, I. Toida, and M. Honda. 2002. Oral recombinant Mycobacterium bovis bacillus Calmette-Guerin expressing HIV-1 antigens as a freeze-dried vaccine induces long-term, HIV-specific mucosal and systemic immunity. Clin. Immunol. 105326-331. [DOI] [PubMed] [Google Scholar]
- 16.Liao, H.-X., L. L. Sutherland, S.-M. Xia, M. E. Brock, R. M. Scearce, S. Vanleeuwen, M.-S. Alam, M. McAdams, E. A. Weaver, Z. T. Camacho, B.-J. Ma, Y. Li, J. M. Decker, G. J. Nabel, D. C. Montefiori, B. H. Hahn, B. T. Korber, F. Gao, and B. F. Haynes. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353268-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasser Eddine, A., and S. H. Kaufmann. 2005. Improved protection by recombinant BCG. Microbes Infect. 7939-946. [DOI] [PubMed] [Google Scholar]
- 18.Northrop, J. K., and H. Shen. 2004. CD8+ T-cell memory: only the good ones last. Curr. Opin. Immunol. 16451-455. [DOI] [PubMed] [Google Scholar]
- 19.Pamer, E., and P. Cresswell. 1998. Mechanisms of MHC class I-restricted antigen processing. Annu. Rev. Immunol. 16323-358. [DOI] [PubMed] [Google Scholar]
- 20.Peacock, J. W., S. K. Nordone, S. S. Jackson, H. X. Liao, N. L. Letvin, A. G. Yafal, L. Gritz, G. P. Mazzara, B. F. Haynes, and H. F. Staats. 2004. Gender differences in human immunodeficiency virus type 1-specific CD8 responses in the reproductive tract and colon following nasal peptide priming and modified vaccinia virus Ankara boosting. J. Virol. 7813163-13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Schaible, U. E., F. Winau, P. A. Sieling, K. Fischer, H. L. Collins, K. Hagens, R. L. Modlin, V. Brinkmann, and S. H. Kaufmann. 2003. Apoptosis facilitates antigen presentation to T lymphocytes through MHC-I and CD1 in tuberculosis. Nat. Med. 91039-1046. [DOI] [PubMed] [Google Scholar]
- 23.Serbina, N. V., V. Lazarevic, and J. L. Flynn. 2001. CD4(+) T cells are required for the development of cytotoxic CD8(+) T cells during Mycobacterium tuberculosis infection. J. Immunol. 1676991-7000. [DOI] [PubMed] [Google Scholar]
- 24.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 41911-1919. [DOI] [PubMed] [Google Scholar]
- 25.Someya, K., D. Cecilia, Y. Ami, T. Nakasone, K. Matsuo, S. Burda, H. Yamamoto, N. Yoshino, M. Kaizu, S. Ando, K. Okuda, S. Zolla-Pazner, S. Yamazaki, N. Yamamoto, and M. Honda. 2005. Vaccination of rhesus macaques with recombinant Mycobacterium bovis bacillus Calmette-Guérin Env V3 elicits neutralizing antibody-mediated protection against simian-human immunodeficiency virus with a homologous but not a heterologous V3 motif. J. Virol. 791452-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351456-460. [DOI] [PubMed] [Google Scholar]
- 27.Wards, B. J., and D. M. Collins. 1996. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol. Lett. 145101-105. [DOI] [PubMed] [Google Scholar]
- 28.Weaver, E. A., Z. Lu, Z. T. Camacho, F. Moukdar, H.-X. Liao, B. Ma, M. Muldon, J. Theiler, G. J. Nabel, N. Letvin, B. T. Korber, B. H. Hahn, B. F. Haynes, and F. Gao. 2006. Cross-subtype T cell immune responses induced by a human immunodeficiency virus type 1 group M consensus Env immunogen. J. Virol. 806745-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winau, F., S. Weber, S. Sad, J. de Diego, S. L. Hoops, B. Breiden, K. Sandhoff, V. Brinkmann, S. H. Kaufmann, and U. E. Schaible. 2006. Apoptotic vesicles crossprime CD8 T cells and protect against tuberculosis. Immunity 24105-117. [DOI] [PubMed] [Google Scholar]
- 30.Yu, J.-S., J. W. Peacock, S. Vanleeuwen, T. Hsu, W. R. Jacobs, Jr., M. J. Cayabyab, N. L. Letvin, R. Frothingham, H. F. Staats, H.-X. Liao, and B. F. Haynes. 2006. Generation of mucosal anti-human immunodeficiency virus type 1 T-cell responses by recombinant Mycobacterium smegmatis. Clin. Vaccine Immunol. 131204-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]