Abstract
The surface protective antigen (Spa) protein of Erysipelothrix rhusiopathiae has been shown to be highly immunogenic and is a potential candidate for a new vaccine against erysipelas. In this study, we cloned and sequenced spa genes from all E. rhusiopathiae serovar reference strains as well as from a serovar 18 strain which was not classified as any species in the genus Erysipelothrix. Sequence analysis revealed that the Spa proteins could be classified into three molecular species, including SpaA, which was previously found in serovars 1a and 2, and the newly designated SpaB and SpaC proteins. The SpaA protein is produced by E. rhusiopathiae serovars 1a, 1b, 2, 5, 8, 9, 12, 15, 16, 17, and N, the SpaB protein is produced by E. rhusiopathiae serovars 4, 6, 11, 19, and 21, and the SpaC protein is produced only by serovar 18. The amino acid sequence similarity was high among members of each Spa type (96 to 99%) but low between different Spa types (∼60%). The greatest diversity in Spa proteins was found in the N-terminal half of the molecule (50 to 57% similarity), which was shown to be involved in immunoprotection. Coinciding with this, immunoblot analysis revealed that rabbit antisera specific to each Spa reacted strongly with the homologous Spa protein but weakly with heterologous Spa proteins. A mouse cross-protection study showed that the three recombinant Spa (rSpa) proteins elicited complete protection against challenge with homologous strains but that the level of protection against challenge with heterologous strains varied depending on the rSpa protein used for immunization. Our study is the first to demonstrate sequence and antigenic diversity in Spa proteins and to indicate that rSpaC may be the most promising antigen for use as a vaccine component because of its broad cross-protectiveness.
Erysipelothrix rhusiopathiae is a small gram-positive rod bacterium that causes erysipelas in swine and a variety of diseases in other animals, as well as erysipeloid, a skin disease of humans (20). E. rhusiopathiae was once thought to be the only species in the genus Erysipelothrix and was classified into 25 serovars based on peptidoglycan antigens of the cell wall. At present, the genus contains at least the following two species: E. rhusiopathiae, including serovars 1a, 1b, 2, 4, 5, 6, 8, 9, 11, 12, 15, 16, 17, 19, and 21 and type N; and Erysipelothrix tonsillarum, including serovars 3, 7, 10, 14, 20, 22, and 23. Serovars 13 and 18 are unclassified but are considered to be assigned to genetically distinct groups from the above two species (15).
E. rhusiopathiae serovars 1 and 2 are most frequently isolated from swine with clinical erysipelas (11, 16), but other serovars of E. rhusiopathiae are occasionally isolated from swine with septicemia, urticaria, arthritis, lymphadenitis, and endocarditis (17). Because of their high frequency of isolation, serovar 1a (Koganei 65-0.15) and serovar 2 (Tama-96) strains have been used to prepare live and killed vaccines, respectively, in Japan. Both vaccines elicit a cross-protective immune response in immunized pigs against challenge with E. rhusiopathiae serovars 1 and 2 (6), but it is not known whether they confer cross-protection against other E. rhusiopathiae serovars.
In swine erysipelas, antibodies against a cell surface component(s) of E. rhusiopathiae have been known to play an important role in protection. A 64- to 66-kDa cell surface antigen in Triton X-100 extracts of bacterial cells has been reported to be a protective molecule (2). Recently, a gene encoding surface protective antigen A (SpaA) was cloned from strains Tama-96 (serovar 2) (9) and Fujisawa (serovar 1a) (14), and its nucleotide sequence was determined. The genetic region responsible for protective immunity in the SpaA molecule has also been identified (5, 14). Southern and immunoblot analyses revealed that E. rhusiopathiae serovars 1a, 1b, 2, 5, 8, 9, 12, 15, 16, and 17 and type N possess the spaA gene and express the SpaA protein (9); however, whether the remaining E. rhusiopathiae serovars, i.e., serovars 4, 6, 11, 19, and 21, can produce Spa proteins or not is still unclear.
In this study, we analyzed spa-related genes of all E. rhusiopathiae serovars and of an unclassified serovar 18 strain in the genus Erysipelothrix and found that three spa-related molecules are present in the genus Erysipelothrix. We then analyzed the immunological properties of the three Spa proteins, mainly focusing on their cross-protectiveness, using a mouse model.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains used in this study included 16 strains of E. rhusiopathiae (Fujisawa, serovar 1a; 422/1E1, serovar 1b; ATCC 19414T, serovar 2; Doggerscharbe, serovar 4; Pe'cs 67, serovar 5; Dolphin E-1, serovar 6; Goda, serovar 8; Kaparek, serovar 9; IV12/8, serovar 11; Pe'cs 9, serovar 12; Pe'cs 3597, serovar 15; Tanzania, serovar 16; 545, serovar 17; 2017, serovar 19; Bãno 36, serovar 21; and MEW 22, type N), 5 strains of E. tonsillarum (ATCC 43339T, serovar 7; ATCC 43338, serovar 7; Lengyel-P, serovar 10; 2553, serovar 20; and Bãno 107, serovar 22), and two unclassified strains in the genus Erysipelothrix (Pe'cs 56, serovar 13; and 715, serovar 18). E. rhusiopathiae strains Fujisawa (serovar 1a), ATCC 19414T (serovar 2), Dolphin E-1 (serovar 6), and 715 (serovar 18) were used to challenge mice. The properties of the strains are described elsewhere (15).
The vector plasmid pGEM-T Easy (Promega, Madison, WI) was used to clone spa genes. Protein expression vectors pQE9 and pQE30 (QIAGEN, Santa Clarita, CA) were used for the construction and expression of histidine-tagged fusion proteins. Escherichia coli XL1-Blue was used as the host strain for replication of these plasmids.
Erysipelothrix strains were grown in tryptose phosphate broth supplemented with 1% proteose peptone no. 3 (Difco Laboratories, Detroit, MI) and 0.1% Tween 80 (pH 7.8). Escherichia coli strains were grown in Luria-Bertani medium. When appropriate, the medium was supplemented with ampicillin (100 μg/ml) or isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM).
PCR amplification.
Chromosomal DNAs of Erysipelothrix strains were prepared as described previously (19). The following primers were custom synthesized (Sawady Technology Co., Ltd., Tokyo, Japan): primer 1, 5′-AGGATCCATGAAAAAGAAAAAACACCTA-3′; primer 2, 5′-GAAGCTTCTATTTTAAACTTCCATCGTT-3′; primer 3, 5′-AGGATCCATGAAAAAGAAAAAACACCTATTTCCGAAAGTA-3′; and primer 4, 5′-GAAGCTTCTATTTTAAACTTCCATCGTTCTTAAATGCATA-3′. Primers 1 and 3 correspond to the sense strand at positions 1 to 21 and 1 to 33, respectively, of the spa gene of E. rhusiopathiae strain Fujisawa (14), with the addition of a new BamHI restriction site at the 5′ end. Primers 2 and 4 correspond to the antisense strand at positions 1881 to 1860 and 1881 to 1848, respectively, of the spa gene of strain Fujisawa, with the addition of a new HindIII restriction site at the 3′ end. Primers 1 and 2 were used for amplification of spaA from serovars 1a, 1b, 2, 5, 8, 9, 12, 15, 16, 17, and N, and primers 3 and 4 were used for amplification of spaB from serovars 4, 6, 11, 19, and 21 and for amplification of spaC from serovar 18. PCR was performed as described previously (10). Briefly, samples were subjected to an initial denaturation step at 94°C for 3 min and then to 25 cycles of denaturation at 94°C for 45 s, annealing at 55°C (primers 1 and 2) or 60°C (primers 3 and 4) for 1 min, and extension at 72°C for 2 min, with a final extension step at 72°C for 5 min.
Cloning and sequencing of spa genes.
PCR products were ligated to the plasmid pGEM-T Easy and then transformed into Escherichia coli XL1-Blue by electroporation with a gene pulser and pulse controller set at 2.5 kV, 25 mF, and 400 Ω (Bio-Rad, Richmond, CA).
Both strands of DNA of cloned PCR products were sequenced on an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA), using a BigDye Terminator cycle sequencing kit (Applied Biosystems). The DNA sequences were determined by a primer-walking procedure, starting with M13 vector primers, and were analyzed with the GENETYX-MAX program, version 12 (SDC, Tokyo, Japan), and the EditSeq and SeqMan programs of the DNAStar software package (DNASTAR Inc., Madison, WI). Alignments and phylogenetic tree analyses of nucleotide and amino acid sequences were performed with the CLUSTAL W program (18) and by the neighbor-joining method of the MEGA program (7), respectively.
Construction, expression, and purification of recombinant fusion proteins.
BamHI-HindIII fragments of 1,881 and 1,893 bp, containing the full coding sequences of SpaA from serovar 1a strain Fujisawa and SpaB from serovar 6 strain Dolphin E-1, respectively, were ligated to BamHI/HindIII-digested pQE9. The 1,995-bp BamHI-SalI fragment containing the full coding sequence of SpaC from serovar 18 strain 715 was ligated to BamHI/SalI-digested pQE30. The ligated DNAs were transformed into Escherichia coli XL1-Blue by electroporation.
The expression of recombinant histidine-tagged Spa proteins (rSpas) in Escherichia coli was induced by adding IPTG to 1 mM. The rSpas were purified by column chromatography with Ni-nitrilotriacetic acid resin (QIAGEN) according to the recommendation of the manufacturer.
Preparation of polyclonal rabbit antisera.
Six inbred New Zealand White rabbits (body weight, 2.5 to 3.0 kg) (line JW-NIBS; Nisseken Co. Ltd., Tokyo, Japan) were used. Groups of two rabbits were injected subcutaneously with 250 μg of purified rSpaA, rSpaB, or rSpaC protein in 1 ml of a 1:1 emulsion of the protein solution and Freund's complete adjuvant. Booster injections were given thrice, in weeks 3, 5, and 7, as described above, except that Freund's incomplete adjuvant was used. Blood samples were collected before immunization and at week 9 to determine serum antibody titers against each rSpa by immunoblotting. Rabbits were exsanguinated at week 10 to collect the sera. Obtained sera showing titers of 1:60,000 or more against the homologous rSpa protein were used.
Preparation of native SpaA, -B, and -C.
Native Spa proteins of E. rhusiopathiae strains Fujisawa (serovar 1a), Dolphin E-1 (serovar 6), and 715 (serovar 18) were extracted by alkaline treatment as described previously (3). Briefly, smooth colonies of the three strains were grown in a medium at 37°C overnight. Cells harvested and washed with phosphate-buffered saline were suspended in 10 mM NaOH at 0.05 g (wet weight)/ml and incubated with constant stirring for 18 h at 4°C. After neutralization, the suspension was centrifuged, concentrated, filter sterilized, and stored at −70°C.
SDS-PAGE and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were performed as described elsewhere (23). Antigenic proteins were probed with anti-Spa rabbit sera and visualized with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulins (Zymed Laboratories, Inc., San Francisco, CA) and a substrate solution containing 4-chloro-1-naphthol and hydrogen peroxide.
Mouse immunization and challenge.
One hundred sixty specific-pathogen-free male ddY mice aged 5 weeks (Japan SLC, Shizuoka, Japan) were used. Each of 40 mice was injected subcutaneously with 40 μg of purified rSpaA, rSpaB, or rSpaC emulsified with an equal volume of an oil-based adjuvant and then was given a booster injection after 3 weeks. The remaining 40 mice served as nonimmunized controls. Two weeks after the second injection, all groups of mice were subdivided into four groups of 10 each, and each group was challenged subcutaneously with 2.9 × 102 CFU of strain Fujisawa (serovar 1a), 3.8 × 102 CFU of strain ATCC 19414T (serovar 2), 4.1 × 104 CFU of strain Dolphin E-1 (serovar 6), or 6.9 × 102 CFU of strain 715 (serovar 18). Mouse mortality was monitored for the following 14 days.
The animals used in the present study were cared for in accordance with the guidelines for animal treatment of the Nippon Institute for Biological Science, which conform to the standard principles of laboratory animal care.
Statistical methods.
Comparisons of live-versus-dead numbers in the mouse experiments were compared by the Fisher exact test, using SAS software (SAS Institute Inc., Cary, NC) (12).
Nucleotide sequence accession numbers.
The spa nucleotide sequences of 17 Erysipelothrix strains have been submitted to the DDBJ/EMBL/GenBank database under accession no. AB259652 (strain Fujisawa [serovar 1a]), AB259653 (442/1E1 [serovar 1b]), AB259654 (ATCC 19414T [serovar 2]), AB259655 (Pe'cs 67 [serovar 5]), AB259656 (Goda [serovar 8]), AB290347 (Kaparek [serovar 9]), AB259657 (Pe'cs 9 [serovar 12]), AB259658 (Pe'cs 3597 [serovar 15]), AB2596529 (Tanzania [serovar 16]), AB259660 (545 [serovar 17]), AB259661 (MEW22 [serovar N]), AB238211 (Doggerscharbe [serovar 4]), AB238212 (Dolphin E-1 [serovar 6]), AB238213 (IV12/8 [serovar 11]), AB238214 (2017 [serovar 19]), AB238215 (Bãno 36 [serovar 21]), and AB238210 (715 [serovar 18]).
RESULTS
PCR amplification, cloning, and sequence analysis of spa genes.
We designed several primer pairs and successfully amplified genes encoding putative Spa proteins from 16 serovars of E. rhusiopathiae and a serovar 18 strain by PCR (data not shown). No products were amplified when DNAs from serovars of E. tonsillarum and serovar 13 were used. The PCR products were cloned into the plasmid pGEM-T Easy to determine their nucleotide sequences. Sequence analysis of the amplified DNA fragments revealed that the sizes of the spa open reading frames (ORFs) ranged from 1,818 to 1,992 nucleotides (encoding 606 to 664 amino acids). The spa ORFs varied in length depending on the E. rhusiopathiae serovar strain. The predicted molecular masses of the Spa proteins ranged from 69.9 to 76.9 kDa.
Based on their deduced amino acid sequence similarities, Spa proteins could be divided into three species of molecules (Table 1). We named the first Spa protein, from E. rhusiopathiae serovars 1a, 1b, 2, 5, 8, 9, 12, 15, 16, 17, and N, “SpaA,” which was used for serovars 1a and 2. We designated the second Spa protein, from serovars 4, 6, 11, 19, and 21, “SpaB” and the third Spa protein, from serovar 18 only, “SpaC.” The amino acid sequence similarities within each Spa type for various serovar strains were 96 to 99% for SpaA and 96 to 99% for SpaB. In contrast, the similarities between different Spa types were 61 to 64% (between SpaA and SpaB), 63 to 65% (between SpaA and SpaC), and 66 to 67% (between SpaB and SpaC), indicating that SpaA, -B, and -C are apparently different molecular species. SpaA proteins contain 626 amino acids, with a deduced molecular mass of 72.3 kDa, with the exception of the SpaA protein of serovar 9, which contains 606 amino acids, with a deduced molecular mass of 69.9 kDa. SpaB and SpaC proteins contain 630 and 664 amino acids, respectively, with deduced molecular masses of 72.9 and 76.9 kDa, respectively.
TABLE 1.
Deduced amino acid sequence similarities among Spas from 17 Erysipelothrix strains
| Serovar of Spa | % Amino acid similarity with Spas from serovara
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 2 | 5 | 8 | 9 | 12 | 15 | 16 | 17 | N | 4 | 6 | 11 | 19 | 21 | 18 | |
| 1a | 100 | ||||||||||||||||
| 1b | 99 | 100 | |||||||||||||||
| 2 | 98 | 98 | 100 | ||||||||||||||
| 5 | 97 | 98 | 99 | 100 | |||||||||||||
| 8 | 99 | 99 | 98 | 98 | 100 | ||||||||||||
| 9 | 98 | 98 | 97 | 96 | 98 | 100 | |||||||||||
| 12 | 99 | 99 | 98 | 98 | 99 | 98 | 100 | ||||||||||
| 15 | 97 | 98 | 99 | 100 | 98 | 96 | 98 | 100 | |||||||||
| 16 | 99 | 99 | 98 | 98 | 99 | 98 | 99 | 98 | 100 | ||||||||
| 17 | 98 | 98 | 97 | 97 | 99 | 97 | 99 | 97 | 98 | 100 | |||||||
| N | 99 | 99 | 98 | 98 | 99 | 98 | 99 | 98 | 99 | 99 | 100 | ||||||
| 4 | 63 | 63 | 63 | 63 | 64 | 61 | 63 | 63 | 63 | 63 | 64 | 100 | |||||
| 6 | 63 | 63 | 63 | 63 | 64 | 61 | 63 | 63 | 63 | 63 | 64 | 99 | 100 | ||||
| 11 | 64 | 64 | 64 | 64 | 64 | 62 | 64 | 64 | 64 | 64 | 64 | 97 | 97 | 100 | |||
| 19 | 62 | 63 | 62 | 63 | 63 | 61 | 63 | 63 | 63 | 63 | 63 | 98 | 99 | 96 | 100 | ||
| 21 | 62 | 63 | 62 | 63 | 63 | 61 | 63 | 63 | 63 | 63 | 63 | 98 | 99 | 96 | 99 | 100 | |
| 18 | 65 | 65 | 65 | 65 | 65 | 63 | 65 | 65 | 65 | 64 | 65 | 67 | 67 | 67 | 66 | 67 | 100 |
Values showing ≥96% amino acid sequence similarity are highlighted in bold.
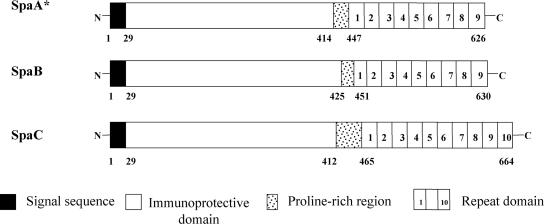
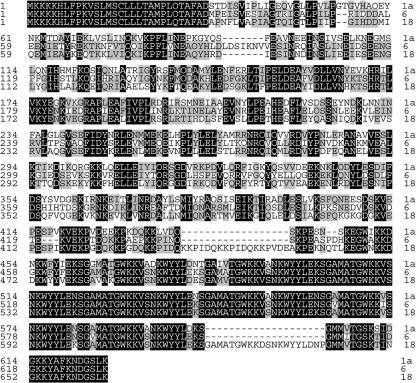
Figure 1 shows a schematic diagram of the domain structures of the three Spa proteins. The signal sequences of 29 amino acids are perfectly conserved among all Spa proteins (100% identity). The following N-terminal half-domain of Spas, which contains predominantly α-helical and amphipathic structures, has been shown to play a role in immunoprotection against E. rhusiopathiae infection in both mice and pigs (5). The protective domain, which is located at amino acid positions 30 to 413, 30 to 424, and 30 to 411 in SpaA, -B, and -C, respectively, and has 50 to 51%, 56 to 57%, and 53 to 54% similarity between SpaA and -B, SpaA and -C, and SpaB and -C, respectively, is hypervariable in both size and sequence among Spa proteins. The proline-rich hydrophobic domain, which is located at amino acid positions 414 to 447, 425 to 451, and 412 to 465 in SpaA, -B, and -C, respectively, and a repeat domain, which consists of 8 to 10 tandem repeats of similar 20-amino-acid sequences and is located at amino acid positions 448 to 626, 452 to 630, and 466 to 664 in SpaA, -B, and -C, respectively, are conserved among the three Spa proteins. The amino acid sequence similarities of the proline-rich hydrophobic domain and the repeat domain among SpaA, -B, and -C are 40 to 61% and 83 to 88%, respectively. In the repeat domain, the number of 20-amino-acid tandem repeats is 9 for SpaA and -B and 10 for SpaC; however, SpaA of E. rhusiopathiae serovar 9 strain Kaparek has only eight tandem repeats, indicating the variation in the number of repeats in SpaA.
FIG. 1.
Schematic diagrams of the domain structure of the three Spa molecules. The 29-amino-acid signal sequence, immunoprotective domain, proline-rich region, and 20-amino-acid repeat domain are indicated by solid (black), open, stippled, and numbered boxes, respectively. *, SpaA of the E. rhusiopathiae serovar 9 strain Kaparek has eight tandem repeats.
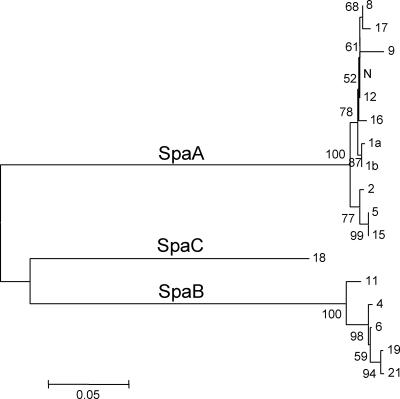
A phylogenetic tree was constructed based on the deduced amino acid sequences of Spa proteins (Fig. 2). The branch lengths between molecules are proportional to the similarities of the sequences. This tree confirms that Spa proteins from 16 E. rhusiopathiae serovars and serovar 18 could be divided into three molecular species, namely, SpaA, SpaB, and SpaC.
FIG. 2.
Phylogenetic tree constructed from the deduced amino acid sequences of Spa proteins from 16 E. rhusiopathiae serovars and one unclassified serovar in the genus Erysipelothrix. The designations shown to the right of the tree show serovars. A scale bar is shown at the bottom. The tree was constructed based on the neighbor-joining method. Bootstrap proportions were plotted at the main internal branches of the phylogram to show support values.
Expression and purification of rSpas.
To produce rSpas, ORFs encoding SpaA, -B, and -C were cloned into the expression vector pQE9 or pQE30. We chose Spa ORFs of serovars 1a and 6 as representatives to produce rSpaA and rSpaB, respectively. A Spa ORF from serovar 18 was used to produce rSpaC. Whether Spa ORFs were correctly inserted downstream of the promoter sequence was confirmed by nucleotide sequence analysis. His-tagged rSpa fusion proteins were expressed in Escherichia coli and purified with column chromatography on Ni-nitrilotriacetic acid resin.
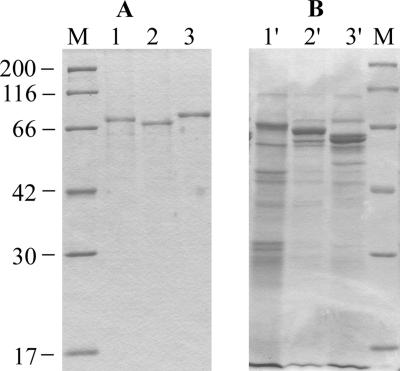
SDS-PAGE analysis of purified rSpaA, rSpaB, and rSpaC showed single bands of approximately 73, 70, and 77 kDa, respectively (Fig. 3A). rSpaA and rSpaC migrated at their expected molecular masses, whereas the rSpaB protein showed a lower molecular mass than predicted. Aberrant migration of rSpaB might result from posttranslational modifications.
FIG. 3.
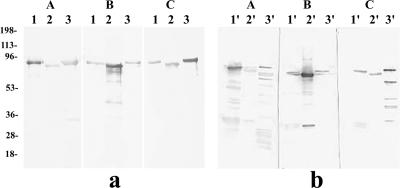
SDS-PAGE profiles of purified rSpa proteins and alkaline extracts of E. rhusiopathiae strains containing native Spa proteins. The gels were stained for protein with Coomassie brilliant blue. (A) Purified recombinant proteins. Lane 1, rSpaA protein prepared from the spaA gene of strain Fujisawa (serovar 1a); lane 2, rSpaB protein prepared from the spaB gene of strain Dolphin E-1 (serovar 6); lane 3, rSpaC protein prepared from the spaC gene of strain 715 (serovar 18); lane M, molecular masses in kilodaltons. (B) Alkaline extracts of E. rhusiopathiae strains Fujisawa (serovar 1a; lane 1′) and Dolphin E-1 (serovar 6; lane 2′) and the unclassified Erysipelothrix strain 715 (serovar 18; lane 3′).
SDS-PAGE profiles of alkaline extracts of E. rhusiopathiae strains, which contained native SpaA, SpaB, and SpaC proteins, are shown in Fig. 3B. Major bands detected in the extracts of E. rhusiopathiae serovars 1a and 6 were located at the same positions as those of rSpaA and rSpaB, respectively. However, in the extract of serovar 18, only a faint band was detected at the same position as that of rSpaC, and a major band was located at a lower-molecular-weight position than that of rSpaC. These results indicated that the alkaline extracts of E. rhusiopathiae strains contained mainly native Spa proteins and that SpaC is easily degraded.
Reactivity of anti-rSpa sera to homologous and heterologous Spa proteins.
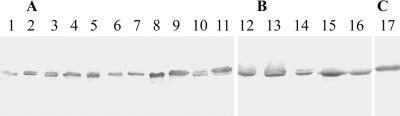
To examine whether all 16 E. rhusiopathiae serovars and the serovar 18 strain express Spas, rabbit antisera specific to rSpaA, rSpaB, and rSpaC were incubated with whole-cell lysates prepared from homologous serovar strains by immunoblotting, i.e., the rSpaA antiserum was incubated with lysates of serovars 1a, 1b, 2, 5, 8, 9, 12, 15, 16, 17, and N; the rSpaB antiserum was incubated with those of serovars 4, 6, 11, 19, and 21; and the rSpaC antiserum was incubated with that of serovar 18. The results showed that all antisera specific to each Spa protein recognized an approximately 70-kDa protein antigen, which corresponds to the expected molecular mass of the Spas, in the lysates of 16 E. rhusiopathiae serovars and the serovar 18 strain (Fig. 4). This result demonstrates that all serovars of E. rhusiopathiae produce at least one of the Spa proteins.
FIG. 4.
Results of immunoblot analysis of E. rhusiopathiae whole-cell proteins from different serovars probed with polyclonal anti-rSpaA (A), anti-rSpaB (B), and anti-rSpaC (C) antibodies. Lanes: 1, MEW 22 (type N); 2, 545 (serovar 17); 3, Tanzania (serovar 16); 4, Pe'cs 3597 (serovar 15); 5, Kaparek (serovar 9); 6, Pe'cs 9 (serovar 12); 7, Goda (serovar 8); 8, Fujisawa (serovar 1a); 9, 422/1E1 (serovar 1b); 10, ATCC 19414T (serovar 2); 11, Pe'cs 67 (serovar 5); 12, Doggerscharbe (serovar 4); 13, Dolphin E-1 (serovar 6); 14, IV12/8 (serovar 11); 15, 2017 (serovar 19); 16, Bãno 36 (serovar 21); and 17, 715 (serovar 18).
Next, to analyze the cross-reactivity among the three Spa proteins, rabbit antisera specific to rSpaA, rSpaB, and rSpaC were incubated with homologous and heterologous Spas (both recombinant and native) by immunoblotting. When rSpas were used as antigens, anti-rSpa sera reacted strongly with homologous rSpas but weakly with heterologous rSpas (Fig. 5a). This lower reactivity to heterologous Spa proteins than to homologous Spa protein was more clearly demonstrated when native Spa proteins were used as antigens. Anti-rSpaA and anti-rSpaB sera reacted strongly with their homologous native SpaA and SpaB proteins (Fig. 5b, panel A, lane 1′, and panel B, lane 2′, respectively), in contrast to the weak reactivity observed with heterologous native Spas; however, anti-rSpaC antiserum reacted strongly with the native SpaC protein (Fig. 5b, panel C, lane 3′) and even reacted with heterologous SpaA and SpaB proteins, with a moderate degree of intensity (Fig. 5b, panel C, lanes 1′ and 2′, respectively). In addition, the native SpaC fraction was found to contain several antigenic proteins, with molecular masses of 60, 45, 36, and 30 kDa, which appeared to be degradation products of the intact SpaC protein. An alignment of deduced amino acid sequences of SpaA (from serovar 1a strain Fujisawa), SpaB (from serovar 6 strain Dolphin E-1), and SpaC (from serovar 18 strain 715) is shown in Fig. 6. These results suggest that the heterogeneity in the primary sequences of the three representative Spas might contribute to differences in cross-reactivity.
FIG. 5.
Results of immunoblot analysis of recombinant (a) and native (b) Spa proteins probed with homologous and heterologous anti-rSpa polyclonal antibodies. Panels A, B, and C show the results using anti-rSpaA, anti-rSpaB, and anti-rSpaC rabbit antisera, respectively. Recombinant and native SpaA (from serovar 1a), SpaB (from serovar 6), and SpaC (from serovar 18) proteins were used as antigens (lanes 1 and 1′, 2 and 2′, and 3 and 3′, respectively). Numbers on the left are molecular masses in kilodaltons.
FIG. 6.
Alignment of deduced amino acid sequences of SpaA, -B, and -C, which originated from three representative strains, namely, Fujisawa (serovar 1a), Dolphin E-1 (serovar 6), and 715 (serovar 18), respectively. The predicted N-terminal signal sequence corresponds to amino acids 1 to 29. Identical and conserved residues are shown as letters shaded with black and gray boxes, respectively. Gaps are shown as dashes.
Cross-protection study using rSpas as immunogens.
To examine whether cross-protection could be induced by immunization with different rSpas as antigens, we injected each rSpa protein twice into mice and challenged the mice with three E. rhusiopathiae strains expressing homologous and heterologous Spas to the immunizing rSpa.
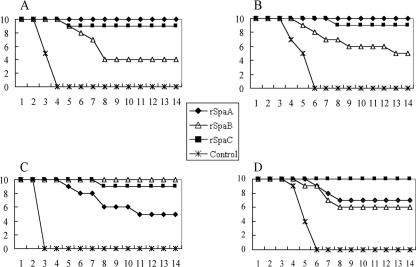
After challenge with the serovar 1a strain Fujisawa, which expresses SpaA, all nonimmunized control mice died within 4 days (Fig. 7A). In contrast, all mice immunized with rSpaA (homologous) survived until 14 days, when the experiment was terminated. The difference between the rSpaA-immunized group and the control group was significant (P = 0.0000). Only 4 of 10 (40%) mice in the rSpaB (heterologous)-immunized group survived (P = 0.0433 [significant]), indicating that the protection conferred by immunization with rSpaB was partial. In the rSpaC (heterologous)-immunized group, 9 of 10 (90%) mice survived, showing that rSpaC induced efficient protection against challenge with strain Fujisawa (P < 0.0001 [significant]). Similar results were observed when mice were challenged with the serovar 2 strain ATCC 19414T, which expresses SpaA: 100%, 50%, and 90% of mice in the rSpaA-, rSpaB-, and rSpaC-immunized groups, respectively, survived under the challenge conditions where all mice in the nonimmunized control group died (Fig. 7B).
FIG. 7.
Cumulative mortality of nonimmunized mice and mice immunized with rSpa proteins after challenge with homologous and heterologous serovar strains. (A) Challenge with serovar 1a strain Fujisawa producing SpaA; (B) challenge with serovar 2 strain ATCC 19414T producing SpaA; (C) challenge with serovar 6 strain Dolphin E-1 producing SpaB; (D) challenge with serovar 18 strain 715 producing SpaC.
When challenged with the serovar 6 strain Dolphin E-1, which expresses SpaB, all of the nonimmunized control mice died within 3 days (Fig. 7C), whereas all mice immunized with rSpaB (homologous) survived (P = 0.0000 [significant]). Only 5 of 10 mice (50%) in the SpaA (heterologous)-immunized group survived (P = 0.0163 [significant]); however, 9 of 10 mice in the SpaC (heterologous)-immunized group survived (P < 0.0001 [significant]).
After challenge with the serovar 18 strain 715, which expresses SpaC, all nonimmunized control mice died within 6 days (Fig. 7D), whereas all mice immunized with rSpaC (homologous) survived. Seven of 10 mice (70%) in the SpaA (heterologous)-immunized group and 6 of 10 mice (60%) in the SpaB (heterologous)-immunized group survived (P = 0.0015 and P = 0.0054, respectively [both significant]). These results indicate that all rSpas have potent protective immunogenicity against challenge with E. rhusiopathiae strains expressing homologous Spas; however, the levels of immunogenicity of the rSpas were variable against E. rhusiopathiae strains producing heterologous Spas. These data also demonstrate that the rSpaC protein is the most potent cross-protective antigen in this mouse challenge model.
DISCUSSION
The Spa protein of E. rhusiopathiae has been shown to be localized at the bacterial cell surface and to be a potent protective antigen against E. rhusiopathiae infection (5, 14). The present study is the first to describe genetic and antigenic diversity in the Spa proteins of 16 E. rhusiopathiae serovars and one unclassified serovar in the genus Erysipelothrix. Our results demonstrate that (i) Spa proteins can be classified into three molecular species, named SpaA, SpaB, and SpaC, based on their amino acid sequence similarities; (ii) the three Spa proteins are antigenically different, as antisera reacted strongly with homologous Spas but reacted moderately or weakly with heterologous Spas; and (iii) that the degree of cross-protection conferred in mouse immunoprotection tests by immunization with each Spa protein varies among Spas and that SpaC is the most broadly cross-protective antigen among the three Spa proteins.
SpaA was previously identified and characterized by Makino et al. (9), using E. rhusiopathiae strain Tama 96 (serovar 2), and by Shimoji et al. (14), using strain Fujisawa (serovar 1a). Makino et al. found that SpaA is produced by only 11 (1a, 1b, 2, 5, 8, 9, 12, 15, 16, 17, and N) of 16 E. rhusiopathiae serovars. However, when the remaining five serovars (4, 6, 11, 19, and 21) were used as samples, a weak positive reaction was shown by enzyme-linked immunosorbent assay with a monoclonal antibody specific to SpaA, but no positive signal was detected by immunoblotting with the monoclonal antibody and Southern blot analysis using the spaA gene as a probe; thus, they concluded that these five serovar strains do not produce SpaA. In the present study, we demonstrated that these five serovar strains produce SpaB, which shows approximately 60% homology to SpaA but is apparently a distinct and novel molecule. Consequently, all E. rhusiopathiae serovar strains produce at least one Spa protein, SpaA or SpaB, as we showed in this study (Fig. 4). In contrast, we could not detect any spa-related genes by PCR or any Spa-related proteins by immunoblot analysis in E. tonsillarum serovar strains (data not shown). This finding suggests that Spa proteins may be virulence factors specifically involved in the pathogenesis of E. rhusiopathiae, and from a practical point of view, the presence or absence of spa may be a suitable marker for the differentiation of E. rhusiopathiae and E. tonsillarum.
Regions or stretches of sequence conservation among Spas are likely to represent domains of protein constrained by a common function. In our alignment of Spas from 17 Erysipelothrix strains, high sequence identity was found within the signal and repetitive amino acid regions. In particular, the amino acid sequences of the signal sequence regions, located at the most N-terminal part of the Spas, were completely identical in the Spa proteins. These data suggest that Spa proteins may cross bacterial cell membranes and eventually be secreted from the bacterial cell by using a secretion machinery common to E. rhusiopathiae strains. The regions containing 20-amino-acid tandem repeats are also structurally similar among the three Spas. The repeat region has been suggested to function as an anchor for binding Spa proteins tightly to the bacterial surface (8), as it was found in other surface proteins of gram-positive bacteria (1, 4, 22). This suggests that the anchoring mechanism of the bacterial cell surface protein may be common not only to E. rhusiopathiae strains but also to other gram-positive pathogenic bacteria.
Sequence diversity among Spa proteins was largely confined to the α-helical N-terminal half, which could be defined as a hypervariable domain (∼50% identity). Importantly, the N-terminal half has been found to play a major role in immunoprotection against E. rhusiopathiae infection (5, 14). In fact, immunoblot analysis showed that antisera raised against rSpaA, rSpaB, or rSpaC reacted strongly with homologous Spas but moderately or weakly with heterologous Spas when native Spas in E. rhusiopathiae whole-cell extracts as well as rSpas purified from recombinant Escherichia coli were used as antigens. Various degrees of cross-protection among different E. rhusiopathiae serovars have been described (13). In our cross-protection study, mice immunized with rSpaA, rSpaB, or rSpaC were completely protected against homologous challenge, but the degree of cross-protection varied for heterologous challenge. However, it was noteworthy that rSpaC induced good protection against even heterologous strains expressing SpaA or SpaB. This high degree of cross-protection induced by immunization with SpaC was unexpected because of the high sequence diversity in the protection domain found between SpaC and other Spa proteins. One possible explanation is that short linear epitopes which are similar between SpaC and other Spas may exist in the protective domain. Another possible explanation is that conformational epitopes in SpaC which work as cross-protective epitopes may exist. Further studies concerning the identification and characterization of epitopes which may be present in Spa molecules should be performed to clarify why the SpaC protein can induce cross-protection in immunized animals.
Our results showed that serovar 18 is a unique serovar which produces SpaC. At present, the genus Erysipelothrix contains at least two species, E. rhusiopathiae and E. tonsillarum. Serovar 18 strains, however, have not been assigned to these two species and are considered to be classified in a genetically distinct group, based on the observation that they hybridize at low levels with the type strains of E. rhusiopathiae and E. tonsillarum (15). The serovar 18 reference strain 715, together with two other serovar 18 strains, has been shown to be virulent for mice and swine (21), raising the possibility that SpaC plays some role in their virulence. The function of SpaC, as well as those of SpaA and SpaB, and its possible role in E. rhusiopathiae virulence should be elucidated by analyzing a Spa gene knockout mutant.
The present findings concerning the diversity of Spa proteins in the genus Erysipelothrix may shed new light on the pathogenesis of Erysipelothrix species and may help to classify various serovar strains in the genus Erysipelothrix. Currently, serovars 18 and 13 are not classified into species. We found that a serovar 13 strain did not have spa-related genes or produce any Spa proteins, as detected by PCR and immunoblot analysis (data not shown). This is an important characteristic for discriminating serovar 13 strains from serovar 18 or other E. rhusiopathiae serovar strains. The finding that the spa-related genes and their products are found only in E. rhusiopathiae species, not in E. tonsillarum species, may be applied to the development of a spa gene-targeted PCR assay and an rSpa antigen-based serodiagnostic test for differential diagnosis. Most importantly, Spa proteins have been shown to be potent immunogens and thus may become good candidates for a vaccine component. This study revealed that rSpaC can induce broad cross-protectiveness against challenge with E. rhusiopathiae serovar strains producing different Spas; therefore, a component vaccine containing rSpaC or a more advanced chimeric vector vaccine producing rSpaC may be useful for the eradication of erysipelas from swine herds.
Acknowledgments
We thank T. Sawada (Nippon Veterinary and Life Science University) for providing the E. rhusiopathiae strains and members of the Second Division of the Nippon Institute for Biological Science for excellent care in the animal experiments.
This work was supported by a grant-in-aid from the Ministry of Agriculture, Forestry and Fisheries of Japan via the Japanese Association of Veterinary Biologics.
Footnotes
Published ahead of print on 2 May 2007.
REFERENCES
- 1.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 676533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galan, J. E., and J. F. Timoney. 1990. Cloning and expression in Escherichia coli of a protective antigen of Erysipelothrix rhusiopathiae. Infect. Immun. 583116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groschup, M. H., K. Cussler, R. Weiss, and J. F. Timoney. 1991. Characterization of a protective protein antigen of Erysipelothrix rhusiopathiae. Epidemiol. Infect. 107637-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammerschmidt, S., S. R. Talay, P. Brandtzaeg, and G. S. Chhatwal. 1997. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol. Microbiol. 251113-1124. [DOI] [PubMed] [Google Scholar]
- 5.Imada, Y., N. Goji, H. Ishikawa, M. Kishima, and T. Sekizaki. 1999. Truncated surface protective antigen (SpaA) of Erysipelothrix rhusiopathiae serotype 1a elicits protection against challenge with serotypes 1a and 2b in pigs. Infect. Immun. 674376-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitajima, T., E. Oishi, K. Amimoto, S. Ui, H. Nakamura, N. Okada, O. Sasaki, and H. Yasuhara. 1997. Protective effect of NaOH-extracted Erysipelothrix rhusiopathiae vaccine in pigs. J. Vet. Med. Sci. 609-14. [DOI] [PubMed] [Google Scholar]
- 7.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 8.Makino, S., K. Yamamoto, H. Asakura, and T. Shirahata. 2000. Surface antigen, SpaA, of Erysipelothrix rhusiopathiae binds to gram-positive bacterial cell surfaces. FEMS Microbiol. Lett. 186313-317. [DOI] [PubMed] [Google Scholar]
- 9.Makino, S., K. Yamamoto, S. Murakami, T. Shirahata, K. Uemura, T. Sawada, H. Wakamoto, and H. Morita. 1998. Properties of repeat domain found in a novel protective antigen, SpaA, of Erysipelothrix rhusiopathiae. Microb. Pathog. 25101-109. [DOI] [PubMed] [Google Scholar]
- 10.Nagai, S., S. Someno, and T. Yagihashi. 1994. Differentiation of toxigenic from nontoxigenic isolates of Pasteurella multocida by PCR. J. Clin. Microbiol. 321004-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opriessnig, T., L. J. Hoffman, D. L. Harris, S. B. Gaul, and P. G. Halbur. 2004. Erysipelothrix rhusiopathiae: genetic characterization of midwest US isolates and live commercial vaccines using pulsed-field gel electrophoresis. J. Vet. Diagn. Investig. 16101-107. [DOI] [PubMed] [Google Scholar]
- 12.SAS Institute Inc. 1988. SAS/SAT user's guide 6.03 ed. SAS Institute Inc., Cary, NC.
- 13.Sawada, T., and T. Takahashi. 1987. Cross protection of mice and swine inoculated with culture filtrate of attenuated Erysipelothrix rhusiopathiae and challenge exposed to strains of various serovars. Am. J. Vet. Res. 48239-242. [PubMed] [Google Scholar]
- 14.Shimoji, Y., Y. Mori, and V. A. Fischetti. 1999. Immunological characterization of a protective antigen of Erysipelothrix rhusiopathiae: identification of the region responsible for protective immunity. Infect. Immun. 671646-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi, T., T. Fujisawa, Y. Tamura, S. Suzuki, M. Muramatsu, T. Sawada, Y. Benno, and T. Mitsuoka. 1992. DNA relatedness among Erysipelothrix rhusiopathiae strains representing all twenty-three serovars and Erysipelothrix tonsillarum. Int. J. Syst. Bacteriol. 42469-473. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi, T., N. Nagamine, M. Kijima, S. Suzuki, M. Takagi, Y. Tamura, M. Nakamura, M. Muramatsu, and T. Sawada. 1996. Serovars of Erysipelothrix strains isolated from pigs affected with erysipelas in Japan. J. Vet. Med. Sci. 58587-589. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi, T., T. Sawada, M. Takagi, K. Seto, M. Kanzaki, and T. Maruyama. 1984. Serotypes of Erysipelothrix rhusiopathiae strains isolated from slaughter pigs affected with chronic erysipelas. Jpn. J. Vet. Sci. 46149-153. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To, H., G. Zhang, H. Otsuka, M. Ogawa, O. Ochiai, S. Nguyen, T. Yamaguchi, N. Nagaoka, M. Akiyama, K. Amano, and K. Hirai. 1996. Q fever pneumonia in children in Japan. J. Clin. Microbiol. 34647-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood, R. L. 1999. Erysipelas, p. 419-430. In B. E. Straw et al. (ed.), Diseases of swine. Iowa State University Press, Ames, IA.
- 21.Wood, R. L., D. R. Haubrich, and R. Harrington. 1978. Isolation of previously unreported serotypes of Erysipelothrix rhusiopathiae from swine. Am. J. Vet. Res. 391958-1961. [PubMed] [Google Scholar]
- 22.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 1762976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, G., H. To, K. E. Russell, L. R. Hendrix, T. Yamaguchi, H. Fukushi, K. Hirai, and J. E. Samuel. 2005. Identification and characterization of an immunodominant 28-kilodalton Coxiella burnetii outer membrane protein specific to isolates associated with acute disease. Infect. Immun. 731561-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]