Abstract
Macaques are the only animal model used to test dengue virus (DENV) vaccine candidates. Nevertheless, the pathogenesis of DENV in macaques is not well understood. In this work, by using Affymetrix oligonucleotide microarrays, we studied the broad transcriptional modifications and cytokine expression profile after infecting rhesus macaques with DENV serotype 1. Five days after infection, these animals produced a potent, innate antiviral immune response by inducing the transcription of signature genes from the interferon (IFN) pathway with demonstrated antiviral activity, such as myxoprotein, 2′,5′-oligoadenylate synthetase, phospholipid scramblase 1, and viperin. Also, IFN regulatory element 7, IFN-stimulated gene 15, and protein ligases linked to the ISGylation process were up-regulated. Unexpectedly, no up-regulation of IFN-α, -β, or -γ genes was detected. Transcription of the genes of interleukin-10 (IL-10), IL-8, IL-6, and tumor necrosis factor alpha was neither up-regulated nor down-regulated. Results were confirmed by real-time PCR and by multiplex cytokine detection in serum samples.
Dengue virus (DENV) is the second most important arthropod-borne tropical disease after malaria, with 50 to 100 million and 500,000 cases of dengue fever (DF) and dengue hemorrhagic fever (DHF)/dengue shock syndrome (DSS), respectively, each year (29). These diseases are induced by DENV, a member of the Flaviviridae family that exists as four different serotypes (DENV-1, -2, -3, and -4). The mechanisms of DENV-induced disease and immune response are not fully understood, but two theories are the most discussed. The theory of antibody-dependent enhancement postulates that higher viremia occurs in secondary infections due to antibody-facilitated viral entry, causing more severe disease (32-35, 71). The cytokine-mediated immunopathology theory proposes a model of plasma leakage in DHF mediated through an interaction between DENV-infected monocytes/macrophages and memory CD4+ and CD8+ DENV-reactive T cells (58-61, 90, 91). This interaction leads to production of type 1 proinflammatory cytokines, mainly gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α), which directly affects the vascular endothelium. This cytokine-mediated proinflammatory response is postulated to be more extended in a secondary infection than in primary infections due to the preferential expansion of memory T cells that recognize cross-reactive epitopes. However, it is well documented that DHF/DSS can occur after a single DENV infection (6, 7, 13, 50, 64, 77, 83, 108). In addition to IFN-γ and TNF-α, other cytokines, such as interleukin-18 (IL-18), IL-6, IL-2, and IL-10, have been detected in the sera of patients with DHF (6, 12, 22, 77, 82, 83). A pathogenic role has been assigned to monocytes/macrophages but also to B cells in humans (67). There is no appropriate animal model for DENV that reproduces DF/DHF/DSS symptoms. Recently, some interesting approaches have been taken using mice as a model (8, 99). However, in spite of its worth and the advance it represents in the study of DENV, macaques continue to be the standard model to test vaccine candidates or treatments against DENV. At present, there is no specific anti-DENV treatment, but several DENV vaccine formulations are being studied. All of them have been or will be tested using macaques before studies are conducted with humans (11, 21, 38, 84, 85, 87, 88, 102, 105). Previous work has been performed to characterize DENV replication in macaques (36, 37). However, little is known about the molecular mechanism of the macaque's immune response to DENV. In particular, the type or level of cytokines produced during DENV infection or the gene expression profile of infected cells is not known in these animals. The efficacy of DENV vaccine trials relies on the production of neutralizing antibodies after vaccination that can counteract viremia after challenge. Macaques do not develop symptoms that mimic the clinical outcomes of DF or DHF/DSS in humans, and only a transient viremia occurs without the development of relevant symptoms. This suggests a natural mechanism that might allow rhesus macaques to antagonize DENV infection better than humans. We have taken preliminary steps to better understand the innate immune response in rhesus macaques. Here, we have tested the profile of gene activation and cytokine production after infecting four rhesus macaques with a low-passage strain of DENV-1. Our studies provide the first evidence that rhesus macaques respond to DENV infection with a potent innate antiviral immune response although without measurable type I or II IFN or proinflammatory cytokine production in peripheral blood mononuclear cells (PBMC). These profiles are quite different from those reported for humans and could explain the absence of symptoms of DF or DHF/DSS in these animals. These findings have implications for vaccine efficacy studies in progress.
MATERIALS AND METHODS
Infection of animals and blood collection.
Six male rhesus macaques that were negative for immunoglobulin G (IgG) and IgM antibodies to DENV were used for the study. Four animals were infected subcutaneously with a 1-ml suspension containing 1 × 104 PFU of a low-passage Western Pacific 74 DV1 strain (L. Markoff, Walter Reed Army Hospital). Two animals were mock infected with supernatant from uninfected LLCMK-2 cells. Sera were collected at days 1, 3, and 5 after infection, quick-frozen, and kept at −80°C until analysis. On day 5, PBMC were also collected using 8-ml Vacutainer CPT tubes (BD, Franklin Lakes, NJ). Tubes were centrifuged at 1,500 × g for 30 min at 20°C. Collected PBMC were washed twice with phosphate-buffered saline, and platelets were removed by centrifugation at 200 × g for 15 min at 20°C. PBMC were used directly for magnetic cell sorting. A fraction was frozen at −80°C for further RNA extraction. All work with animals was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to the use of animals in research. In addition, all procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the Medical Sciences Campus, University of Puerto Rico, and performed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Serologic tests.
All animals were tested for the presence of anti-DENV IgM by the IgM antibody capture enzyme-linked immunosorbent assay in order to preclude recent natural infections in the monkeys. To preclude past infections, IgG antibody tests were performed using a quantitative enzyme-linked immunosorbent assay. Neutralizing antibodies to DENV-1 were quantified using a flow cytometry-based neutralization (FNT) assay on Vero cells. This neutralization assay is based on a fluorescence-activated cell sorter (FACS)-based dengue virus titration assay as described previously (63). In the flow cytometry-based neutralization assay, the ability of immune serum to neutralize the infectivity of DENV on Vero cells is measured at 24 h postinfection by enumerating cells that are positive for intracellular staining of DENV E protein by flow cytometry.
The neutralization titer for each serum sample was expressed as the reciprocal of the highest dilution of serum that neutralized the challenge virus by 50% (FNT50) (63). Alanine aminotransferase and aspartate aminotransferase levels were determined using a commercial system (Dimension Expand; Dade Behring, Deerfield, IL).
Isolation of B cells and macrophages from PBMC.
PBMC were obtained as described above and separated by magnetic sorting using superparamagnetic microbeads according to the manufacturer's instructions (all reagents and equipment for magnetic cell separation were purchased from Miltenyi Biotec, Auburn, CA). Macrophages were purified with anti-CD14 microbeads using LS columns. PBMC were CD3 depleted with the CD3 MicroBead kit using LD columns. B cells were subsequently purified with anti-CD20 microbeads using LS columns. The remaining fraction of PBMC and the eluted fraction from the CD3 depletion (which includes CD4, CD8, NK, and other cells) were labeled as other cells (OC) and kept frozen at −80°C for further analysis. The cells' purity was determined by FACS analysis using conjugated mouse anti-human monoclonal antibodies known to cross-react with rhesus monkey antigens. CD20 fluorescein isothiocyanate (2H7) and CD14 fluorescein isothiocyanate (M5E2) were used to label purified B cells or macrophages, respectively. In both cases, specific antibodies were combined with CD3 allophycocyanin (SP34-2), and CD20+/CD3− or CD14+/CD3+ cells were quantified. Antibodies for FACS analysis were obtained from BD Biosciences (San Diego, CA).
Total cellular RNA preparation.
RNA was extracted from 3 × 105 PBMC or from 3 × 106 isolated B cells, macrophages, or OC by using an RNeasy Mini kit (QIAGEN, Valencia, CA) and eluted in 30 μl of RNase-free double-distilled H2O. For real-time PCR (RT-PCR), RNA was also extracted from frozen PBMC. The RNA was resuspended in diethyl pyrocarbonate-treated 0.01% distilled H2O (Ambion, Austin, TX). The quality and quantity of RNA were estimated by the Agilent Bioanalyzer RNA Nanochip methodology. RNA samples from macrophages, B cells, and OC were used for microarray, and RNA from macrophages, B cells, and PBMC was used for RT-PCR.
Affymetrix GeneChip analysis.
The Affymetrix protocol used was essentially described previously (109). Briefly, 100 picomoles of a T7-(dT)24 primer was added to 10 μg total RNA. The primer and RNA were denatured at 70°C for 10 min and placed on ice, and cDNA was then synthesized using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Reactions were terminated by the addition of EDTA to a final concentration of 30 mM, and samples were placed on ice. An equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added to each cDNA sample and centrifuged at 16,000 × g for 2 min. The aqueous upper layer was precipitated with 2.5 M ammonium acetate and a 2.5× final volume of 100% ethanol. Samples were centrifuged, and the resulting pellets were extensively washed with 80% ethanol, air dried, and resuspended in 12 μl diethyl pyrocarbonate-treated water. The in vitro transcription labeling reactions were done using ENZO BioArray (Affymetrix) to synthesize biotin-labeled cRNA targets. The labeled cRNA was cleaned using RNeasy Mini kits, and 15 μg was hybridized to rhesus macaque GeneChip arrays (Affymetrix, Santa Clara, CA) containing 56,867 probe sets, 47,400 transcripts, and 38,500 genes. The microarray procedure was performed at the Expression Analysis Laboratory (Durham, NC) using a GeneChip Scanner 3000 (Affymetrix). Probe intensity values were extracted from the array image using GCOS software (Affymetrix).
Data analysis.
Genes were normalized to the mean of the controls, and data from corresponding negative controls were selected as a reference to allow the calculation of infected/uninfected gene expression ratios. Arrays were median normalized. Genes with low signals (defined as a signal of <100 above the background) were not considered. The remaining signals were log2 transformed prior to analysis so that relative increases and decreases in gene expression were represented on a linear scale. Gene expression profiles (txt version of the CHP files) were loaded into the GeneSifter microarray data analysis system (VizX Labs, Seattle, WA). The data were filtered by using the criteria absent (A), marginal (M), and present (P). To identify highly expressed genes, only genes with at least a fourfold increase in expression with a P value of <0.05 after t test and Benjamini and Hochberg correction were selected and displayed as scatter plots. For hierarchical clustering, only genes differentially expressed with a P value of <0.005 and called P were included. Analysis was performed individually for B cells, macrophages, or OC. When data from all three kinds of samples were grouped in a single project, the sample was considered to be PBMC.
Relative quantification of cytokine mRNA expression levels.
Rhesus macaque cytokines and other gene mRNA levels were determined by RT-PCR as described previously (1). For this experiment, RNA isolated from PBMC, B cells, and macrophages was used. Briefly, samples were tested in duplicate, and the PCR for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene and the target gene from each sample were run in parallel on the same plate. The reaction was carried out on a 96-well optical plate (Applied Biosystems, Foster City, CA) in a 25-μl reaction volume containing 5 μl cDNA plus 20 μl Mastermix (Applied Biosystems). All sequences were amplified using the 7900 default amplification program. PCR conditions and cytokine and other protein mRNA expression levels were calculated from normalized ΔCT values according to methods reported previously (1, 2). In the present study, a minimum of two normal samples for each kind of studied cell (B cells, macrophages, and PBMC) were analyzed to determine baseline mRNA levels for each cytokine and protein. The mean value for each cytokine mRNA level of all animals in each tissue (mean ΔCT) and the statistical analysis were performed as previously described (2).
DENV RT-PCR.
DENV RNA was isolated from aliquots of serum using the QIAamp Viral RNA Mini kit (QIAGEN, Valencia, CA) to a final volume of 50 μl. One hundred nanograms of RNA was used to amplify a 170-bp product in the capsid gene region of DENV by RT-PCR according to a methodology described previously (67). PCR products were separated by 1.5% agarose gel electrophoresis and visualized in a GelDoc station using Quantity One software (Bio-Rad, Hercules, CA).
Rhesus macaque cytokine detection.
The detection of 20 nonhuman primate chemokines and cytokines was performed in a single sample using the Luminex100 system as previously described (25). Serum was collected at days 1, 3, and 5 after infection. The panel included granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, IFN-γ, IFN-α, IL-1β, IL-1Ra, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12 (p40), IL-17, IL-18, monocyte chemoattractant protein 1, macrophage inflammatory protein la, macrophage inflammatory protein 1β, RANTES, and TNF-α. The raw data (mean fluorescence intensity) from all the bead combinations tested were analyzed with Master-Plex QT quantification software (MiraiBio Inc., Alameda, CA) in order to obtain concentration values.
RESULTS
Animal infections and serological tests.
To better understand immune responses in rhesus macaques, four animals were infected with a low-passage strain of DENV-1, and two animals were mock infected with supernatant from uninfected cells. All infected animals developed specific anti-DENV-1 neutralizing antibodies as measured by FNT. In Fig. 1, the titers of neutralizing antibodies at 5 and 20 days and then at 5 months after the infection are shown. Twenty days after infection, all infected animals had neutralizing antibody titers (FNT50) above 1:80. At 5 months after infection, titers were higher in three out of four animals. One animal (animal 27S) showed a decrease in the antibody titer. Viral RNA was detected in serum by PCR 5 days after infection in all except one animal (Fig. 1). This animal, animal 92R, showed the highest levels of transcriptional induction of the genes involved in the innate immune response (data not shown). In particular, the aspartate aminotransferase level was increased in all animals, and in one of them (animal AI73), the alanine aminotransferase level was also increased (data not shown), possibly indicating different levels of hepatic injury. No other significant signs of illness were detected after DENV infection in these animals.
FIG. 1.
Titers of neutralizing antibodies and serum DENV PCR. Specific antibodies to DENV-1 were measured at different points using a flow cytometry-based neutralization assay. Titers represent the FNT50 values. Twenty days after infection, all infected animals developed titers over 1:80. Five months later, all infected animals except one had neutralizing antibodies over 1:320. PCR for DENV was performed 5 days after the infection; the presence of the virus in serum was evidenced by this method in all infected animals except animal 92R. This animal showed a higher level of induction of antiviral genes (see the text for details). None of the mock-infected animals developed neutralizing antibodies or showed specific DENV PCR bands in serum.
Cell separation.
The magnetic microbead system was used to separate the cells. Cells were labeled with antibodies with known cross-reactivity to rhesus macaques according to the manufacturer's instructions. After separation, B cells (CD3−/CD20+) from infected animals were 89 to 95.4% pure (average, 92.75%), while the purity of the same cells isolated from control animals was 86% to 88.5% (average, 87.2%). Macrophage (CD3+/CD14+) purity was 90.6% to 95.6% (average, 93.1%) and 76% to 90.1 (average, 83.5%) when isolated from infected and uninfected animals, respectively.
Differential gene expression is cell specific.
For microarray studies, we used RNA collected from different cells 5 days after infection. RNA from B cells, macrophages, and OC was run using rhesus macaque chips. Data obtained from all three kinds of samples were analyzed together and considered to be PBMC. Figure 2 shows the scattered plots obtained with samples from different kinds of cells. The overall pattern was similar for PBMC and macrophages, with more genes down-regulated than up-regulated (72.29% versus 27.7% and 74.49% versus 25.50%, respectively), while B cells showed three times more up-regulated than down-regulated genes (63.06% versus 36.93%). These results showed a cell type-specific effect in vivo, which can be coherent with the specific role for each type of cell. However, a common aspect in all cell types was the intensities of the differential modifications, with a stronger up-regulation and a limited down-regulation of the affected genes. Common ontologic pathways such as immune responses, inflammatory responses, antiviral responses, and defense were activated in all samples (data not shown) (a complete list of ontologic pathways can be found at http://ucm.rcm.upr.edu/geneexpression.html).
FIG. 2.
Scatter plot of signal intensity comparisons of baselines and infected samples. The log of the mean intensity for the groups is plotted. Genes with fourfold up- or down-regulation and P values of <0.05 are represented. A cell type-specific response is noticeable by the number of modified genes (771, 3,721, and 3,341 for PBMC, macrophages, and B cells, respectively) and by its distribution according to the profile of modification (numbers and percentages are indicated). PBMC and macrophages showed a similar pattern of modification, with a higher percentage of genes being down-regulated. In contrast, in B cells, the higher number of modified genes was up-regulated. Only macrophages showed down-regulated genes with signals above 1 log. Genes with positive or negative changes are represented by red and green dots, respectively.
Analysis of gene expression in DENV-1-infected rhesus macaques.
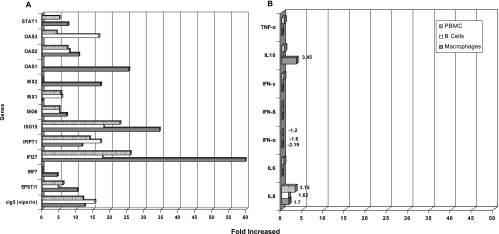
Gene expression profiles for rhesus macaques after experimental infection have not been studied previously. Following the procedure described above, a total of 812, 3,341, and 3,671 genes were differentially regulated in PBMC, B cells, and macrophages, respectively (Fig. 2). The complete gene lists for each type of cell can be reviewed at http://ucm.rcm.upr.edu/geneexpression.html. To identify specific genes differentially regulated with higher significance, conditions were set to P values of <0.005 and with signal calls detected as present (P). With these conditions, the number of up-regulated genes was set down to 20, 30, and 40 in PBMC, B cells, and macrophages, respectively. No down-regulated genes met these stringent conditions. A summary of all these genes is displayed in Table 1. The newly identified genes can be set apart into three categories: IFN-α/β-stimulated genes (IFN-induced proteins, IFN-stimulated gene 15 [ISG15], IFN-α-inducible protein 6-16 [G1P3 or ISG6-16], IFN-α-inducible transmembrane protein 27 [IFI27], IFN regulatory element 7 [IRF7], and phospholipid scramblase 1 [PLSCR1]), IFN-α/β-induced and virally induced genes (2′,5′-oligoadenylate synthetase 1 [OAS1], OAS2, OAS3, Mx1, Mx2, and PRKR), and other genes with known or unknown antiviral function (cig5 or viperin, CKCL10, and epithelial-stromal interactor 1 [EPSTI1]). The increase (n-fold) of selected genes of each group is shown in Fig. 3A. Some of these genes (ISG15, ISG6, IFI27, EPSTI1, IFIPT-1, OAS, and viperin) were up-regulated in all three tested samples. ISG15 is an IFN-α/β-induced protein implicated in a process known as ISGylation (5, 17, 55). Its role in the antiviral response was first suggested because human influenza B virus was able to inhibit its conjugation to target proteins (112). Its direct antiviral activity against Sindbis virus, a member of the Alphaviridae family, has recently been demonstrated (66). Its role as an antiflavivirus has been documented in vitro for West Nile virus (23) and for hepatitis C virus (HCV) both in vitro and in vivo using chimpanzees as the animal model (10, 49). However, there was no previous evidence of ISG15 up-regulation in response to DENV in vitro or in vivo. The role of this protein in the anti-DENV immune response is reinforced in this work by the co-up-regulation of two members of the HERC protein ligase family (RLD5 and RLD6) linked to the ISGylation process (see references 24 and 43 and references therein) (Table 1). The up-regulation of ISG6-16 in response to DENV infection is documented for the first time in this work. This member of the ISG group has been shown to have an antiapoptotic function through inhibiting caspase-3 in cancer cell lines (106). Its role as anti-HCV (9, 113) and its induction after West Nile virus infection (23) have also been recently demonstrated. The induction of high levels of IFI27 (ISG12) and its antiviral role in response to HCV both in vivo and in vitro (9, 49) as well as its protective role in the development of lethal alphavirus encephalitis in mice have been well recognized (62). It has been quite interesting to identify the transcriptional activation of the EPSTI1 gene (Table 1), which has not been linked so far to the IFN signaling pathway. This gene was first identified as being highly upregulated in invasive breast carcinomas compared with normal breast cells (25, 78). More recently, it has been related to the pathogenesis of systemic lupus erythematosus (48). In concordance with this work, EPSTI1 and IFI27 were up-regulated in patients with systemic lupus erythematosus. To our knowledge, there were no previous reports of EPSTI1 activation after viral infection. Its induction after DENV infection in rhesus macaques will be further investigated.
TABLE 1.
Genes up regulated after DENV-1 infectiona
| Gene | Description | Ratio (fold)
|
Function | ||
|---|---|---|---|---|---|
| PBMC (20 genes) | MO (40 genes) | BC (30 genes) | |||
| 1 | CBP/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain | NC | 7.83 | NC | Protein binding, transcription factor activity |
| 2 | 10-kDa IFN-γ-inducible protein (CXCL10_1179) | NC | NC | 5.69 | Pleiotropic effects, stimulation of monocytes |
| 3 | 5′ nucleotidase, cytosolic III | NC | 4.12 | NC | Nucleotide binding, hydrolase activity |
| 4 | CCL3 (chemokine ligand 3) | NC | 11.93 | NC | Immune response |
| 5 | Chromosome 17 open reading frame 27 | NC | 4.44 | 4 | Protein binding |
| 6 | C-type lectin domain family 1, member B | NC | 5.53 | NC | Defense response, transmembrane receptor activity, response to virus |
| 7 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | 5.97 | NC | NC | Innate immune response, protein binding |
| 8 | DNA polymerase-transactivated protein 6 | NC | 8.11 | NC | DNA replication |
| 9 | DTFT5783 (hypothetical protein LOC388352) | NC | 4.37 | NC | ? |
| 10 | Endothelial cell growth factor 1 (platelet derived) | NC | 6.39 | NC | Growth factor activity, thymidine phosphorylase activity |
| 11 | EPSTI1 | 6.13 | 10.29 | 4.64 | ? |
| 12 | FBJ murine osteosarcoma viral oncogene homolog B | NC | 7.89 | NC | Sequence-specific DNA binding, transcription factor activity |
| 13 | Full-length cDNA clone CS0DK002YF13 of HeLa cells, Cot25 normalized | NC | NC | 5.53 | ? |
| 14 | G1P3, IFN-α-induced protein (ISG6-16) | 5.01 | 7.17 | 5.01 | Exonuclease activity |
| 15 | gb:AY044446.1 DB_XREF = gi:22074157 = Mmu.11363.1 (similar to glutathione) | NC | NC | 4.22 | ? |
| 16 | GEN = PRIC285 peroxisome proliferator-activated receptor alpha-interacting complex protein 28S | 4.29 | NC | NC | ? |
| 17 | gi:60301523 DEF = USP18_9254 rhesus (ubiquitin-specific protease) | NC | NC | 5.08 | Ubiquitin-specific protease |
| 18 | Hect domain and RLD5 (HERC family) | 11.52 | NC | 9.63 | Ligase activity, ubiquitin-protein ligase activity |
| 19 | Hect domain and RLD6 (HERC family) | 5.7 | NC | NC | Ubiquitin-protein ligase activity |
| 20 | HS-5-8-S rRNA ribosomal DNA complete repeating unit | NC | 5.23 | NC | ? |
| 21 | Hypothetical protein FLJ20035 (no assigned function) | 5.22 | NC | 5.08 | Nucleic acid binding |
| 22 | Hypothetical protein LOC129607 (thymidylate kinase) | 15.6 | NC | 22.17 | Kinase activity |
| 23 | IBRDC3 (zinc ion binding metal ion binding protein binding) | NC | 4.68 | NC | ? |
| 24 | Immunoglobulin heavy-chain variable region | NC | 11.74 | NC | Nucleotide binding, transferase activity, protein serine/threonine kinase |
| 25 | Immunoglobulin kappa constant | NC | 10.1 | NC | Antigen binding |
| 26 | IFI27 | 25.95 | 59.78 | 17.61 | Immune response |
| 27 | IFN-induced protein 44 | NC | NC | 12.58 | Response to virus |
| 28 | INF-induced transmembrane protein 1 | 13.96 | 11.73 | 17.15 | Receptor signaling protein |
| 29 | IRF7 | NC | 4.37 | NC | Transcription factor |
| 30 | IFN-induced protein with tetratricopeptide repeat 1 (IFIT1_6442) | NC | NC | 17.15 | Immune response |
| 31 | IFN-induced protein with tetratricopeptide repeat 2 | 10.52 | NC | 6.69 | Immune response |
| 32 | IFN-induced protein with tetratricopeptide repeat 3 | 6.35 | NC | 10.27 | Immune response |
| 33 | IFN-induced protein with tetratricopeptide repeat 4 | 6.17 | NC | 6.51 | Immune response |
| 34 | IFN-induced protein with tetratricopeptide repeat 5 (IFIT5_6444) | NC | NC | 5.6 | Immune response |
| 35 | ISG15 ubiquitin-like modifier | 22.85 | 34.5 | 18 | Immune response |
| 36 | Kruppel-like factor 6 | NC | 4.51 | NC | Nucleic acid binding |
| 37 | Lectin, galactoside-binding soluble 3 binding protein | NC | 6.8 | NC | Cell adhesion, activation of immune cells |
| 38 | Likely ortholog of mouse d11lgp2 | 4.64 | NC | 6.95 | Nucleotide binding, hydrolase activity |
| 39 | Lymphocyte antigen 6 complex, locus E | NC | 5.24 | NC | Defense response, glycosylphosphatidylinositol anchor binding |
| 40 | Mx1 | 5.44 | NC | 5.7 | Immune response, response to virus, GTP-binding protein |
| 41 | Mx2 | NC | 4.9 | 5.98 | Immune response, response to virus, GTP-binding protein |
| 42 | Mx2 human-like protein | NC | 4.92 | NC | Immune response, response to virus, nucleotide binding, transferase activity |
| 43 | OAS1 | NC | 25.48 | NC | Nucleotidyltransferase |
| 44 | OAS2 | 7.29 | 10.76 | 7.92 | Immune response |
| 45 | OAS2 (69/71 kDa) | NC | NC | 6.83 | Immune response, nucleotidyltransferase, ATP/RNA binding |
| 46 | OAS3 | 4.2 | 16.62 | NC | Immune response |
| 47 | Platelet factor 4 (CXCL4 variant 1) | NC | 10.51 | NC | Immune response, chemokine activity |
| 48 | Platelet factor 4 (CXCL4) | NC | 5.57 | NC | Leukocyte chemotaxis, immune response, chemokine activity |
| 49 | Platelet-derived endothelial cell growth factor/thymidine phosphorylase | NC | 4.61 | NC | ? |
| 50 | PRKR | NC | NC | 4.16 | Immune response, response to virus, negative regulation of cell proliferation |
| 51 | PTX3_4 pentaxin-related gene | NC | 5.73 | NC | Inflammatory response |
| 52 | Schlafen family member 5 | NC | NC | 8.74 | DNA/RNA helicase |
| 53 | Scramblase 1 | NC | 4.11 | NC | Response to virus, protein binding |
| 54 | Serum/glucocorticoid-regulated kinase | NC | 5.95 | 4.5 | Nucleotide binding, protein/serine/threonine kinase activity |
| 55 | STAT, 1.91 kDa (STAT1) | NC | NC | 5.01 | Transcription factor |
| 56 | Similar to MGC52679 | NC | 4.61 | NC | Phosphorylase activity |
| 57 | Sterile alpha motif domain-containing 9 | NC | 6.14 | NC | Binding |
| 58 | Syndecan 3 (N-syndecan) | NC | 4.11 | NC | Cytoskeletal protein binding |
| 59 | Thymidylate kinase activity | 15.6 | NC | NC | Kinase/transferase activity |
| 60 | Tribbles homolog 2 (Drosophila melanogaster) | NC | 4.84 | NC | Nucleotide binding, protein kinase activity, protein kinase inhibitor activity |
| 61 | Tripartite motif-containing 14 (TRIM14) | NC | NC | 4.24 | Protein binding, metal/zinc ion binding |
| 62 | Tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10) | NC | 5 | NC | Immune response, apoptosis, TNF receptor binding |
| 63 | Unc-93 homolog B1 (Caenorhabditis elegans) | NC | 5.27 | NC | ? |
| 64 | Radical S-adenosyl methionine domain-containing 2 (cig5, viperin) | 11.96 | 12.49 | 15.47 | Catalytic activity, iron ion binding/antiviral |
Genes that were increased by 4.0-fold or greater were included. Conditions for this set of genes were a P value of <0.005 and call P. Twenty, 40, and 30 genes were detected under these conditions in PBMC, macrophages (MO), and B cells (BC), respectively. Gene names are given at the left and are organized alphabetically. The function attributable to each gene is given on the right. Information on how to access the full list of all genes, specific P values, and ontologic pathways is provided in the text. NC, no changes.
FIG. 3.
Transcriptional increase (n-fold) detected by microarray. Selected genes were plotted for comparative purposes among samples and among genes. (A) Increment of the ISGs. The IFI27 and ISG15 genes were the most highly up-regulated, followed by the OAS genes Mx, viperin, and EPSTI1. (B) Transcriptional modifications of the cytokine genes. Only a limited increase in the transcription levels of IL-10 and IL-8 was detected. IFN-α was detected as being down-regulated in all kinds of samples. However, none of these changes were statistically significant, reflecting no changes in the transcriptional activity of these genes. The transcriptional increase (n-fold) was plotted on the abscissa. The names of the genes are shown on the ordinate.
In contrast to what has been reported for human cells (12, 109), we were unable to detect a significant modification in the level of cytokine gene transcription after a primary DENV infection. As shown in Fig. 3B, only a limited level of transcriptional activation was detected for IL-10 and IL-8. These data were obtained 5 days after infection. However, serum detection, using antibodies with known cross-reactivity to rhesus macaque cytokines, did not detect the presence of such cytokines at day 1, 3, or 5 after infection (data not shown).
RT-PCR confirmation of microarray results.
From the microarray data, we attempted to confirm three central genes associated with IFN induction by RT-PCR: IRF7, OAS, and Mx. All of them were shown to be up-regulated by this method, confirming their increased induction found in the microarrays (Fig. 4). IRF7 was selected for confirmation because of its crucial role in the induction of the type I IFN response after viral infection (44-46). In addition to macrophages, this gene was found to be up-regulated in B cells. The transcriptional increase (n-fold) was higher by RT-PCR (compare Fig. 3A to Fig. 4). These variations could be attributable to differences in sample processing.
FIG. 4.
Real-time confirmation of ISG gene but not of cytokine gene up-regulation. Three ISG genes (IRF7, OAS, and Mx) and three cytokine genes (IFN-γ, TNF-α, and IFN-β) were selected to be amplified by RT-PCR to confirm the microarray data. Figures represent increases (n-fold) in up-regulated genes compared to baseline samples. All three ISGs were up regulated except IRF7 in PBMCs. No increase in the transcription level of the studied cytokine genes was detected.
Both OAS and Mx proteins were amplified with generic primers that were unable to discriminate among different subtypes of these proteins. However, in both cases, an increase in gene transcription was also detected. As expected from the microarray data, both genes were activated in all three kinds of cells compared to baseline. OAS showed a peak of increase that duplicated the value detected by microarray. In this case, the detection of a whole pool of genes could account for these marked differences. As shown in Table 1, different members of this family were up-regulated in each type of cell (OAS1 in macrophages, OAS3 in macrophages and PBMC, and OAS2 in macrophages, PBMC, and B cells), suggesting a possible cell type-specific mechanism of activation of this pathway. The severalfold increases in Mx proteins detected by RT-PCR were comparable to those detected by microarray, possibly due to the existence of fewer variants of the proteins of this family. In summary, the three selected genes (Mx, OAS, and IRF) were found to be up-regulated upon DENV-1 infection, confirming their increased induction originally detected by the microarray method.
The induction of IRF7 was also confirmed by RT-PCR. In contrast to IRF3, the other key player in the type I IFN pathway, IRF7 is not constitutively expressed in most cell types, and it has a short half-life (69, 79, 93, 110). After activation by phosphorylation, IRF7 is translocated to the nucleus, where it induces the transcription of IFN-α/β genes (69, 93, 94). These aspects are very important in our work, because it was detected 5 days after infection with DENV, supporting the presence of a very recent stimulus. In addition, by the use of RT-PCR, we were able to confirm the lack of induction of cytokine genes like IFN-β, IFN-γ, and TNF-α 5 days after in vivo infection with DENV-1 (Fig. 4). This result was confirmed by cytokine detection in serum at days 1, 3, and 5 after infection (data not shown).
DISCUSSION
Presently, there is not an adequate animal model to study DENV pathogenesis or to test immune responses to DENV candidate vaccines. Interesting approaches using mice have been done in recent years (8, 99). It is well known that macaques respond to DENV infection with quite limited signs and symptoms compared to humans; however, they are the ultimate model of choice to test DENV vaccine candidates and their elicited immune responses. Nevertheless, the molecular basis of the immune response to DENV in macaques is remarkably unknown, and the continued use of this model requires a better understanding of underlying differences between human and macaque immune systems. We infected four rhesus macaques with a low-passage strain of DENV-1, and 5 days thereafter, by using microarrays and RT-PCR, we observed the gene transcriptional pattern. Also, the profile of serum cytokines was studied by multiplex cytokine detection after 1, 3, and 5 days of infection.
We found a robust innate immune response characterized by the induction of several genes implicated in the immune response to viral infections. Most of those genes were ISGs and virus-induced genes. A third group included some genes with no relation to the IFN signaling pathway. By RT-PCR, we confirmed that at least three of those genes (IRF7, OAS, and Mx) were indeed up-regulated.
2′,5′-Oligoadenlyate is a system that is well documented as an endogenous player in the antiviral pathway (16, 89, 92). This protein is activated by dsRNA to produce 5′-phosphorylated, 2′,5′-linked oligoadenylates, whose function is to activate RNase L with subsequent RNA degradation. The implication of this system to control flavivirus infections has been demonstrated before in vitro and in vivo for West Nile virus (54, 70, 95) and HCV (56, 68, 75) and in vitro for DENV (109). Our finding in vivo of increased levels of OAS transcripts confirms the role of this system in controlling DENV replication in rhesus macaques.
The Mx protein family is well known for its role in antiviral responses in vivo and in vitro (31, 65, 80, 81), and the proteins are preferentially induced by IFN-α/β (20). Evidence from animal studies established that Mx alone is sufficient to block the replication of virus in the absence of any other IFN-α/β-inducible proteins (3, 30). The induction of Mx1, Mx2, and MxA genes was produced after in vitro infection of HUVEC cells with DENV-2 (110); however, in our study, only Mx1 and Mx2 were detected as being up-regulated by microarrays. These results together confirm the specific anti-DENV activity of Mx proteins.
In contrast with other key players in the type I IFN pathway such as IRF3, IRF7 is not constitutively expressed in most cell types, and it has a short half-life (69, 79, 93, 110). After activation by phosphorylation, IRF7 is translocated to the nucleus, where it induces the transcription of IFN-α/β genes (69, 93, 94). The detection of high levels of IRF7 5 days after the infection with DENV-1 in rhesus monkeys could then be interpreted as evidence of the presence of a very recent stimulus. This is particularly remarkable in the lack of confirmed cytokine induction, including IFN-β, IFN-γ, and TNF-α, 5 days after DENV-1 infection (Fig. 4). These results were confirmed by cytokine detection in serum at days 1, 3, and 5 after infection (data not shown).
One of the key and unexpected outcomes of this study was the apparent lack of induction of three key cytokines in human innate immunity: IFN-β, IFN-γ, and TNF-α (Fig. 3 and 4). In addition, only limited but not significant up-regulation of IL-10 and IL-8 was detected by microarray, (Fig. 3). This is highly contrasting with what has been reported for humans. The high-level presence of IL-10, IL-8, IL-6, TNF-α, and IFN-γ is a hallmark of the development of DHF/DSS and has been associated with an increased risk of developing severe forms of disease in primary infections, which may be accentuated during secondary infections (7, 13, 18, 28, 40-42, 50, 52, 53, 64, 82, 86). Several groups documented an increased production of these proinflammatory cytokines after a primary infection (6, 7, 13, 50, 64, 77, 83, 108), even in cases without DHF/DSS manifestations (6, 64, 77, 83, 108).
Because our study was performed 5 days after infection, we cannot rule out later cytokine gene induction and cytokine production. One of the advantages of the macaque model is that we have the opportunity to analyze the very early virus-host interaction, which is often not possible for humans. DENV is able to induce the activation of PBMC in mice as early as 3 days after infection (97) and in humans with less than 72 h of fever/viremia (27). Also, the production of cytokines has been detected in children presenting less than 72 h of fever (26-28, 60). Considering that disease outcome is likely to be decided by those early events, it is remarkable that we were unable to find the inflammatory response that is so typical for disease in humans. Nevertheless, in the future, we plan to conduct similar studies at later time points following infection.
Our new findings may explain why rhesus macaques do not develop DF or DHF/DSS after a primary DENV infection. Indirectly, these results confirm the proposed role of TNF-α and IFN-γ in the development of DHF/DSS. Thus, it might be argued that a therapy aimed at the suppression of inflammation/activation may reduce the incidence of DHF/DSS in humans. It can be affirmed that a different profile could be expected after a secondary infection of these animals. However, our data show that the cytokine profile after secondary infection with DENV-2 is very similar to the one described after primary infection (C. A. Sariol et al., unpublished results).
The up-regulation of OAS and Mx proteins shown by microarrays and RT-PCR was expected, as they constitute a standard response after infection with several viruses (3, 30, 47, 54, 92, 103, 109, 110) and are a confirmation of the role of the innate immune response generated after DENV infection in macaques. The increased detection by both methods reinforces the importance of this type of immune response in controlling DENV replication in rhesus macaques. Additionally, several ISGs such as ISG15 and ISG16-56, which are acting during the innate immune response, were found to be highly up-regulated by microarrays.
Induction of ISGs without increased levels of type I or type II IFN has been reported for HCV-infected chimpanzees (10). Similar to data from that report, we found an apparent contradiction in that the induction of ISGs may not necessarily be accompanied by a higher induction of IFN. Additionally, increases in serum levels of IFN-α or IFN-γ were not detected by the multiplex cytokine array even as early as 24 h after the infection (data not shown). However, the following explanations for the apparent discrepancy may account for this fact. An early transcriptional activation of these genes before the day of the sampling for microarray and RT-PCR study (5 days after infection) with a restricted level of expression below the limit of detection of our assay but still able to stimulate the ISG transcription detected 5 days later may have occurred. Second, only the first wave of the IFN response, characterized by the production of IFN-β (69, 93, 94), was allowed to occur (animal or virus restriction). Because of the lack of a reagent with reactivity for this rhesus macaque cytokine (IFN-β), we were unable to test its levels in serum, and we were missing a crucial and early-phase activation of the innate immune response. For the same reasons, we may be missing the possible expression of IFN-λ, which has been shown to have an active role in the activation of the immune response (57, 96). Third, anti-IFN activity of DENV protein NS4B through the inhibition of the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway has been documented in vitro using monkey kidney cells (72, 73). This mechanism may lead to the partial impairment of the IFN pathway. Fourth, novel virus-host interactions may limit the transcriptional activation and expression of IFN-α and IFN-γ and other cytokines, such as TNF-α, IL-6, IL-8, and IL-10, linked to the immunopathogenesis of DENV.
The detection of IRF7 up-regulation by microarray and confirmation by RT-PCR 5 days after infection also support the existence of alternative mechanisms in this animal model to regulate type I IFNs and activate ISGs. The role of IRF7 as a master regulator in the induction of type I IFN is well known (44-46). It was long believed that the activation of IRF7 was dependent exclusively on basal levels of IFN-α/β or in response to an early wave of IFNs following virus infection, which in turn amplified IFN-α/β production through the increase in the intracellular level of IRF7 (69, 76, 93, 94, 107). However, it has recently been shown that the regulation of IRF7 transcription includes a virus-dependent but IFN-independent signaling pathway (79). In such alternative mechanisms, the viral infection induces virus-activated factor complex formation (including IRF7/IRF3), which directly binds to specific regions of the IRF7 promoter. One essential component of this pathway is the recruitment of cyclic AMP response element binding protein (CREB)-binding protein (CBP)/p300 for IRF3 activation (79). However, it has also been shown that this activator has an opposing activity by negatively modulating the IRF7 DNA binding (14). Supporting this mechanism in our model, as an alternative induction of IRF7 without detectable transcriptional activation or serum levels of type I IFN, is the transcriptional up-regulation of the CBP/300 gene detected by microarray (Table 1). It could act in synergy with the anti-IFN activity of the nonstructural proteins of DENV, particularly NS4B through the JAK/STAT pathway (39, 51, 72, 73). Although we have no data to support the occurrence of this mechanism in vivo in our model, it is an alternative to be addressed in further studies.
Other genes such as PLSCR1 or EPSTI1, here detected as being up-regulated by microarray, are also interesting targets for further studies. PLSCR1 is a membrane protein implicated in the synthesis and translocation of phospholipids to the cell surface in response to cell activation, injury, or apoptotic stimulus (111), and it is able to strengthen the antiviral activity of IFNs (19). This gene was previously found to be activated after DENV-2 infection in HUVEC cells (109). Our in vivo results support the anti-DENV activity of this protein. Confirmation of the up-regulation of EPSTI1 by other methods will be performed.
The immune response to DENV described in this work is quite different from the immune response characterized in mice (4, 97-100) and in humans in vivo or in vitro using human cells (12, 13, 15, 18, 41, 83, 99, 104, 109). This strongly suggests interspecies differences in the complex molecular mechanisms of the IFN response. This reinforces the need to search for a proper animal model to study the pathogenesis and immune response to vaccines against DENV.
This balance of innate immune response without cytokine induction can be an example of a long-term host-parasite evolutionary process. Similar situations have been demonstrated for sooty mangabeys infected with simian immunodeficiency virus. These animals are naturally infected without developing symptoms. The key feature of that process is a controlled activation of the immune response (74, 101).
Results documented in this work prompted us to consider other interventions in addition to vaccine approaches, like the modulation of the innate immunity, to control the development of severe forms of this disease.
Acknowledgments
We thank the entire Staff of the Animal Resources Center and SSFS for taking excellent care of the monkeys and for the use of the facilities. We also thank Roberto Medina for his very useful help as a laboratory assistant.
This work was supported by a Northeast Biodefense Center Developmental grant (NBC-Lipkin AI57158) to C.A.S. and partially by NIH grants U42 RR16021 and U24 RR18108.
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Abel, K., M. J. Alegria-Hartman, K. Rothaeusler, M. Marthas, and C. J. Miller. 2002. The relationship between simian immunodeficiency virus RNA levels and the mRNA levels of alpha/beta interferons (IFN-α/β) and IFN-α/β-inducible Mx in lymphoid tissues of rhesus macaques during acute and chronic infection. J. Virol. 76:8433-8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abel, K., B. Pahar, K. K. Van Rompay, L. Fritts, C. Sin, K. Schmidt, R. Colon, M. McChesney, and M. L. Marthas. 2006. Rapid virus dissemination in infant macaques after oral simian immunodeficiency virus exposure in the presence of local innate immune responses. J. Virol. 80:6357-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnheiter, H., M. Frese, R. Kambadur, E. Meier, and O. Haller. 1996. Mx transgenic mice—animal models of health. Curr. Top. Microbiol. Immunol. 206:119-147. [DOI] [PubMed] [Google Scholar]
- 4.Atrasheuskaya, A., P. Petzelbauer, T. M. Fredeking, and G. Ignatyev. 2003. Anti-TNF antibody treatment reduces mortality in experimental dengue virus infection. FEMS Immunol. Med. Microbiol. 35:33-42. [DOI] [PubMed] [Google Scholar]
- 5.Au, W. C., P. A. Moore, W. Lowther, Y. T. Juang, and P. M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. USA 92:11657-11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avila-Aguero, M. L., C. R. Avila-Aguero, S. L. Um, A. Soriano-Fallas, A. Canas-Coto, and S. B. Yan. 2004. Systemic host inflammatory and coagulation response in the dengue virus primo-infection. Cytokine 27:173-179. [DOI] [PubMed] [Google Scholar]
- 7.Azeredo, E. L., S. M. Zagne, M. A. Santiago, A. S. Gouvea, A. A. Santana, P. C. Neves-Souza, R. M. Nogueira, M. P. Miagostovich, and C. F. Kubelka. 2001. Characterisation of lymphocyte response and cytokine patterns in patients with dengue fever. Immunobiology 204:494-507. [DOI] [PubMed] [Google Scholar]
- 8.Bente, D. A., M. W. Melkus, J. V. Garcia, and R. Rico-Hesse. 2005. Dengue fever in humanized NOD/SCID mice. J. Virol. 79:13797-13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieche, I., T. Asselah, I. Laurendeau, D. Vidaud, C. Degot, V. Paradis, P. Bedossa, D. C. Valla, P. Marcellin, and M. Vidaud. 2005. Molecular profiling of early stage liver fibrosis in patients with chronic hepatitis C virus infection. Virology 332:130-144. [DOI] [PubMed] [Google Scholar]
- 10.Bigger, C. B., B. Guerra, K. M. Brasky, G. Hubbard, M. R. Beard, B. A. Luxon, S. M. Lemon, and R. E. Lanford. 2004. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J. Virol. 78:13779-13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaney, J. E., Jr., J. M. Matro, B. R. Murphy, and S. S. Whitehead. 2005. Recombinant, live-attenuated tetravalent dengue virus vaccine formulations induce a balanced, broad, and protective neutralizing antibody response against each of the four serotypes in rhesus monkeys. J. Virol. 79:5516-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosch, I., K. Xhaja, L. Estevez, G. Raines, H. Melichar, R. V. Warke, M. V. Fournier, F. A. Ennis, and A. L. Rothman. 2002. Increased production of interleukin-8 in primary human monocytes and in human epithelial and endothelial cell lines after dengue virus challenge. J. Virol. 76:5588-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braga, E. L., P. Moura, L. M. Pinto, S. R. Ignacio, M. J. Oliveira, M. T. Cordeiro, and C. F. Kubelka. 2001. Detection of circulant tumor necrosis factor-alpha, soluble tumor necrosis factor p75 and interferon-gamma in Brazilian patients with dengue fever and dengue hemorrhagic fever. Mem. Inst. Oswaldo Cruz 96:229-232. [DOI] [PubMed] [Google Scholar]
- 14.Caillaud, A., A. Prakash, E. Smith, A. Masumi, A. G. Hovanessian, D. E. Levy, and I. Marie. 2002. Acetylation of interferon regulatory factor-7 by p300/CREB-binding protein (CBP)-associated factor (PCAF) impairs its DNA binding. J. Biol. Chem. 277:49417-49421. [DOI] [PubMed] [Google Scholar]
- 15.Cardier, J. E., E. Marino, E. Romano, P. Taylor, F. Liprandi, N. Bosch, and A. L. Rothman. 2005. Proinflammatory factors present in sera from patients with acute dengue infection induce activation and apoptosis of human microvascular endothelial cells: possible role of TNF-alpha in endothelial cell damage in dengue. Cytokine 30:359-365. [DOI] [PubMed] [Google Scholar]
- 16.Castelli, J., K. A. Wood, and R. J. Youle. 1998. The 2-5A system in viral infection and apoptosis. Biomed. Pharmacother. 52:386-390. [DOI] [PubMed] [Google Scholar]
- 17.D'Cunha, J., E. Knight, Jr., A. L. Haas, R. L. Truitt, and E. C. Borden. 1996. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. USA 93:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dewi, B. E., T. Takasaki, and I. Kurane. 2004. In vitro assessment of human endothelial cell permeability: effects of inflammatory cytokines and dengue virus infection. J. Virol. Methods 121:171-180. [DOI] [PubMed] [Google Scholar]
- 19.Dong, B., Q. Zhou, J. Zhao, A. Zhou, R. N. Harty, S. Bose, A. Banerjee, R. Slee, J. Guenther, B. R. Williams, T. Wiedmer, P. J. Sims, and R. H. Silverman. 2004. Phospholipid scramblase 1 potentiates the antiviral activity of interferon. J. Virol. 78:8983-8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupuis, S., E. Jouanguy, S. Al-Hajjar, C. Fieschi, I. Z. Al-Mohsen, S. Al-Jumaah, K. Yang, A. Chapgier, C. Eidenschenk, P. Eid, A. Al Ghonaium, H. Tufenkeji, H. Frayha, S. Al-Gazlan, H. Al-Rayes, R. D. Schreiber, I. Gresser, and J. L. Casanova. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388-391. [DOI] [PubMed] [Google Scholar]
- 21.Eckels, K. H., D. R. Dubois, R. Putnak, D. W. Vaughn, B. L. Innis, E. A. Henchal, and C. H. Hoke, Jr. 2003. Modification of dengue virus strains by passage in primary dog kidney cells: preparation of candidate vaccines and immunization of monkeys. Am. J. Trop. Med. Hyg. 69:12-16. [DOI] [PubMed] [Google Scholar]
- 22.Fink, J., F. Gu, and S. G. Vasudevan. 2006. Role of T cells, cytokines and antibody in dengue fever and dengue haemorrhagic fever. Rev. Med. Virol. 16:263-275. [DOI] [PubMed] [Google Scholar]
- 23.Fredericksen, B. L., M. Smith, M. G. Katze, P. Y. Shi, and M. Gale, Jr. 2004. The host response to West Nile virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737-7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Gonzalo, F. R., and J. L. Rosa. 2005. The HERC proteins: functional and evolutionary insights. Cell. Mol. Life Sci. 62:1826-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giavedoni, L. D. 2005. Simultaneous detection of multiple cytokines and chemokines from nonhuman primates using Luminex technology. J. Immunol. Methods 301:89-101. [DOI] [PubMed] [Google Scholar]
- 26.Green, S., S. Pichyangkul, D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, A. Nisalak, I. Kurane, A. L. Rothman, and F. A. Ennis. 1999. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J. Infect. Dis. 180:1429-1435. [DOI] [PubMed] [Google Scholar]
- 27.Green, S., D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, S. Suntayakorn, A. Nisalak, R. Lew, B. L. Innis, I. Kurane, A. L. Rothman, and F. A. Ennis. 1999. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J. Infect. Dis. 179:755-762. [DOI] [PubMed] [Google Scholar]
- 28.Green, S., D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, S. Suntayakorn, A. Nisalak, A. L. Rothman, and F. A. Ennis. 1999. Elevated plasma interleukin-10 levels in acute dengue correlate with disease severity. J. Med. Virol. 59:329-334. [PubMed] [Google Scholar]
- 29.Gubler, D. J. 2002. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 10:100-103. [DOI] [PubMed] [Google Scholar]
- 30.Haller, O., M. Frese, and G. Kochs. 1998. Mx proteins: mediators of innate resistance to RNA viruses. Rev. Sci. Tech. 17:220-230. [DOI] [PubMed] [Google Scholar]
- 31.Haller, O., and G. Kochs. 2002. Interferon-induced Mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710-717. [DOI] [PubMed] [Google Scholar]
- 32.Halstead, S. B. 1982. Immune enhancement of viral infection. Prog. Allergy 31:301-364. [PubMed] [Google Scholar]
- 33.Halstead, S. B. 2003. Neutralization and antibody-dependent enhancement of dengue viruses. Adv. Virus Res. 60:421-467. [DOI] [PubMed] [Google Scholar]
- 34.Halstead, S. B. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239:476-481. [DOI] [PubMed] [Google Scholar]
- 35.Halstead, S. B., S. Rojanasuphot, and N. Sangkawibha. 1983. Original antigenic sin in dengue. Am. J. Trop. Med. Hyg. 32:154-156. [DOI] [PubMed] [Google Scholar]
- 36.Halstead, S. B., H. Shotwell, and J. Casals. 1973. Studies on the pathogenesis of dengue infection in monkeys. II. Clinical laboratory responses to heterologous infection. J. Infect. Dis. 128:15-22. [DOI] [PubMed] [Google Scholar]
- 37.Halstead, S. B., H. Shotwell, J. Casals, N. J. Marchette, W. A. Falkler, Jr., A. Stenhouse, and D. Nash. 1973. Studies on the pathogenesis of dengue infection in monkeys. II. Clinical laboratory responses to heterologous infection. J. Infect. Dis. 128:7-14. [DOI] [PubMed] [Google Scholar]
- 38.Hanley, K. A., L. R. Manlucu, G. G. Manipon, C. T. Hanson, S. S. Whitehead, B. R. Murphy, and J. E. Blaney, Jr. 2004. Introduction of mutations into the non-structural genes or 3′ untranslated region of an attenuated dengue virus type 4 vaccine candidate further decreases replication in rhesus monkeys while retaining protective immunity. Vaccine 22:3440-3448. [DOI] [PubMed] [Google Scholar]
- 39.Ho, L. J., L. F. Hung, C. Y. Weng, W. L. Wu, P. Chou, Y. L. Lin, D. M. Chang, T. Y. Tai, and J. H. Lai. 2005. Dengue virus type 2 antagonizes IFN-alpha but not IFN-gamma antiviral effect via down-regulating Tyk2-STAT signaling in the human dendritic cell. J. Immunol. 174:8163-8172. [DOI] [PubMed] [Google Scholar]
- 40.Hober, D., A. S. Delannoy, S. Benyoucef, D. De Groote, and P. Wattre. 1996. High levels of sTNFR p75 and TNF alpha in dengue-infected patients. Microbiol. Immunol. 40:569-573. [DOI] [PubMed] [Google Scholar]
- 41.Hober, D., T. L. Nguyen, L. Shen, D. Q. Ha, V. T. Huong, S. Benyoucef, T. H. Nguyen, T. M. Bui, H. K. Loan, B. L. Le, A. Bouzidi, D. De Groote, M. T. Drouet, V. Deubel, and P. Wattre. 1998. Tumor necrosis factor alpha levels in plasma and whole-blood culture in dengue-infected patients: relationship between virus detection and pre-existing specific antibodies. J. Med. Virol. 54:210-218. [DOI] [PubMed] [Google Scholar]
- 42.Hober, D., L. Poli, B. Roblin, P. Gestas, E. Chungue, G. Granic, P. Imbert, J. L. Pecarere, R. Vergez-Pascal, P. Wattre, et al. 1993. Serum levels of tumor necrosis factor-alpha (TNF-alpha), interleukin-6 (IL-6), and interleukin-1 beta (IL-1 beta) in dengue-infected patients. Am. J. Trop. Med. Hyg. 48:324-331. [DOI] [PubMed] [Google Scholar]
- 43.Hochrainer, K., H. Mayer, U. Baranyi, B. Binder, J. Lipp, and R. Kroismayr. 2005. The human HERC family of ubiquitin ligases: novel members, genomic organization, expression profiling, and evolutionary aspects. Genomics 85:153-164. [DOI] [PubMed] [Google Scholar]
- 44.Honda, K., Y. Ohba, H. Yanai, H. Negishi, T. Mizutani, A. Takaoka, C. Taya, and T. Taniguchi. 2005. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434:1035-1040. [DOI] [PubMed] [Google Scholar]
- 45.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 46.Honda, K., H. Yanai, A. Takaoka, and T. Taniguchi. 2005. Regulation of the type I IFN induction: a current view. Int. Immunol. 17:1367-1378. [DOI] [PubMed] [Google Scholar]
- 47.Horisberger, M. A. 1995. Interferons, Mx genes, and resistance to influenza virus. Am. J. Respir. Crit. Care Med. 152:S67-71. [DOI] [PubMed] [Google Scholar]
- 48.Ishii, T., H. Onda, A. Tanigawa, S. Ohshima, H. Fujiwara, T. Mima, Y. Katada, H. Deguchi, M. Suemura, T. Miyake, K. Miyatake, I. Kawase, H. Zhao, Y. Tomiyama, Y. Saeki, and H. Nojima. 2005. Isolation and expression profiling of genes upregulated in the peripheral blood cells of systemic lupus erythematosus patients. DNA Res. 12:429-439. [DOI] [PubMed] [Google Scholar]
- 49.Itsui, Y., N. Sakamoto, M. Kurosaki, N. Kanazawa, Y. Tanabe, T. Koyama, Y. Takeda, M. Nakagawa, S. Kakinuma, Y. Sekine, S. Maekawa, N. Enomoto, and M. Watanabe. 2006. Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J. Viral Hepat. 13:690-700. [DOI] [PubMed] [Google Scholar]
- 50.Iyngkaran, N., M. Yadav, and M. Sinniah. 1995. Augmented inflammatory cytokines in primary dengue infection progressing to shock. Singapore Med. J. 36:218-221. [PubMed] [Google Scholar]
- 51.Jones, M., A. Davidson, L. Hibbert, P. Gruenwald, J. Schlaak, S. Ball, G. R. Foster, and M. Jacobs. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 79:5414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Juffrie, M., G. M. Meer, C. E. Hack, K. Haasnoot, Sutaryo, A. J. Veerman, and L. G. Thijs. 2001. Inflammatory mediators in dengue virus infection in children: interleukin-6 and its relation to C-reactive protein and secretory phospholipase A2. Am. J. Trop. Med. Hyg. 65:70-75. [DOI] [PubMed] [Google Scholar]
- 53.Juffrie, M., G. M. van Der Meer, C. E. Hack, K. Haasnoot, Sutaryo, A. J. Veerman, and L. G. Thijs. 2000. Inflammatory mediators in dengue virus infection in children: interleukin-8 and its relationship to neutrophil degranulation. Infect. Immun. 68:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kajaste-Rudnitski, A., T. Mashimo, M. P. Frenkiel, J. L. Guenet, M. Lucas, and P. Despres. 2006. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J. Biol. Chem. 281:4624-4637. [DOI] [PubMed] [Google Scholar]
- 55.Kessler, D. S., D. E. Levy, and J. E. Darnell, Jr. 1988. Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. USA 85:8521-8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knapp, S., L. J. Yee, A. J. Frodsham, B. J. Hennig, S. Hellier, L. Zhang, M. Wright, M. Chiaramonte, M. Graves, H. C. Thomas, A. V. Hill, and M. R. Thursz. 2003. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 4:411-419. [DOI] [PubMed] [Google Scholar]
- 57.Kotenko, S. V., G. Gallagher, V. V. Baurin, A. Lewis-Antes, M. Shen, N. K. Shah, J. A. Langer, F. Sheikh, H. Dickensheets, and R. P. Donnelly. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69-77. [DOI] [PubMed] [Google Scholar]
- 58.Kurane, I., D. Hebblewaite, W. E. Brandt, and F. A. Ennis. 1984. Lysis of dengue virus-infected cells by natural cell-mediated cytotoxicity and antibody-dependent cell-mediated cytotoxicity. J. Virol. 52:223-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurane, I., B. L. Innis, C. H. Hoke, Jr., K. H. Eckels, A. Meager, J. Janus, and F. A. Ennis. 1995. T cell activation in vivo by dengue virus infection. J. Clin. Lab. Immunol. 46:35-40. [PubMed] [Google Scholar]
- 60.Kurane, I., B. L. Innis, S. Nimmannitya, A. Nisalak, A. Meager, J. Janus, and F. A. Ennis. 1991. Activation of T lymphocytes in dengue virus infections. High levels of soluble interleukin 2 receptor, soluble CD4, soluble CD8, interleukin 2, and interferon-gamma in sera of children with dengue. J. Clin. Investig. 88:1473-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurane, I., B. L. Innis, S. Nimmannitya, A. Nisalak, A. L. Rothman, P. G. Livingston, J. Janus, and F. A. Ennis. 1990. Human immune responses to dengue viruses. Southeast Asian J. Trop. Med. Public Health 21:658-662. [PubMed] [Google Scholar]
- 62.Labrada, L., X. H. Liang, W. Zheng, C. Johnston, and B. Levine. 2002. Age-dependent resistance to lethal alphavirus encephalitis in mice: analysis of gene expression in the central nervous system and identification of a novel interferon-inducible protective gene, mouse ISG12. J. Virol. 76:11688-11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lambeth, C. R., L. J. White, R. E. Johnston, and A. M. de Silva. 2005. Flow cytometry-based assay for titrating dengue virus. J. Clin. Microbiol. 43:3267-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laur, F., B. Murgue, X. Deparis, C. Roche, O. Cassar, and E. Chungue. 1998. Plasma levels of tumour necrosis factor alpha and transforming growth factor beta-1 in children with dengue 2 virus infection in French Polynesia. Trans. R. Soc. Trop. Med. Hyg. 92:654-656. [DOI] [PubMed] [Google Scholar]
- 65.Lee, S. H., and S. M. Vidal. 2002. Functional diversity of Mx proteins: variations on a theme of host resistance to infection. Genome Res. 12:527-530. [DOI] [PubMed] [Google Scholar]
- 66.Lenschow, D. J., N. V. Giannakopoulos, L. J. Gunn, C. Johnston, A. K. O'Guin, R. E. Schmidt, B. Levine, and H. W. Virgin IV. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin, Y. W., K. J. Wang, H. Y. Lei, Y. S. Lin, T. M. Yeh, H. S. Liu, C. C. Liu, and S. H. Chen. 2002. Virus replication and cytokine production in dengue virus-infected human B lymphocytes. J. Virol. 76:12242-12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacQuillan, G. C., C. Mamotte, W. D. Reed, G. P. Jeffrey, and J. E. Allan. 2003. Upregulation of endogenous intrahepatic interferon stimulated genes during chronic hepatitis C virus infection. J. Med. Virol. 70:219-227. [DOI] [PubMed] [Google Scholar]
- 69.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mashimo, T., M. Lucas, D. Simon-Chazottes, M. P. Frenkiel, X. Montagutelli, P. E. Ceccaldi, V. Deubel, J. L. Guenet, and P. Despres. 2002. A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc. Natl. Acad. Sci. USA 99:11311-11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morens, D. M., and S. B. Halstead. 1990. Measurement of antibody-dependent infection enhancement of four dengue virus serotypes by monoclonal and polyclonal antibodies. J Gen. Virol. 71:2909-2914. [DOI] [PubMed] [Google Scholar]
- 72.Muñoz-Jordán, J. L., M. Laurent-Rolle, J. Ashour, L. Martínez-Sobrido, M. Ashok, W. I. Lipkin, and A. García-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muthukumar, A., D. Zhou, M. Paiardini, A. P. Barry, K. S. Cole, H. M. McClure, S. I. Staprans, G. Silvestri, and D. L. Sodora. 2005. Timely triggering of homeostatic mechanisms involved in the regulation of T-cell levels in SIVsm-infected sooty mangabeys. Blood 106:3839-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naganuma, A., A. Nozaki, T. Tanaka, K. Sugiyama, H. Takagi, M. Mori, K. Shimotohno, and N. Kato. 2000. Activation of the interferon-inducible 2′-5′-oligoadenylate synthetase gene by hepatitis C virus core protein. J. Virol. 74:8744-8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nguyen, H., J. Hiscott, and P. M. Pitha. 1997. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 8:293-312. [DOI] [PubMed] [Google Scholar]
- 77.Nguyen, T. H., H. Y. Lei, T. L. Nguyen, Y. S. Lin, K. J. Huang, B. L. Le, C. F. Lin, T. M. Yeh, Q. H. Do, T. Q. Vu, L. C. Chen, J. H. Huang, T. M. Lam, C. C. Liu, and S. B. Halstead. 2004. Dengue hemorrhagic fever in infants: a study of clinical and cytokine profiles. J. Infect. Dis. 189:221-232. [DOI] [PubMed] [Google Scholar]
- 78.Nielsen, H. L., L. Ronnov-Jessen, R. Villadsen, and O. W. Petersen. 2002. Identification of EPSTI1, a novel gene induced by epithelial-stromal interaction in human breast cancer. Genomics 79:703-710. [DOI] [PubMed] [Google Scholar]
- 79.Ning, S., L. E. Huye, and J. S. Pagano. 2005. Regulation of the transcriptional activity of the IRF7 promoter by a pathway independent of interferon signaling. J. Biol. Chem. 280:12262-12270. [DOI] [PubMed] [Google Scholar]
- 80.Pavlovic, J., A. Schroder, A. Blank, F. Pitossi, and P. Staeheli. 1993. Mx proteins: GTPases involved in the interferon-induced antiviral state. Ciba Found. Symp. 176:233-247. [DOI] [PubMed] [Google Scholar]
- 81.Pavlovic, J., and P. Staeheli. 1991. The antiviral potentials of Mx proteins. J. Interf. Res. 11:215-219. [DOI] [PubMed] [Google Scholar]
- 82.Perez, A. B., G. Garcia, B. Sierra, M. Alvarez, S. Vazquez, M. V. Cabrera, R. Rodriguez, D. Rosario, E. Martinez, T. Denny, and M. G. Guzman. 2004. IL-10 levels in dengue patients: some findings from the exceptional epidemiological conditions in Cuba. J. Med. Virol. 73:230-234. [DOI] [PubMed] [Google Scholar]
- 83.Pinto, L. M., S. A. Oliveira, E. L. Braga, R. M. Nogueira, and C. F. Kubelka. 1999. Increased pro-inflammatory cytokines (TNF-alpha and IL-6) and anti-inflammatory compounds (sTNFRp55 and sTNFRp75) in Brazilian patients during exanthematic dengue fever. Mem. Inst. Oswaldo Cruz 94:387-394. [DOI] [PubMed] [Google Scholar]
- 84.Putnak, J. R., B. A. Coller, G. Voss, D. W. Vaughn, D. Clements, I. Peters, G. Bignami, H. S. Houng, R. C. Chen, D. A. Barvir, J. Seriwatana, S. Cayphas, N. Garcon, D. Gheysen, N. Kanesa-Thasan, M. McDonell, T. Humphreys, K. H. Eckels, J. P. Prieels, and B. L. Innis. 2005. An evaluation of dengue type-2 inactivated, recombinant subunit, and live-attenuated vaccine candidates in the rhesus macaque model. Vaccine 23:4442-4452. [DOI] [PubMed] [Google Scholar]
- 85.Putnak, R., J. Fuller, L. VanderZanden, B. L. Innis, and D. W. Vaughn. 2003. Vaccination of rhesus macaques against dengue-2 virus with a plasmid DNA vaccine encoding the viral pre-membrane and envelope genes. Am. J. Trop. Med. Hyg. 68:469-476. [PubMed] [Google Scholar]
- 86.Raghupathy, R., U. C. Chaturvedi, H. Al-Sayer, E. A. Elbishbishi, R. Agarwal, R. Nagar, S. Kapoor, A. Misra, A. Mathur, H. Nusrat, F. Azizieh, M. A. Khan, and A. S. Mustafa. 1998. Elevated levels of IL-8 in dengue hemorrhagic fever. J. Med. Virol. 56:280-285. [DOI] [PubMed] [Google Scholar]
- 87.Ratterree, M. S., R. A. Gutierrez, A. P. Travassos da Rosa, B. J. Dille, D. W. Beasley, R. P. Bohm, S. M. Desai, P. J. Didier, L. G. Bikenmeyer, G. J. Dawson, T. P. Leary, G. Schochetman, K. Phillippi-Falkenstein, J. Arroyo, A. D. Barrett, R. B. Tesh, A. P. da Rosa, R. P. Bohm, Jr., F. B. Cogswell, K. M. Phillippi, K. Caillouet, S. Schwanberger, R. E. Shope, V. V. Pogodina, M. P. Frolova, G. V. Malenko, G. I. Fokina, G. V. Koreshkova, L. L. Kiseleva, N. G. Bochkova, and N. M. Ralph. 2004. Experimental infection of rhesus macaques with West Nile virus: level and duration of viremia and kinetics of the antibody response after infection. J. Infect. Dis. 189:669-676. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 88.Raviprakash, K., D. Apt, A. Brinkman, C. Skinner, S. Yang, G. Dawes, D. Ewing, S. J. Wu, S. Bass, J. Punnonen, and K. Porter. 2006. A chimeric tetravalent dengue DNA vaccine elicits neutralizing antibody to all four virus serotypes in rhesus macaques. Virology 353:166-173. [DOI] [PubMed] [Google Scholar]
- 89.Ronni, T., S. Matikainen, T. Sareneva, K. Melen, J. Pirhonen, P. Keskinen, and I. Julkunen. 1997. Regulation of IFN-alpha/beta, MxA, 2′,5′-oligoadenylate synthetase, and HLA gene expression in influenza A-infected human lung epithelial cells. J. Immunol. 158:2363-2374. [PubMed] [Google Scholar]
- 90.Rothman, A. L. 2003. Immunology and immunopathogenesis of dengue disease. Adv. Virus Res. 60:397-419. [DOI] [PubMed] [Google Scholar]
- 91.Rothman, A. L., and F. A. Ennis. 1999. Immunopathogenesis of dengue hemorrhagic fever. Virology 257:1-6. [DOI] [PubMed] [Google Scholar]
- 92.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 94.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 95.Scherbik, S. V., J. M. Paranjape, B. M. Stockman, R. H. Silverman, and M. A. Brinton. 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80:2987-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sheppard, P., W. Kindsvogel, W. Xu, K. Henderson, S. Schlutsmeyer, T. E. Whitmore, R. Kuestner, U. Garrigues, C. Birks, J. Roraback, C. Ostrander, D. Dong, J. Shin, S. Presnell, B. Fox, B. Haldeman, E. Cooper, D. Taft, T. Gilbert, F. J. Grant, M. Tackett, W. Krivan, G. McKnight, C. Clegg, D. Foster, and K. M. Klucher. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63-68. [DOI] [PubMed] [Google Scholar]
- 97.Shresta, S., J. L. Kyle, P. R. Beatty, and E. Harris. 2004. Early activation of natural killer and B cells in response to primary dengue virus infection in A/J mice. Virology 319:262-273. [DOI] [PubMed] [Google Scholar]
- 98.Shresta, S., J. L. Kyle, H. M. Snider, M. Basavapatna, P. R. Beatty, and E. Harris. 2004. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J. Virol. 78:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shresta, S., K. L. Sharar, D. M. Prigozhin, P. R. Beatty, and E. Harris. 2006. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J. Virol. 80:10208-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shresta, S., K. L. Sharar, D. M. Prigozhin, H. M. Snider, P. R. Beatty, and E. Harris. 2005. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J. Immunol. 175:3946-3954. [DOI] [PubMed] [Google Scholar]
- 101.Silvestri, G., D. L. Sodora, R. A. Koup, M. Paiardini, S. P. O'Neil, H. M. McClure, S. I. Staprans, and M. B. Feinberg. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441-452. [DOI] [PubMed] [Google Scholar]
- 102.Simmons, M., K. R. Porter, C. G. Hayes, D. W. Vaughn, and R. Putnak. 2006. Characterization of antibody responses to combinations of a dengue virus type 2 DNA vaccine and two dengue virus type 2 protein vaccines in rhesus macaques. J. Virol. 80:9577-9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith, P. L., G. Lombardi, and G. R. Foster. 2005. Type I interferons and the innate immune response—more than just antiviral cytokines. Mol. Immunol. 42:869-877. [DOI] [PubMed] [Google Scholar]
- 104.Suharti, C., E. C. van Gorp, W. M. Dolmans, T. E. Setiati, C. E. Hack, R. Djokomoeljanto, and J. W. van der Meer. 2003. Cytokine patterns during dengue shock syndrome. Eur. Cytok. Netw. 14:172-177. [PubMed] [Google Scholar]
- 105.Sun, W., A. Nisalak, M. Gettayacamin, K. H. Eckels, J. R. Putnak, D. W. Vaughn, B. L. Innis, S. J. Thomas, and T. P. Endy. 2006. Protection of rhesus monkeys against dengue virus challenge after tetravalent live attenuated dengue virus vaccination. J. Infect. Dis. 193:1658-1665. [DOI] [PubMed] [Google Scholar]
- 106.Tahara, E., Jr., H. Tahara, M. Kanno, K. Naka, Y. Takeda, T. Matsuzaki, R. Yamazaki, H. Ishihara, W. Yasui, J. C. Barrett, T. Ide, and E. Tahara. 2005. G1P3, an interferon inducible gene 6-16, is expressed in gastric cancers and inhibits mitochondrial-mediated apoptosis in gastric cancer cell line TMK-1 cell. Cancer Immunol. Immunother. 54:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell. Microbiol. 8:907-922. [DOI] [PubMed] [Google Scholar]
- 108.Vitarana, T., H. de Silva, N. Withana, and C. Gunasekera. 1991. Elevated tumour necrosis factor in dengue fever and dengue haemorrhagic fever. Ceylon Med. J. 36:63-65. [PubMed] [Google Scholar]
- 109.Warke, R. V., K. Xhaja, K. J. Martin, M. F. Fournier, S. K. Shaw, N. Brizuela, N. de Bosch, D. Lapointe, F. A. Ennis, A. L. Rothman, I. Bosch, L. Estevez, G. Raines, H. Melichar, and M. V. Fournier. 2003. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J. Virol. 77:11822-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weber, F., G. Kochs, and O. Haller. 2004. Inverse interference: how viruses fight the interferon system. Viral Immunol. 17:498-515. [DOI] [PubMed] [Google Scholar]
- 111.Wiedmer, T., Q. Zhou, D. Y. Kwoh, and P. J. Sims. 2000. Identification of three new members of the phospholipid scramblase gene family. Biochim. Biophys. Acta 1467:244-253. [DOI] [PubMed] [Google Scholar]
- 112.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu, H., H. Zhao, C. D. Collins, S. E. Eckenrode, Q. Run, R. A. McIndoe, J. M. Crawford, D. R. Nelson, J. X. She, and C. Liu. 2003. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology 37:1180-1188. [DOI] [PubMed] [Google Scholar]