Abstract
The parasitic protozoan Trypanosoma brucei contains two type III phosphatidylinositol 4-kinases (α and β). We have cloned the gene encoding the T. brucei type III phosphatidylinositol 4-kinase β (TbPI4KIII-β), expressed the protein in COS-7 cells, and confirmed that the protein catalyzes the phosphorylation of phosphatidylinositol. Depletion of TbPI4KIII-β in procyclic T. brucei by RNA interference (RNAi) resulted in inhibition of cell growth and a distorted cellular morphology. RNAi cells had a distorted Golgi apparatus, and lysosomal and flagellar pocket proteins were mislocalized. Ultrastructural analysis revealed the internal accumulation of a heterogeneous population of vesicles, abnormal positioning of organelles, and a loss of cell polarity. Scanning electron microcopy revealed a twisted phenotype, and dividing cells often exhibited a detached daughter flagellum and lacked a cleavage furrow. Cell cycle analysis confirmed that cells depleted of TbPI4KIII-β have a postmitotic cytokinesis block that occurs after a single round of mitosis, suggestive of a specific cell cycle block. In summary, TbPI4KIII-β is an essential protein in procyclic T. brucei, required for maintenance of Golgi structure, protein trafficking, normal cellular shape, and cytokinesis.
Phosphoinositides (PIs), although minor constituents of eukaryotic cell membranes, play a crucial role in cellular regulation. PIs are phosphorylated derivatives of phosphatidylinositol (PtdIns), which can be differentially phosphorylated at the 3′, 4′, or 5′ position of the inositol ring, leading to seven possible PIs (16). Their synthesis, mediated by specific kinases and/or phosphatases, is partitioned to distinct subcellular compartments. The PIs then serve to recruit effector proteins by interacting with lipid binding domains in these effectors (37).
PtdIns 4-kinases phosphorylate PtdIns at the 4′ position to yield PtdIns 4-monophosphate (PI4P). Subsequent phosphorylation at the 3′ and/or 5′ position results in the synthesis of PI 3,4-P2, PI 4,5-P2, and PI 3,4,5-P3, which participate in a wide variety of cell signaling pathways. Recently it has been shown that PI4P has an essential role in Golgi budding, and hence, its role extends beyond serving as a precursor (45). The D4-phosphorylated PIs are essential lipid mediators for the regulation of multiple cellular functions, including endocytosis, vesicular trafficking, actin cytoskeletal organization, and Ca2+ signaling (41).
Eukaryotes express two classes of PtdIns 4-kinase, termed type II and type III. These two classes are distinguished by their sequences, biochemical properties, enzymatic parameters, and inhibitor profiles (4, 18). Most eukaryotes express at least one type II enzyme and have two type III enzymes (PI4KIII-α and PI4KIII-β). Although the specific functions of each isoform are still poorly understood, it appears that the type II kinases and PI4KIII-β are important for membrane trafficking in the secretory/endocytotic pathways (4), whereas PI4KIII-α may serve to generate pools of PI4P required for phospholipase C-dependent signaling (5).
Pik1, the yeast ortholog of PI4KIII-β, localizes to the nucleus and to the Golgi apparatus, where it is involved in the regulation of secretion (3, 23, 44). The mammalian enzyme also localizes to the Golgi apparatus (20, 48) and the nucleus (12) and regulates trafficking from the Golgi apparatus to the plasma membrane (7, 19). PI4KIII-β-dependent synthesis of PI4P at the Golgi apparatus is also important for ceramide transport from the endoplasmic reticulum (42), which is mediated by the PI4P-binding protein CERT (30). More recently, PI4KIII-β has been implicated in the priming of neurosecretory vesicles in regulated exocytosis (11).
Trypanosoma brucei, an extracellular protozoan parasite, is the causative agent of African sleeping sickness in humans. The parasite exists in two distinct life forms, the mammalian bloodstream form and the insect procyclic form. T. brucei has a highly polarized exocytotic and endocytotic system, which is located between the nucleus and a specialized organelle called the flagellar pocket. This invagination of the plasma membrane near the posterior of the cell is the only site of exocytosis and endocytosis (21). Both forms of the parasite express a stage-specific glycosylphosphatidylinositol-anchored surface protein: variable surface glycoprotein (VSG) in the bloodstream form and procyclin in the procyclic form (10, 40). Surface coat proteins comprise the bulk of protein cargo moving through the secretory system (15). Bloodstream form parasites also have a high rate of VSG endocytosis and recycling at the flagellar pocket. This process is also important for immune evasion, allowing for degradation of bound host immune complexes. Endocytosis at the flagellar pocket is important in both forms for uptake of nutrients and for host-parasite interactions. Trafficking, endocytosis, and Golgi maintenance are all PI-mediated events in other eukaryotes (4). Many of the components of the exocytotic and endocytotic systems in T. brucei have been identified, including clathrin, adapter proteins, and Rab GTPases (15), some of which are known effector proteins of D4-phosphorylated PIs in other eukaryotes. These observations suggest that the synthesis of D4-phosphorylated PIs may play a key regulatory role in the secretory pathway in T. brucei.
The T. brucei database contains the sequences for two putative PtdIns 4-kinases, TbPI4KIII-α and TbPI4KIII-β. Unlike other eukaryotes, it appears that T. brucei lacks a type II PtdIns 4-kinase. A class III PtdIns 3-kinase was recently identified in T. brucei, and its depletion by RNA interference (RNAi) implicated it in Golgi segregation and endocytotic trafficking (22). Like yeast, T. brucei does not have class I or II PtdIns 3-kinases, suggesting that T. brucei does not have PtdIns 3-kinase-dependent signaling pathways. Four putative PtdIns monophosphate (PIP) kinases have been identified in the T. brucei genome: a type I PI4P 5-kinase, a type III PI3P 5-kinase (PIKFYVE), and two type II enzymes, both of which are most similar to the type II PIP kinase-α isoforms. The presence of these enzymes indicates that T. brucei can, in principle, synthesize all of the mono- and bisphosphorylated PIs.
In the present study, we have cloned and functionally characterized the T. brucei PI4KIII-β. Using RNAi, we show that synthesis of PI4P by TbPI4KIII-β is essential for normal growth and morphology in procyclic T. brucei. Further analysis revealed that TbPI4KIII-β is required for maintenance of Golgi structure, protein trafficking, normal cellular shape, and cytokinesis.
MATERIALS AND METHODS
Identification and cloning of Tb PI4KIII-β.
TbPI4KIII-β and TbPI4KIII-α were identified by BLAST searching of the TIGR T. brucei database with either the human PI4KIII-β protein sequence (Q9UBF8) or the human PI4KIII-α protein sequence (P42356). An open reading frame for TbPI4KIII-β (accession number XP 844264) was found on bacterial artificial chromosome (BAC) clone RPCI93-5E12 from chromosome 4, and TbPI4KIII-α (accession number XP 843994) was found on BAC clone RPCI93-28C22 from chromosome 3. The full-length gene for TbPI4KIII-β was amplified by PCR from 427 genomic DNA and subcloned into the mammalian expression vector pCMV-tag2b (Stratagene) using the primers 5′-GTTTCCTTTTTCCGGATCCATGTCGAATGCTTTGTTTTG-3′ and 5′-CATTCACCACATCCCCTCGAGCTAGAGTATACCATT-3′. A fragment of the TbPI4KIII-α gene (nucleotides 5097 to 6936) was likewise amplified by PCR from 427 genomic DNA and subcloned into the Zero Blunt TOPO PCR cloning vector (Invitrogen) using the primers 5′-GCCGGGAACGGCTAATGAGCCTCATCC-3′ and 5′-CCCTTCACTCACCGGGTACCCC-3′.
Transfections, immunoprecipitations, and Western blot analysis.
COS-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). Purified plasmid, either empty vector or pCMV-TbPI4KIII-β, was transfected into COS-7 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. Cells were incubated at 37°C in 5% CO2 for 18 h. Transfected COS-7 cells were washed with phosphate-buffered saline (PBS), lysed on ice in buffer A (20 mM Tris-HCl [pH 7.5], 25 mM sucrose, 100 mM NaCl, 1.0 mM EDTA, 1 mM NaVO3, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 20 μg/ml antipain, 100 μg/ml benzamidine, 10 μg/ml pepstatin, and 10 μg/ml chymostatin), and sonicated. After centrifugation (70,000 rpm for 15 min at 4°C), Triton X-100 was added to the supernatant for a final concentration of 0.1%. Cleared lysate (300 μl) was incubated overnight with 75 μl anti-Flag M2 resin (Sigma). Beads were then washed three times with buffer A plus 0.1% Triton X-100 and then resuspended in 75 μl buffer A plus 0.1% Triton X-100, yielding a 50% bead slurry.
For Western analysis, 10 μl of bead slurry was separated on a 10% sodium dodecyl sulfate gel and transferred to Hybond-P polyvinylidene difluoride membranes (Amersham). Membranes were blocked in 5% milk (wt/vol) in TBS-T (20 mM Tris-HCl, 137 mM NaCl, 0.1% [vol/vol] Tween 20, pH 7.6) for 1 h at room temperature. Primary and secondary antibodies were diluted in blocking buffer and were incubated for 1 h at room temperature. Antibody dilutions were as follows: anti-Flag M2 antibody (Sigma), 1:1,000, and sheep anti-mouse immunoglobulin G (IgG) horseradish peroxidase conjugate secondary (Amersham), 1:5,000.
Immune complex kinase assays.
PtdIns 4-kinase activity of immunoprecipitates was measured by phosphorylation of PtdIns-Triton X-100 micelles using [γ-32P]ATP (10 mCi/mmol) as the phosphate donor. PtdIns-Triton X-100 micelles were prepared by sonicating PtdIns (Avanti) in buffer C (50 mM Tris-HCl [pH 7.5], 1.0 mM EGTA, 0.4% Triton X-100, and 0.5 mg/ml bovine serum albumin [BSA]). Reaction mixtures contained 10 μl bead slurry and 38 μl PtdIns-Triton X-100 micelles and were initiated with the addition of 2 μl of ATP mix (50 mM Tris-HCl [pH 7.5], 375 mM MgCl2, 5 mM ATP, and [γ-32P]ATP [10 mCi/mmol]). The reaction mixtures contained 200 μM PtdIns, 0.3% Triton X-100, 400 μM ATP, and 15 mM MgCl, and reactions were carried out at room temperature for 30 min. Reactions were stopped by addition of 3.75 volumes of chloroform-methanol-HCl (100:200:1), followed by extraction with 1.25 volumes of chloroform and 0.1 N HCl. The lipids were separated by thin-layer chromatography (TLC) on Whatman Partisil LK5D TLC plates using an n-propyl alcohol-H2O-NH4OH (65:20:15) solvent system, and radioactive spots were detected by autoradiography, scraped, and quantified by scintillation counting.
RNAi plasmid construction.
Primers were designed to amplify the coding region between positions 421 and 1002 of the DNA sequence. Primers 5′-GGAACTGCGACTTAAGCTTTTCAACGATGAGAG-3′ and 5′-GCTAGCTCCTCTTGCCGCAAAT-3′ contain restriction sites HindIII and NheI, respectively. This fragment was subcloned into the HindIII and NheI sites of pJM326 (a generous gift of Paul Englund, Johns Hopkins University). The complementary strand was amplified using primers 5′-GGAACTGCGACTTACGCGTTTCAACGATGAGAG-3′ and 5′-TCTAGATCCTCTTGCCGCAAAT-3′, which contain restriction sites for MluI and Xba, respectively. This fragment was subcloned into the MluI and XbaI sites in pLew100 (a generous gift of Paul Englund, Johns Hopkins University). The pJM326 construct containing our insert was digested with HindIII and XbaI, releasing a larger fragment containing the target gene fragment fused to a fragment which will form the loop of the stem-loop transcript. This fragment was then subcloned into the HindIII and XbaI sites in the pLew100 construct containing the complement of the target gene.
Trypanosome cell culture and transfection.
Procyclic culture form 29-13 cells (a generous gift of George Cross, Rockefeller University), which stably express T7 polymerase and a tetracycline repressor, were used for creating RNAi cell lines (47). 29-13 parental line cultures were grown in SDM-79 supplemented with 10% FBS in the presence of 25 μg/ml G418 and 25 μg/ml hygromycin. 29-13 cells were transfected with EcoRV-linearized RNAi plasmid, using a Bio-Rad Gene Pulser II at 1.5 kV and 25 μF in 0.4-cm electrocuvettes (Bio-Rad). After electroporation, transfectants were selected for by culturing cells in the presence of 2.5 μg/ml phleomycin in addition to G418 and hygromycin. Synthesis of RNA was induced by the addition of tetracycline at 1 μg/ml (46).
RNA isolation and Northern blot analysis.
Total RNA was extracted from log-phase cultures using the RNAqueous kit (Ambion). Total RNA samples were collected from an uninduced culture and from tetracycline-induced cultures every 24 h, over 6 days. Two micrograms of total RNA was separated on a 1% agarose-formaldehyde gel and transferred to BrightStar-Plus nylon membranes (Ambion). Hybridization was performed overnight at 48°C in ULTRAhyb hybridization buffer (Ambion). 32P-labeled probes for TbPI4KIII-β, TbPI4KIII-α, and α-tubulin were synthesized using the StripEZ-PCR probe synthesis kit (Ambion). Blots were stripped and reprobed according to the manufacturer's protocols. The DNA probes were designed to hybridize with the following regions of the transcripts; TbPI4KIII-β, 1 to 675; TbPI4KIII-α, 5097 to 5770, and α-tubulin, 1 to 650.
Growth curves.
For growth curves, cell counts were performed using a hemocytometer. Cells from three independent experiments were counted, and the averages are reported.
Inositol labeling.
Mid-log-phase trypanosomes from uninduced cultures and from cultures induced for 3 and 5 days were centrifuged at 400 × g and washed once with PBS. Trypanosomes were resuspended in 5 ml fresh SDM-79 supplemented with 10% FBS at 2 × 107 cells/ml, and 50 μl of 10 μCi/μl [3H]myoinositol (Amersham) was added to the mixture and incubated for 24 h at 25°C. Cells were centrifuged at 400 × g and washed with PBS. Lipids were extracted with 375 μl of methanol-HCl (1:1) and 190 μl of chloroform. The lipids were separated by TLC on Whatman Partisil LK5D TLC plates using an n-propyl alcohol-H2O-NH4OH (65:20:15) solvent system, and radioactive spots were detected by autoradiography, scraped, and quantified by scintillation counting. The PIP/PtdIns ratio was calculated for five independent experiments at each time point, and results were averaged.
Immunofluorescence.
Cells for immunofluorescence were fixed with 3.7% formaldehyde (EMS) in PBS and permeabilized with 0.1% Triton X-100-PBS. Cells were blocked for 1 h with 10% normal goat serum-PBS. Incubations with primary antibody(ies) and secondary antibody(ies) were performed in 10% normal goat serum-PBS for 1 h at room temperature. Primary antibody dilutions were as follows: mouse anti-p67, 1:500 (1), rat-anti YL1/2, 1:500 (Chemicon) (39), and rabbit anti-CRAM, 1:300 (28). To visualize the Golgi apparatus, cells were fixed and permeabilized with ice-cold 100% methanol. Cells were blocked for 1 h with 3% BSA-PBS, and incubations with primary antibodies and secondary antibodies were performed in 3% BSA-PBS for 1 h at room temperature. Primary antibody dilutions were as follows: rabbit anti-TbGrasp antibody, 1:500 (24), and mouse anti-p67, 1:500 (1). Secondary antibody dilutions were as follows: Dylight560-labeled goat anti-rabbit IgG (Pierce), 1:200; AlexaFluor488-labeled goat anti-rat IgG (Molecular Probes), 1:200; and AlexaFluor488-labeled goat anti-mouse IgG (Molecular Probes), 1:200. All slides were mounted in Vectashield mounting medium containing DAPI (4′,6′-diamidino-2-phenylindole) (Vector Labs). All fluorescence microscopy was performed with a Zeiss Axiovert 200 M inverted microscope equipped with a Zeiss 100× 1.30-numerical-aperture oil immersion objective and a charge-coupled device camera. Digital images were captured and analyzed using Slidebook 4.0 software (Intelligent Imaging Innovations). For the preparation of the final figures, signal enhancement and background reduction were performed on the images using Adobe Photoshop (5.5).
Electron microscopy.
Cell fixation followed the procedure of Allen et al. (2) with some modifications. Double-strength fixative containing 5% glutaraldehyde and 6.4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.4) was added to an equal volume of cell suspension. After 10 min, small aliquots were removed for scanning electron microscopy. The remaining cell suspension was centrifuged and the supernatant replaced with single-strength fixative. Cells were stored overnight in fixative.
Transmission electron microscopy.
Fixed, pelleted cells were rinsed in sodium phosphate buffer, embedded in 2% agarose, postfixed in 1% osmium tetroxide, dehydrated in a graded ethanol series, and embedded in EMbed-812 resin. Thin sections were cut on a Leica EM UC6 microtome, poststained with uranyl acetate and lead citrate, and viewed in a JEOL JEM-1200EX transmission electron microscope operating at 80 kV.
Scanning electron microscopy.
Small drops of fixed cell suspension were incubated on poly-l-lysine-coated coverslips in a humidity chamber for 30 minutes. The coverslips were then rinsed, and adhered cells were postfixed in 1% osmium tetroxide, dehydrated in a graded ethanol series, transferred through several changes of hexamethyldisilazane, and allowed to air dry overnight. Cells on coverslips were then sputter coated with gold and viewed in a JEOL JSM-840A scanning electron microscope at 10 kV.
Analysis of DNA content.
Log-phase cells were collected over a 6-day period, and 2 × 107 cells were washed once with PBS, resuspended in 1.0 ml PBS, and adhered to poly-l-lysine-coated slides. Cells were fixed for 5 min in ice-cold 100% methanol and washed with PBS. Slides were then mounted in Vectashield mounting medium containing DAPI (Vector Labs).
Individual cells were examined by fluorescence microscopy in order to determine the percentages of cells in different phases of the cell cycle. A population of 100 cells at each time point (days 0, 4, 5, and 6) was counted, and four different cell cycle phenotypes were scored. The four phenotypes correspond to kinetoplast and nuclear contents. The first phenotype, which corresponds to G1, has one kinetoplast and one nucleus. The second phenotype has two kinetoplasts and one nucleus. The third phenotype, which has two kinetoplasts and two nuclei, corresponds to cells in G2/M. The fourth phenotype corresponded to any other combination of kinetoplast and nuclear contents. The counts from three independent experiments were averaged, and the standard deviation was calculated.
RESULTS
Identification and characterization of T. brucei PI4KIII-β.
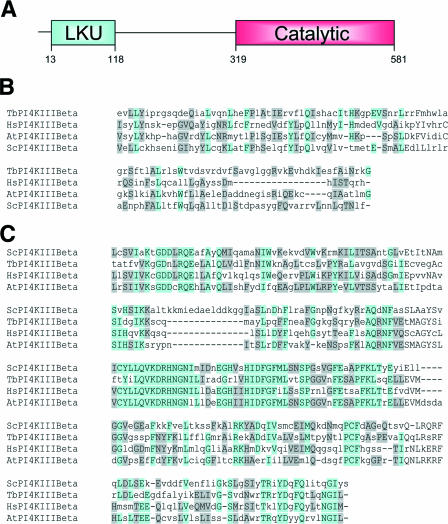
The gene for T. brucei PI4KIII-β was identified through a BLAST search of the TIGR T. brucei database. The search originally identified an open reading frame of 1,746 nucleotides in a BAC clone from chromosome 4. The deduced amino acid sequence corresponded to a 581-amino-acid protein with a predicted molecular mass of 66 kDa. CLUSTALW analysis revealed that the T. brucei enzyme shares 20.7% identity and 49.2% similarity with the human enzyme. The domain structure of the predicted protein was characteristic of the type III PtdIns 4-kinases, with a highly conserved C-terminal PtdIns 4-kinase catalytic domain and a less conserved N-terminal lipid kinase unique (LKU) domain (Fig. 1A). The catalytic domain, residues 319 to 581, shares 41.1% identity and 68.8% similarity with the human enzyme, whereas the LKU domain, residues 13 to 118, shares only 14.0% identity and 47.6% similarity (Fig. 1B and C).
FIG. 1.
Domain structure of TbPI4KIII-β and sequence alignments with known type III-β kinases. (A) TbPI4KIII-β, like other PI4KIII-βs, has an N-terminal LKU domain and a C-terminal catalytic domain. (B) Sequence alignment of the LKU domain of TbPI4KIII-β with those of Homo sapiens (Q9UBF8), A. thaliana (NP_201212), and S. cerevisiae (NP_014132). Alignments were prepared using the MUSCLE alignment program (http://www.bioinformatics.nl/tools/muscle.html). (C) Alignment of the catalytic domain according to the MUSCLE program.
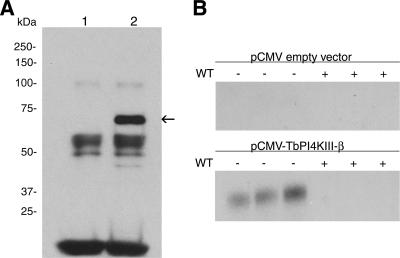
In order to confirm the identity of the gene, it was overexpressed in COS-7 cells, using the pCMV-tag2b (N-terminally Flag-tagged) mammalian expression vector. The overexpressed protein was immunoprecipitated from COS-7 cell lysates using anti-Flag M2 resin. The Flag-tagged product was detected only in lysates from cells transfected with the pCMV-TbPI4KIII-β vector and not in cells transfected with empty vector (Fig. 2A). The Flag-tagged product migrated between the 50-kDa and 75-kDa markers on sodium dodecyl sulfate gels, consistent with its predicted molecular mass of 66 kDa. PtdIns kinase assays of the immunoprecipitated recombinant protein confirmed that the expressed protein was able to phosphorylate PtdIns (Fig. 2B). Similarly to type III PtdIns 4-kinases from other sources, the T. brucei enzyme activity is wortmannin sensitive and is completely inhibited by 100 μM wortmannin. No PtdIns kinase activity was detected in the empty vector control immunoprecipitations.
FIG. 2.
Expression and enzymatic characterization of TbPI4KIII-β. (A) Immunoblot of Flag-tagged TbPI4KIII-β expressed in COS-7 cells and immunoprecipitated with anti-Flag M2 resin. Anti-Flag antibody detects a band migrating at between 50 and 75 kDa in immunoprecipitates from cells expressing TbPI4KIII-β (lane 2) but not in those from control cells expressing empty vector (lane 1). (B) Kinase assay of the immune complex of Flag-tagged recombinant TbPI4KIII-β. Immunoprecipitated proteins were tested for PtdIns kinase activity in the presence and absence of 100 μM wortmannin (WT), which inhibits mammalian PI4KIII-β activity. Reaction mixtures contained 200 μM PtdIns, 0.3% Triton X-100, 400 μM ATP, and 15 mM MgCl2, and reactions were carried out at room temperature for 30 min. Radioactive PI4P is detected by TLC only in immunoprecipitates expressing TbPI4KIII-β and is prevented from forming by WT. Triplicate measurements are shown.
TbPI4KIII-β is essential for the growth of procyclic T. brucei.
In order to investigate possible functional roles of TbPI4KIII-β in procyclic T. brucei, a stable tetracycline-inducible RNAi procyclic cell line was generated. An RNAi vector which produces double-stranded RNA as a stem-loop structure in procyclic form T. brucei was utilized (46). This construct contains a region of our target gene (nucleotides 421 to 1002) and is complement separated by a ∼550-bp region, which forms the loop of the stem-loop transcript. The linearized vector was transfected into 29-13 procyclic T. brucei by electroporation, and transfectants were selected using phleomycin.
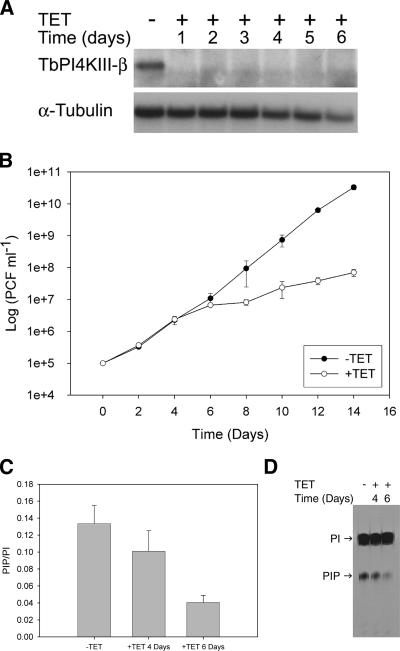
Northern blot analysis was employed to ascertain that expression of TbPI4KIII-β double-stranded RNA results in degradation of TbPI4KIII-β mRNA. After 24 h of tetracycline induction, transcript levels were virtually undetectable, compared to those in the uninduced cells, whereas α-tubulin levels remain relatively unchanged (Fig. 3A). The transcript levels remain undetectable throughout 6 days of induction. TbPI4KIII-β and TbPI4KIII-α share only 8% sequence identity at the amino acid level, and thus it is highly unlikely that knockdown of TbPI4KIII-β by RNAi would affect the levels of TbPI4KIII-α. However, as a control transcript levels for TbPI4KIII-α were also examined, and they are not significantly changed throughout 6 days of induction (data not shown), indicating that the gene knockdown is specific for TbPI4KIII-β.
FIG. 3.
Characterization of TbPI4KIII-β RNAi cell line. (A) Northern blot analysis of TbPI4KIII-β RNAi cells. Transcript levels from samples collected over 6 days of tetracycline (TET) induction were determined. Two micrograms of total RNA was separated on a 1% agarose-formaldehyde gel and transferred to BrightStar-Plus nylon membranes. The blot was then probed for TbPI4KIII-β and α-tubulin. After 24 h of induction, transcripts for TbPI4KIII-β were undetectable, and they remained so through day 6. (B) Effect of TbPI4KIII-β depletion on growth. Cell growth of uninduced (−TET) and induced (+TET) cells was monitored over 2 weeks. Induced cells show a defect in growth beginning at day 5. Error bars indicate standard deviations. (C) Effect of TbPI4KIII-β depletion on PIP levels. PIP levels were monitored by inositol labeling. Uninduced cells and cells induced for 3 and 5 days were incubated with [3H]inositol for 24 h. The lipids were then extracted, separated by TLC, and quantitated by scintillation counting. PIP levels were normalized against PtdIns (PI) levels to account for differences in cell number. Each bar represents the average from five independent experiments; error bars represent standard deviations. After 4 days of induction PIP levels were reduced by 25%, and by 6 days levels were reduced by 70%. (D) Autoradiograph of TLC-separated lipids.
A cell growth defect is observed beginning 4 days after tetracycline induction of TbPI4KIII-β RNAi (Fig. 3B). The depletion of TbPI4KIII-β led to a concomitant decrease in PIP levels (Fig. 3C). PIP levels were decreased by 25% after 4 days of induction and by 70% after 6 days. The decrease in PIP levels correlates with the timing of the appearance of the growth defect, indicating that the synthesis of PI4P by TbPI4KIII-β is required for cell growth. T. brucei has two PtdIns 4-kinases, TbPI4KIII-α and TbPI4KIII-β, both of which presumably contribute to total PIP levels. The decrease of PI4P by 70% at 6 days postinduction indicates that TbPI4KIII-β is responsible for the majority of PtdIns kinase activity in procyclic form T. brucei.
Depletion of TbPI4KIII-β results in a severe morphological defect.
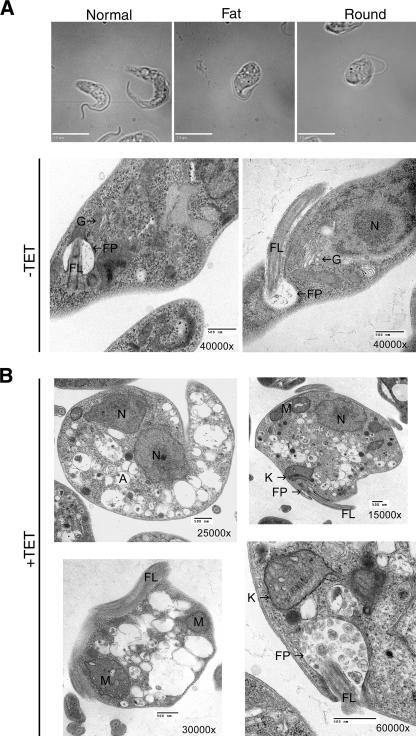
Light microscopic analysis of induced cultures showed the appearance of an abnormal phenotype, consisting of fat and round cells approximately 4 days after induction (Fig. 4A). The proportion of cells having this phenotype increases with time. At day 4 only 6% of the 100 cells counted displayed an abnormal morphology, at day 5 12% are abnormal, and at day 6 approximately 43% of the population is abnormal. The morphologically abnormal cells also exhibit defects in motility. Instead of swimming through the medium, they remain in one place, spinning in circles.
FIG. 4.
Requirement of TbPI4KIII-β for normal morphology in procyclic T. brucei. (A) Bright-field differential interference contrast microcopy reveals an abnormal phenotype in cells depleted of TbPI4KIII-β. The normal phenotype (uninduced cells), fat phenotype, and round phenotype are shown (both fat and round phenotype images are from cultures induced for 6 days). Bars, 10 μm. (B) Ultrastructural analysis of TbPI4KIII-β-depleted cells. Transmission electron microscopy was used to examine the ultrastructure of uninduced cells (−TET) and cells induced for 6 days (+TET). Cells depleted of TbPI4KIII-β are abnormally shaped, and their cytoplasm is filled with vesicles. There is also an accumulation of vesicles in the flagellar pocket (bottom right). Abbreviations: FP, flagellar pocket; Fl, flagellum; G, Golgi apparatus; M, mitochondria; N, nucleus; A, acidocalcisome. Bars, 500 nm.
Ultrastructural analysis of TbPI4KIII-β-depleted cells by transmission electron microscopy revealed that the cytoplasm of these cells was filled with numerous vesicles of various sizes, ranging from approximately 50 nm to 750 nm (Fig. 4B). Most of the vesicles were translucent, although some contained electron-dense material that morphologically is reminiscent of acidocalcisomes (36). The morphology of the major organelles, including the kinetoplast, flagellum, flagellar pocket, and nucleus, appeared to be normal, but their polarized organization within the cell was lost. As in the light micrographs, the shapes of the cells varied, with most having lost their original slender morphologies. In addition, abnormal membrane structures accumulated within the cytoplasm, and many cells had an abnormal accumulation of vesicles within the flagellar pocket.
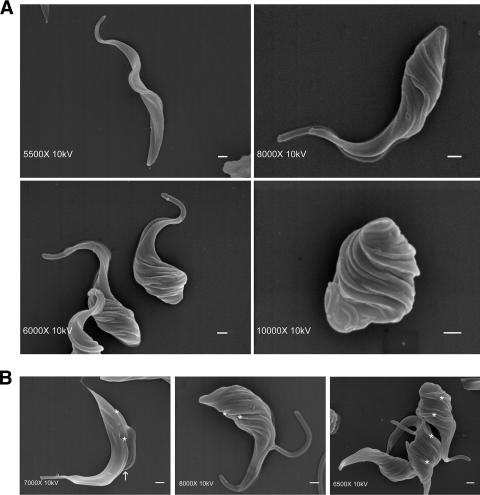
Scanning electron microscopy was employed to examine the surface structure of the induced cells. The long and slender shape of wild-type T. brucei is defined by a highly polarized microtubule cytoskeleton (31). The microtubules are organized lengthwise, with their minus ends at the anterior and their plus ends at the posterior of the cell (35). The flagellum originates from the flagellar pocket at the posterior end and runs the length of the organism (Fig. 5A). In contrast, scanning electron microscopy images of induced cells revealed an aberrant morphology with a unique characteristic. Depletion of TbPI4KIII-β resulted in a twisted phenotype, which appears to begin at the posterior end of the parasite and, over time, results in a twisted round cell (Fig. 5). This twisted phenotype is seen in thin and round cells and in dividing and nondividing cells (Fig. 5A and B). Uninduced cells, which are progressing through the cell cycle, can be detected by the presence of the daughter flagellum/flagellar pocket and the cleavage furrow (Fig. 5B). Induced cells progressing through the cell cycle also have a second flagellum/flagellar pocket, but the daughter flagellum is sometimes detached (Fig. 5B). The presence of a cleavage furrow has not been detected in the TbPI4KIII-β-depleted cells, indicating that cytokinesis may be defective in these cells.
FIG. 5.
Scanning electron microscopic analysis of TbPI4KIII-β-depleted T. brucei. (A) Scanning electron micrographs of cells depleted of TbPI4KIII-β by 6-day RNAi induction, showing an abnormal twisted shape compared to uninduced cells (top left). (B) Dividing TbPI4KIII-β-depleted cells lack a cleavage furrow. Normally, dividing T. brucei cells contain an evident cleavage furrow (arrow) and two flagella (*) (left panel). Dividing cells in induced cultures (center and right panels) also contain two flagella (*) but contain no obvious cleavage furrow. Some induced cells display a detached flagellum. Bars, 1 μm.
Effect of TbPI4KIII-β depletion on Golgi morphology.
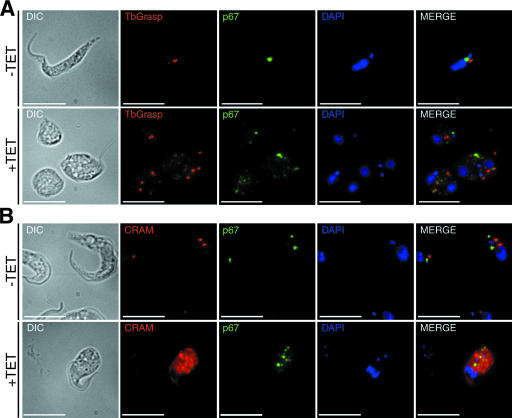
PI4KIII-β plays a major role in Golgi maintenance and secretion in other organisms. The accumulation of vesicles and abnormal membrane structures seen in the ultrastructural analysis of TbPI4KIII-β-depleted cells indicated a possible defect at the Golgi apparatus. We used immunofluorescence microscopy to further explore the abnormal phenotype seen in cells depleted of TbPI4KIII-β. We used TbGRASP as a marker for the Golgi apparatus (24). In uninduced cells (Fig. 6A), TbGRASP stains a single Golgi structure located between the nucleus and the kinetoplast. Cells depleted of TbPI4KIII-β displayed an abnormal staining pattern. In these cells, TbGRASP stained multiple punctate structures, and there was an increase in background staining, indicating that cells depleted of TbPI4KIII-β have a defect in the Golgi structure. Temperature-sensitive mutants of the yeast ortholog, Pik1, also display an aberrant Golgi structure. This indicates that TbPI4KIII-β, like Pik1, is required for maintenance of the Golgi apparatus.
FIG. 6.
Abnormal localization of protein markers for the Golgi apparatus, lysosome, and flagellar pocket in cells depleted of TbPI4KIII-β. Immunofluorescence analysis of markers for the Golgi apparatus (TbGrasp) (red) and lysosome (p67) (green) (A) and for the flagellar pocket (CRAM) (red) and lysosome (p67) (green) (B) are shown. The kinetoplast and nucleus are stained with DAPI (blue). Cells depleted of TbPI4KIII-β display an abnormal staining pattern for all markers examined, indicating a defect in Golgi trafficking. +TET, cells were induced for 6 days. Bars, 10 μm. DIC, differential interference contrast.
TbPI4KIII-β is required for proper localization of protein markers for the flagellar pocket and lysosome.
The effect of depleting TbPI4KIII-β on the localization of lysosomal and flagellar pocket markers was also examined. The p67 protein, a type I membrane protein, was used as a marker for the lysosome (1). In uninduced cells, p67 stains a region between the kinetoplast and the nucleus (Fig. 6A and B). After induction, p67 is no longer localized to one region but stains several punctate structures throughout the cytoplasm. Similarly to the staining of TbGRASP, there is an increase in background p67 distribution. The CRAM protein, which is a highly expressed type I membrane protein in procyclic cells, was used as a marker for the flagellar pocket. It is a putative lipoprotein receptor, which localizes to the flagellar pocket (28). Typically CRAM stains one or two structures in the posterior region of the cell, near the kinetoplast (Fig. 6B). TbPI4KIII-β depleted cells display an abnormal staining pattern. CRAM was mislocalized throughout the cytoplasm, with diffuse staining and staining of multiple punctate structures, indicating that CRAM is no longer localized to the flagellar pocket (Fig. 6B). Both CRAM and p67 are distributed throughout the cytoplasm in the TbPI4KIII-β depleted cells, but CRAM displays a much more extreme phenotype, with intense staining throughout the cytoplasm (Fig. 6B). Because p67 and CRAM are integral membrane proteins, they should be targeted to their respective locations via the secretory pathway. The mislocalization of these proteins may therefore reflect a defect in sorting and/or trafficking from the Golgi apparatus or may be due to a defect in the Golgi apparatus itself.
Effect of TbPI4III-β depletion on cell cycle progression in procyclic form T. brucei.
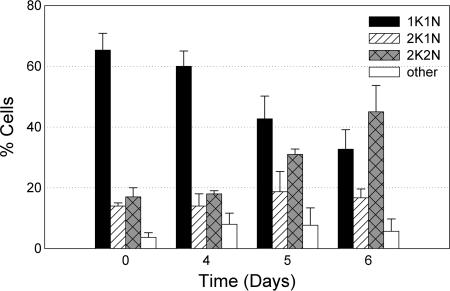
Scanning electron microscopy reveals that dividing cells depleted of TbPI4KIII-β lacked a cleavage furrow, indicating a possible defect in cytokinesis. In order to determine the effect of TbPI4KIII-β depletion on cell cycle progression, the kinetoplast and nuclear contents of uninduced and induced cells were analyzed in DAPI-stained cells. The percentages of cells, in four different stages of the cell cycle, were calculated from a population of 100 cells for each sample. Analysis of the uninduced cells, in log-phase growth, reveals that approximately 65% of the cells were in G1 and approximately 15 to 20% of the cells were in G2/M at all times examined (Fig. 7). After 5 days of tetracycline induction, the percentage of cells in G1 decreased from 68% to 43% and the percentage of cells in G2/M increased from 16% to 31%. By 6 days of induction, the percentage of cells in G1 had decreased to 33% and the percentage of cells in G2/M had increased to 45%. Although a few cells with abnormal DNA content were found in both uninduced and induced cultures, depletion of TbPI4KIII-β did not result in an increase in the number of anucleated cells (zoids) or in cells with a DNA content of greater than 2K2N. Cells depleted of TbPI4KIII-β replicated both the kinetoplast and the nucleus but were blocked at cytokinesis.
FIG. 7.
Cell cycle analysis of TbPI4KIII-β-depleted T. brucei. The cell cycle was followed in induced RNAi cells over time. The number of kinetoplasts and nuclei in 100 individual DAPI-stained cells were counted at different time points after induction. Cells in three independent experiments were counted and averaged for each day. Error bars represent the standard deviations. In cells depleted of TbPI4KIII-β, the percentage of cells in G2/M begins to increase at day 5, indicating a cytokinesis block.
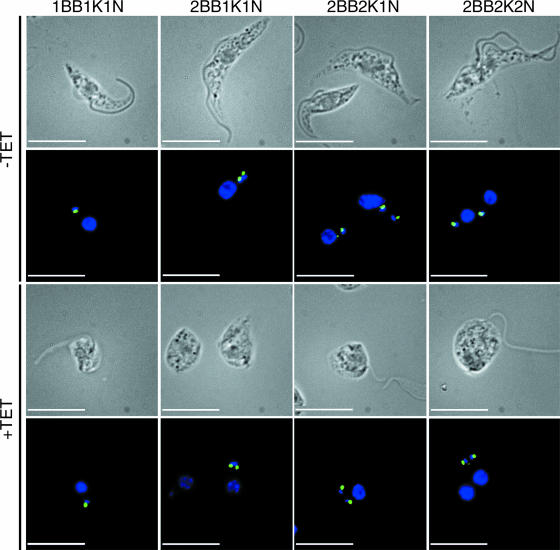
In order to investigate further the effects of depletion of TbPI4KIII-β on the cell cycle, we followed kinetoplast segregation and nuclear division throughout the cell cycle (Fig. 8). Uninduced and induced cells were stained with DAPI to stain the kinetoplast and the nucleus and with YL1/2 to stain the basal body. The basal body of T. brucei is physically attached to the kinetoplast through the tripartite attachment region, which extends through both the cell and mitochondrial membranes (33). Elongation and maturation of the pro-basal body and nucleation of the new flagellum are the first events in the cell cycle of T. brucei (32). Kinetoplast replication and division occur soon after, followed by nuclear division (49). Depletion of TbPI4KIII-β did not affect duplication of the basal bodies or the kinetoplasts. Both the basal body and kinetoplast duplicated and segregated together (Fig. 8), although the direction of segregation was aberrant. Normally, the new basal body/kinetoplast is positioned posterior to the old basal body/kinetoplast. As the cell cycle progresses, the new basal body/kinetoplast moves toward the posterior end of the cell. During mitosis the daughter nucleus moves towards the posterior so that eventually the cell has the characteristic 2K2N configuration, with the old basal body/kinetoplast between the two nuclei. The induced cells no longer maintain this strict spatial organization. The basal bodies/kinetoplasts segregate perpendicular to the normal anterior-to-posterior directional axis (Fig. 8). Nuclear division proceeds at the appropriate time, although the positioning of the kinetoplasts and nuclei are abnormal in the induced cells. These cells do not have the characteristic 2K2N configuration, with the old basal body/kinetoplast between the two nuclei.
FIG. 8.
Analysis of morphological cell cycle events in TbPI4KIII-β-depleted T. brucei. The number and position of the basal body (BB), kinetoplast (K), and nucleus (N) were examined by fluorescence microscopy in uninduced cells (−TET) and cells induced for 6 days (+TET). Cells were stained with DAPI (blue) and YL1/2 (green), which identifies the basal body. In cells depleted of TbPI4KIII-β, the basal bodies/kinetoplasts segregate perpendicular to the normal anterior-to-posterior directional axis. Nuclear division proceeds at the appropriate time, although the positioning of the kinetoplasts and nuclei is abnormal in the induced cells. These cells do not have characteristic 2K2N configuration, with the old basal body/kinetoplast between the two nuclei as seen in uninduced cells (top right). Bars, 10 μm.
DISCUSSION
Based on amino acid sequence analysis and enzymatic characterization of the expressed protein, we have succeeded in cloning PI4KIII-β from T. brucei. Although the T. brucei kinase is only 20.7% identical overall to human PI4KIII-β, it contains a relatively conserved catalytic domain (41.1% identity) and a somewhat less conserved LKU domain (14.0% identity).
TbPI4KIII-β is an essential protein in T. brucei, as its depletion by RNAi causes severe growth and morphological defects. There is a 3-day lag between RNAi-mediated loss of transcripts (undetectable 24 h after induction) and the appearance of abnormally shaped fat/round cells (beginning 4 days after induction). There is a corresponding lag between RNAi induction and decreased PI4P levels, which are diminished by ∼25% at day 4 and by ∼70% at day 6 after induction.
Synthesis of PI4P is important for maintenance of the structural and functional organization of the Golgi apparatus (13). Loss of PI4KIII-β in other organisms results in morphological defects in the Golgi apparatus and defects in the secretory pathway (4). Immunofluorescence analysis of TbPI4KIII-β-depleted cells reveals mislocalization of a Golgi marker and markers for the lysosome and flagellar pocket. These markers, TbGrasp, p67, and CRAM, redistribute predominantly to what appear to be membrane-bounded vesicles throughout the cytoplasm. Ultrastructural analysis confirms that the cytoplasm of abnormally shaped cells is filled with a heterogeneous population of vesicles. These abnormal structures may be vesicle intermediates, which originate from affected organelles, such as the endoplasmic reticulum, Golgi apparatus, or endosomes. Ablation of clathrin heavy chain in procyclic T. brucei results in similar morphological abnormalities, including rounded shape, accumulation of cytoplasmic vesicles, and mislocalization of CRAM (2). Trafficking of CRAM to the flagellar pocket, but not of p67 to the lysosome or of EP procyclin to the surface, is clathrin dependent in procyclic T. brucei (25). In mammalian cells, the clathrin adapter protein AP-1 is recruited to the trans-Golgi network by a pool of PI4P synthesized by PI4KII-α (45). T. brucei lacks the type II isoforms, and it is possible that this function in T. brucei is performed by TbPI4KIII-β. Loss of PI4P synthesis apparently causes a more global defect in secretion, affecting trafficking to the lysosome as well as transport to the flagellar pocket. The importance of PIs in trafficking in T. brucei is also illustrated by the finding that depletion of TbVps34, a class III PtdIns 3-kinase, in bloodstream form T. brucei caused a defect in export of VSG to the cell surface, in addition to its effect on Golgi segregation (22).
Impairment of PI4P synthesis in T. brucei may also influence lipid homeostasis in these organisms. For example, synthesis of sphingomyelin in the Golgi apparatus in COS-7 cells requires ceramide transfer protein (CERT)-dependent transport of ceramide from the endoplasmic reticulum (42). CERT localizes to Golgi membranes through its pleckstrin homology domain, which binds selectively to PI4P. Oxysterol-binding proteins also bind specifically to PI4P through their pleckstrin homology domains. Oxysterol-binding protein localizes to the Golgi apparatus in response to oxysterols, in a PI4P- and Arf1-dependent manner (19, 27, 29). Localization to the Golgi apparatus activates CERT-dependent transport of ceramide to the Golgi apparatus, thereby integrating sterol homeostasis and sphingomyelin synthesis (34). Procyclic T. brucei is able to synthesize ergosterol but also uses exogenous low-density lipoprotein as a source of cholesterol and lipids (9). Inhibition of sterol biosynthesis in Leishmania amazonesis causes a similar alteration of membrane-bound compartments as seen with depletion of TbPI4KIII-β (43). Inhibition of 3-hydroxy-3-methylglutaryl-coenzyme A reductase in procyclic T. brucei also results in a cytokinesis block and in the accumulation of abnormal membrane compartments (9). The similarities with the morphological defects in PI4P-depleted cells indicate that loss of TbPI4KIII-β may affect lipid homeostasis in addition to its effects on the secretory pathway.
As evident in scanning electron micrographs, the fat and round cells that result from TbPI4KIII-β depletion are actually twisted, and this structural abnormality progresses over time. This twisted phenotype may be caused by a defect in the microtubule cytoskeleton, which maintains the shape of the cell. The twist is seen in both nondividing and dividing cells, indicating that it is not cell cycle dependent. Some twisted cells are not fat or round and otherwise appear relatively normal, suggesting that the twist may begin before the cells become fat or round. Twisting begins at the posterior of the cell, where the flagellum originates from the flagellar pocket. It is possible that a defect in the structure, attachment, or movement of the flagellum causes the abnormal surface structure. Because the flagellum is physically attached to the surface of the cell in a helical pattern (38), abnormal flagellar attachment and subsequent movement may induce this helical pattern in the cell surface.
Depletion of TbPI4KIII-β in procyclic T. brucei results in the accumulation of cells in the G2/M stage of mitosis, suggesting that TbPI4KIII-β is required for cytokinesis. Unlike most of the RNAi phenotypes that disrupt cytokinesis, TbPI4KIII-β-depleted cells do not continue to replicate their DNA, indicating the possible activation of a cell cycle checkpoint. Knockdown of dynamin-like protein in procyclic parasites also led to a cell cycle block after a single round of mitosis, which is suggestive of a cytokinesis checkpoint associated with mitochondrial fission (8). Although TbPI4KIII-β RNAi-induced cells can replicate their DNA, they are unable to correctly position the kinetoplasts and the nuclei. Correct positioning of cytoskeletal elements and organelles is important for the progression through the cell cycle and cytokinesis (32). After nuclear division, the daughter kinetoplast and nucleus normally segregate towards the cell posterior, resulting in one kinetoplast being situated between the two nuclei (38). In TbPI4KIII-β-depleted cells, the kinetoplasts and the nuclei often segregate perpendicular to the normal lengthwise axis. Moreover, these cells often exhibit a detached daughter flagellum, indicative of a possible abnormality in the flagellar attachment zone. Establishment of the cleavage furrow requires correct positioning of the daughter flagellum and/or the flagellar attachment zone (21). Loss or impairment of these structural cues may explain the block in cytokinesis observed in TbPI4KIII-β-depleted cells.
PIs have been implicated in the reorganization of the cortical cytoskeleton during cytokinesis (26). In particular, the type III-β isoform of PtdIns 4-kinase was shown to be essential for cytokinesis in Saccharomyces cerevisiae (17, 44) and for cytokinesis during male meiosis in Drosophila melanogaster (6). At present, the mechanism of cleavage furrow formation in T. brucei is unclear, although it appears not to require an actin/myosin II contractile ring (14). The lack of protein markers for the T. brucei cleavage furrow prevents localization studies. However, our results indicate that PI synthesis is also likely to be important for cell division in T. brucei, and they provide some of the first evidence for PI signaling pathways involved in regulating cytokinesis in this parasite.
PIs have long been recognized as key regulators of membrane trafficking in yeast and mammalian cells, yet there are very few studies addressing the roles of these signaling molecules in protozoans. Our studies reveal essential roles for PI4P synthesis in Golgi maintenance, secretion, and cytokinesis of T. brucei, providing insight into the function of these signaling pathways in this parasite. Unexpected effects on morphology were also observed, leading to the observation of a twisted phenotype in cells lacking TbPI4KIII-β.
Acknowledgments
This work was supported by National Institutes of Health grant R01 AI34432 (to M.A.P.) and Welch Foundation grant I-1257 (to M.A.P.).
We thank Jay Bangs, Gwo-Shu Mary Lee, and Graham Warren for antibodies used in this study and Laurie Mueller for assistance with the electron microscopy.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Alexander, D. L., K. J. Schwartz, A. E. Balber, and J. D. Bangs. 2002. Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J. Cell Sci. 115:3253-3263. [DOI] [PubMed] [Google Scholar]
- 2.Allen, C. L., D. Goulding, and M. C. Field. 2003. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 22:4991-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Audhya, A., M. Foti, and S. D. Emr. 2000. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11:2673-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balla, A., and T. Balla. 2006. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 16:351-361. [DOI] [PubMed] [Google Scholar]
- 5.Balla, A., G. Tuymetova, A. Tsiomenko, P. Varnai, and T. Balla. 2005. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell 16:1282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brill, J. A., G. R. Hime, M. Scharer-Schuksz, and M. T. Fuller. 2000. A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development 127:3855-3864. [DOI] [PubMed] [Google Scholar]
- 7.Bruns, J. R., M. A. Ellis, A. Jeromin, and O. A. Weisz. 2002. Multiple roles for phosphatidylinositol 4-kinase in biosynthetic transport in polarized Madin-Darby canine kidney cells. J. Biol. Chem. 277:2012-2018. [DOI] [PubMed] [Google Scholar]
- 8.Chanez, A. L., A. B. Hehl, M. Engstler, and A. Schneider. 2006. Ablation of the single dynamin of T. brucei blocks mitochondrial fission and endocytosis and leads to a precise cytokinesis arrest. J. Cell Sci. 119:2968-2974. [DOI] [PubMed] [Google Scholar]
- 9.Coppens, I., and P. J. Courtoy. 1995. Exogenous and endogenous sources of sterols in the culture-adapted procyclic trypomastigotes of Trypanosoma brucei. Mol. Biochem. Parasitol. 73:179-188. [DOI] [PubMed] [Google Scholar]
- 10.Cross, G. A. 1975. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 71:393-417. [DOI] [PubMed] [Google Scholar]
- 11.de Barry, J., A. Janoshazi, J. L. Dupont, O. Procksch, S. Chasserot-Golaz, A. Jeromin, and N. Vitale. 2006. Functional implication of neuronal calcium sensor-1 and phosphoinositol 4-kinase-beta interaction in regulated exocytosis of PC12 cells. J. Biol. Chem. 281:18098-18111. [DOI] [PubMed] [Google Scholar]
- 12.de Graaf, P., E. E. Klapisz, T. K. Schulz, A. F. Cremers, A. J. Verkleij, and P. M. van Bergen en Henegouwen. 2002. Nuclear localization of phosphatidylinositol 4-kinase beta. J. Cell Sci. 115:1769-1775. [DOI] [PubMed] [Google Scholar]
- 13.De Matteis, M. A., A. Di Campli, and A. Godi. 2005. The role of the phosphoinositides at the Golgi complex. Biochim. Biophys. Acta 1744:396-405. [DOI] [PubMed] [Google Scholar]
- 14.Dutcher, S. K., B. Huang, and D. J. Luck. 1984. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 98:229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field, M. C., and M. Carrington. 2004. Intracellular membrane transport systems in Trypanosoma brucei. Traffic 5:905-913. [DOI] [PubMed] [Google Scholar]
- 16.Fruman, D. A., R. E. Meyers, and L. C. Cantley. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67:481-507. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Bustos, J. F., F. Marini, I. Stevenson, C. Frei, and M. N. Hall. 1994. PIK1, an essential phosphatidylinositol 4-kinase associated with the yeast nucleus. EMBO J. 13:2352-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gehrmann, T., and L. M. Heilmeyer, Jr. 1998. Phosphatidylinositol 4-kinases. Eur. J. Biochem. 253:357-370. [DOI] [PubMed] [Google Scholar]
- 19.Godi, A., A. Di Campli, A. Konstantakopoulos, G. Di Tullio, D. R. Alessi, G. S. Kular, T. Daniele, P. Marra, J. M. Lucocq, and M. A. De Matteis. 2004. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6:393-404. [DOI] [PubMed] [Google Scholar]
- 20.Godi, A., P. Pertile, R. Meyers, P. Marra, G. Di Tullio, C. Iurisci, A. Luini, D. Corda, and M. A. De Matteis. 1999. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1:280-287. [DOI] [PubMed] [Google Scholar]
- 21.Gull, K. 2003. Host-parasite interactions and trypanosome morphogenesis: a flagellar pocketful of goodies. Curr. Opin. Microbiol. 6:365-370. [DOI] [PubMed] [Google Scholar]
- 22.Hall, B. S., C. Gabernet-Castello, A. Voak, D. Goulding, S. K. Natesan, and M. C. Field. 2006. TbVps34, the trypanosome orthologue of Vps34, is required for Golgi complex segregation. J. Biol. Chem. 281:27600-27612. [DOI] [PubMed] [Google Scholar]
- 23.Hama, H., E. A. Schnieders, J. Thorner, J. Y. Takemoto, and D. B. DeWald. 1999. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274:34294-34300. [DOI] [PubMed] [Google Scholar]
- 24.He, C. Y., H. H. Ho, J. Malsam, C. Chalouni, C. M. West, E. Ullu, D. Toomre, and G. Warren. 2004. Golgi duplication in Trypanosoma brucei. J. Cell Biol. 165:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung, C. H., X. Qiao, P. T. Lee, and M. G. Lee. 2004. Clathrin-dependent targeting of receptors to the flagellar pocket of procyclic-form Trypanosoma brucei. Eukaryot. Cell 3:1004-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janetopoulos, C., and P. Devreotes. 2006. Phosphoinositide signaling plays a key role in cytokinesis. J. Cell Biol. 174:485-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagace, T. A., D. M. Byers, H. W. Cook, and N. D. Ridgway. 1997. Altered regulation of cholesterol and cholesteryl ester synthesis in Chinese-hamster ovary cells overexpressing the oxysterol-binding protein is dependent on the pleckstrin homology domain. Biochem. J. 326:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, M. G., B. E. Bihain, D. G. Russell, R. J. Deckelbaum, and L. H. Van der Ploeg. 1990. Characterization of a cDNA encoding a cysteine-rich cell surface protein located in the flagellar pocket of the protozoan Trypanosoma brucei. Mol. Cell. Biol. 10:4506-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine, T. P., and S. Munro. 1998. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 8:729-739. [DOI] [PubMed] [Google Scholar]
- 30.Levine, T. P., and S. Munro. 2002. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 12:695-704. [DOI] [PubMed] [Google Scholar]
- 31.Matthews, K. R. 2005. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 118:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKean, P. G., A. Baines, S. Vaughan, and K. Gull. 2003. Gamma-tubulin functions in the nucleation of a discrete subset of microtubules in the eukaryotic flagellum. Curr. Biol. 13:598-602. [DOI] [PubMed] [Google Scholar]
- 33.Ogbadoyi, E. O., D. R. Robinson, and K. Gull. 2003. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell 14:1769-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry, R. J., and N. D. Ridgway. 2006. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol. Biol. Cell 17:2604-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson, D. R., T. Sherwin, A. Ploubidou, E. H. Byard, and K. Gull. 1995. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J. Cell Biol. 128:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodrigues, C. O., D. A. Scott, and R. Docampo. 1999. Characterization of a vacuolar pyrophosphatase in Trypanosoma brucei and its localization to acidocalcisomes. Mol. Cell. Biol. 19:7712-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth, M. G. 2004. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 84:699-730. [DOI] [PubMed] [Google Scholar]
- 38.Sherwin, T., and K. Gull. 1989. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. London B 323:573-588. [DOI] [PubMed] [Google Scholar]
- 39.Sherwin, T., and K. Gull. 1989. Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell 57:211-221. [DOI] [PubMed] [Google Scholar]
- 40.Stebeck, C. E., and T. W. Pearson. 1994. Major surface glycoproteins of procyclic stage African trypanosomes. Exp. Parasitol. 78:432-436. [DOI] [PubMed] [Google Scholar]
- 41.Toker, A. 2002. Phosphoinositides and signal transduction. Cell Mol. Life Sci. 59:761-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toth, B., A. Balla, H. Ma, Z. A. Knight, K. M. Shokat, and T. Balla. 2006. Phosphatidylinositol 4-kinase IIIbeta regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J. Biol. Chem. 281:36369-36377. [DOI] [PubMed] [Google Scholar]
- 43.Vannier-Santos, M. A., A. Martiny, U. Lins, J. A. Urbina, V. M. Borges, and W. de Souza. 1999. Impairment of sterol biosynthesis leads to phosphorus and calcium accumulation in Leishmania acidocalcisomes. Microbiology 145:3213-3220. [DOI] [PubMed] [Google Scholar]
- 44.Walch-Solimena, C., and P. Novick. 1999. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1:523-525. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Y. J., J. Wang, H. Q. Sun, M. Martinez, Y. X. Sun, E. Macia, T. Kirchhausen, J. P. Albanesi, M. G. Roth, and H. L. Yin. 2003. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell 114:299-310. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 47.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 48.Wong, K., R. Meyers, and L. C. Cantley. 1997. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 272:13236-13241. [DOI] [PubMed] [Google Scholar]
- 49.Woodward, R., and K. Gull. 1990. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 95:49-57. [DOI] [PubMed] [Google Scholar]