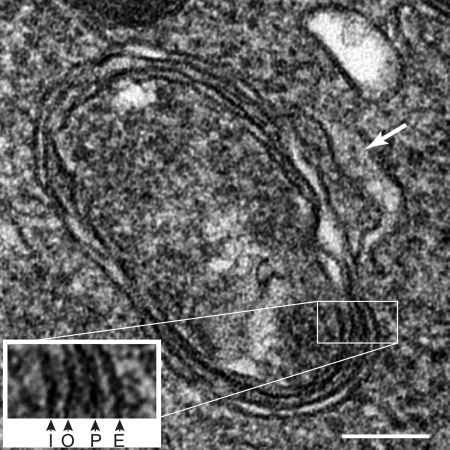
Almost 20 years ago, the first sequence was published from a 35-kb circular molecule found in Plasmodium falciparum, the most virulent of human malaria parasites (20). Consistent with the expected mitochondrial origin of the small genome, the sequence showed strong similarities to eubacterial small-subunit rRNA. But further sequencing found open reading frames similar to subunits of eubacterial RNA polymerases (22) and a large set of ribosomal proteins as well as large inverted repeats that carry rRNAs and additional genes, all features that are common for chloroplast genomes but uncommon for mitochondrial genomes (93). Clearly, the circular genome was something quite unexpected, but what was it? And if indeed it was a chloroplast-like genome, what was it doing in an obligate intracellular parasite? Ten years ago, the subcellular location of the mysterious DNA was identified in Toxoplasma gondii, a parasite related to Plasmodium (41, 50). It resides in an organelle surrounded by four membranes (three or four in Plasmodium) (reviewed in reference 89). The unexpected organelle genome now had a home. Similar multimembraned organelles had previously been described from electron micrographs of many members of phylum Apicomplexa (73), which includes the human pathogens Plasmodium and Toxoplasma as well as animal pathogens Eimeria and Theileria. A similar extrachromosomal DNA is present in each of these species that has been examined to date (35, 60). The apicomplexan Cryptosporidium parvum lacks such a structure and also lacks the corresponding genome, further strengthening the correlation (1). The organelle in which the circular genome resides has been dubbed the apicoplast, for apicomplexan plastid. Figure 1 shows an electron micrograph of the T. gondii apicoplast with its four membranes.
FIG. 1.
Transmission electron micrograph of a T. gondii apicoplast. The inset shows an enlargement of the four membranes visible in the image, marked according to their origin. I, derived from inner chloroplast membrane of cyanobacterial origin; O, derived from outer chloroplast membrane of cyanobacterial origin; P, periplastid membrane of algal plasma membrane origin; and E, outer membrane of apicomplexan endomembrane system origin. The bulge on the side of the outer membrane of the apicoplast is marked with an arrow. Bar = 100 nm.
The combination of the genome's chloroplast-like features and the multiple membranes of its resident organelle strongly suggests that apicomplexan parasites derived their apicoplasts from a secondary endosymbiosis and that the 35-kb DNA is a remnant chloroplast genome. Endosymbiosis has become a familiar concept in biology; one organism engulfs another, and rather than digesting the intruder, the host adopts it. Over time, the “visitor” loses many of its genes; some are lost to the nucleus of the host and others are lost altogether. The visitor becomes dependent on the host for many functions and is incapable of independent action. At the same time, the host becomes dependent on activities carried out by the visitor, now degenerated to an organelle. In primary endosymbiosis, the visitor is a prokaryote, while in secondary endosymbiosis, it is another eukaryote. In the case of the ancestral apicomplexan, it appears that an algal cell was engulfed and the chloroplast was retained. The algal nucleus has disappeared, although some of its genes likely have been transferred to the apicomplexan nucleus. The most easily recognized of such genes are those that encode plastid proteins, but genes encoding cytosolic proteins likely were also appropriated; several candidates have been proposed on the basis of “plant-like” features of the encoded proteins (32). Of the four membranes that bound the organelle, the inner two are thought to correspond to the algal chloroplast membranes. The next membrane, the periplastid membrane, arose from the plasma membrane of the algal cell, and the outermost membrane arose from the vacuole membrane formed during engulfment (Fig. 1, inset). The most direct evidence for secondary endosymbiosis comes from two groups of algae, the chlorarachniophytes and cryptomonads (61). In these organisms, a remnant yet functional nucleus (called the nucleomorph) still exists between the second and third membranes of the complex plastid.
Plasmodium falciparum (93), Toxoplasma gondii (www.toxodb.org; data provided by J. Kissinger and D. Roos), Eimeria (10), and Theileria (21) apicoplast genomes have been sequenced in their entirety, and gene content and organization are well conserved. But these sequences provide few clues as to the role of the apicoplast. Among the protein coding genes, only two have ascribed functions not associated with transcription and translation: ClpC, a molecular chaperone, and ORF470 (also known as ycf24 and sufB), which encodes a putative orthologue of a protein required for iron sulfur cluster biogenesis (17). The apicoplast is an essential organelle (13, 18). As discussed below, studies of apicoplast protein trafficking have allowed the identification of chloroplast-like proteins that are localized to the apicoplast, leading to predicted functions of the apicoplast (19, 65). These studies, along with further experimental analyses, indicate that enzymes for heme biosynthesis, type II fatty acid biosynthesis, and the deoxyxylulose-5-phosphate pathway for isoprenoid synthesis as well as accessory pathways are localized to the apicoplast (for examples, see references 36, 49, 85, and 88). Many apicoplast proteins and pathways have “plant-like” characteristics by virtue of their common cyanobacterial ancestor and are either absent or very diverged in the human host. Furthermore, some of these molecules have proven druggable in other systems. Hence, they are promising drug targets (24, 91, 92). The importance of the apicoplast is illustrated by the finding that over 500 P. falciparum proteins, about 9% of the predicted proteins encoded by the organism's genome, are suggested to be localized to the apicoplast (19, 65). Most are of unknown function.
Chloroplasts and secondary plastids: models and precedents for apicoplast targeting.
Studies of protein trafficking to chloroplasts and secondary plastids have proven to be valuable models for investigations into the mechanisms of protein import into the apicomplexan plastid. Import of proteins into the stroma (lumen) of higher plant chloroplasts has been studied extensively (reviewed in references 38, 66, and 75). Most chloroplast proteins are encoded in the nucleus and synthesized on cytosolic free ribosomes. The proteins are posttranslationally translocated across the chloroplast double membrane. Proteins destined for the chloroplast stroma possess a transit peptide at their amino terminus that is both necessary and sufficient for import into the chloroplast. This domain, which varies from 25 to over 100 amino acids, is cleaved when exposed to the stroma (67). Although plastid transit peptides do not bear specific motifs, they are rich in basic and hydroxylated amino acids, while being poor in acidic amino acids (8). They have little or no secondary structure. Overall, chloroplast transit peptides resemble mitochondrial targeting sequences, except that the latter typically form amphiphilic alpha helices. The ability of some transit peptides to direct import into the chloroplast is boosted by phosphorylation (87), which may also assist in discrimination from mitochondrial targeting sequences, and is enhanced by association with HSP70 (34) and 14-3-3 proteins (48).
The translocons of the inner and outer membranes of the chloroplast, Tic and Toc, respectively, mediate the import of stromal proteins. Their components include well-characterized receptors, transmembrane channel proteins, and accessory molecules (reviewed in references 4 and 75). Some of the key players are briefly described here. Transit peptides mediate import by interacting with the GTP binding proteins Toc159 and then Toc 34 (3, 71). The proteins traverse the outer membrane through a beta barrel channel formed by Toc75 (30). To cross the inner membrane, proteins must engage the Tic complex, a step thought to be facilitated by Tic22, a protein of the intermembrane space (75). The channel itself appears to be formed by Tic20 and Tic110 (29). Tic110 also provides a scaffold for stromal chaperones (42, 47). ATP hydrolysis by the stromal chaperone ClpC provides motive force for protein translocation (2, 58); the transit peptide is cleaved by the signal processing protease (67) and subsequently degraded. Together, these steps comprise the general import pathway of chloroplasts. Although the vast majority of stromal chloroplast proteins of plants appear to be directly imported from the cytosol via this pathway, there have been a few recent reports of plant proteins traversing the endomembrane system en route to the chloroplast stroma (55, 86). The trafficking of chloroplast membrane proteins will be considered below.
Organisms with complex plastids employ several different strategies to import proteins across their multiple membranes and into the lumen (reviewed in references 57 and 83). One feature found across diverse organisms is that the plastid targeting sequences is bipartite, comprised of a signal sequence and transit peptide. Thus, the first step in the trafficking of nucleus-encoded, plastid-targeted proteins is entry into the endoplasmic reticulum (ER). This fits well with the presumed origin of these plastids as organelles captured by and residing in the endocytic system. Indeed, in some organisms with four-membraned plastids, such as heterokonts, the outermost membrane of the organelle is studded with ribosomes (23). However, plastids from many organisms, including Apicomplexa, lack ribosomes on the outer membrane. In the case of Euglena and Gonyaulax, which nabbed plastids in separate endosymbiotic events, proteins reach the three-membraned organelles via vesicular trafficking through the Golgi (56, 78). In an interesting case of convergent evolution, most transit peptides that route proteins to plastids in these two species contain a hydrophobic stop-transfer sequence (16, 56). The topology is such that the majority of each protein is cytosolic as it traffics (78). How this orientation is reversed to generate a luminal protein is not known. Mechanisms addressing proteins into different compartments of secondary plastids have been explored in only a few organisms, such as the diatoms Thalassiosira and Phaeodactylum. In these species, sequences targeting proteins to the four-membraned plastid contain a six-amino-acid motif (ASA↓FAP) at the junction between the signal sequence and the transit peptide (39). Interestingly, when the invariant phenylalanine was mutated, reporter proteins bearing the targeting sequences localized to a blob-like structure (39) likely to be the periplastid space between the two central membranes (25). In organisms bearing secondary plastids, some candidate components of the Tic/Toc apparatus have been identified, although the complement is far from complete.
Trafficking of luminal proteins to the apicoplast.
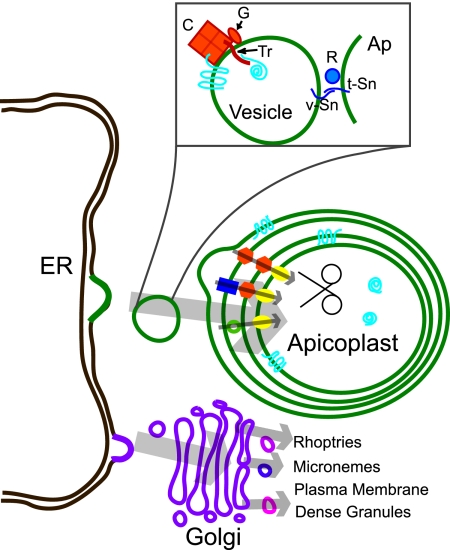
How do nucleus-encoded, apicoplast-targeted (NEAT) proteins make their way to the apicoplast and across its multiple membranes? The plastids described above provide many clues, but the apicoplast clearly has its own peculiarities. Since the discovery of the first apicoplast-targeted proteins, several models have been proposed to account for how proteins traffic to the plastid, including the apicoplast residing within the ER, vesicular trafficking through the Golgi to the apicoplast, and direct vesicular trafficking from ER to apicoplast, either terminally or as the first destination in the secretory system (see references 69, 83, and 89 and references therein). Figure 2 shows an updated model for the life journey of a luminal NEAT protein, which is based in part on experiments summarized in this review as well as previous models. Among the studies that led to this model were bioinformatic analysis coupled with transfection technology employing reporter proteins, such as green fluorescent protein (GFP) or epitope-tagged proteins. Unfortunately, no system has been developed which allows the recapitulation of apicoplast targeting in vitro.
FIG. 2.
Model for protein trafficking to the apicoplast. In the proposed model, vesicles bound for the apicoplast (green) bud from the ER or nuclear envelope (brown) and are distinct from vesicles targeted to the Golgi (violet). Vesicles derived from the latter move to other secretory destinations. The boxed enlargement shows some potential features of the apicoplast-destined vesicles, cargo, and apicoplast. Molecules involved in ER exit (orange) are proposed here to be a protein coat (C), a GTPase (G), and the putative transit peptide receptor (Tr). Molecules mediating fusion to the apicoplast (blue) are suggested to include an apicoplast-specific Rab (R) and v-SNAREs (v-Sn) and t-SNAREs (t-Sn) as well as others not shown. Apicoplast-destined proteins (turquoise) are proposed to interact directly with the coat or indirectly via the transit peptide receptor. Once the vesicles reach the apicoplast, they fuse with the outer membrane, releasing their contents to the space between the outer two membranes. Within the apicoplast, three models are shown to explain the translocation across the apicoplast membranes. The mediators in these models include Toc (orange hexagon) and Tic complexes (yellow ovals), ER-derived or mitochondrially derived translocators (blue rectangle), and vesicles between the periplastid and chloroplast outer membranes (green circles). Once proteins achieve the lumen of the apicoplast, the transit peptide domain is cleaved by the processing protease.
Shortly after the apicoplast was first identified as a plastid-derived organelle of apicomplexans, searches of draft genome sequences of T. gondii and P. falciparum identified proteins related to those found in chloroplasts. These included ribosomal proteins L28 and S9 as well as several enzymes involved in fatty acid biosynthesis (88). As with proteins destined for other secondary plastids, each of the predicted proteins contained a bipartite N-terminal extension that commenced with a 20- to 30-amino-acid signal peptide, followed by a region rich in basic amino acids, reminiscent of a chloroplast transit peptide. As expected, fusions of the bipartite extensions to GFP resulted in the localization of the reporter to the apicoplast in both P. falciparum (90) and T. gondii (15, 88) (Fig. 3, red).
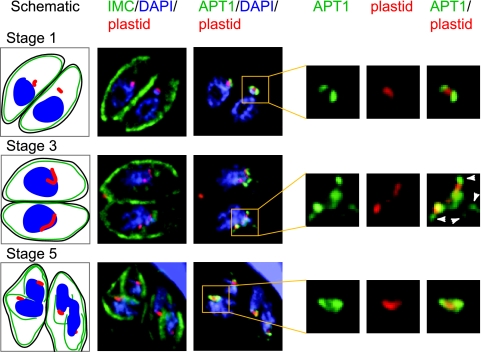
FIG. 3.
Regulation of membrane protein trafficking during the T. gondii cell cycle. Toxoplasma isolates were stably transfected with genes encoding HcRed bearing the signal and transit peptide of ACP (plastid lumen, red) or with the apicoplast membrane protein APT1 with four repeats of the HA tag appended to the C terminus. The parasites were fixed, probed with antisera directed against inner membrane complex protein 1 (revealed by anti-rabbit immunoglobulin G, shown in green) and anti-HA (also shown in green but detected with a different filter set), and stained with DAPI (4′,6′-diamidino-2-phenylindole) (blue). Samples were viewed on a DeltaVision deconvolution microscope. The schematics show the relative location of the plasma membrane (black), inner membrane complex (green), apicoplast (red), and nucleus (blue). Enlargements of the apicoplast are shown at right. Arrowheads indicate structures near the apicoplast that contain APT1-HA.
The signal sequences of NEAT proteins show no unusual features. When placed at the N terminus of GFP or other reporters in the absence of a transit peptide, these signal sequences can localize a normally cytosolic protein to the secretory pathway, typically resulting in secretion into the parasitophorous vacuole surrounding the organism (15, 27, 90). Conversely, the signal sequences from two different secreted proteins (erythrocyte binding antigen 175 or knob-associated, histidine-rich protein) can function in lieu of the signal sequence from an apicoplast protein to target proteins to the plastid (81). Furthermore, the signal sequence is necessary for the trafficking of these reporters to the apicoplast. When deleted from constructs, the reporter typically remains cytosolic (27, 90). Thus, both the signal and transit regions of the N-terminal extensions of apicoplast proteins are required for proper localization.
In one interesting case, the transit peptide of ribosomal protein S9 routed GFP to the mitochondrion (15, 94). Unlike most transit peptides, the S9 transit peptide is predicted to form an amphipathic alpha helix that could be recognized by the mitochondrial import machinery. As noted above, the phosphorylation of hydroxylated residues in at least some plant transit peptides is thought to provide further discrimination between the two types of targeting sequences (87). However, in contrast to plant transit peptides, hydroxylated residues (serine and threonine) are not essential for targeting mediated by the transit peptide of P. falciparum β-ketoacyl-acyl carrier protein synthase III (90). Such discrimination is provided in Apicomplexa by the presence of a signal peptide.
From the ER to the apicoplast.
Following the nascent NEAT protein on its journey, the signal sequence is rapidly cleaved upon import into the ER, revealing the apicoplast transit peptide at the new N terminus, like the transit peptides of chloroplasts. The exposed transit peptide is thought to be crucial for interaction with a hypothetical “receptor” in the ER lumen, which then allows selective trafficking to the apicoplast. Apicoplast transit peptides are considerably more complex than signal sequences. First, they show considerable variation in length, ranging from 24 to 150 amino acids. Second, large sections of some of the longer T. gondii transit peptides can be deleted without blocking the trafficking of proteins to the apicoplast (15, 27, 94). Nonetheless, they must all be specifically recognized to enable proper transport, much as the diverse transit peptides of plant chloroplast proteins must be recognized by the Toc complex. The amino acid composition of transit peptides in apicomplexans differ from one another in detail, with lysines being common in P. falciparum transit peptides, and arginines being common in T. gondii transit peptides. However, this difference does not appear to relate to the function of the transit peptide but rather reflects a dramatic difference in A/T bias of the two genomes (64).
Some elegant studies have defined essential characteristics of apicoplast transit peptides in P. falciparum and T. gondii. These studies demonstrate that basic residues within the transit peptide are important for faithful targeting. In P. falciparum, mutational analysis of the 24-amino-acid transit peptide of acyl carrier protein (ACP) showed that the substitution of acidic residues for two basic residues near the N terminus of the transit peptide abrogated targeting to the apicoplast, even though downstream basic residues were maintained (19). When alanines were substituted at the same positions, apicoplast targeting was not affected. In T. gondii, studies of the 63-amino-acid transit peptide of ACP showed that an overall positive charge of the transit peptide is essential for trafficking but that the exact position of the basic residues is not important (80). However, changes affecting basic residues near the N terminus of the transit peptide had a more significant impact on targeting than did those affecting basic residues located more distally. It appears that either lysine or arginine can provide the positive charges that are essential for apicoplast targeting (80). A second important feature of the P. falciparum ACP transit peptide is a putative Hsp70 binding site (19).
At least three hypothetical routes of transit from the ER to the apicoplast have been proposed, vesicular trafficking routed through the Golgi, direct vesicular trafficking from the ER, and nonvesicular trafficking (69, 83, 89). Several experiments argue against the Golgi model. First, the treatment of parasites with the Golgi inhibitor brefeldin A does not lead to a change in the steady-state distribution of NEAT reporters in either T. gondii (14) or P. falciparum (81). The exploitation of a ligand-regulated system that allows the accumulation and release of proteins from the ER (68) generated more convincing conclusions. Ligand-induced release of a NEAT reporter from the ER led to rapid localization to the apicoplast, which was not blocked by brefeldin A (14). Furthermore, low-temperature incubation, which blocks Golgi trafficking, did not block the apicoplast localization of the reporter upon ligand-induced release. With respect to the other models, we are unaware of any evidence that directly addresses trafficking of luminal proteins. However, as discussed below, recent data show that apicoplast membrane proteins undergo vesicular trafficking. Such a process would provide a means for proteins to exit the ER and enter the outermost intermembrane space of the plastid (Fig. 2).
If proteins trafficking to the apicoplast do not pass through the Golgi, the site of the canonical ER retrieval receptor, can mistargeted ER resident proteins be retrieved? Interestingly, when an ER retrieval sequence is appended to reporters bearing wild-type bipartite targeting sequences in T. gondii or in P. falciparum, the reporters are not retained in the ER but rather show solely an apicoplast localization (15, 81). However, when a mutated targeting sequence was used, the addition of the retrieval sequence allowed retention in the ER (9). These data suggest that a retrieval mechanism may be present, but wild-type apicoplast targeting is dominant over that retrieval. Such a retrieval mechanism, if it exists, likely acts at the outermost compartment of the apicoplast. Apicomplexans possess multiple isoforms of ER retrieval-like proteins, which could potentially function in different compartments (62).
As the properties of apicoplast proteins were recognized, bioinformatic tools were developed to predict NEAT proteins in P. falciparum (19, 95). Such algorithms have identified a large complement of proteins as likely to be localized to the apicoplast, although experimental verification exists for relatively few. In P. falciparum, microarray studies show that many of the transcripts corresponding to these predicted apicoplast proteins are expressed at a specific point in the erythrocytic cycle (7). Furthermore, if a NEAT-GFP fusion protein is expressed under the control of a heterologous promoter that drives expression early in the cycle (e.g., the ring stage), the fusion protein is not localized to the apicoplast, but rather is secreted into the parasitophorous vacuole. These data indicate that the machinery for the trafficking of apicoplast proteins from the ER to the plastid is not fully operational early in the erythrocytic cycle of P. falciparum (12). In these studies, the authors suggested that the secreted NEAT-GFP fusion protein can be subsequently localized from the vacuole to the apicoplast. Such a pathway has not been confirmed. To date, cell cycle-regulated targeting of apicoplast luminal reporters has not been observed in T. gondii.
Into the lumen.
A large gap in our knowledge concerns how proteins are translocated across the multiple membranes of the apicoplast. It has been assumed, based on the evolutionary origin of the organelle, that the translocon apparatus of the inner two membranes should resemble those of chloroplasts. However, the multiple polypeptides which make up the Tic and Toc complexes must be quite divergent in Apicomplexa, since only two candidates have been identified through homology-based searches, Tic22 and Toc34 (89). The relationships are relatively weak, and no studies localizing the candidate molecules to the apicoplast have yet been published.
If vesicular trafficking delivers proteins to the outermost intermembrane space (see below) and a Tic/Toc-like apparatus delivers them through the inner two membranes, mechanisms for crossing the periplastid membrane remain more obscure. One possibility is that the Toc apparatus is also present in this membrane (11); another is that vesicular trafficking occurs between the periplastid and “chloroplast” outer membrane (23, 83). It has also been suggested that the periplastid membrane has large pores or nonspecific protein transporters that allow passage across the membrane (43). A fourth suggestion is that a mitochondrial channel was usurped from the endosymbiont and relocalized to the apicoplast (6). However, recently an additional intriguing hypothesis has emerged (76). In ER-associated degradation (ERAD), proteins targeted for degradation are extruded out of the ER by complex machinery. Recently, multiple distantly related homologues of ERAD components were identified in several species bearing secondary plastids, Guillardia, Phaeodactylum, and P. falciparum (76). Several of the predicted proteins bear plastid targeting sequences, which in Phaeodactylum can target a reporter to the periplastid space. These intriguing data suggest that a system homologous to the ERAD ER export process serves to translocate proteins across the periplastid membrane.
Following import into the apicoplast, the transit peptide is clipped from the mature protein by a stromal processing peptidase related to those found in chloroplasts (84). Furthermore, cleavage of the transit peptide is quite delayed compared to the time of synthesis (14, 84). The lag in transit peptide processing could represent inefficient cleavage of the imported protein or a lengthy time course for import across all membranes (because of a generally inefficient import process or perhaps cell cycle regulation of certain steps). Some experiments using NEAT reporters show not only the presence of the transit peptide bearing reporter and the fully processed form but also the presence of an additional, intermediately sized species (27, 94). These data suggest that some processing may occur prior to complete import. Recently, falcilysin, a protease previously thought to be localized to the food vacuole of P. falciparum, was demonstrated to be predominantly localized to the apicoplast, where it appears to participate in the degradation of cleaved transit peptides (63).
Protein targeting to chloroplast membranes.
Unlike the single highway that proteins travel to the lumen of the chloroplast, protein targeting to the chloroplast membrane is like a branched and winding network of pathways, with distinctive features of trafficking for different proteins. For example, several proteins require GTP or ATP for insertion into the membrane, while others do not (31). Membrane insertion can require the Toc translocon or the assistance of other as-yet-unidentified proteins. Some proteins can simply insert into protein-free liposomes (72). Some proteins reach the membranes via the general import pathway, but others lack a cleavable N-terminal transit peptide. Generally, the former are inner membrane proteins and the latter are outer membrane proteins (45, 46, 74), but exceptions do occur (52, 54, 82). Only one outer membrane protein with an N-terminal transit peptide has been described, Toc75. It uses the general import pathway, and the first section of the transit peptide is cleaved in the lumen (82). Cleavage at a second site by a different membrane-associated protease releases Toc75 to the outer membrane (33). In outer membrane proteins, targeting information is typically contained within or adjacent to the transmembrane domains of the proteins (31). In a further twist, a methyltransferase and a ferrochelatase are targeted to two sites in the chloroplast, the inner membrane and the thylakoid membrane (the site of photosynthesis) (5, 79).
Targeting of apicoplast membrane proteins.
Analysis of the targeting of NEAT membrane proteins has lagged behind that of luminal proteins. Initial efforts to target a marker protein to the apicoplast membrane were unsuccessful (28), few candidate membrane proteins have been identified in homology-based searches, and attempts to directly identify membrane proteins have been hampered by the inability to isolate the apicoplast in T. gondii (28). Nonetheless, the four membranes that surround the apicoplast should have unique and essential proteins that are necessary for the proper functioning of the organelle. Based on the secondary endosymbiosis theory, the inner two membranes of the apicoplast should be populated with homologues of chloroplast membrane proteins. Such proteins should include components of the Tic/Toc machinery, transporters, and possibly proteins with metabolic functions. As noted above, BLAST searches of the apicomplexan databases have not yielded clear homologues of Tic or Toc proteins, with the possible exceptions of Tic22 (in P. falciparum) and Toc34 (89).
Certain transporters are postulated to be essential for apicoplast function based on known pathways within the organelle. The apicoplast phosphate translocator (APT) family was identified by high homology scores to chloroplast sugar phosphate translocators, and APT proteins are localized to apicoplast membranes in both T. gondii and P. falciparum (37, 53). These proteins belong to the plastidic phosphate translocator (pPT) family, integral membrane proteins that exchange inorganic phosphate for phosphorylated sugars or their derivatives (40). Based on the known metabolic pathways of apicoplasts, these molecules are likely to be involved in the import of 3-carbon phosphorylated compounds, such as phosphoenolpyruvate, required for the type II fatty acid synthesis, or glyceraldehyde-3-phosphate, a substrate for the isoprenoid synthesis pathway (65).
Plasmodium species possess two pPTs, whereas T. gondii has only one (37, 53). Like most proteins of the inner chloroplast membrane, P. falciparum APT2, also known as PfiPPT, possesses a predicted transit peptide, which is adjacent to its N-terminal signal sequence. As with such sequences from luminal NEAT proteins, this bipartite extension can route a reporter protein to the apicoplast of P. falciparum or T. gondii (37, 53). The transit sequence of endogenous PfAPT2 is processed in vivo, suggesting exposure to the stromal processing peptidase (53). Additionally, protease protection experiments indicate that PfAPT2 is protected by a membrane. The authors conclude that this molecule most likely resides in the innermost apicoplast membrane (53).
In contrast to APT2s, none of the APT1s identified thus far in a variety of Apicomplexa possess a canonical apicoplast targeting sequence. Additionally, no evidence of processing has been seen. In studies of P. falciparum organellar preparations, the molecule was found to be protease sensitive, suggesting that it is displayed on the outer membrane of the apicoplast (53). Thus, the two different APTs of P. falciparum appear to reside in different membranes. However, it is difficult to exclude the possibility that one or more membranes contain both transporters or that each may populate more than one membrane. In fact, in T. gondii, which has only a single APT gene, immunoelectron microscopy of epitope-tagged TgAPT1 showed that it resides in multiple membranes, most likely at least three and possibly all four (37). The mechanisms by which the protein is targeted to multiple membranes is not known, although precedent exists for targeting to multiple chloroplast membranes (see above).
The lack of an N-terminal signal sequence on APT1 raises the question of whether it traffics through the ER, as do luminal proteins and most likely APT2. The addition of the ACP signal sequence to the N terminus of TgAPT1 resulted in proper targeting to the apicoplast. This indicates that TgAPT1 is capable of trafficking to the apicoplast after cotranslational import into the ER (37). Some proteins rely on a transmembrane domain as an internal signal sequence to enter the ER, and APT1, with its multiple transmembrane domains, is likely to do the same. Upon entry into the ER, it is probable that apicoplast-targeted membrane proteins are embedded in the ER membrane where they can interact with either luminal or cytosolic proteins that may be important for their trafficking. The sequences of APT1, which correspond functionally to a transit peptide, have not been identified. APT1 might bind the putative transit peptide receptor in the ER lumen. However, based on the mechanisms of targeting of membrane proteins from the ER to other destinations in the secretory system (51, 59, 70), it seems just as feasible that APT1 is targeted to the apicoplast via interactions with cytosolic factors rather than the luminally disposed putative transit peptide receptor.
Recent work shows that apicoplast targeting of TgAPT1 bearing a hemagglutinin (HA) epitope tag is regulated during the cell cycle (37). This contrasts with targeting of luminal proteins in T. gondii, which thus far appears to be cell cycle independent (14, 15, 27, 88). However, as noted above, the expression and trafficking of apicoplast luminal proteins in P. falciparum are modulated during the erythrocytic cycle (12). The T. gondii cell cycle can be divided into six stages based on the apicoplast shape and its location relative to the nucleus as well as the apicoplast and nuclear division and inner membrane complex formation (77). The relationship of these stages to classical stages of the cell cycle has not been fully elucidated. In stage 1, TgAPT1-HA mostly surrounds the apicoplast luminal marker (Fig. 3). As the apicoplast elongates, TgAPT1-HA is localized to beads and tubules along the length of the apicoplast as well as extending from it (stage 3). Following plastid division, TgAPT1-HA staining is again circumplastid. When the heterologous DHFR promoter was used in lieu of the endogenous promoter, much more extraplastid staining was observed (37), although protein abundance was not increased. These data imply that the timing of expression is critical for appropriate trafficking. We suggest that the expression or function of the machinery for the targeting of membrane proteins to the plastid is enhanced during the time of plastid enlargement. Whether similar cell cycle-associated changes are seen in trafficking of apicoplast membrane proteins routed by virtue of a signal and transit sequence remains to be established.
The “bead and tubule” staining pattern of TgAPT1-HA was further explored by immunoelectron microscopy (37). These studies showed, for the first time, the presence of vesicles bearing an apicoplast protein. Labeled protrusions of the apicoplast outer membrane that are similar in size and electron density to the vesicles bearing TgAPT1 were also observed, suggestive of vesicular fusion with the outer apicoplast membrane (small bulges can also be seen on transmission electron microscopy, such as in Fig. 1). These studies also showed some labeling of the nuclear envelope, which is the intermediate compartment for ER to Golgi trafficking in T. gondii (26). However, no labeling of the Golgi itself was seen, suggesting parallels to the targeting of luminal proteins. These APT proteins are the first apicoplast membrane proteins that have been identified; clearly, an analysis of additional membrane proteins will help us better understand the mechanisms of targeting to the apicoplast membranes.
Model for apicoplast targeting.
Based on the above observations, previous models, and inferences from other eukaryotic systems and to provide testable predictions, we developed the following “straw man” model of protein trafficking to the apicoplast (Fig. 2). Apicoplast proteins are cotranslationally imported into the ER, and their import can be directed either by an N-terminal signal sequence or, in the case of some membrane proteins, by an internal signal sequence. The transit peptide domain of luminal proteins is bound by a specific (hypothetical) receptor protein at the ER exit sites and the receptor directs packaging of these proteins into vesicles, possibly via an interaction with a vesicle coat protein. Signals directing appropriate packaging of apicoplast membrane proteins are facing the cytosol, where they too interact with such a coat protein. In this model, the trafficking of both soluble and membrane proteins is independent of the Golgi and vesicles bearing such proteins bud off the ER and are directly escorted to the apicoplast. The vesicles then fuse with the outer membrane of the apicoplast, utilizing proteins homologous to those involved in trafficking to other destinations in the secretory system, e.g., SNAREs, SNAP25, and Rabs (44). The trafficking of membrane proteins to the apicoplast occurs predominantly at the time of plastid growth, but a modest level of soluble proteins can be mobilized to the apicoplast at other times due to the trafficking system's larger capacity for soluble proteins. From the initial vesicular fusion event, specific mechanisms which remain unknown serve to distribute proteins to the various membranes and to the lumen of the organelle (several possibilities are diagrammed on the internal apicoplast membranes in Fig. 2). Once the protein arrives in the lumen, its transit peptide is cleaved by the processing protease.
Like most models, this model raises more questions than it answers. Among the most important are the following. What is the transit peptide receptor? How do the vesicles carrying apicoplast membrane proteins (and luminal proteins) form? How do proteins cross the periplastid membrane? Have the pore proteins of the Toc and Tic complexes been displaced by unrelated transmembrane proteins or simply diverged to a point where they are not recognized in bioinformatics searches? Many more questions can be listed, but all await future experimentation. Clearly, we have only begun to understand protein transport into this Achilles' heel of apicomplexan parasites.
Acknowledgments
This work was supported by NIH R01 AI50506.
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441-445. [DOI] [PubMed] [Google Scholar]
- 2.Akita, M., E. Nielsen, and K. Keegstra. 1997. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136:983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, T., M. Jelic, A. Vojta, A. Radunz, J. Soll, and E. Schleiff. 2004. Preprotein recognition by the Toc complex. EMBO J. 23:520-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bédard, J., and P. Jarvis. 2005. Recognition and envelope translocation of chloroplast preproteins. J. Exp. Bot. 56:2287-2320. [DOI] [PubMed] [Google Scholar]
- 5.Block, M. A., A. K. Tewari, C. Albrieux, E. Marechal, and J. Joyard. 2002. The plant S-adenosyl-l-methionine:Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. Eur. J. Biochem. 269:240-248. [DOI] [PubMed] [Google Scholar]
- 6.Bodyl, A. 2004. Evolutionary origin of a preprotein translocase in the periplastid membrane of complex plastids: a hypothesis. Plant Biol. (Stuttgart) 6:513-518. [DOI] [PubMed] [Google Scholar]
- 7.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruce, B. D. 2001. The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim. Biophys. Acta 1541:2-21. [DOI] [PubMed] [Google Scholar]
- 9.Brydges, S. D., and V. B. Carruthers. 2003. Mutation of an unusual mitochondrial targeting sequence of SODB2 produces multiple targeting fates in Toxoplasma gondii. J. Cell Sci. 116:4675-4685. [DOI] [PubMed] [Google Scholar]
- 10.Cai, X., A. L. Fuller, L. R. McDougald, and G. Zhu. 2003. Apicoplast genome of the coccidian Eimeria tenella. Gene 321:39-46. [DOI] [PubMed] [Google Scholar]
- 11.Cavalier-Smith, T. 2003. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae). Philos. Trans. R. Soc. Lond. B 358:109-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheresh, P., T. Harrison, H. Fujioka, and K. Haldar. 2002. Targeting the malarial plastid via the parasitophorous vacuole. J. Biol. Chem. 277:16265-16277. [DOI] [PubMed] [Google Scholar]
- 13.Dahl, E. L., J. L. Shock, B. R. Shenai, J. Gut, J. L. DeRisi, and P. J. Rosenthal. 2006. Tetracyclines specifically target the apicoplast of the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 50:3124-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeRocher, A., B. Gilbert, J. E. Feagin, and M. Parsons. 2005. Dissection of brefeldin A-sensitive and -insensitive steps in apicoplast protein targeting. J. Cell Sci. 118:565-574. [DOI] [PubMed] [Google Scholar]
- 15.DeRocher, A., C. B. Hagen, J. E. Froehlich, J. E. Feagin, and M. Parsons. 2000. Analysis of targeting sequences demonstrates that trafficking to the Toxoplasma gondii plastid branches off the secretory system. J. Cell Sci. 113:3969-3977. [DOI] [PubMed] [Google Scholar]
- 16.Durnford, D. G., and M. W. Gray. 2006. Analysis of Euglena gracilis plastid-targeted proteins reveals different classes of transit sequences. Eukaryot. Cell 5:2079-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis, K. E., B. Clough, J. W. Saldanha, and R. J. Wilson. 2001. Nifs and Sufs in malaria. Mol. Microbiol. 41:973-981. [DOI] [PubMed] [Google Scholar]
- 18.Fichera, M. E., and D. S. Roos. 1997. A plastid organelle as a drug target in apicomplexan parasites. Nature 390:407-409. [DOI] [PubMed] [Google Scholar]
- 19.Foth, B. J., S. A. Ralph, C. J. Tonkin, N. S. Struck, M. Fraunholz, D. S. Roos, A. F. Cowman, and G. I. McFadden. 2003. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science 299:705-708. [DOI] [PubMed] [Google Scholar]
- 20.Gardner, M. J., P. A. Bates, I. T. Ling, D. J. Moore, S. McCready, M. B. Gunasekera, R. J. Wilson, and D. H. Williamson. 1988. Mitochondrial DNA of the human malarial parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 31:11-17. [DOI] [PubMed] [Google Scholar]
- 21.Gardner, M. J., R. Bishop, T. Shah, E. P. de Villiers, J. M. Carlton, N. Hall, Q. Ren, I. T. Paulsen, A. Pain, M. Berriman, R. J. Wilson, S. Sato, S. A. Ralph, D. J. Mann, Z. Xiong, S. J. Shallom, J. Weidman, L. Jiang, J. Lynn, B. Weaver, A. Shoaibi, A. R. Domingo, D. Wasawo, J. Crabtree, J. R. Wortman, B. Haas, S. V. Angiuoli, T. H. Creasy, C. Lu, B. Suh, J. C. Silva, T. R. Utterback, T. V. Feldblyum, M. Pertea, J. Allen, W. C. Nierman, E. L. Taracha, S. L. Salzberg, O. R. White, H. A. Fitzhugh, S. Morzaria, J. C. Venter, C. M. Fraser, and V. Nene. 2005. Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science 309:134-137. [DOI] [PubMed] [Google Scholar]
- 22.Gardner, M. J., D. H. Williamson, and R. J. M. Wilson. 1991. A circular DNA in malaria parasites encodes an RNA polymerase like that of prokaryotes and chloroplasts. Mol. Biochem. Parasitol. 44:115-124. [DOI] [PubMed] [Google Scholar]
- 23.Gibbs, S. P. 1979. The route of entry of cytoplasmically synthesized proteins into chloroplasts of algae possessing chloroplast ER. J. Cell Sci. 35:253-266. [DOI] [PubMed] [Google Scholar]
- 24.Goodman, C. D., and G. I. McFadden. 2007. Fatty acid biosynthesis as a drug target in apicomplexan parasites. Curr. Drug Targets 8:15-30. [DOI] [PubMed] [Google Scholar]
- 25.Gould, S. B., M. S. Sommer, P. G. Kroth, G. H. Gile, P. J. Keeling, and U. G. Maier. 2006. Nucleus-to-nucleus gene transfer and protein retargeting into a remnant cytoplasm of cryptophytes and diatoms. Mol. Biol. Evol. 23:2413-2422. [DOI] [PubMed] [Google Scholar]
- 26.Hager, K. M., B. Striepen, L. G. Tilney, and D. S. Roos. 1999. The nuclear envelope serves as an intermediary between the ER and Golgi complex in the intracellular parasite Toxoplasma gondii. J. Cell Sci. 112:2631-2638. [DOI] [PubMed] [Google Scholar]
- 27.Harb, O. S., B. Chatterjee, M. J. Fraunholz, M. J. Crawford, M. Nishi, and D. S. Roos. 2004. Multiple functionally redundant signals mediate targeting to the apicoplast in the apicomplexan parasite Toxoplasma gondii. Eukaryot. Cell 3:663-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He, C. Y., B. Striepen, C. H. Pletcher, J. M. Murray, and D. S. Roos. 2001. Targeting and processing of nuclear-encoded apicoplast proteins in plastid segregation mutants of Toxoplasma gondii. J. Biol. Chem. 276:28436-28442. [DOI] [PubMed] [Google Scholar]
- 29.Heins, L., A. Mehrle, R. Hemmler, R. Wagner, M. Kuchler, F. Hormann, D. Sveshnikov, and J. Soll. 2002. The preprotein conducting channel at the inner envelope membrane of plastids. EMBO J. 21:2616-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinnah, S. C., K. Hill, R. Wagner, T. Schlicher, and J. Soll. 1997. Reconstitution of a chloroplast protein import channel. EMBO J. 16:7351-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann, N. R., and S. M. Theg. 2005. Chloroplast outer membrane protein targeting and insertion. Trends Plant Sci. 10:450-457. [DOI] [PubMed] [Google Scholar]
- 32.Huang, J., N. Mullapudi, T. Sicheritz-Ponten, and J. C. Kissinger. 2004. A first glimpse into the pattern and scale of gene transfer in Apicomplexa. Int. J. Parasitol. 34:265-274. [DOI] [PubMed] [Google Scholar]
- 33.Inoue, K., A. J. Baldwin, R. L. Shipman, K. Matsui, S. M. Theg, and M. Ohme-Takagi. 2005. Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J. Cell Biol. 171:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivey, R. A., III, C. Subramanian, and B. D. Bruce. 2000. Identification of a Hsp70 recognition domain within the rubisco small subunit transit peptide. Plant Physiol. 122:1289-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeffries, A. C., and A. M. Johnson. 1996. The growing importance of the plastid-like DNAs of the Apicomplexa. Int. J. Parasitol. 26:1139-1150. [DOI] [PubMed] [Google Scholar]
- 36.Jomaa, H., J. Wiesner, S. Sanderbrand, B. Altincicek, C. Weidemeyer, M. Hintz, I. Türbachova, M. Eberl, J. Zeidler, H. K. Lichtenthaler, D. Soldati, and E. Beck. 1999. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalerial drugs. Science 285:1573-1576. [DOI] [PubMed] [Google Scholar]
- 37.Karnataki, A., A. DeRocher, I. Coppens, C. Nash, J. E. Feagin, and M. Parsons. 2007. Cell-cycle regulated targeting of an apicoplast membrane protein that lacks a canonical targeting sequence. Mol. Microbiol. 63:1653-1668. [DOI] [PubMed] [Google Scholar]
- 38.Kessler, F., and D. J. Schnell. 2006. The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic 7:248-257. [DOI] [PubMed] [Google Scholar]
- 39.Kilian, O., and P. G. Kroth. 2005. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 41:175-183. [DOI] [PubMed] [Google Scholar]
- 40.Knappe, S., U. I. Flugge, and K. Fischer. 2003. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 131:1178-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Köhler, S., C. F. Delwiche, P. W. Denny, L. G. Tilney, P. Webster, R. J. Wilson, J. D. Palmer, and D. S. Roos. 1997. A plastid of probable green algal origin in apicomplexan parasites. Science 275:1485-1489. [DOI] [PubMed] [Google Scholar]
- 42.Kouranov, A., and D. J. Schnell. 1997. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139:1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kroth, P., and H. Strotman. 1999. Diatom plastids: secondary endocytobiosis, plastid genome, and protein import. Physiol. Plant. 107:136-141. [Google Scholar]
- 44.Lee, M. C., E. A. Miller, J. Goldberg, L. Orci, and R. Schekman. 2004. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20:87-123. [DOI] [PubMed] [Google Scholar]
- 45.Li, M., and D. J. Schnell. 2006. Reconstitution of protein targeting to the inner envelope membrane of chloroplasts. J. Cell Biol. 175:249-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lübeck, J., L. Heins, and J. Soll. 1997. A nuclear-coded chloroplastic inner envelope membrane protein uses a soluble sorting intermediate upon import into the organelle. J. Cell Biol. 137:1279-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lübeck, J., J. Soll, M. Akita, E. Nielsen, and K. Keegstra. 1996. Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J. 15:4230-4238. [PMC free article] [PubMed] [Google Scholar]
- 48.May, T., and J. Soll. 2000. 14-3-3 Proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazumdar, J., H. Wilson, K. Masek, A. Hunter, and B. Striepen. 2006. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. USA 103:13192-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McFadden, G. I., M. E. Reith, J. Munholland, and N. Lang-Unnasch. 1996. Plastid in human parasites. Nature 381:482. [DOI] [PubMed] [Google Scholar]
- 51.Michelsen, K., H. Yuan, and B. Schwappach. 2005. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 6:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miras, S., D. Salvi, M. Ferro, D. Grunwald, J. Garin, J. Joyard, and N. Rolland. 2002. Non-canonical transit peptide for import into the chloroplast. J. Biol. Chem. 277:47770-47778. [DOI] [PubMed] [Google Scholar]
- 53.Mullin, K. A., L. Lim, S. A. Ralph, T. P. Spurck, E. Handman, and G. I. McFadden. 2006. Membrane transporters in the relict plastid of malaria parasites. Proc. Natl. Acad. Sci. USA 103:9572-9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nada, A., and J. Soll. 2004. Inner envelope protein 32 is imported into chloroplasts by a novel pathway. J. Cell Sci. 117:3975-3982. [DOI] [PubMed] [Google Scholar]
- 55.Nanjo, Y., H. Oka, N. Ikarashi, K. Kaneko, A. Kitajima, T. Mitsui, F. J. Munoz, M. Rodriguez-Lopez, E. Baroja-Fernandez, and J. Pozueta-Romero. 2006. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell 18:2582-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nassoury, N., M. Cappadocia, and D. Morse. 2003. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J. Cell Sci. 116:2867-2874. [DOI] [PubMed] [Google Scholar]
- 57.Nassoury, N., and D. Morse. 2005. Protein targeting to the chloroplasts of photosynthetic eukaryotes: getting there is half the fun. Biochim. Biophys. Acta 1743:5-19. [DOI] [PubMed] [Google Scholar]
- 58.Nielsen, E., M. Akita, J. Davila-Aponte, and K. Keegstra. 1997. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16:935-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishimura, N., S. Bannykh, S. Slabough, J. Matteson, Y. Altschuler, K. Hahn, and W. E. Balch. 1999. A di-acidic (DXE) code directs concentration of cargo during export from the endoplasmic reticulum. J. Biol. Chem. 274:15937-15946. [DOI] [PubMed] [Google Scholar]
- 60.Oborník, M., M. Jirku, J. R. Slapeta, D. Modry, B. Koudela, and J. Lukes. 2002. Notes on coccidian phylogeny, based on the apicoplast small subunit ribosomal DNA. Parasitol. Res. 88:360-363. [DOI] [PubMed] [Google Scholar]
- 61.Palmer, J. D., and C. F. Delwiche. 1996. Second-hand chloroplasts and the case of the disappearing nucleus. Proc. Natl. Acad. Sci. USA 93:7432-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfluger, S. L., H. V. Goodson, J. M. Moran, C. J. Ruggiero, X. Ye, K. M. Emmons, and K. M. Hager. 2005. Receptor for retrograde transport in the apicomplexan parasite Toxoplasma gondii. Eukaryot. Cell 4:432-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ponpuak, M., M. Klemba, M. Park, I. Y. Gluzman, G. K. Lamppa, and D. E. Goldberg. 2007. A role for falcilysin in transit peptide degradation in the Plasmodium falciparum apicoplast. Mol. Microbiol. 63:314-334. [DOI] [PubMed] [Google Scholar]
- 64.Ralph, S. A., B. J. Foth, N. Hall, and G. I. McFadden. 2004. Evolutionary pressures on apicoplast transit peptides. Mol. Biol. Evol. 21:2183-2194. [DOI] [PubMed] [Google Scholar]
- 65.Ralph, S. A., G. G. van Dooren, R. F. Waller, M. J. Crawford, M. J. Fraunholz, B. J. Foth, C. J. Tonkin, D. S. Roos, and G. I. McFadden. 2004. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2:203-216. [DOI] [PubMed] [Google Scholar]
- 66.Reumann, S., K. Inoue, and K. Keegstra. 2005. Evolution of the general protein import pathway of plastids. Mol. Membr. Biol. 22:73-86. [DOI] [PubMed] [Google Scholar]
- 67.Richter, S., and G. K. Lamppa. 1998. A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA 95:7463-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rivera, V. M., X. Wang, S. Wardwell, N. L. Courage, A. Volchuk, T. Keenan, D. A. Holt, M. Gilman, L. Orci, F. J. Cerasoli, J. E. Rothman, and T. Clackson. 2000. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science 287:826-830. [DOI] [PubMed] [Google Scholar]
- 69.Roos, D. S., M. J. Crawford, R. G. Donald, M. Fraunholz, O. S. Harb, C. Y. He, J. C. Kissinger, M. K. Shaw, and B. Striepen. 2002. Mining the Plasmodium genome database to define organellar function: what does the apicoplast do? Philos. Trans. R. Soc. Lond. B 357:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sato, K., and A. Nakano. 2002. Emp47p and its close homolog Emp46p have a tyrosine-containing endoplasmic reticulum exit signal and function in glycoprotein secretion in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2518-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schleiff, E., J. Soll, M. Kuchler, W. Kuhlbrandt, and R. Harrer. 2003. Characterization of the translocon of the outer envelope of chloroplasts. J. Cell Biol. 160:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schleiff, E., R. Tien, M. Salomon, and J. Soll. 2001. Lipid composition of outer leaflet of chloroplast outer envelope determines topology of OEP7. Mol. Biol. Cell 12:4090-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siddall, M. E. 1992. Hohlzylinders. Parasitol. Today 8:90-91. [DOI] [PubMed] [Google Scholar]
- 74.Silva-Filho, M. D., M. C. Wieërs, U. I. Flügge, F. Chaumont, and M. Boutry. 1997. Different in vitro and in vivo targeting properties of the transit peptide of a chloroplast envelope inner membrane protein. J. Biol. Chem. 272:15264-15269. [DOI] [PubMed] [Google Scholar]
- 75.Soll, J., and E. Schleiff. 2004. Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 5:198-208. [DOI] [PubMed] [Google Scholar]
- 76.Sommer, M. S., S. B. Gould, P. Lehmann, A. Gruber, J. M. Przyborski, and U. G. Maier. 2007. Der1-mediated pre-protein import into the periplastid compartment of chromalveolates? Mol. Biol. Evol. 24:918-928. [DOI] [PubMed] [Google Scholar]
- 77.Striepen, B., M. J. Crawford, M. K. Shaw, L. G. Tilney, F. Seeber, and D. S. Roos. 2000. The plastid of Toxoplasma gondii is divided by association with the centrosomes. J. Cell Biol. 151:1423-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sulli, C., Z. Fang, U. Muchhal, and S. D. Schwartzbach. 1999. Topology of Euglena chloroplast protein precursors within endoplasmic reticulum to Golgi to chloroplast transport vesicles. J. Biol. Chem. 274:457-463. [DOI] [PubMed] [Google Scholar]
- 79.Suzuki, T., T. Masuda, D. P. Singh, F. C. Tan, T. Tsuchiya, H. Shimada, H. Ohta, A. G. Smith, and K. Takamiya. 2002. Two types of ferrochelatase in photosynthetic and nonphotosynthetic tissues of cucumber: their difference in phylogeny, gene expression, and localization. J. Biol. Chem. 277:4731-4737. [DOI] [PubMed] [Google Scholar]
- 80.Tonkin, C. J., D. S. Roos, and G. I. McFadden. 2006. N-terminal positively charged amino acids, but not their exact position, are important for apicoplast transit peptide fidelity in Toxoplasma gondii. Mol. Biochem. Parasitol. 150:192-200. [DOI] [PubMed] [Google Scholar]
- 81.Tonkin, C. J., N. S. Struck, K. A. Mullin, L. M. Stimmler, and G. I. McFadden. 2006. Evidence for Golgi-independent transport from the early secretory pathway to the plastid in malaria parasites. Mol. Microbiol. 61:614-630. [DOI] [PubMed] [Google Scholar]
- 82.Tranel, P. J., and K. Keegstra. 1996. A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell 8:2093-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Dooren, G. G., S. D. Schwartzbach, T. Osafune, and G. I. McFadden. 2001. Translocation of proteins across the multiple membranes of complex plastids. Biochim. Biophys. Acta 1541:34-53. [DOI] [PubMed] [Google Scholar]
- 84.van Dooren, G. G., V. Su, M. C. D'Ombrain, and G. I. McFadden. 2002. Processing of an apicoplast leader sequence in Plasmodium falciparum and the identification of a putative leader cleavage enzyme. J. Biol. Chem. 277:23612-23619. [DOI] [PubMed] [Google Scholar]
- 85.Varadharajan, S., B. K. Sagar, P. N. Rangarajan, and G. Padmanaban. 2004. Localization of ferrochelatase in Plasmodium falciparum. Biochem. J. 384:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Villarejo, A., S. Buren, S. Larsson, A. Dejardin, M. Monne, C. Rudhe, J. Karlsson, S. Jansson, P. Lerouge, N. Rolland, G. von Heijne, M. Grebe, L. Bako, and G. Samuelsson. 2005. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 12:1224-1231. [DOI] [PubMed] [Google Scholar]
- 87.Waegemann, K., and J. Soll. 1996. Phosphorylation of the transit sequence of chloroplast precursor proteins. J. Biol. Chem. 271:6545-6554. [DOI] [PubMed] [Google Scholar]
- 88.Waller, R. F., P. J. Keeling, R. G. K. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A. F. Cowman, G. S. Besra, D. S. Roos, and G. McFadden. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 95:12352-12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Waller, R. F., and G. I. McFadden. 2005. The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr. Issues Mol. Biol. 7:57-79. [PubMed] [Google Scholar]
- 90.Waller, R. F., M. B. Reed, A. F. Cowman, and G. I. McFadden. 2000. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 19:1794-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wiesner, J., and H. Jomaa. 2007. Isoprenoid biosynthesis of the apicoplast as drug target. Curr. Drug Targets 8:3-13. [DOI] [PubMed] [Google Scholar]
- 92.Wiesner, J., and F. Seeber. 2005. The plastid-derived organelle of protozoan human parasites as a target of established and emerging drugs. Expert Opin. Ther. Targets 9:23-44. [DOI] [PubMed] [Google Scholar]
- 93.Wilson, R. J., P. W. Denny, P. R. Preiser, K. Rangachari, K. Roberts, A. Roy, A. Whyte, M. Strath, D. J. Moore, P. W. Moore, and D. H. Williamson. 1996. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 261:155-172. [DOI] [PubMed] [Google Scholar]
- 94.Yung, S., T. R. Unnasch, and N. Lang-Unnasch. 2001. Analysis of apicoplast targeting and transit peptide processing in Toxoplasma gondii by deletional and insertional mutagenesis. Mol. Biochem. Parasitol. 118:11-21. [DOI] [PubMed] [Google Scholar]
- 95.Zuegge, J., S. Ralph, M. Schmuker, G. I. McFadden, and G. Schneider. 2001. Deciphering apicoplast targeting signals—feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene 280:19-26. [DOI] [PubMed] [Google Scholar]