Retroviruses comprise a distinct group of enveloped RNA viruses that replicate by reverse transcribing their RNA genomes to form a DNA copy within a viral core particle. The DNA copy, or cDNA, is then integrated into the host genome. The integrated proviral DNA is transcribed by RNA polymerase II (Pol II) to produce polyadenylated mRNAs that are translated into viral proteins and also packaged into assembling core particles in the cytoplasm or at the plasma membrane. Core particles acquire a host-derived envelope as they bud out of the cell. The membrane of retroviral virions fuses with the membrane of new host cells, and the replication cycle begins again (Fig. 1). Retroviruses are obligate parasites with small genomes and a complex mode of replication; thus, they are reliant on a multitude of host factors for replication. At the same time, mammalian cells have evolved a variety of mechanisms to impede retroviruses, which are potentially pathogenic or mutagenic to their hosts. Significant progress toward identifying mammalian host factors that regulate the activities of retroviruses has been made in recent years. A number of dominant retroviral resistance factors, including the APOBEC3 family of cytosine deaminases, the mouse Fv1 restriction factor, and the primate antiviral factor TRIM5α, have been uncovered using genetic approaches (reviewed in references 7 and 49). In addition, a diverse collection of recessive genes that promote replication at a variety of steps in the retroviral life cycle have been identified through the analysis of biochemical and two-hybrid interactions and dominant-negative mutants (reviewed in references 47 and 48). This body of work has deepened our appreciation of the complexity of the host-retrovirus relationship and illustrated how much remains to be understood about the intricate interplay between host and pathogen. In this review, we explore the value of using a simple model organism to systematically identify functional orthologs of host factors involved in retroviral replication.
FIG. 1.
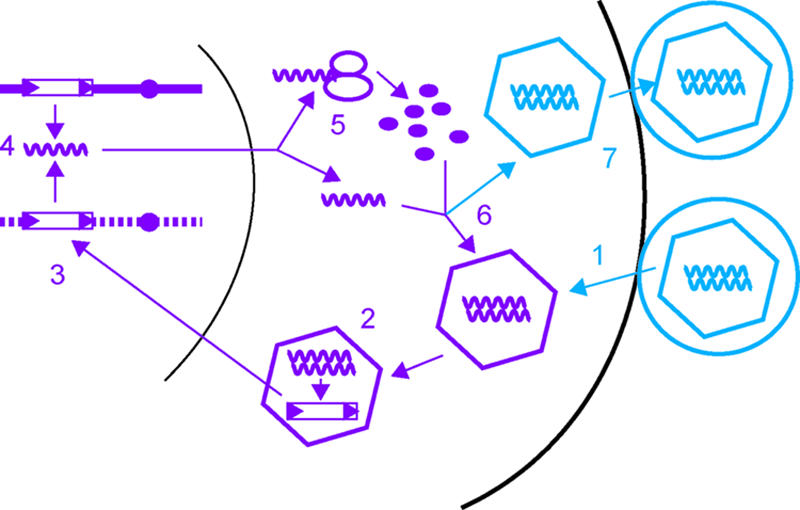
General steps of retrovirus and retrovirus-like transposable element replication. The drawings and numbers are ordered to indicate stages of retrovirus replication. The wavy lines represent retrovirus or retrovirus-like transposon RNA; open rectangles with black triangles at each end represent retrovirus or retrovirus-like transposon DNA. Stages that are common to retroviruses and retrovirus-like transposons are indicated in purple, and stages specific to retroviruses are indicated in blue. The stages of retroviral replication include viral entry (1); reverse transcription of the viral genome to form a linear double-stranded cDNA (2); integration of the cDNA into the host genome (3); transcription of the provirus by RNA polymerase II (4); translation of viral mRNA (5); viral core particle assembly, which includes encapsidation of viral mRNA (6); and viral-particle exit (7). The stages of retrotransposon replication include transcription of a genomic element by RNA polymerase II (4), translation of retrotransposon mRNA (5), virus-like particle assembly and packaging of the retrotransposon mRNA (6), reverse transcription of retroelement RNA to form a linear double-stranded cDNA (2), and insertion of cDNA into the genome by integration or homologous recombination with preexisting retrotransposons or solo LTRs in the genome (3).
A facile approach to the identification of mammalian genes that participate positively or negatively in retroviral propagation is first to identify genes that regulate retrovirus-like transposons in a model organism and then to test the effect on retroviral replication of mutating or reducing the expression of the corresponding mammalian protein. The development of tools to specifically reduce the expression of individual genes through RNA interference in many mammalian species has dramatically enhanced the feasibility of this approach. Retrovirus-like transposons are ubiquitous in eukaryotes and constitute a significant percentage of the host genome, from 3% of the genome of the budding yeast Saccharomyces cerevisiae to approximately 8% of the human genome (12, 36). While retrovirus-like transposons are not pathogenic, they are potent insertional mutagens. Many of the steps in transposition, with the notable exception of viral-particle budding and infection of new cells, are analogous to steps involved in the replication of retroviruses (Fig. 1). Consequently, the Ty1 and Ty3 retrovirus-like transposons in S. cerevisiae have been studied extensively as models to explore the influence of the host cell on retroviral propagation (71, 87, 106). The stable haploid phase of growth, the availability of genetic and genomics tools, and the feasibility of biochemical studies of S. cerevisiae are some of the features that permit rapid identification of host factors and analysis of their effects on the replication of retrovirus-like elements.
Retroviruses and retrovirus-like transposons consist of terminal direct-repeat sequences known as long terminal repeats (LTRs) flanking a central coding region, which at a minimum consists of gag, pol, and, in some cases, prt genes. The gag gene encodes the structural protein or proteins necessary to form the viral core particle or the virus-like particle (VLP). The pol domain encodes reverse transcriptase (RT) and integrase (IN) enzymatic activities and sometimes protease (PR) activity, if PR is not encoded by a separate prt gene. Retroviruses and retrovirus-like transposons, also known as LTR retroelements, can be divided into three major families on the basis of their amino acid sequences in RT: the retroviruses (Retroviridae), the Ty3/gypsy/BEL family (Metaviridae), and the Ty1/copia family (Pseudoviridae) (52). Retroviral genomes harbor an additional open reading frame (ORF) called env, which encodes an envelope glycoprotein that is incorporated into the cell-derived lipid bilayer of the retroviral virion when it exits the cell. The presence of a functional env gene is correlated with the infectivity of retroviruses, and acquisition of env is thought to be a key step in the evolution of retroviruses from LTR retrotransposons (63). While most members of the Ty3/gypsy/BEL and Ty1/copia families lack both an env gene and an extracellular phase of replication, the two families do include several examples of elements that have obtained env-like ORFs (52), and the Drosophila metavirus gypsy has been shown to be an infectious retrovirus (97). Regardless of whether they carry an env gene, metaviruses, such as Ty3, are more closely related to retroviruses, and they have a common structural organization of the pol domain. Pseudoviruses, such as Ty1, comprise the most ancient family of LTR retroelements.
The identification of conserved host genes that inhibit or contribute to Ty1 and/or Ty3 retrotransposition provides candidates for genes that influence retrovirus replication. Recent genetic screens for host factors that control Ty1 or Ty3 retrotransposition have identified large sets of genes involved in a variety of basic cellular processes (4, 50, 56, 90). Some regulatory themes are evident among these collections of host factors and restriction factors, such as regulation through RNA-processing factors and DNA damage-signaling/repair proteins. The utility of the yeast model systems is emphasized by the recent identification of a small number of genes involved in RNA processing and DNA repair as regulators of both retrovirus replication in mammalian cells and Ty1 and/or Ty3 elements in yeast (67, 110, 111). Moreover, the human antiviral protein, APOBEC3G, when expressed in yeast potently inhibits Ty1 retrotransposition by a mechanism similar to that by which human immunodeficiency virus type 1 (HIV-1) replication is restricted in human cells (40, 92). These recent findings suggest that not only the mechanisms of retroelement replication, but also the various means by which retroelements interact with their hosts, can be conserved in eukaryotes.
GENOME-WIDE SCREENS IN YEAST: DIFFERENT SCREENS, DIFFERENT GENES?
During the past six years, four genome-wide screens in S. cerevisiae have identified large sets of putative regulators of Ty1 and Ty3 retrotransposition (4, 50, 56, 90), but little overlap was observed among the sets (Table 1). In fact, even different screens for regulators of the same element type (either Ty1 or Ty3) uncovered largely nonoverlapping sets of host factors. Some of the variability among screens could be due to the large number of host proteins involved in modulating the activities of Ty1 and Ty3 retrotransposons and the fact that some, or perhaps all, of the screens were not exhaustive. The initial screen that was performed in each of these four studies identified 1.17% to 5.65% of mutants as having an altered transposition phenotype (Table 2). Both hypertransposition and hypotransposition mutants were found in high numbers if the design of the screen permitted their identification. The large, nonoverlapping gene sets identified in these screens could reflect the presence of differences between regulatory pathways that control Ty1 elements and those that control Ty3 elements, regulation of multiple steps in retrotransposition through numerous cellular pathways, and/or the influence of many genes on a few major pathways that regulate retrotransposon mobility. Below, we consider differences in experimental design among the four genetic screens that could account for these divergent results (Table 2).
TABLE 1.
Genes identified by multiple genetic screens for host factors that regulate Ty elements
| Gene | Identification ina:
|
|||
|---|---|---|---|---|
| Scholes et al. (90) | Griffith et al. (50) | Aye et al. (4) | Irwin et al. (56) | |
| BRO1 | — | — | Activator | Activator |
| CPR7 | — | Activator | — | Activator |
| DEP1 | — | Activator | — | Activator |
| DBP3 | — | Activator | — | Activator |
| DBR1 | — | Activator | — | Activator |
| ELG1 | Repressor | — | — | Repressor |
| NUP84 | — | Activator | — | Repressor |
| NUP133 | — | Activator | — | Repressor |
| POP2 | — | Activator | — | Repressor |
| RIM13 | — | — | Activator | Activator |
| RPL6A | — | Activator | — | Activator |
| RRM3 | Repressor | — | — | Repressor |
| RTT101 | Repressor | — | — | Repressor |
| RTT103 | Repressor | — | — | Repressor |
| RTT109 | Repressor | — | — | Repressor |
| SEC22 | — | Activator | — | Repressor |
| SGS1 | Repressor | — | — | Repressor |
| SIN4 | — | Activator | Activator | Activator |
| SWD1 | — | — | Activator | Repressor |
| TPS2 | — | Activator | — | Repressor |
| VAC8 | Repressor | — | — | Repressor |
| VPH1 | — | Activator | — | Activator |
| VPS9 | — | Activator | — | Activator |
| YBR284W | — | — | Activator | Repressor |
| Fractionb | 7/29 | 13/101 | 5/22 | 24/130 |
A dash indicates that the gene was not identified as a regulator of Ty in the screen. Activators are genes that, when mutated, decrease Ty mobility, and repressors are genes that, when mutated, increase Ty mobility.
The number of Ty regulatory genes identified in the screen that were also identified in one or more additional screens divided by the total number of Ty1 regulatory genes identified in the screen.
TABLE 2.
Comparison of genetic screens for host factors that regulate Ty elements
| Screen parameter | Value in:
|
|||
|---|---|---|---|---|
| Scholes et al. (90) | Griffith et al. (50) | Aye et al. (4) | Irwin et al. (56) | |
| Primary | ||||
| Ty Element | Ty1 | Ty1 | Ty3 | Ty3 |
| Location/promoter | Chromosome/native LTR | Low-copy-number plasmid/GAL1 | Low-copy-number plasmid/GAL1 | High-copy-number plasmid/GAL1 |
| Selection | Marker gene (his3AI) | Marker gene (HIS3) | Target locus | Marker gene (HIS3) |
| Mutant cell type | Haploid | Homozygous diploid | Haploid | Haploid |
| Mutagenesis | Mini-Tn3 insertion | Systematic ORF deletion strains | Mini-Tn3 insertion | Systematic ORF deletion strains |
| Candidate mutants (% up or down) | 1.66 | 2.25 | 1.17 | 5.65 |
| Secondary | ||||
| Features | Identification of single gene mutations causing transposition phenotype in genetic crosses | Elimination of mutations affecting GAL1 promoter or HIS3 marker | Test of corresponding ORF deletion alleles for the same phenotype using a high-copy pGAL1Ty1HIS3 element. | Repeated test of ORF deletion mutants using a low-copy pGAL1Ty1HIS3 element |
| Regulators identified | 29 repressors | 2 repressors, 99 activators | 1 repressor, 21 activators | 64 repressors, 66 activators |
On the most basic level, two screens identified regulators of Ty1 elements (50, 90) while the other two identified regulators of Ty3 elements (4, 56); hence, element-specific regulators could have numbered among the proteins identified in each screen. Aside from the fact that Ty1 and Ty3 belong to different families of LTR retroelements, a number of differences in the two elements' relationships with their hosts support the notion that element-specific regulators exist. For instance, Ty1 elements are present at about 30 copies in haploid laboratory strains; Ty1 RNA is very abundant, but transposition is repressed at posttranscriptional and posttranslational steps (28, 29, 31, 44); also, Ty1 integrates into a ∼750-bp region upstream of genes transcribed by RNA Pol III or into promoters of genes transcribed by RNA Pol II (37, 59, 79, 108). In contrast, Ty3 elements are present in one to five copies in haploid laboratory strains; their transcription and transposition both occur at very low levels but are induced to high levels by mating or treatment of cells with mating pheromone (11, 22, 64, 105), and Ty3 integrates 1 to 2 bp upstream of start sites for RNA Pol III transcription (17, 18). These differences in expression levels and integration specificities suggest that the sets of host factors that activate or inhibit Ty1 and Ty3 retrotransposition do not completely overlap.
A second difference among the four screens was related to whether expression of a Ty1 or Ty3 element was induced to high levels to aid in the detection of transposition. Because the frequency of retromobility is so low (≤10−5 per Ty element per generation in yeast), transposition can be detected only if an individual element is expressed from a strong heterologous promoter (13) or if a sensitive genetic assay for retrotransposition is employed (30). Only the screen for Ty1 regulators reported by Scholes et al. (90) measured the activity of a chromosomal Ty element expressed from its native promoter (Table 2). While the mobility of chromosomal Ty1 elements is measurable, it is too low to permit screening of a large number of mutants for reduced mobility. Consequently, Scholes et al. (90) exclusively identified mutations that elevate Ty1 mobility, which correspond to genes that encode repressors of Ty1 retromobility. In contrast, the other screen for regulators of Ty1 transposition measured the activity of a plasmid-borne Ty1 element expressed from the inducible GAL1 promoter, referred to as a pGAL1Ty1 element (50). Expression of a pGAL1Ty1 element overrides transpositional dormancy, a strong posttranslational repression of Ty1 mobility, and results in an ∼1,000-fold increase in retrotransposition (29, 44). Therefore, the Griffith et al. (50) screen for regulators of pGAL1Ty1 yielded primarily those mutants that reduce retrotransposition, which correspond to genes that encode activators of Ty1 retromobility (Table 2). To isolate regulators of Ty3 mobility, both screens used pGAL1Ty3 elements as a means of circumventing the problem of low levels of Ty3 expression and transposition (4, 56). Notably, Ty3 driven by its native promoter transposes efficiently in mating populations (64); therefore, it is likely that transposition of a chromosomal, LTR-driven Ty3 element could be measured by combining the use of mating pheromone for induction of Ty3 expression with the sensitive genetic assay for retrotransposition (see below). An assay for mobility of chromosomal Ty3 elements would be useful as a secondary screen for mutants that have been isolated as regulators of pGAL1Ty3 elements.
Third, the screens employed different methods of quantifying Ty transposition in host mutants. Three of the four screens measured the frequency with which a copy of a genetically marked Ty1 or Ty3 element was incorporated into the genome (Table 2). In the Scholes et al. screen (90), a chromosomal Ty1 element was marked with the retrotranscript indicator gene his3AI, thereby allowing HIS3-marked retromobility events that occurred at a frequency of >10−9 to be detected in a simple genetic assay (Fig. 2). In Griffith et al. (50) and Irwin et al. (56), the pGAL1Ty1 or pGAL1Ty3 element was marked with HIS3 and transfer of HIS3 from the plasmid to the chromosome was assayed. In all three cases, insertion of TyHIS3 cDNA could occur by transposition or by recombination of TyHIS3 cDNA with Ty elements or LTRs preexisting in the genome, both of which are quite abundant (Fig. 2). In contrast, the initial screen performed by Aye et al. (4) made use of a clever plasmid target assay for Ty3 transposition (Fig. 2). Approximately 90% of phenotypically selectable events in this system result from Ty3 transposition into the target (64). Hence, different mutants may have been identified in the study of Aye et al. (4), because the assay that was used measured only cDNA integration, whereas the other assays measured both cDNA integration and recombination events. The notion that some host factors exert their influence primarily on cDNA recombination is supported by the observation that the RecQ family helicase Sgs1 reduces the level of Ty1 transposition events measured in genetic assays, specifically by blocking the formation of multimeric cDNAs by recombination (16).
FIG. 2.
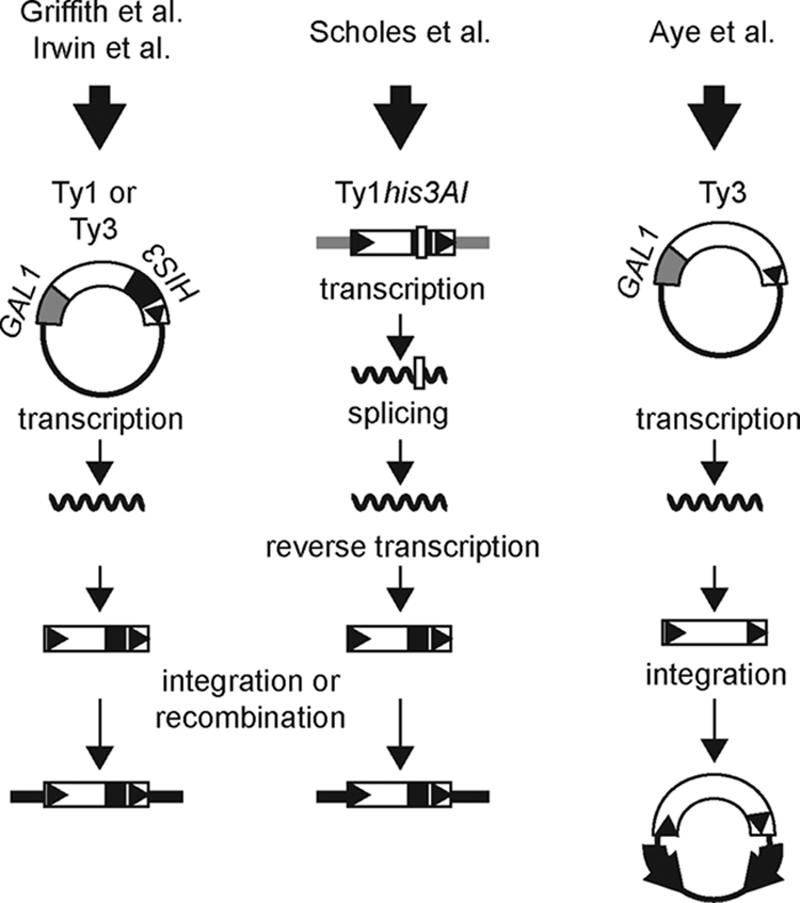
Comparison of Ty mobility assays used in the four high-throughput screens for Ty regulators. The three columns are schematic representations of the different experimental approaches used to measure Ty mobility in the primary screen of each of four studies. The column on the left depicts a plasmid copy of a Ty element expressed from the GAL1 promoter and marked with a HIS3 reporter gene. The TyHIS3 element is transcribed and then reverse transcribed to produce a TyHIS3 cDNA that is inserted into the genome through Ty IN-mediated integration or host-mediated recombination with genomic Ty elements or LTRs. New TyHIS3 mobility events are detected by selecting for plasmid loss and the presence of a genomic copy of HIS3. The middle column depicts a genomic copy of a Ty1his3AI element in which a HIS3 marker gene is disrupted by an artificial intron (AI). Following transcription from the Ty1 promoter, the AI is spliced out of the Ty1his3AI transcript. After reverse transcription and insertion of the resulting cDNA molecule into the genome through integration or recombination with genomic Ty1 sequences, a functional HIS3 gene is restored, allowing new Ty1HIS3 mobility events to be detected. The column on the right depicts a plasmid copy of Ty3 expressed from the GAL1 promoter. The Ty3 element is transcribed, and the Ty3 transcript is reverse transcribed to produce a cDNA. Integration of the cDNA into a target locus, which consists of two closely spaced, divergently transcribed tRNA genes (block arrows) on a plasmid, is detected by relief of transcriptional repression of the tRNA genes, resulting in expression of reporter genes that require the corresponding tRNA suppressors.
Fourth, the nature of the yeast strain used for each screen could also have contributed to the identification of different regulators. The Griffith et al. (50) screen was the only one that used homozygous diploid mutants, while the other three screens employed haploid mutants (Table 2). Since the expression of some genes, including the Ty1 and Ty3 elements themselves, is differentially regulated between haploid and diploid yeast strains, certain mutations may have been more or less likely to be identified in each cell type. In addition, mutant strains can harbor a second, unselected mutation sustained during the transformation step used to construct the mutant library. When present in a haploid strain, an unselected mutation could influence the activity of the retrotransposon being assayed. Therefore, candidate host factors identified in haploid strains have a greater risk of being false positives. Scholes et al. (90) addressed the latter caveat by testing whether the hypertransposition phenotype of haploid mutants segregated as a single gene trait in genetic crosses.
Fifth, the methods used to generate mutations varied among screens. Mutant strains screened by Scholes et al. (90) and Aye et al. (4) contained random insertions of a mini-Tn3 transposon (65), while strains from the systematic ORF gene deletion collection (46, 109) were tested by Griffith et al. (50) and Irwin et al. (56) (Table 2). Neither screen performed using transposon mutagenesis was likely to have been saturating, since well-characterized host mutants were not identified. Furthermore, mutant alleles generated by transposon mutagenesis are not necessarily null alleles, and the phenotypes of hypomorphic alleles could differ from those of the corresponding deletion alleles. Indeed, the altered Ty1 and Ty3 transposition phenotypes of some mini-Tn3-tagged alleles differed considerably from those of the corresponding deletion alleles (4, 90). However, this feature of mini-Tn3 mutagenesis also permitted the identification of essential genes in both the Aye et al. and Scholes et al. screens, whereas essential genes were not tested in the Griffith et al. or Irwin et al. screens.
Sixth, vastly disparate secondary screens were employed among the studies to confirm the altered transposition phenotype (Table 2). As noted above, Scholes et al. (90) tested mutants with increased mobility of a chromosomal Ty1his3AI for segregation of the hypertransposition phenotype as a single gene trait. Griffith et al. (50) eliminated mutants that had defects in GAL1 promoter or HIS3 marker function and those that failed to grow on medium containing galactose. In the Aye et al. screen (4), mutants identified by their activity on a selectable target gene were confirmed by measuring the activity of a pGAL1Ty3HIS3 element in the corresponding ORF deletion mutants (Fig. 2). Irwin et al. (56) used a low-copy-number pGAL1Ty3HIS3 element to measure the transposition phenotype in mutants identified using a high-copy-number pGAL1Ty3HIS3 element assay. Each of these secondary screens eliminated a large number of the initial mutants identified. Overall, the substantial variation in screening methods was probably the most significant factor in the lack of overlap among gene sets identified in each study.
MANY REGULATORY PATHWAYS OR FEW PATHWAYS WITH MANY COMPONENTS?
The four screens for Ty regulators likely provide complementary information in terms of retrotransposon regulation, considering that each of them had the potential to identify distinct gene sets. However, the challenge of fully interpreting and incorporating these findings into a coherent model for Ty regulation still remains. Given the complexity of LTR element mobility and its dependence on the host machinery at many steps (Fig. 1), there are some obvious potential roles for regulators involved in diverse cellular processes. For example, host factors involved in RNA elongation, processing, transport, or translation could regulate the expression of Ty proteins or the packaging of RNA into VLPs. A large number of the host factors identified have roles in vesicular trafficking, vacuolar function, secretion, stress response, and protein modification. These regulators could participate in the modification of Ty proteins, Ty protein processing, protein turnover, VLP assembly, and/or VLP maturation. Genes involved in nuclear transport may regulate the import of cDNA into the nucleus, which is a necessary step in Ty retrotransposition, since the yeast nuclear envelope does not break down during mitosis. Genes with roles in DNA repair, replication, recombination, or other aspects of genome maintenance could influence the synthesis, stability, or fate of Ty cDNA. Since Ty cDNA integrates into a chromatin substrate, genes that influence chromatin structure and transcriptional processes could alter target site choice or integration efficiency. In three of the screens, Ty elements were expressed from the GAL1 promoter, so it is unlikely that the transcription and chromatin factors identified directly regulate native Ty promoters. An alternative explanation for the role of any of the classes of host factors mentioned above is that they act indirectly to alter the expression or function of a gene product that regulates Ty transposition.
This straightforward interpretation of the data is consistent with regulation of retrotransposition at many steps and does not place strong emphasis on the regulatory role of any particular cellular pathway. However, some observations are in conflict with such a simple perspective on retrotransposition regulation. For example, Griffith et al. (50) measured the efficiency of Ty1 targeting to the upstream regions of tRNA genes in 10 mutants that had deletions of genes involved in chromatin structure. The authors anticipated that some or all of these mutations would alter the efficiency of targeting to tRNA genes, but they found only marginal differences in targeting efficiency between wild-type and mutant strains. In contrast, Ty1 cDNA levels were reduced in all 10 mutants. Moreover, Ty1 cDNA levels were decreased in 44 of the 99 hypotransposition mutants found by Griffith et al. We examined the genetic interaction data for these 44 genes in the Saccharomyces Genome Database (http://www.yeastgenome.org) to determine whether they would interact in common with genes in particular cellular pathways or processes. While the 44 genes are involved in diverse cellular processes, 33 of them show synthetic genetic interactions with genes that are components of DNA damage-signaling networks or that have roles in DNA replication or repair (2, 62, 73, 81, 103). Perhaps a central pathway that is influenced by many genes, such as a DNA damage-signaling pathway, is responsible for the common phenotype of many of these diverse genes.
It is also useful to consider the numbers and functions of those genes that were identified in more than one screen. While the percentages of genes that were identified as host regulators in multiple screens were similar for each individual screen, most of the overlap in identification of Ty activators was found between the studies by Griffith et al. (50) and Irwin et al. (56) and most of the overlap in Ty repressors was found between the Scholes et al. (90) and Irwin et al. studies. Only 24 genes were identified in more than one screen; of these, 10 genes encoded Ty activators and 7 encoded Ty inhibitors, while the other 7 genes yielded opposing results in each of two screens (Table 1). The 10 Ty activators have quite diverse functions. For example, Sin4, which was identified in three screens, is a component of the RNA Pol II mediator complex, and a global chromatin modifier; Rpl6A, which is a candidate activator of Ty1 and Ty3, is a ribosomal protein; Vps9 and Vph1, putative activators of Ty1 and Ty3 transposition, function in vacuole trafficking; and Bro1, identified in two screens as an activator of Ty3 mobility, is a vacuolar-protein-sorting factor required in the multivesicular body pathway. Interestingly, the human homolog of Bro1 interacts with the p6-Gag protein of HIV-1 retroviruses and functions in virion budding (98). In contrast to Ty activators, Ty1 and Ty3 repressors identified in more than one screen (Table 1) primarily have roles in DNA replication and replication-dependent repair. For example, Elg1/Rtt110 is a replication factor C homolog important for recovery from DNA damage arising during replication (9, 10, 60). The Rrm3 helicase promotes replication through nonnucleosomal protein-DNA complexes, while the cullin Rtt101 is involved in replication through natural and methyl methanesulfonate-induced pause sites (57, 74). The Sgs1 helicase influences the resolution of repair intermediates that form during repair of damage during replication (6, 23). Rtt109 is a histone acetyltransferase with a role in replication-dependent DNA repair (39, 51, 89, 104). Perhaps DNA damage that results in the absence of these factors stimulates an S-phase-specific signaling pathway that has a common activating influence on Ty1 and Ty3 mobility.
Below, we consider in detail host factors identified as regulators of retrotransposition that have characterized or putative roles in either of two basic areas of host cell biology: mRNA metabolism and the maintenance of genome integrity. We use these two sets of host factors to illustrate the potential mechanisms of interaction between Ty elements and their hosts and consider the implications of these findings for the regulation of retroviral replication in mammalian cells.
RNA METABOLISM AND THE DUAL ROLE OF RETROELEMENT RNA
The Griffith et al. (50) screen for regulators of Ty1 and the Irwin et al. (56) screen for regulators of Ty3 each identified numerous factors involved in RNA processing. Surprisingly, though, only a mutation in DBP3 (which encodes an RNA helicase involved in ribosome biogenesis), RPL6A (which encodes a ribosomal protein), or DBR1 (which encodes an RNA lariat-debranching enzyme) produced the equivalent phenotype in the two screens. Dbp3 and Rpl6 are putative activators of Ty transposition that could affect translation of the Ty RNA; alternatively, they could facilitate transposition indirectly by altering the translation of another host protein. Dbr1 is a 2′-5′ phosphodiesterase required to remove the phosphodiester bond within an intron lariat RNA that is formed in the process of mRNA splicing (19). Chapman and Boeke (19) discovered more than 15 years ago that Ty1 retrotransposition is reduced ∼85% in the absence of lariat-debranching enzyme. Remarkably, not only Ty1 and Ty3 retrotransposition, but also replication of the HIV-1 retrovirus, requires Dbr1 (61, 110). Furthermore, all three LTR retroelements appear to have a defect in the accumulation of cDNA when Dbr1 expression is reduced or abolished (61, 86, 110). However, the exact mechanism by which Dbr1 promotes LTR retroelement activity remains unknown. Cheng and Menees (20) found evidence for a lariat form of the Ty1 RNA containing a phosphodiester bond between the 5′ phosphate on the first nucleotide of Ty1 mRNA and a 2′ hydroxyl group of an internal nucleotide at the U3-R boundary of the 3′ LTR in dbr1 mutants. They hypothesized that the lariat promotes transfer of minus-strand strong-stop DNA (the first segment of cDNA synthesized) from the 5′ LTR to the 3′ LTR and that Dbr1 was necessary to resolve the lariat intermediate so that cDNA synthesis could resume. Consistent with these authors' observations, Ye et al. (110) presented evidence that small-interfering-RNA-mediated RNA interference of DBR1 inhibits HIV-1 replication by blocking cDNA synthesis at a step subsequent to minus-strand strong-stop DNA formation. On the other hand, Coombes and Boeke (27) were unable to confirm the presence of a Ty1 RNA lariat species using RNase H digestion, RNA ligase-mediated rapid amplification of cDNA ends, or RNase protection. Moreover, they demonstrated that RT-PCR could produce artifactual evidence for the existence of a lariat Ty1 RNA. Thus, the exact role of Dbr1 in retroelement replication remains to be defined.
An interesting observation made by Cheng and Menees (20) and Coombes and Boeke (27) is that both capped and uncapped Ty1 mRNAs are detected in nucleic acids isolated from VLPs of wild-type cells. In dbr1 mutants, Cheng and Menees could not detect capped or uncapped Ty1 mRNA by RNA ligase-mediated rapid amplification of cDNA ends, supporting their hypothesis that the 5′ end of Ty1 mRNA was protected within a lariat. In contrast, Coombes and Boeke found both the capped and uncapped species in dbr1 mutants. The presence of stable, uncapped Ty1 mRNA in wild-type cells is surprising, since Ty1 mRNA is capped during transcription by RNA Pol II and mRNA decapping leads to rapid mRNA degradation (76). If Ty1 mRNA is not decapped by lariat formation and subsequent debranching, then how does decapping of Ty1 RNA occur, and what is its role?
Decapping of mRNAs is a critical regulatory step in a major pathway of mRNA degradation in eukaryotes. In this pathway, deadenylation of the mRNA leads to decapping by the Dcp1-Dcp2 complex, which in turns allows 5′-to-3′ exonucleolytic degradation by Kem1/Xrn1 (the yeast homolog of the mammalian Pacman protein). Decapping of mRNAs occurs in dynamic ribonucleoprotein complexes known as mRNA-processing bodies, or “P bodies” in yeast and mammalian cells. P bodies contain untranslated mRNAs, and they are sites of mRNA storage or mRNA decapping and degradation (24). Interestingly, although no individual P-body component was identified as a regulator of both Ty1 and Ty3 retrotransposition, the Griffith et al. (50) and Irwin et al. (56) screens each identified different P-body components as activators of retrotransposition. Griffith et al. found Lsm1 (the homolog of human LSm1) and Pat1, which both stimulate decapping, to be activators of Ty1 transposition, while Irwin et al. identified Kem1/Xrn1 and Dhh1 (mammalian p54/RCK), a DExD/H-box RNA helicase that promotes decapping (25), as activators of Ty3 transposition. Not surprisingly, though, subsequent testing of individual mutants revealed that retrotransposition of Ty3 is also reduced in lsm1Δ and pat1Δ mutants (8). These findings are consistent with a role for P bodies and possibly decapping in Ty1 and Ty3 transposition, although the effect of P-body proteins on retrotransposition could be indirect.
A large body of evidence suggests that P-body formation competes with translation initiation and that P bodies sequester mRNA away from translation initiation factors so that the RNA can be degraded or stored for later use (15, 24, 25, 42, 93, 95, 101). Thus, Beliakova-Bethell et al. (8) investigated the possibility that P bodies play a role in VLP assembly by sequestering the Ty3 RNA away from translation so that it can be packaged in VLPs. They found that not only Ty3 RNA, but also Ty3 proteins and VLPs colocalized with P-body components. Moreover, expression of Ty3 from the GAL1 promoter led to the hyperaccumulation of P-body components in cytoplasmic foci, suggesting that elevated Ty3 RNA and protein levels result in an increase in P-body size. In xrn1Δ mutants, which have increases in the size and abundance of P bodies (96), Ty3 proteins accumulated in large P bodies (8). In contrast, Ty3 proteins were more diffusely distributed in dhh1Δ mutants, which have a defect in P-body formation (25). Together, these observations indicate that the association of Ty3 VLPs with P bodies is likely to have functional significance.
If the association of Ty RNA with P bodies promotes VLP assembly by sequestering Ty RNA away from translation, then mutations in genes that encode translation initiation factors should enhance Ty VLP formation and transposition, since decapping and P-body formation are enhanced when translation initiation is repressed (25, 93, 94). In support of this model, Scholes et al. (90) found that a mutation in TIF4632, one of two eukaryotic initiation factor 4G (a component of the cap binding complex) homologs in yeast, resulted in a modest increase in Ty1 mobility. None of the other screens found mutations in translation initiation factor genes that derepressed Ty1 or Ty3 transposition, but this could reflect the fact that most translation initiation factors are essential for viability of yeast cells.
Three of the four P-body components identified as Ty activators in the yeast screens have mammalian homologs with analogous functions, and mammalian P bodies resemble yeast P bodies in composition and function (42). Thus, it is possible that P bodies play a similar role in the early stages of retroviral core particle assembly in mammalian cells as well. While this possibility has not yet been explored, the human APOBEC3F and APOBEC3G proteins, which inhibit the activities of a variety of retroviruses and endogenous retrotransposons, have been shown in biochemical analyses and localization studies to be associated with components of P bodies and mRNA storage granules (21, 43, 107). It remains to be determined whether retrovirus and LTR retrotransposon-ribonucleoprotein complexes are associated with mammalian P bodies and, if this is the case, whether their localization is dependent on APOBEC3 proteins. In yeast, however, the association of LTR retrotransposon VLPs with P bodies is clearly independent of APOBEC3 proteins, as this family of cytidine deaminases is found only in mammals.
GENOME INTEGRITY AND REPRESSION OF Ty TRANSPOSITION—THREE MODELS
The studies by Irwin et al. (56) and Scholes et al. (90), as well as other studies focused on selected host factors, have identified a large number of proteins that play roles in genome maintenance and also repress Ty retrotransposition (16, 69, 70, 83, 91, 99). Irwin et al. (56) identified 11 DNA maintenance factors as repressors of Ty3. In most instances, the absence of the repressor resulted in elevated levels of Ty3 Gag protein and/or Ty3 cDNA. Studies with endogenous Ty1 elements have shown that mutations in DNA maintenance genes result in an increase in Ty1 cDNA levels, but not Ty1 RNA or Gag protein levels (16, 69, 70, 83, 90, 91, 99). Therefore, the inhibitory effects of such genes are exerted at posttranslational steps, such as Ty1 VLP assembly/maturation; cDNA synthesis, stability, or transport to the nucleus; or successful incorporation of cDNA into the genome (through recombination or integration). DNA maintenance factors that repress retrotransposition could work by distinct or common mechanisms. Studies illustrating the effects on Ty1 mobility of mutations in individual DNA maintenance factors, such as Est2 (the catalytic subunit of telomerase), Rad27 (a homolog of the human FEN-1 endonuclease, involved in Okazaki fragment processing and base excision repair), Ssl2 and Rad3 (TFIIH-associated helicases), and Sgs1 (a RecQ family helicase), support the notion that both the level and destination of Ty1 cDNA are modulated by a variety of different mechanisms (16, 69, 70, 91, 99).
Since more is known about host regulation of Ty1 retrotransposition than Ty3 retrotransposition, we will focus on three hypotheses for the mechanism that underlies the posttranslational increases in Ty1 cDNA and mobility in genome maintenance mutants. These hypotheses are not mutually exclusive, and it is conceivable that more than one mechanism affects the levels of cDNA and retromobility in genome integrity mutants. The first hypothesis, which is based on studies of Ty1 transposition intermediates in rad27, ssl2, and rad3 mutants, is that Ty1 cDNA is stabilized in genome maintenance mutants (69, 70, 99). Following exposure to an RT inhibitor, Ty1 cDNA levels remain higher in these mutants than in wild-type strains, supporting a role for Rad27, Ssl2, and Rad3 in promoting destabilization of cDNA. Rad27 possesses 5′-flap endonuclease and 5′-3′ exonuclease activities that are required to process Okazaki fragments during replication. Ty1 RNA-cDNA hybrids, as well as cDNA containing a DNA flap structure, are intermediates in transposition (45, 53, 54), so Rad27 could interact directly with partially synthesized Ty1 cDNA intermediates, promoting degradation. Ssl2 and Rad3 are essential DNA helicases and components of the RNA Pol II transcription initiation factor, TFIIH. It has been proposed that these proteins bind free Ty cDNA ends and facilitate unwinding of Ty1 cDNA, leading to its degradation (69).
While the model that genome maintenance factors directly promote degradation of Ty1 cDNA fits with their roles in preserving genome integrity, some observations are inconsistent with this model. First, host factors that influence Ty1 cDNA stability should presumably have the same effect on Ty1 cDNA produced from pGAL1Ty1 elements, but this is not the case. None of the approximately 20 nonessential DNA maintenance factors that inhibit genomic Ty1his3AI element mobility, including Rad27, was identified as a factor that restricts pGAL1Ty1 transposition (50). Although pGAL1Ty1 elements produce much more cDNA than do genomic elements, pGAL1Ty1 mobility still occurs at frequencies of less than one event per cell; consequently, it is unlikely that expression of pGAL1Ty1 elements would be able to overwhelm host factors that degrade cDNA. Second, the half-life of Ty1 cDNA is very long in wild-type strains, varying between 93 and 252 min in different studies (69, 91, 99). Moreover, the half-lives were equivalent to or a few times shorter than the doubling times of the yeast cells in these experiments, so it is questionable whether cDNA degradation could be a primary means of inhibiting Ty1 mobility. A testable hypothesis that could explain both the striking stability of Ty1 cDNA in wild-type strains and the apparent increase in cDNA stability in ssl2, rad3, and rad27 mutants is that synthesis of double-stranded Ty1 cDNA is completed by an RT-independent mechanism in these mutants. The synthesis of the plus strand of Ty1 cDNA has been reported to be inefficient and to result in the accumulation of Ty1 RNA-cDNA hybrid molecules (82). RNA-cDNA hybrids would not be scored as cDNA molecules in quantitative Ty cDNA assays, which rely on restriction digestion and the formation of discrete bands on agarose gels (26, 70). Therefore, we propose that partially synthesized Ty1 RNA-cDNA molecules are converted to double-stranded cDNAs by host repair enzymes and that this process occurs more efficiently in certain genome maintenance mutants, such as ssl2, rad3, and rad27 mutants. Thus, cDNA levels might remain elevated in these mutants when they are treated with an RT inhibitor, because partially synthesized cDNAs are still being converted to double-stranded molecules and not because cDNA is more stable. Perhaps this effect is due to the induction of a specific family of DNA repair enzymes in the absence of certain genome maintenance factors.
A second hypothesis to explain the increased Ty1 cDNA and retromobility in genome maintenance mutants is that reverse transcription of Ty1 cDNA is stimulated as a result of the activation of a DNA damage-signaling pathway. For example, the substantial increase in Ty1 cDNA observed in est2 mutants was interpreted as being the result of increased cDNA synthesis, because the half-life of cDNA in est2 mutants was comparable to that in a wild-type strain, after correcting for differences in the rate of cell doubling (91). The increase in Ty1 cDNA levels was correlated with the severity of telomere erosion, indicating that the DNA lesion created in the absence of telomerase, rather than the absence of telomerase per se, was responsible for inducing the accumulation of Ty1 cDNA. Moreover, cDNA levels were reduced in both est2 rad9 and est2 rad24 double mutants relative to est2 single mutants, implying a role for DNA damage signaling through RAD9 and RAD24 in the elevation of cDNA levels. The possibility that DNA damage-signaling pathways activate Ty1 reverse transcription and mobility has fascinating implications for the host-Ty1 relationship, but there is not yet any evidence for a specific mechanism of activation. One model is that DNA damage-signaling pathways act by relieving posttranslational inhibition of Ty1. This hypothesis is consistent with the observation that many genome maintenance factors that repress the mobility of chromosomal Ty1 elements fail to inhibit the mobility of pGAL1Ty1 elements, since pGAL1Ty1 expression already overrides the posttranslational control of Ty1 mediated by the host cell (29, 44). Activation of Ty1 RT activity through a signaling pathway raises the possibility that elevated retrotransposition is part of the cellular response to DNA damage. It has been demonstrated that fragments of Ty1 cDNA, particularly minus strand strong-stop DNA, can be captured at sites of DNA double-strand break repair (77, 102). In addition, cDNA copies of an unrelated subtelomeric repeat element, Y′, which are reverse transcribed in Ty1 VLPs using Ty1 cDNA as a primer, are introduced into the genome at high levels in telomerase-negative survivors. This process is efficient enough to play a significant role in telomere elongation in the absence of telomerase (75). Hence, elevated retrotransposition triggered by DNA damage could play a functional role in yeast.
The third hypothesis is that delay of the cell cycle in the G2/M phase, which is characteristic of many genome maintenance mutants due to the increased levels of DNA lesions, promotes accumulation of Ty1 cDNA. This is an alternative interpretation of the data just presented for the second hypothesis, since est2 mutants exhibit a G2/M delay that is dependent on a functional DNA damage-signaling pathway (41, 55). Since there is currently no evidence that DNA repair or damage-signaling proteins specifically interact with Ty1 proteins or nucleic acids or directly influence a particular step of transposition, it is possible that these factors act indirectly. One hypothesis for indirect regulation is that G2/M arrest promotes Ty1 mobility because of some characteristic of the cellular environment, such as an increase in the pools of available deoxynucleotides or the presence of a newly replicated genome. To distinguish between the last two hypotheses, it will be necessary to obtain evidence that a genome maintenance factor or damage-signaling protein directly influences a specific step of transposition or evidence that a specific characteristic of cells in G2/M promotes transposition.
MAMMALIAN HOMOLOGS OF YEAST Ty REGULATORS/GENOME INTEGRITY FACTORS
Mammalian homologs of a number of yeast genome maintenance factors that regulate Ty elements have been reported to regulate steps in retrovirus life cycles. For example, mutations in XPB, XPD, RAD18, and RAD52 increase the efficiency of retroviral transduction and the levels of retroviral cDNA (67, 72, 111). Mutations in the yeast homologs of these four genes, SSL2, RAD3, RAD18, and RAD52, respectively, also increase the levels of Ty1 cDNA and the mobility of native chromosomal Ty1 elements (69, 70, 84; M. J. Curcio, A. E. Kenny, S. Moore, D. J. Garfinkel, M. Weintraub, E. Gamache and D. T. Scholes, submitted for publication). Based on the results of experiments in which cDNA levels were measured following treatment with RT inhibitors, it was proposed that the XPB and XPD proteins facilitate retroviral cDNA degradation, just as their yeast homologs have been proposed to promote Ty1 cDNA degradation (69, 70, 111). However, in XPB and XPD mutant cell lines, the rate of HIV-1 cDNA degradation was measured by comparing cDNA levels in cells treated with an RT inhibitor to cDNA levels in untreated cells at the same time points (111) rather than by comparing cDNA levels to genomic DNA levels, as was done for ssl2 and rad3 mutants in yeast (69). Reductions over time in the ratio of cDNA in treated cells to cDNA in untreated cells could result from either decay of cDNA in the treated cells or synthesis of cDNA in the untreated cells. Thus, the methodology employed in this study by Yoder et al. (111) complicates the determination of exactly how much of the relative decrease in cDNA levels is due to degradation.
Rad18 is a conserved E3 ubiquitin ligase that functions in postreplicative DNA repair in yeast and mammals. The mechanisms by which Rad18 represses Ty1 retrotransposition in yeast and retroviral transduction in mammalian cells are not yet understood. Elevated Ty1 retrotransposition in rad18Δ mutants is suppressed by deletion of RAD9, which encodes a component of the DNA damage checkpoint pathway (Curcio et al., submitted). This observation is consistent with the hypothesis that Rad18 represses Ty1 mobility indirectly by preventing the formation of DNA lesions that trigger the DNA damage checkpoint. Mammalian Rad18 interacts with HIV-1 IN (78), and mutations in RAD18 lead to increased retroviral transduction efficiencies. Therefore, Rad18 could play a role in maintaining the integrity of the genome by inhibiting viral integration in mammalian cells (72). Perhaps these findings indicate that Rad18 represses LTR retrotransposon activity in yeast and retroviral transduction in mammalian cells by distinct mechanisms, but further studies are necessary to elucidate the role of Rad18 in restricting the replication of retroelements. The homologous repair protein Rad52, on the other hand, appears to bind directly to HIV-1 cDNA and to repress HIV-1 infection and integration via binding to cDNA (67). Similarly, Ty1 cDNA levels and integration are increased in the absence of an increase in RT activity in yeast rad52 mutants (84).
Regulatory roles for nonhomologous end-joining (NHEJ) proteins in retroviral integration and Ty transposition have been proposed but remain controversial. Some authors have suggested that NHEJ proteins function positively in retroviral integration, possibly by promoting repair of the DNA gaps that are created during integration of cDNA (32, 34, 58), but other authors have reported no effect of NHEJ proteins on retroviral transduction (3, 5). Contradictory conclusions have also been reached in studies addressing a role for NHEJ proteins in Ty transposition. A few NHEJ genes, including XRS2 and NEJ1, are evolving under positive selection, leading to the suggestion that the NHEJ factors they encode are in genetic conflict with Ty elements (88). Some support for this model is provided by studies indicating that both Ku and components of the Mre11-Rad50-Xrs2 complex repress endogenous Ty1 element transposition (38, 84, 90, 91); however, Ku and Xrs2 have also been reported to promote pGAL1Ty1 retromobility (38, 50). Considering these disparate observations, it seems unlikely that NHEJ factors play a direct role in the regulation of Ty1 transposition.
DNA damage signaling and arrest in the G2 stage of the cell cycle have also been proposed to play roles in modulating the activities of mammalian retroviruses and yeast retrotransposons. The relationship between DNA damage signaling, G2 arrest, and productive retroviral infection has been considered from two perspectives: first, single-stranded DNA gaps present following retroviral integration may activate DNA damage-signaling proteins, and second, some viral proteins, including HIV-1 viral protein R (Vpr), induce G2 arrest (113). If retroviral integration stimulates DNA damage checkpoints, then the signaling would be expected to involve the ATM and/or ATR kinase. These kinases initiate signaling cascades as a result of double-stranded DNA breaks, replication stress, UV damage, and other forms of DNA damage to promote cell cycle arrest and DNA repair activities. A number of groups have investigated a role for these kinases, particularly ATM, in retroviral transduction using a variety of approaches and retroviral reporter systems (including wild-type retroviruses). These kinases are not required for efficient transduction in all systems (3, 35), but loss or reduction of their function greatly reduces transduction and/or cell survival following infection in several systems (32, 33, 68, 80). DNA damage signaling through ATM or ATR may, therefore, facilitate retroviral infection.
HIV-1 Vpr protein can cause G2 arrest through an ATR-dependent DNA damage-signaling pathway and through a mitogen-activated protein kinase signaling pathway (1, 85, 112). In addition, Vpr induces apoptosis, and signaling through ATR also appears to play a role in this effect (1). The specific consequences of G2 arrest for retroviral infection are not fully understood, but presumably, G2 arrest provides a cellular environment or condition that favors retroviral infection. There are conflicting reports as to whether the activation of ATR by Vpr is the result of Vpr indirectly inducing double-stranded DNA breaks (100) or whether the activation is a result of Vpr binding chromatin and promoting the formation of single-stranded DNA that is bound by RPA protein (66). Rad17 and Rad9-Rad1-Hus1 (9-1-1) complex proteins work together with Atr to activate the G2 checkpoint signal, and Rad17 and Hus1 have both been shown to be involved in Vpr-mediated G2 arrest (114).
Ty retrotransposition in yeast has not been observed to stimulate DNA damage checkpoints. However, as discussed above, the increased mobility of endogenous Ty1 elements in some DNA maintenance mutants involves the DNA damage-signaling pathway. The activation of Ty1 mobility by telomere erosion depends at least partially on yeast RAD24, RAD17, and RAD9, which signal for activation of the G2 checkpoint (91). Yeast Rad24 and the Rad17-Mec3-Ddc1 complex are homologs of mammalian Rad17 and the 9-1-1 complex, which are involved in Vpr-mediated G2 arrest. Yeast MEC1 and TEL1, homologs of ATR and ATM, respectively, are not required for Ty1 mobility; however, activation of Ty1 mobility in rtt101Δ mutants, which have defects in replication fork progression, depends on MEC1, as well RAD9, RAD24, and the RAD53 gene, which encodes the checkpoint transducer kinase in the DNA damage checkpoint (90; Curcio et al., submitted). Conservation of components of the Vpr-induced checkpoint pathway and the checkpoint pathway that activates Ty1 could reflect conserved mechanisms through which DNA damage signaling and G2 arrest promote both retroviral infection and retrotransposition. Therefore, further characterization of the activation of Ty1 through DNA damage signaling is likely to provide information useful for understanding mechanisms that influence retroviral infection.
SUMMARY
In the past several years, significant progress has been made in understanding how retroviruses and LTR retrotransposons commandeer the host cell machinery to promote their replication and how the host cell restricts the activities of retroelements at multiple steps in replication. In this review, we have discussed the results of genetic screens employing endogenous LTR retrotransposons in S. cerevisiae, in which thousands of yeast genes have been probed for roles in LTR retrotransposon mobility and over two hundred putative regulators have been identified (Table 2). These studies have revealed roles for highly conserved components of fundamental cellular processes ranging from transcription elongation to DNA replication. Secondary genetic and biochemical analyses of host regulatory proteins are beginning to elucidate their mechanisms of action, and the findings of these studies suggest that the levels of host regulation of yeast retrotransposons are numerous and varied. One of many challenges that lie ahead is to integrate the results derived from various screens and other studies in budding yeast into a comprehensive picture of the interchange between retroelements and the host cell. Researchers are beginning to use the information gained by studying LTR retrotransposons in budding yeast as a paradigm for the regulation of retroviruses in mammalian cells (14, 67, 110, 111). Despite the small number of such investigations that have been performed to date, the findings suggest that many pathways that control retroelements are conserved between yeast and mammals and illustrate the strength of yeast as a model system. Further exploitation of this approach is likely to quicken the pace of progress toward an understanding of how mammalian host cells modulate the activities of retroviruses.
Acknowledgments
We thank Suzanne Sandmeyer for helpful comments on the manuscript.
Work in our laboratory is supported by National Institutes of Health grants GM52072 and AI65301.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Andersen, J. L., E. S. Zimmerman, J. L. DeHart, S. Murala, O. Ardon, J. Blackett, J. Chen, and V. Planelles. 2005. ATR and GADD45α mediate HIV-1 Vpr-induced apoptosis. Cell Death Differ. 12:326-334. [DOI] [PubMed] [Google Scholar]
- 2.Archambault, V., A. E. Ikui, B. J. Drapkin, and F. R. Cross. 2005. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 25:6707-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariumi, Y., P. Turelli, M. Masutani, and D. Trono. 2005. DNA damage sensors ATM, ATR, DNA-PKcs, and PARP-1 are dispensable for human immunodeficiency virus type 1 integration. J. Virol. 79:2973-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aye, M., B. Irwin, N. Beliakova-Bethell, E. Chen, J. Garrus, and S. Sandmeyer. 2004. Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics 168:1159-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baekelandt, V., A. Claeys, P. Cherepanov, E. De Clercq, B. De Strooper, B. Nuttin, and Z. Debyser. 2000. DNA-Dependent protein kinase is not required for efficient lentivirus integration. J. Virol. 74:11278-11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour, L., and W. Xiao. 2003. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat. Res. 532:137-155. [DOI] [PubMed] [Google Scholar]
- 7.Baumann, J. G. 2006. Intracellular restriction factors in mammalian cells—an ancient defense system finds a modern foe. Curr. HIV Res. 4:141-168. [DOI] [PubMed] [Google Scholar]
- 8.Beliakova-Bethell, N., C. Beckham, T. H. Giddings, Jr., M. Winey, R. Parker, and S. Sandmeyer. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellaoui, M., M. Chang, J. Ou, H. Xu, C. Boone, and G. W. Brown. 2003. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 22:4304-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Aroya, S., A. Koren, B. Liefshitz, R. Steinlauf, and M. Kupiec. 2003. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. USA 100:9906-9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilanchone, V. W., J. A. Claypool, P. T. Kinsey, and S. B. Sandmeyer. 1993. Positive and negative regulatory elements control expression of the yeast retrotransposon Ty3. Genetics 134:685-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boeke, J. D., and S. E. Devine. 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93:1087-1089. [DOI] [PubMed] [Google Scholar]
- 13.Boeke, J. D., D. J. Garfinkel, C. A. Styles, and G. R. Fink. 1985. Ty elements transpose through an RNA intermediate. Cell 40:491-500. [DOI] [PubMed] [Google Scholar]
- 14.Bolton, E. C., A. S. Mildvan, and J. D. Boeke. 2002. Inhibition of reverse transcription in vivo by elevated manganese ion concentration. Mol. Cell. 9:879-889. [DOI] [PubMed] [Google Scholar]
- 15.Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryk, M., M. Banerjee, D. Conte, Jr., and M. J. Curcio. 2001. The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol. Cell. Biol. 21:5374-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chalker, D. L., and S. B. Sandmeyer. 1993. Sites of RNA polymerase III transcription initiation and Ty3 integration at the U6 gene are positioned by the TATA box. Proc. Natl. Acad. Sci. USA 90:4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalker, D. L., and S. B. Sandmeyer. 1992. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 6:117-128. [DOI] [PubMed] [Google Scholar]
- 19.Chapman, K. B., and J. D. Boeke. 1991. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65:483-492. [DOI] [PubMed] [Google Scholar]
- 20.Cheng, Z., and T. M. Menees. 2004. RNA branching and debranching in the yeast retrovirus-like element Ty1. Science 303:240-243. [DOI] [PubMed] [Google Scholar]
- 21.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:15588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark, D. J., V. W. Bilanchone, L. J. Haywood, S. L. Dildine, and S. B. Sandmeyer. 1988. A yeast sigma composite element, TY3, has properties of a retrotransposon. J. Biol. Chem. 263:1413-1423. [PubMed] [Google Scholar]
- 23.Cobb, J. A., L. Bjergbaek, and S. M. Gasser. 2002. RecQ helicases: at the heart of genetic stability. FEBS Lett. 529:43-48. [DOI] [PubMed] [Google Scholar]
- 24.Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861-890. [DOI] [PubMed] [Google Scholar]
- 25.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conte, D., E. Barber, M. Banerjee, D. J. Garfinkel, and M. J. Curcio. 1998. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol. Cell. Biol. 18:2502-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coombes, C. E., and J. D. Boeke. 2005. An evaluation of detection methods for large lariat RNAs. RNA 11:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curcio, M. J., and D. J. Garfinkel. 1994. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics 136:1245-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curcio, M. J., and D. J. Garfinkel. 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell. Biol. 12:2813-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curcio, M. J., A. M. Hedge, J. D. Boeke, and D. J. Garfinkel. 1990. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol. Gen. Genet. 220:213-221. [DOI] [PubMed] [Google Scholar]
- 32.Daniel, R., G. Kao, K. Taganov, J. G. Greger, O. Favorova, G. Merkel, T. J. Yen, R. A. Katz, and A. M. Skalka. 2003. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc. Natl. Acad. Sci. USA 100:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniel, R., R. A. Katz, G. Merkel, J. C. Hittle, T. J. Yen, and A. M. Skalka. 2001. Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficiency of stable transduction by retroviruses. Mol. Cell. Biol. 21:1164-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 35.Dehart, J. L., J. L. Andersen, E. S. Zimmerman, O. Ardon, D. S. An, J. Blackett, B. Kim, and V. Planelles. 2005. The ataxia telangiectasia-mutated and Rad3-related protein is dispensable for retroviral integration. J. Virol. 79:1389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deininger, P. L., and M. A. Batzer. 2002. Mammalian retroelements. Genome Res. 12:1455-1465. [DOI] [PubMed] [Google Scholar]
- 37.Devine, S. E., and J. D. Boeke. 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10:620-633. [DOI] [PubMed] [Google Scholar]
- 38.Downs, J. A., and S. P. Jackson. 1999. Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol. Cell. Biol. 19:6260-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Driscoll, R., A. Hudson, and S. P. Jackson. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315:649-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enomoto, S., L. Glowczewski, and J. Berman. 2002. Mec3, Mec1 and Ddc2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2626-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell. Biol. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 43.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81:2165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garfinkel, D. J., K. Nyswaner, J. Wang, and J. Y. Cho. 2003. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165:83-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garfinkel, D. J., K. M. Stefanisko, K. M. Nyswaner, S. P. Moore, J. Oh, and S. H. Hughes. 2006. Retrotransposon suicide: formation of Ty1 circles and autointegration via a central DNA flap. J. Virol. 80:11920-11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 47.Goff, S. P. 2004. Genetic control of retrovirus susceptibility in mammalian cells. Annu. Rev. Genet. 38:61-85. [DOI] [PubMed] [Google Scholar]
- 48.Goff, S. P. 2007. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 5:253-263. [DOI] [PubMed] [Google Scholar]
- 49.Goff, S. P. 2004. Retrovirus restriction factors. Mol. Cell 16:849-859. [DOI] [PubMed] [Google Scholar]
- 50.Griffith, J. L., L. E. Coleman, A. S. Raymond, S. G. Goodson, W. S. Pittard, C. Tsui, and S. E. Devine. 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164:867-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han, J., H. Zhou, B. Horazdovsky, K. Zhang, R. M. Xu, and Z. Zhang. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315:653-655. [DOI] [PubMed] [Google Scholar]
- 52.Havecker, E. R., X. Gao, and D. F. Voytas. 2004. The diversity of LTR retrotransposons. Genome Biol. 5:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heyman, T., B. Agoutin, S. Friant, F. X. Wilhelm, and M. L. Wilhelm. 1995. Plus-strand DNA synthesis of the yeast retrotransposon Ty1 is initiated at two sites, PPT1 next to the 3′ LTR and PPT2 within the pol gene. PPT1 is sufficient for Ty1 transposition. J. Mol. Biol. 253:291-303. [DOI] [PubMed] [Google Scholar]
- 54.Heyman, T., M. Wilhelm, and F. X. Wilhelm. 2003. The central PPT of the yeast retrotransposon Ty1 is not essential for transposition. J. Mol. Biol. 331:315-320. [DOI] [PubMed] [Google Scholar]
- 55.Ijpma, A. S., and C. W. Greider. 2003. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14:987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irwin, B., M. Aye, P. Baldi, N. Beliakova-Bethell, H. Cheng, Y. Dou, W. Liou, and S. Sandmeyer. 2005. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 15:641-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivessa, A. S., B. A. Lenzmeier, J. B. Bessler, L. K. Goudsouzian, S. L. Schnakenberg, and V. A. Zakian. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12:1525-1536. [DOI] [PubMed] [Google Scholar]
- 58.Jeanson, L., F. Subra, S. Vaganay, M. Hervy, E. Marangoni, J. Bourhis, and J. F. Mouscadet. 2002. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology 300:100-108. [DOI] [PubMed] [Google Scholar]
- 59.Ji, H., D. P. Moore, M. A. Blomberg, L. T. Braiterman, D. F. Voytas, G. Natsoulis, and J. D. Boeke. 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73:1007-1018. [DOI] [PubMed] [Google Scholar]
- 60.Kanellis, P., R. Agyei, and D. Durocher. 2003. Elg1 Forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 13:1583-1595. [DOI] [PubMed] [Google Scholar]
- 61.Karst, S. M., M. L. Rutz, and T. M. Menees. 2000. The yeast retrotransposons Ty1 and Ty3 require the RNA lariat debranching enzyme, Dbr1p, for efficient accumulation of reverse transcripts. Biochem. Biophys. Res. Commun. 268:112-117. [DOI] [PubMed] [Google Scholar]
- 62.Karumbati, A. S., and T. E. Wilson. 2005. Abrogation of the Chk1-Pds1 checkpoint leads to tolerance of persistent single-strand breaks in Saccharomyces cerevisiae. Genetics 169:1833-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim, F. J., J. L. Battini, N. Manel, and M. Sitbon. 2004. Emergence of vertebrate retroviruses and envelope capture. Virology 318:183-191. [DOI] [PubMed] [Google Scholar]
- 64.Kinsey, P. T., and S. B. Sandmeyer. 1995. Ty3 transposes in mating populations of yeast: a novel transposition assay for Ty3. Genetics 139:81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar, A., S. Vidan, and M. Snyder. 2002. Insertional mutagenesis: transposon-insertion libraries as mutagens in yeast. Methods Enzymol. 350:219-229. [DOI] [PubMed] [Google Scholar]
- 66.Lai, M., E. S. Zimmerman, V. Planelles, and J. Chen. 2005. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J. Virol. 79:15443-15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lau, A., R. Kanaar, S. P. Jackson, and M. J. O'Connor. 2004. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 23:3421-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lau, A., K. M. Swinbank, P. S. Ahmed, D. L. Taylor, S. P. Jackson, G. C. Smith, and M. J. O'Connor. 2005. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat. Cell Biol. 7:493-500. [DOI] [PubMed] [Google Scholar]
- 69.Lee, B. S., L. Bi, D. J. Garfinkel, and A. M. Bailis. 2000. Nucleotide excision repair/TFIIH helicases RAD3 and SSL2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol. Cell. Biol. 20:2436-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee, B. S., C. P. Lichtenstein, B. Faiola, L. A. Rinckel, W. Wysock, M. J. Curcio, and D. J. Garfinkel. 1998. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair transcription factor TFIIH subunits Ss12p and Rad3p. Genetics 148:1743-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lesage, P., and A. L. Todeschini. 2005. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 110:70-90. [DOI] [PubMed] [Google Scholar]
- 72.Lloyd, A. G., S. Tateishi, P. D. Bieniasz, M. A. Muesing, M. Yamaizumi, and L. C. Mulder. 2006. Effect of DNA repair protein Rad18 on viral infection. PLoS Pathog. 2:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loeillet, S., B. Palancade, M. Cartron, A. Thierry, G. F. Richard, B. Dujon, V. Doye, and A. Nicolas. 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair 4:459-468. [DOI] [PubMed] [Google Scholar]
- 74.Luke, B., G. Versini, M. Jaquenoud, I. W. Zaidi, T. Kurz, L. Pintard, P. Pasero, and M. Peter. 2006. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr. Biol. 16:786-792. [DOI] [PubMed] [Google Scholar]
- 75.Maxwell, P. H., C. Coombes, A. E. Kenny, J. F. Lawler, J. D. Boeke, and M. J. Curcio. 2004. Ty1 mobilizes subtelomeric Y′ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol. Cell. Biol. 24:9887-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer, S., C. Temme, and E. Wahle. 2004. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 39:197-216. [DOI] [PubMed] [Google Scholar]
- 77.Moore, J. K., and J. E. Haber. 1996. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature 383:644-646. [DOI] [PubMed] [Google Scholar]
- 78.Mulder, L. C., L. A. Chakrabarti, and M. A. Muesing. 2002. Interaction of HIV-1 integrase with DNA repair protein hRad18. J. Biol. Chem. 277:27489-27493. [DOI] [PubMed] [Google Scholar]
- 79.Natsoulis, G., W. Thomas, M. C. Roghmann, F. Winston, and J. D. Boeke. 1989. Ty1 transposition in Saccharomyces cerevisiae is nonrandom. Genetics 123:269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nunnari, G., E. Argyris, J. Fang, K. E. Mehlman, R. J. Pomerantz, and R. Daniel. 2005. Inhibition of HIV-1 replication by caffeine and caffeine-related methylxanthines. Virology 335:177-184. [DOI] [PubMed] [Google Scholar]
- 81.Pan, X., P. Ye, D. S. Yuan, X. Wang, J. S. Bader, and J. D. Boeke. 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124:1069-1081. [DOI] [PubMed] [Google Scholar]
- 82.Pochart, P., B. Agoutin, S. Rousset, R. Chanet, V. Doroszkiewicz, and T. Heyman. 1993. Biochemical and electron microscope analyses of the DNA reverse transcripts present in the virus-like particles of the yeast transposon Ty1. Identification of a second origin of Ty1 DNA plus strand synthesis. Nucleic Acids Res. 21:3513-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rattray, A. J., C. B. McGill, B. K. Shafer, and J. N. Strathern. 2001. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae. A role for SAE2/COM1. Genetics 158:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rattray, A. J., B. K. Shafer, and D. J. Garfinkel. 2000. The Saccharomyces cerevisiae DNA recombination and repair functions of the RAD52 epistasis group inhibit Ty1 transposition. Genetics 154:543-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roshal, M., B. Kim, Y. Zhu, P. Nghiem, and V. Planelles. 2003. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 278:25879-25886. [DOI] [PubMed] [Google Scholar]
- 86.Salem, L. A., C. L. Boucher, and T. M. Menees. 2003. Relationship between RNA lariat debranching and Ty1 element retrotransposition. J. Virol. 77:12795-12806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sandmeyer, S. B., M. Aye, and T. Menees. 2002. Ty3, a position specific, gypsy-like element in Saccharomyces cerevisiae, p. 663-683. In N. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 88.Sawyer, S. L., and H. S. Malik. 2006. Positive selection of yeast nonhomologous end-joining genes and a retrotransposon conflict hypothesis. Proc. Natl. Acad. Sci. USA 103:17614-17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schneider, J., P. Bajwa, F. C. Johnson, S. R. Bhaumik, and A. Shilatifard. 2006. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 281:37270-37274. [DOI] [PubMed] [Google Scholar]
- 90.Scholes, D. T., M. Banerjee, B. Bowen, and M. J. Curcio. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159:1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scholes, D. T., A. E. Kenny, E. R. Gamache, Z. Mou, and M. J. Curcio. 2003. Activation of a LTR-retrotransposon by telomere erosion. Proc. Natl. Acad. Sci. USA 100. [DOI] [PMC free article] [PubMed]
- 92.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 102:9854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwartz, D. C., and R. Parker. 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 20:7933-7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwartz, D. C., and R. Parker. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5247-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Segal, S. P., T. Dunckley, and R. Parker. 2006. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:5120-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Song, S. U., T. Gerasimova, M. Kurkulos, J. D. Boeke, and V. G. Corces. 1994. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 8:2046-2057. [DOI] [PubMed] [Google Scholar]
- 98.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 99.Sundararajan, A., B. S. Lee, and D. J. Garfinkel. 2003. The Rad27 (Fen-1) nuclease inhibits Ty1 mobility in Saccharomyces cerevisiae. Genetics 163:55-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tachiwana, H., M. Shimura, C. Nakai-Murakami, K. Tokunaga, Y. Takizawa, T. Sata, H. Kurumizaka, and Y. Ishizaka. 2006. HIV-1 Vpr induces DNA double-strand breaks. Cancer Res. 66:627-631. [DOI] [PubMed] [Google Scholar]
- 101.Teixeira, D., U. Sheth, M. A. Valencia-Sanchez, M. Brengues, and R. Parker. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Teng, S. C., B. Kim, and A. Gabriel. 1996. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature 383:641-644. [DOI] [PubMed] [Google Scholar]
- 103.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 104.Tsubota, T., C. E. Berndsen, J. A. Erkmann, C. L. Smith, L. Yang, M. A. Freitas, J. M. Denu, and P. D. Kaufman. 2007. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25:703-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Arsdell, S. W., G. L. Stetler, and J. Thorner. 1987. The yeast repeated element sigma contains a hormone-inducible promoter. Mol. Cell. Biol. 7:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Voytas, D. F., and J. D. Boeke. 2002. Ty1 and Ty5 of Saccharomyces cerevisiae, p. 631-662. In N. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 107.Wichroski, M. J., G. B. Robb, and T. M. Rana. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wilke, C. M., S. H. Heidler, N. Brown, and S. W. Liebman. 1989. Analysis of yeast retrotransposon Ty insertions at the CAN1 locus. Genetics 123:655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittman, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philipssen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 110.Ye, Y., J. De Leon, N. Yokoyama, Y. Naidu, and D. Camerini. 2005. DBR1 siRNA inhibition of HIV-1 replication. Retrovirology 2:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoder, K., A. Sarasin, K. Kraemer, M. McIlhatton, F. Bushman, and R. Fishel. 2006. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proc. Natl. Acad. Sci. USA 103:4622-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yoshizuka, N., Y. Yoshizuka-Chadani, V. Krishnan, and S. L. Zeichner. 2005. Human immunodeficiency virus type 1 Vpr-dependent cell cycle arrest through a mitogen-activated protein kinase signal transduction pathway. J. Virol. 79:11366-11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao, R. Y., and R. T. Elder. 2005. Viral infections and cell cycle G2/M regulation. Cell Res. 15:143-149. [DOI] [PubMed] [Google Scholar]
- 114.Zimmerman, E. S., J. Chen, J. L. Andersen, O. Ardon, J. L. Dehart, J. Blackett, S. K. Choudhary, D. Camerini, P. Nghiem, and V. Planelles. 2004. Human immunodeficiency virus type 1 Vpr-mediated G2 arrest requires Rad17 and Hus1 and induces nuclear BRCA1 and gamma-H2AX focus formation. Mol. Cell. Biol. 24:9286-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]


