Abstract
Cysteine proteinases are key virulence factors of the protozoan parasite Entamoeba histolytica. We have shown that cysteine proteinases play a central role in tissue invasion and disruption of host defenses by digesting components of the extracellular matrix, immunoglobulins, complement, and cytokines. Analysis of the E. histolytica genome project has revealed more than 40 genes encoding cysteine proteinases. We have focused on E. histolytica cysteine proteinase 1 (EhCP1) because it is one of two cysteine proteinases unique to invasive E. histolytica and is highly expressed and released. Recombinant EhCP1 was expressed in Escherichia coli and refolded to an active enzyme with a pH optimum of 6.0. We used positional-scanning synthetic tetrapeptide combinatorial libraries to map the specificity of the P1 to P4 subsites of the active site cleft. Arginine was strongly preferred at P2, an unusual specificity among clan CA proteinases. A new vinyl sulfone inhibitor, WRR483, was synthesized based on this specificity to target EhCP1. Recombinant EhCP1 cleaved key components of the host immune system, C3, immunoglobulin G, and pro-interleukin-18, in a time- and dose-dependent manner. EhCP1 localized to large cytoplasmic vesicles, distinct from the sites of other proteinases. To gain insight into the role of secreted cysteine proteinases in amebic invasion, we tested the effect of the vinyl sulfone cysteine proteinase inhibitors K11777 and WRR483 on invasion of human colonic xenografts. The resultant dramatic inhibition of invasion by both inhibitors in this human colonic model of amebiasis strongly suggests a significant role of secreted amebic proteinases, such as EhCP1, in the pathogenesis of amebiasis.
The intestinal protozoan parasite Entamoeba histolytica is the etiologic agent of amebic colitis and liver abscess, which cause high rates of morbidity and mortality worldwide (49). The mechanism by which Entamoeba histolytica is able to invade and damage the host's target tissues has been the subject of intense research. Several virulence factors have been identified, including secreted cysteine proteinases (39, 42). These amebic enzymes have been implicated in the in vitro cytopathology of cell monolayers (20, 23), which correlates with the observed separation of colonic epithelial cells before invasion (51). Other correlates with invasion include the ability of cysteine proteinases to degrade extracellular matrix components (19) and colonic mucin (31, 32). Furthermore, cysteine proteinases enable E. histolytica to evade the host's immune defenses by activating and locally depleting complement (43), and by degrading anaphylotoxins C3a and C5a (41), human immunoglobulin G (IgG) (53), human IgA (21), and interleukin-18 (IL-18) (37).
The recent completion of the Entamoeba histolytica genome project has revealed the presence of at least 40 genes encoding cysteine proteinases (25). Of all the cysteine proteinase genes, only ehcp1 and ehcp5 are unique to E. histolytica, as their orthologs are either absent (ehcp1) or nonfunctional (ehcp5) in Entamoeba dispar, a morphologically identical but noninvasive Entamoeba species (5, 6, 54). Surprisingly, only a small subset of these genes are expressed in cultured Entamoeba histolytica trophozoites (6), and only three, ehcp1, ehcp2, and ehcp5, account for more than 90% of the cysteine proteinase-specific transcripts in culture (5, 6). Quantitative studies of the expression of the major Entamoeba histolytica cysteine proteinases have shown that E. histolytica cysteine proteinase 1 (EhCP1) is one of the most highly expressed and released cysteine proteinases in cultured trophozoites (6, 8, 17). In a recent study of gene expression in a mouse model of amebic colitis, EhCP1 expression was increased almost twofold following invasion, while expression of EhCP5, the other E. histolytica-specific proteinase, was not (11). To further characterize this important amebic cysteine proteinase, we have cloned, expressed, and refolded recombinant EhCP1 (rEhCP1) to obtain active proteinase. We now show that rEhCP1 can cleave physiologic substrates, such as the third component of complement, pro-IL-18, and IgG, components of the host immune response which must be circumvented for the amebae to invade. We designed a new inhibitor based on the marked preference of EhCP1 for arginine in the P2 position. We now show for the first time that specific inhibitors of cysteine proteinases block invasion in the human colon.
MATERIALS AND METHODS
Entamoeba cultures and purification of genomic DNA.
E. histolytica strain HM1:IMSS was grown axenically in TYI-S-33 medium (9) and subcultured every 48 to 72 h. E. histolytica genomic DNA was purified from trophozoite nuclei with the DNeasy Kit (QIAGEN, Valencia, CA).
Isolation and purification of released amebic cysteine proteinases.
Released proteinases in conditioned medium (CM) from Entamoeba histolytica trophozoites were prepared as previously described in phosphate-buffered saline supplemented with additional cysteine and divalent cations which maintained >95% viability (by trypan blue exclusion) (17).
Cloning, expression, and purification of EhCP1 in Escherichia coli.
The sequence encoding the pro- and mature region of the E. histolytica EhCP1 gene (accession no. M94164) was amplified from E. histolytica genomic DNA by PCR (HotstartTaq Master Mix kit; QIAGEN, Valencia, CA) and cloned into the pBAD/Thio-TOPO plasmid (Invitrogen, Carlsbad, CA), resulting in expression of the recombinant protein as a thioredoxin fusion (amino terminus) with a six-residue histidine tail (carboxy terminus). Expression was induced as previously described (36), except that 0.2% arabinose was used for 4 h. For antibody production, rEhCP1 was expressed without the thioredoxin tail in the pRSETA vector (Invitrogen, Carlsbad, CA) and purified under denaturing conditions over nickel-nitrilotriacetic acid (36). Recombinant thio-pro-EhCP1 was purified from frozen pellets as previously described (37) except that we used 6 M urea and a 20 to 500 mM imidazole gradient for elution from the HiTrap-Ni-chelating column.
Refolding and purification of thio-pro-EhCP1.
The refolding of denatured thio-pro-EhCP1 was tested under the screening conditions elucidated by Armstrong et al. (3) as we have previously modified them (38). The highest proteinase activity was obtained by adding 1 μM thio-pro-EhCP1 to 50 mM morpholineethanesulfonic acid, pH 6.0, 30 mM NaCl, 0.1% polyethylene glycol 4000, 10 mM EDTA, 750 mM arginine, 5 mM reduced glutathione, and 0.5 mM oxidized glutathione for 48 to 72 h at 4°C. The proteinase was dialyzed against Tris-buffered saline, concentrated, and separated by ion-exchange fast-protein liquid chromatography on a Hi-Trap-Q column (Amersham Biosciences, United Kingdom).
Proteinase activity assay.
The proteinase activity was determined by measuring the release of the fluorescent leaving group, 4-amino-7-methylcoumarin (AMC), from synthetic peptide substrates (Bachem, Torrance, CA) in a Fluoroskan-Ascent fluorometer (Labsystems) (17). Enzyme activity, initial velocity, and relative fluorescence units (RFU; the amount of proteinase activity needed for the release of 1 pmol of AMC per minute) were calculated with Ascent software. The Michaelis constant (Km) of rEhCP1 for the synthetic peptide substrates Z-Arg-Arg-AMC and Z-Ala-Arg-Arg-AMC was determined with increasing concentrations of synthetic peptide substrates (0.5 to 20 μM) using the Enzfitter software (Biosoft, Cambridge, United Kingdom).
Production of Ab and immunoblot assays.
Polyclonal antibodies (Abs) were raised by immunizing rabbits subcutaneously three times with gel-purified rEhCP1 (100 μg) mixed with TiterMax (Sigma, St. Louis, MO). The resulting immune serum was absorbed with proteins from lysed E. coli cells coupled to cyanogen bromide-activated Sepharose 4B. The absorbed serum IgG fraction was then purified by protein A/G affinity chromatography (GE Health Care, Amersham Biosciences, Pharmacia).
For immunoblots, rEhCP1 was electrophoresed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred as previously described (40). Membranes were probed with monoclonal antithioredoxin Ab (1:10,000; Invitrogen, Carlsbad, CA) or rabbit anti-rEhCP1 Ab (1:500) and visualized with goat anti-mouse or anti-rabbit IgG-alkaline phosphatase (1:10,000 dilution; Zymed, San Francisco, CA) (40).
Gelatin substrate gels.
Refolded rEhCP1 was analyzed with the use of gelatin substrate gels (12% polyacrylamide-SDS copolymerized with 0.1% porcine gelatin) as previously described (37). Samples were incubated for 15 min in buffer or with 100 μM E-64 prior to electrophoresis.
Substrate specificity of rEhCP1.
Two synthetic combinatorial libraries were used to determine the substrate specificities of the S1 to S4 subsites of active, purified EhCP1 using a bifunctional fluorophore, 7-amino-4-carbamoylmethylcoumarin, coupled to the peptides as previously described (15, 27). The P1 diverse library consisted of 20 sublibraries with one native amino acid (cysteine was replaced by norleucine) in the P1 position and the P2, P3, and P4 positions randomized with equimolar mixtures of amino acids. To determine P2, P3, and P4 specificity, a P1-lysine fixed library was used; the P2, P3, and P4 positions were spatially addressed with 19 amino acids; and the remaining two positions were randomized.
pH profile of purified active rEhCP1.
The pH optimum of rEhCP1 was determined by preincubating rEhCP1 (50 pmol) in standard assay buffer (17) for pH 7.0 to 9.0 or with 25 mM sodium acetate for pH 4.0 to 6.5, and the remaining enzymatic activity was determined as described above.
Cleavage of biologic substrates by purified, active rEhCP1.
Human C3 (1 μg; Quidel, San Diego, CA) was incubated with increasing amounts of active purified rEhCP1 or CM which was preincubated for 15 min at room temperature in buffer alone or containing 80 μM E-64 (an irreversible inhibitor of all cysteine proteinases) for 60 min at 37°C, and the cleavage products were analyzed by SDS-PAGE as previously described (43). Cleavage of native human IgG (2 μg; Sigma, St. Louis, MO) was performed under similar conditions except for 16-h incubation. The resulting immunoblots were probed with goat anti-human IgG-horseradish peroxidase conjugate (1:20,000; Zymed, San Francisco, CA) and developed with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL). Human pro-IL-18 was expressed in E. coli as previously detailed (22, 37) and incubated with increasing amounts of purified active rEhCP1, active rEhCP5 (expressed in Pichia pastoris [37]), or E. histolytica released proteinases for 1 to 3 h at 37°C, and the cleavage products were detected on immunoblots as previously described (37).
Intracellular localization of EhCP1.
For confocal microscopy, E. histolytica trophozoites were fixed in 10% formalin, permeabilized in 0.1% Triton X-100, and detected with a mouse monoclonal Ab to EhCP3 (1:200) and a polyclonal rabbit Ab to EhCP1 (1:200), as previously described (36). For immunoelectron microscopy, trophozoites were fixed in 1% glutaraldehyde-4% paraformaldehyde and then processed and embedded in glycol-metacrylate (24). Ultrathin sections were reacted with rabbit polyclonal EhCP1 Ab (1:200) and mouse monoclonal anti-EhCP3 (1:200) Ab and detected with anti-rabbit IgG conjugated with gold particles (15-nm diameter; Ted Pella Inc., Redding, CA) and/or anti-mouse IgG conjugated with gold particles (20-nm diameter). Sections were contrasted with uranyl acetate and lead citrate and viewed under a Zeiss EM910 transmission electron microscope.
Inhibition of rEhCP1 and released proteinases by cysteine proteinase inhibitors.
K11777 is a vinyl sulfone inhibitor (N-methylpiperazine-urea-phenylalanyl-homophenylalanyl-vinylsulfone-benzene), which has been shown to inhibit a number of protozoan cysteine proteinases (10, 26) and has undergone extensive toxicity testing (1). WRR483 was synthesized as a K11777 derivative with an arginine substituted for phenylalanine at the P2 position. Kinetic analyses of the irreversible cysteine protease inhibitors (4, 52) were performed by adding rEHCP1 (20 to 30 nM) to 10-fold dilutions of inhibitor (500 nM to 100 μM) with 10 μM Z-Arg-Arg-AMC (Km = 2 μM) on a Flex Station with robotics (Molecular Devices) and an 0.7-s read time. The value of kobs, the rate constant for loss of enzyme activity, was determined from an equation for pseudo-first-order dynamics using Prism 4 (GraphPad). When kobs varied linearly with inhibitor concentration, Kass was determined by linear regression analysis (4). If the variation was hyperbolic, indicating saturation inhibition kinetics, kinact and Ki were determined from an equation describing a two-step irreversible inhibitor mechanism {kobs = kinact [I]o/([I]o + Ki·(1 + [S]o/Km) } and nonlinear regression analysis using Prism (4).
Amebic infection of a human intestinal xenograft model.
Human intestinal xenografts were transplanted subcutaneously into the backs of SCID mice and infected with E. histolytica trophozoites as previously detailed (28, 46, 55). Xenografts were injected with medium alone, E. histolytica trophozoites (1 × 106) in medium, or trophozoites preincubated with K11777 (20 μM final concentration) or WRR483 (10 μM final concentration). These conditions blocked all detectable cysteine proteinase activity in trophozoites for >24 h (data not shown). A total dose of 1.5 mg of either inhibitor was administered intraperitoneally to each mouse (10). The xenografts were harvested at 24 h, the mucus plug was removed, and sections were frozen in liquid nitrogen for DNA extraction or fixed for histology. The presence of an intact muscularis layer and complete removal of the mucus plug were confirmed in each experimental xenograft.
Quantification of amebic invasion in human intestinal xenografts.
We developed a sensitive, quantitative real-time PCR assay to measure amebic invasion in xenografts infected with E. histolytica. A standard curve to measure the number of E. histolytica trophozoites invading human intestinal xenografts was generated by adding trophozoites (102 to 106) to 25 to 50 mg of human intestinal xenograft tissue and extracting the DNA with the PUREGENE DNA purification kit (Gentra Systems, Minneapolis, MN). Real-time PCRs were carried out using the SYBR green PCR Master Mix (Applied Biosystems, Foster City, CA) with 100 ng of the extracted DNA (from xenografts and standards) and 100 nM (final concentration) of both the forward primer (5′-AAATCAATTGTGAAGTTATTGGAGTGA-3′) and the reverse primer (5′-TCCTACTCCTCCTTTACTTTTATCTGCT-3′), to amplify a 92-bp region of the E. histolytica peroxiredoxin gene (7). Real-time PCR was performed under conditions previously described (40) with a standard curve and negative controls consisting of reaction mixtures without template or containing noninfected xenograft DNA (100 ng).
The potential presence of any PCR inhibitor in each of the human intestinal xenograft DNA samples was tested by the quantitative measure of amplifiable human genomic DNA with primers specific for the detection of human Alu sequences (56). Xenograft DNA (0.1 ng) was amplified with 100 nM of sense (5′ ACG CCT GTA ATC CCA GCA CTT 3′) and antisense (5′ TCG CCC AGG CTG GAG TGC A 3′) human Alu primers, under the same PCR conditions as described above. Negative controls, positive controls, and the experimental samples were run in duplicate. The threshold cycle values from each sample were averaged, and the standard deviation was calculated.
RESULTS
Expression, purification, and refolding of thio-pro-EhCP1.
Expression of the thio-pro-EhCP1 recombinant protein in E. coli resulted in an overexpressed protein of ∼50 kDa, corresponding to the predicted molecular mass for thio-pro-EhCP1 of 50.3 kDa (36.3 kDa for the zymogen form of EhCP1, 13 kDa for the thioredoxin peptide, and 0.9 kDa for the six-His tag) (Fig. 1). rEhCP1 was purified by nickel affinity chromatography and reacted with rabbit polyclonal anti-EhCP1 Ab and monoclonal antithioredoxin Ab (Fig. 1).
FIG. 1.
Purification of recombinant thio-pro-EhCP1 and refolded, active rEhCP1. Recombinant thio-pro-EhCP1 was purified by Ni affinity chromatography, analyzed by 12% SDS-PAGE, and visualized by Coomassie blue staining (lane 1), probing of an immunoblot with polyclonal rabbit anti-EhCP1 Ab (lane 2), and probing of an immunoblot with mouse monoclonal antithioredoxin Ab (lane 3). Hi-Trap-Q-purified, refolded, and active EhCP1 was analyzed by SDS-PAGE (12% gels, silver stained) (lane 4), gelatin zymography (12% SDS-PAGE, 0.1% gelatin, run under nonreducing conditions) (lane 5), and immunoblotting (rabbit polyclonal anti-EhCP1 Ab) (lane 6). Numbers at left are molecular masses in kilodaltons.
The optimal refolding conditions to produce active rEhCP1 were determined by using a 16-condition screen as previously reported (3, 38). The most effective conditions were identified by cleavage of the synthetic peptide substrate Z-Arg-Arg-AMC. Refolded rEhCP1 was unique in several ways. (i) Refolded rEhCP1 did not require preincubation at low pH, which is usually required for the self-activation from the proform to the mature form of eukaryotic cysteine proteinases. (ii) Refolded rEhCP1 could be activated in a Tris-EDTA, phosphate-EDTA, or citrate-EDTA buffer at neutral or slightly alkaline pH (7.0 to 7.5) with 2 to 5 mM dithiothreitol (DTT). (iii) Refolded rEhCP1 could be activated with 5 to 10 mM cysteine alone. rEhCP1 activation by cysteine is of importance as it would not affect the in vitro cleavage of several of its putative biological substrates, particularly C3, which is readily denatured in the presence of other thiol-reducing agents such as DTT (43) or 2-mercaptoethanol, each of which is widely used for the in vitro activation of cysteine proteinases.
Refolded active rEhCP1 was purified by ion-exchange chromatography with a Hi-Trap-Q column (Amersham Biosciences, United Kingdom). As shown in Fig. 1, analysis of the peak fractions revealed the presence of one polypeptide band with a molecular mass of ∼31 kDa shown by silver staining of 12% SDS gels, gelatin zymography, and probing of Western blots with polyclonal anti-EhCP1 Ab. The active recombinant enzyme ran true to Mr, even on gelatin gels, in contrast to findings by Hellberg et al. (16), who found that a gelatinolytic band of ∼48 kDa had the same N-terminal peptide sequence as did mature EhCP1.
Substrate specificity of rEhCP1.
The preliminary substrate specificity of refolded, active rEhCP1 was determined with fluorogenic peptide substrates. Z-Arg-Arg-AMC and Z-Ala-Arg-Arg-AMC, typical cathepsin B substrates, were readily cleaved, whereas Z-Phe-Arg-AMC, a typical cathepsin L substrate, was not (data not shown). To further define the substrate requirements of rEhCP1, we mapped the specificity of the rEhCP1 active site using two synthetic tetrapeptide combinatorial libraries (Fig. 2). An almost absolute requirement for arginine at the P2 position was found.
FIG. 2.
Substrate specificity determined by synthetic combinatorial libraries. A P1 complete diverse library and P2, P3, and P4 sublibraries of the P1-lysine fixed library were used to determine the substrate specificities of EhCP1. Activities are displayed as percentages of the maximum at each position. Amino acids are represented by the single-letter code (“n” is norleucine). Error bars represent the standard deviations from the results of duplicate experiments.
pH optimum of rEhCP1.
rEhCP1 was found to have a pH optimum of 6.0, retaining most of its catalytic activity in the pH range of 5.5 to 7.5 (data not shown). A 50% or higher reduction in the catalytic activity is seen at pH values of ≤5.0 or ≥8.0.
Biological substrate specificity of rEhCP1.
To determine if rEhCP1 had the same biological substrate specificity as the secreted native proteinases, human C3, IgG, and pro-IL-18 were incubated with equal amounts of active purified rEhCP1 or native Eh-secreted proteinases (CM). rEhCP1 cleaves C3 in a fashion identical to that of the secreted native proteinases (43), generating the α′ subunit (Fig. 3A). rEhCP1 also cleaves the human IgG heavy chain in a dose- (Fig. 3B) and time-dependent manner (53) (data not shown), similar to native proteinases. Human pro-IL-18 is also cleaved by rEhCP1, similar to the degradation seen by released amebic proteinases (CM) and rEhCP5 (Fig. 3C) (37).
FIG. 3.
Cleavage of biological substrates by rEhCP1. (A) Human complement component C3 (1 μg) was incubated with rEhCP1 (0 to 600 RFU) or Hi-Trap-Q-purified HM1 CM (50 RFU) for 60 min at 37°C, analyzed by SDS-PAGE, and stained with Coomassie blue. Both native and recombinant proteinases cleaved the α to an α′ chain, which was inhibited by preincubation with E-64. (B) Human IgG (2 μg) was digested with CM or purified rEhCP1 (1,400 RFU) for 16 h at 37°C, and the resulting immunoblots were probed with goat anti-human IgG-horseradish peroxidase conjugate. The cleavage was inhibited by preincubating the proteinases with E-64. The heavy (H) and light (L) chains of IgG are indicated on the right. (C) Recombinant pro-IL-18 (0.4 μg) was incubated for 1 h at 37°C in buffer alone, native proteinases from CM (750 RFU), or 200 to 800 RFU of rEhCP1. The degradation of pro-IL-18 was completely inhibited by preincubation with E-64. Molecular weights (in thousands) are labeled at the left of each panel.
Intracellular localization of EhCP1.
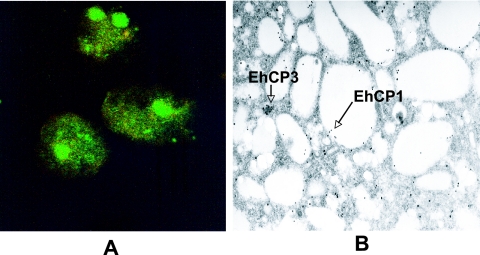
In order to determine the intracellular localization of EhCP1, Entamoeba histolytica trophozoites were labeled with rabbit polyclonal anti-EhCP1 Ab and imaged by fluorescence, confocal, and electron microscopy. EhCP1 localizes within large cytoplasmic vesicles, which are distinct from those containing EhCP3 (Fig. 4A). These results were confirmed by immunoelectron microscopy (Fig. 4B).
FIG. 4.
Localization of EhCP1. EhCP1 was localized in amebic trophozoites by confocal microscopy (A) using polyclonal Ab to EhCP1 (green) and monoclonal Ab to EhCP3 (red). These results were confirmed by immunoelectron microscopy (B) where EhCP1 (15-nm gold particles) was present in large vesicles, distinct from EhCP3 (20-nm gold particles).
Inhibition of EhCP1 by vinyl sulfone inhibitors.
We first tested the inhibition of purified rEhCP1 by K11777, a vinyl sulfone inhibitor which has been shown to cure Trypanosoma cruzi infection in animals (10), has undergone extensive toxicity testing (1), and is approaching clinical trials for Chagas' disease. The Kass (kinact/Ki app) of K11777 against EhCP1 was 350 (1/M·1/s). We next synthesized a peptidomimetic vinyl sulfone with an arginine substituted for phenylalanine in the P2 position based on the distinct specificity for arginine in P2 detected by active site mapping (Fig. 2). The resulting Kass for WRR483 was 849,000 (1/M·1/s), an increase of ∼2,500-fold.
Effect of the cysteine proteinase inhibitors K11777 and WRR483 on amebic invasion in the intestinal xenograft model.
We next tested the efficacy of the inhibitors in an in vivo model of invasion of human intestine. The pathology of human colon xenografts infected for 24 h with virulent E. histolytica trophozoites closely mimics human disease, with undermined ulcers containing trophozoites interspersed with normal mucosa (Fig. 5, left). When colon xenografts were infected with E. histolytica trophozoites preincubated with the cysteine proteinase inhibitor K11777 (1) (data not shown) or WRR483 (Fig. 5, right), no invasion was seen. Amebic invasion in human colon xenografts was quantified by a real-time PCR assay that could detect as few as 10 trophozoites (data not shown). The amplification of human Alu sequences in all of the xenografts was equivalent (15.93 ± 1.3) (56). Xenografts infected with E. histolytica trophozoites preincubated with the specific irreversible vinyl sulfone cysteine proteinase inhibitor K11777 had a >80% reduction in the number of detectable E. histolytica trophozoites (Fig. 6A). Inhibition of amebic invasion with the vinyl sulfone cysteine proteinase inhibitor WRR483 was even more dramatic, with a >95% reduction in the number of detectable trophozoites in the xenograft tissue (Fig. 6B).
FIG. 5.
Pathology of invasion of E. histolytica in human intestinal xenografts. Sections of intestinal xenografts of human colon in SCID mice 24 h following luminal injection of 1 × 106 Entamoeba histolytica (HM1) trophozoites are shown. (Left) Acute inflammatory infiltrate and undermining of the mucosa in infected xenografts. The lumen is filled with proteinaceous debris and clumps of trophozoites (closed arrows). Neutrophils (mouse) are shown with open arrowheads. (Right) Xenografts from mice treated with the cysteine proteinase inhibitor WRR483 showed no evidence of invasion (hematoxylin and eosin stain).
FIG. 6.
Quantification of amebic invasion in colonic xenografts. Human intestinal xenografts were infected with 1 × 106 Entamoeba histolytica trophozoites which had been preincubated in the presence or absence of 20 μM specific irreversible cysteine proteinase inhibitor K11777 (A) or 10 μM of WRR483 (B). At 24 h of incubation the xenografts were harvested and DNA was extracted from 100 mg of xenograft tissue. The amount of E. histolytica trophozoites present in each sample of tissue was determined by a real-time PCR assay using a standard curve which related the number of E. histolytica trophozoites to the cycle threshold. Values represent the means ± standard errors of duplicates from three xenograft experiments.
DISCUSSION
Released cysteine proteinases play a fundamental role in invasion of target tissues by E. histolytica trophozoites. Analysis of the E. histolytica genome reveals that 40 genes encode cysteine proteinases (25). Studies of the expression of the cysteine proteinase genes have shown that only three, ehcp1, ehcp2, and ehcp5, account for more than 90% of the cysteine proteinase-specific transcripts (5, 6) and more than 95% of secreted amebic proteinases in vitro (6, 8, 17). We focused on EhCP1 because it is unique to E. histolytica, there being no homologous gene in the closely related but noninvasive E. dispar (5). It is also one of the most highly expressed and released cysteine proteinases (16, 17). Because of the difficulty in separating the large number of similar native cysteine proteinases, we focused on the expression of active, recombinant enzyme. We have previously expressed active EhCP2 and EhCP3 in baculovirus (36) and EhCP5 in Pichia (37). We expressed the proenzyme of EhCP1 in E. coli as a thioredoxin fusion with a 3′ histidine tag which autocatalytically processed itself to the mature, active enzyme during refolding (Fig. 1). Surprisingly, purified, refolded rEhCP1 did not require activation by preincubation at low pH or with thiol reagents, such as DTT, 2-mercaptoethanol, or cysteine, which distinguishes rEhCP1 from previously purified recombinant parasite cysteine proteinases (16, 33, 44, 45, 47, 48). Purified refolded rEhCP1 has a pH optimum of 6.0, although 50% or more of its catalytic activity is retained across a pH range of 5.0 to 8.5. This broad pH range is consistent with the release of native EhCP1 in the neutral environment of the large bowel (29).
We have shown previously by homology modeling that the structures of EhCP2, EhCP3, and EhCP5 are characteristic of cathepsin L proteins with the ERFNIN motif, but their ability to accept positively charged amino acids at P2 resembles cathepsin B proteins (36). To further investigate this peptide substrate preference, we mapped the specificity of the active site cleft of rEhCP1 using two positional-scanning synthetic tetrapeptide combinatorial libraries (15, 27). We found an almost absolute preference for arginine at the P2 position (Fig. 2), an unusual specificity among clan CA proteinases. We took advantage of this unique substrate specificity to design a vinyl sulfone inhibitor, WRR483, from the scaffold compound K11777.
rEhCP1 cleaves a number of key physiologic substrates equivalently to the native enzymes. For example, active rEhCP1 readily cleaves the α chain of human complement component C3 (Fig. 3A), which we have shown mimics the cleavage of C3 convertases, producing an active C3b molecule (43). The cleavage site is actually one amino acid residue distal to native convertases, placing an arginine in the P2 position of EhCP1 (43). The released C3a anaphylatoxin fragment is further degraded by the amebic cysteine proteinases (41), limiting the host inflammatory response. Human IgG is also cleaved by refolded active rEhCP1 (Fig. 3B) as previously shown with purified amebic proteinases (53). This observation is consistent with recent clinical findings that systemic antiamebic IgG responses do not protect people from reinfection with E. histolytica (12, 13, 14). Previously, we have demonstrated that rEhCP5, the only other cysteine proteinase unique to E. histolytica, degraded pro-IL-18, a finding that implicates amebic cysteine proteinases in the observed lack of neutrophils in amebic lesions (37). Now we have found that rEhCP1 produces a similar dose- (Fig. 3C) and time-dependent degradation of pro-IL-18.
The intracellular localization is known for only a few amebic cysteine proteinases. EhCP5 (18), EhCP2 (35, 36), and EhCP112 (34) are membrane associated, while EhCP3 is intracellular (17, 36). By using confocal and immunoelectron microscopy, we found that EhCP1 localizes to large cytoplasmic vesicles, sites distinct from the vesicles containing EhCP3 (Fig. 4). These are likely the larger, prephagocytic, nonacidic vesicles, which were shown to contain EhCP1 in proteomic studies (35). This localization is consistent with the higher pH optimum (6.0) of EhCP1 as well. We had previously shown that EhCP1, EhCP2, and EhCP5 are passively released during phagocytosis (17). This release is likely to be part of the newly described EhRab11B-associated secretory pathway, as overexpression of EhRab11B led to increased release of all three proteinases (30).
Animal models of amebiasis have been problematic as only humans and higher primates are naturally susceptible to infection. Cysteine proteinase inhibitors such as E-64 and an EhCP5 antisense construct, which blocked expression of multiple cysteine proteinases, inhibited amebic liver abscess formation in SCID mice (50) and hamsters (2). The trophozoites had to be injected directly into the liver, however, which bypasses the normal route of infection through the colon. To avoid this limitation, we closely mimicked human infection by using the human xenograft model (55). When fetal intestinal loops are implanted under the skin of SCID mice, they develop physiologically normal colonic mucosa. Following intraluminal injection of invasive E. histolytica trophozoites, classic flask-shaped ulcers formed in the human graft (Fig. 5).
To test the proof of principle that EhCP1 and released cysteine proteinases are critical for invasion of the human bowel, we evaluated the effect of inhibition of EhCP1 by the vinyl sulfone inhibitor K11777, which has a Kass of 350 (1/M·1/s) for rEhCP1. We initially chose this inhibitor because it has undergone extensive pharmacokinetic, bioavailability, and toxicity testing (1) and is approaching phase I clinical trials for treatment of Chagas' disease. We quantified invasion by real time-PCR, with the specific amplification of a short sequence from the amebic peroxiredoxin gene (7, 40). Our initial results show for the first time that a specific cysteine proteinase inhibitor blocks amebic invasion of the human colon by more than 80% (Fig. 6A).
We next tested the new vinyl sulfone inhibitor WRR483, which was synthesized with arginine at the P2 position instead of phenylalanine to specifically target EhCP1. This specific inhibitor was almost 2,500-fold more potent than K11777 against rEhCP1 (849,000 1/M·1/s). WRR483 is even more effective than K11777 at blocking amebic invasion in the human intestinal xenograft model (Fig. 5, right), as it reduced invasion by more than 95% (Fig. 6B).
Cysteine proteinases are an attractive target for agents designed to disrupt invasion by E. histolytica. The unique requirement of EhCP1 for arginine in the P2 position and the demonstrated ability of a specific cysteine proteinase inhibitor with these particular structural features (WRR483) to block amebic invasion of the bowel support the feasibility of this approach. We have now shown that the expression of active recombinant enzymes will enable us to dissect the role of individual cysteine proteinases in the virulence of this important protozoan parasite. Also, these recombinant enzymes will be a fundamental tool as we address the question of whether E. histolytica releases a higher level of cysteine proteinases facilitating invasion, in contrast to noninvasive E. dispar, or whether the key determinants of its invasiveness are the unique proteinases, EhCP1 and EhCP5. We expect that our continuing efforts in specific inhibition of these unique proteinases will shed yet more light on their roles in the pathogenesis of amebiasis. In the meantime, our current studies have proven that inhibition of amebic cysteine proteinases blocks invasion of the human bowel and have identified a promising, orally available scaffold inhibitor.
Acknowledgments
This work was supported in part by United States Public Health Service grants AI49531 (S.L.R.), DK35108 (S.L.R.), AI35707 (J.M.), and AI30084 (S.L.S.) and the Sandler Center for Basic Research in Parasitic Diseases. S.G.M.-L. was a recipient of a PROMEP-UABC Scholarship (UABC-123, Clave P/PROMEP: UABC-2000-12-01; Ministry of Public Education, México).
We thank Fran Gillin and Charles Davis for their helpful comments.
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Abdulla, M. H., K.-C. Lim, M. Sajid, J. H. McKerrow, and C. R. Caffrey. 2007. Schistosomiasis mansoni: novel chemotherapy using a cysteine protease inhibitor. PLoS Med. 4:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankri, S., T. Stolarsky, R. Bracha, F. Padilla-Vaca, and D. Mirelman. 1999. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect. Immun. 67:421-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, N., A. De Lencastre, and E. Gouaux. 1999. A new protein folding screen: application to the ligand binding domains of glutamate and kinate receptor and to lysozyme and carbonic anhydrase. Protein Sci. 8:1475-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieth, J. G. 1995. Theoretical and practical aspects of proteinase inhibition kinetics. Methods Enzymol. 248:59-84. [DOI] [PubMed] [Google Scholar]
- 5.Bruchhaus, I., T. Jacobs, M. Leippe, and E. Tannich. 1996. Entamoeba histolytica and Entamoeba dispar: differences in numbers and expression of cysteine proteinase genes. Mol. Microbiol. 22:255-263. [DOI] [PubMed] [Google Scholar]
- 6.Bruchhaus, I., B. J. Loftus, N. Hall, and E. Tannich. 2003. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2:501-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, M., D. Sajed, L. Poole, K. Hirata, D. S. Herdman, B. E. Torian, and S. L. Reed. 2005. An unusual surface peroxiredoxin protects invasive Entamoeba histolytica from oxidant attack. Mol. Biochem. Parasitol. 143:80-89. [DOI] [PubMed] [Google Scholar]
- 8.Davis, P. H., J. Schulze, and S. L. Stanley, Jr. 2007. Transcriptomic comparison of two Entamoeba histolytica strains with defined virulence phenotypes identifies new virulence factor candidates and key differences in the expression patterns of cysteine proteases, lectin light chains, and calmodulin. Mol. Biochem. Parasitol. 151:118-128. [DOI] [PubMed] [Google Scholar]
- 9.Diamond, L. S., D. R. Harlow, and C. C. Cunnick. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431-432. [DOI] [PubMed] [Google Scholar]
- 10.Engel, J. C., P. S. Doyle, I. Hsieh, and J. H. McKerrow. 1998. Cysteine proteinase inhibitors cure an experimental Trypanosoma cruzi infection. J. Exp. Med. 188:725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilchrist, C. A., E. Houpt, N. Trapaidze, Z. Fei, O. Crasta, A. Asgharpour, C. Evans, S. Martino-Catt, D. J. Baba, S. Stroup, S. Hamano, G. Ehrenkaufer, M. Okada, U. Singh, T. Nozaki, B. J. Mann, and W. A. Petri, Jr. 2006. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol. Biochem. Parasitol. 147:163-176. [DOI] [PubMed] [Google Scholar]
- 12.Haque, R., I. M. Ali, R. B. Sack, B. M. Farr, G. Ramakrishnan, and W. A. Petri. 2001. Amebiasis and mucosal IgA antibody against the Entamoeba histolytica adherence lectin in Bangladeshi children. J. Infect. Dis. 183:1787-1793. [DOI] [PubMed] [Google Scholar]
- 13.Haque, R., P. Dugal, I. M. Ali, M. B. Hossain, D. Mondal, R. B. Sack, B. M. Farr, T. H. Beaty, and W. A. Petri. 2002. Innate and acquired resistance to amebiasis in Bangladeshi children. J. Infect. Dis. 186:547-552. [DOI] [PubMed] [Google Scholar]
- 14.Haque, R., D. Mondal, P. Duggal, M. Kabir, S. Roy, B. M. Farr, R. B. Sack, and W. A. Petri. 2006. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect. Immun. 74:904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, J. L., B. J. Backes, F. Leonetti, S. Mahrus, J. A. Ellman, and C. S. Craik. 2000. A rapid and general profiling of protease specificity by using combinational fluorogenic substrate libraries. Proc. Natl. Acad. Sci. USA 97:7754-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellberg, A., M. Leippe, and I. Bruchhaus. 2000. Two major ‘higher molecular mass proteinases’ of Entamoeba histolytica are identified as cysteine proteinases 1 and 2. Mol. Biochem. Parasitol. 105:305-309. [DOI] [PubMed] [Google Scholar]
- 17.Hirata, K., X. Que, S. G. Meléndez-López, A. Debnath, S. Myers, S. Herdman, E. Orozco, A. Bhattacharya, J. McKerrow, and S. L. Reed. 2007. A phagocytosis mutant of Entamoeba histolytica is less virulent due to deficient proteinase expression and release. Exp. Parasitol. 115:192-199. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, T., I. Bruchhaus, T. Dandekar, E. Tannich, and M. Leippe. 1998. Isolation and molecular characterization of a surface-bound proteinase of Entamoeba histolytica. Mol. Microbiol. 27:269-276. [DOI] [PubMed] [Google Scholar]
- 19.Keene, W. E., M. G. Pettit, S. Allen, and J. H. McKerrow. 1986. The major neutral proteinase of Entamoeba histolytica. J. Exp. Med. 163:536-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keene, W. E., M. E. Hidalgo, E. Orozco, and J. H. McKerrow. 1990. Entamoeba histolytica: correlation of the cytopathic effect of virulent trophozoites with secretion of cysteine proteinase. Exp. Parasitol. 71:199-206. [DOI] [PubMed] [Google Scholar]
- 21.Kelsall, B. L., and J. I. Ravdin. 1993. Degradation of human IgA by Entamoeba histolytica. J. Infect. Dis. 168:1319-1322. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. H., T. Azam, D. Y. Yoon, L. L. Reznikov, D. Novick, M. Rubenstein, and C. A. Dinarello. 2001. Site specific mutations in the mature form of human IL-18 with enhanced biological activity and decreased neutralization by IL-18 binding protein. Proc. Natl. Acad. Sci. USA 98:3304-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauwaet, T., M. J. Oliveira, B. Callewaert, G. De Bruyne, M. Mareel, and A. Leroy. 2004. Proteinase inhibitors TPCK and TLCK prevent Entamoeba histolytica disturbance of tight junctions and microvilli in enteric cell layers in vitro. Int. J. Parasitol. 34:785-794. [DOI] [PubMed] [Google Scholar]
- 24.Leduc, E. H., and W. Bernhard. 1967. Recent modifications of the glycol methacrylate embedding procedure. J. Ultrastruct. Res. 19:196-199. [DOI] [PubMed] [Google Scholar]
- 25.Loftus, B., I. Anderson, R. Davies, U. C. M. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillén, C. Gilchrist, S. E. Stroup, S. Bhattacharya, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, Jr., C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser, and N. Hall. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865-868. [DOI] [PubMed] [Google Scholar]
- 26.Mahmoudzadeh-Niknam, H., and J. H. McKerrow. 2004. Leishmania tropica: cysteine proteases are essential for growth and pathogenicity. Exp. Parasitol. 106:158-163. [DOI] [PubMed] [Google Scholar]
- 27.Maly, D., F. Leonetti, B. J. Backes, D. S. Dauber, J. L. Harris, C. S. Craik, and J. A. Ellman. 2002. Expedient solid phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J. Org. Chem. 67:910-915. [DOI] [PubMed] [Google Scholar]
- 28.Masser, C., L. Eckmann, G. Paesold, H. S. Kim, and M. F. Kagnoff. 2002. Ubiquitous production of macrophage migration inhibitory factor by human gastric and intestinal epithelium. Gastroenterology 122:667-680. [DOI] [PubMed] [Google Scholar]
- 29.McNeil, N. I., K. L. Ling, and J. Wager. 1987. Mucosal surface pH of the large intestine of the rat and of normal and inflamed large intestine in man. Gut 28:707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitra, B. N., Y. Saito-Nakano, K. Nakada-Tsukui, D. Sato, and T. Nozaki. 17 April 2007. RAB11B small GTPase regulates secretion of cysteine proteases in the enteric protozoan parasite Entamoeba histolytica. Cell. Microbiol. [Epub ahead of print.] [DOI] [PubMed]
- 31.Moncada, D., K. Keller, and K. Chadee. 2003. Entamoeba histolytica cysteine proteinases disrupt the polymeric structure of colonic mucin and alter its protective function. Infect. Immun. 71:838-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moncada, D., K. Keller, S. Ankir, D. Mirelman, and K. Chadee. 2006. Antisense inhibition of Entamoeba histolytica cysteine proteases inhibits colonic mucus degradation. Gastroenterology 130:721-730. [DOI] [PubMed] [Google Scholar]
- 33.Na, B. K., B. R. Shenai, P. S. Sijwali, Y. Choe, K. C. Pandey, A. Singh, C. S. Craik, and P. J. Rosenthal. 2004. Identification and biochemical characterization of vivapains, cysteine proteases of the malaria parasite Plasmodium vivax. Biochem. J. 378:529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ocadiz, R., E. Orozco, E. Carrillo, L. Quintas, J. Ortega-Lopez, R. García-Pérez, T. Sánchez, B. A. Castillo-Juárez, G. García-Rivera, and M. A. Rodríguez. 2005. EhCP112 is an Entamoeba histolytica secreted cysteine proteinase that maybe involved in the parasite virulence. Cell. Microbiol. 7:221-232. [DOI] [PubMed] [Google Scholar]
- 35.Okada, M., C. D. Huston, B. J. Mann, W. A. Petri, K. Kita, and T. Nozaki. 2005. Proteomic analysis of phagocytosis of the protozoan parasite Entamoeba histolytica. Eukaryot. Cell 4:827-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Que, X., L. S. Brinen, P. Perkins, D. S. Herdman, K. Hirata, B. E. Torian, H. M. Rubin, J. H. McKerrow, and S. L. Reed. 2002. Cysteine proteinases from distinct cellular compartments are recruited to phagocytic vesicles by Entamoeba histolytica. Mol. Biochem. Parasitol. 119:23-32. [DOI] [PubMed] [Google Scholar]
- 37.Que, X., S. H. Kim, M. Sajid, L. Eckmann, C. A. Dinarello, J. H. McKerrow, and S. L. Reed. 2003. A surface amebic cysteine proteinase inactivates interleukin-18. Infect. Immun. 71:1274-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Que, X., H. Ngo, J. Lawton, M. Gray, Q. Liu, J. Engel, L. Brinen, P. Ghosh, K. Joiner, and S. L. Reed. 2002. The cathepsin B of Toxoplasma gondii, toxopain-1, is critical for parasite invasion and rhoptry protein processing. J. Biol. Chem. 277:25791-25797. [DOI] [PubMed] [Google Scholar]
- 39.Que, X., and S. L. Reed. 2000. Cysteine proteinases and the pathogenesis of amebiasis. Clin. Microbiol. Rev. 13:196-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Que, X., A. Wunderlich, K. A. Joiner, and S. L. Reed. 2004. Toxopain-1 is critical for infection in a novel chicken embryo model of congenital toxoplasmosis. Infect. Immun. 72:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed, S. L., J. A. Ember, D. S. Herdman, R. G. DiScipio, T. E. Hugli, and I. Gigli. 1995. The extracellular neutral cysteine proteinase of Entamoeba histolytica degrades anaphylatoxins C3a and C5a. J. Immunol. 155:266-274. [PubMed] [Google Scholar]
- 42.Reed, S. L., W. E. Keene, and J. H. McKerrow. 1989. Thiol proteinase expression and pathogenicity of Entamoeba histolytica. J. Clin. Microbiol. 27:2772-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed, S. L., W. E. Keene, J. H. McKerrow, and I. Gigli. 1989. Cleavage of C3 by a neutral cysteine proteinase of Entamoeba histolytica. J. Immunol. 143:189-195. [PubMed] [Google Scholar]
- 44.Sajid, M., and J. H. McKerrow. 2002. Cysteine proteases of parasitic organisms. Mol. Biochem. Parasitol. 120:1-21. [DOI] [PubMed] [Google Scholar]
- 45.Sanderson, S. J., K. J. Pollock, J. D. Hilley, M. Meldal, P. S. Hilaire, M. A. Juliano, L. Juliano, J. C. Mottram, and G. H. Coombs. 2000. Expression and characterization of a recombinant cysteine proteinase of Leishmania mexicana. Biochem. J. 347:383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seydel, K. B., E. Li, Z. Zhang, and S. L. Stanley. 1998. Epithelial cell-initiated inflammation plays a crucial role in early tissue damage in amebic infection of human intestine. Gastroenterology 115:1446-1453. [DOI] [PubMed] [Google Scholar]
- 47.Sijwali, P. S., L. S. Brinen, and P. J. Rosenthal. 2001. Systematic optimization of expression and refolding of the Plasmodium falciparum cysteine protease falcipain-2. Protein Expr. Purif. 22:128-134. [DOI] [PubMed] [Google Scholar]
- 48.Sijwali, P. S., B. R. Shenai, J. Gut, A. Singh, and P. J. Rosenthal. 2001. Expression and characterization of the Plasmodium falciparum haemoglobinase falcipain-3. Biochem. J. 360:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanley, S. L. 2003. Amoebiasis. Lancet 361:1025-1034. [DOI] [PubMed] [Google Scholar]
- 50.Stanley, S. L., T. Zhang, D. Rubin, and E. Li. 1995. Role of the Entamoeba histolytica cysteine proteinase in amebic liver abscess formation in severe combined immunodeficient mice. Infect. Immun. 63:1587-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeuchi, A., and B. P. Phillips. 1975. Electron microscope studies of experimental Entamoeba histolytica infections in the guinea pig. I. Penetration of the intestinal epithelium by trophozoites. Am. J. Trop. Med. Hyg. 24:34-48. [DOI] [PubMed] [Google Scholar]
- 52.Tian, W.-X., and C.-L. Tsou. 1982. Determination of the rate constant of enzyme by measuring the substrate reaction in the presence of modifier. Biochemistry 21:1028-1032. [DOI] [PubMed] [Google Scholar]
- 53.Tran, V. Q., D. S. Herdman, B. E. Torian, and S. L. Reed. 1998. The neutral cysteine proteinase of Entamoeba histolytica degrades IgG and prevents its binding. J. Infect. Dis. 177:508-511. [DOI] [PubMed] [Google Scholar]
- 54.Willhoeft, U., L. Hamann, and E. Tannich. 1999. A DNA sequence corresponding to the gene encoding cysteine proteinase 5 in Entamoeba histolytica is present and positionally conserved but highly degenerated in Entamoeba dispar. Infect. Immun. 67:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Z., L. Yan, L. Wang, K. B. Seydel, E. Li, S. Ankri, D. Mirelman, and S. L. Stanley. 2000. Entamoeba histolytica cysteine proteinases with interleukin-1 beta converting enzyme (ICE) activity cause intestinal inflammation and tissue damage in amoebiasis. Mol. Microbiol. 37:542-548. [DOI] [PubMed] [Google Scholar]
- 56.Zijlstra, A., R. Mellor, G. Panzarella, R. T. Aimes, J. D. Hooper, N. D. Marchenko, and J. P. Quigley. 2002. A quantitative analysis of rate-limiting steps in the metastatic cascade using human-specific real-time polymerase chain reaction. Cancer Res. 62:7083-7092. [PubMed] [Google Scholar]