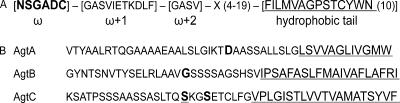Abstract
In the genome sequence of Aspergillus niger CBS 513.88, three genes were identified with high similarity to fungal α-amylases. The protein sequences derived from these genes were different in two ways from all described fungal α-amylases: they were predicted to be glycosylphosphatidylinositol anchored, and some highly conserved amino acids of enzymes in the α-amylase family were absent. We expressed two of these enzymes in a suitable A. niger strain and characterized the purified proteins. Both enzymes showed transglycosylation activity on donor substrates with α-(1,4)-glycosidic bonds and at least five anhydroglucose units. The enzymes, designated AgtA and AgtB, produced new α-(1,4)-glycosidic bonds and therefore belong to the group of the 4-α-glucanotransferases (EC 2.4.1.25). Their reaction products reached a degree of polymerization of at least 30. Maltose and larger maltooligosaccharides were the most efficient acceptor substrates, although AgtA also used small nigerooligosaccharides containing α-(1,3)-glycosidic bonds as acceptor substrate. An agtA knockout of A. niger showed an increased susceptibility towards the cell wall-disrupting compound calcofluor white, indicating a cell wall integrity defect in this strain. Homologues of AgtA and AgtB are present in other fungal species with α-glucans in their cell walls, but not in yeast species lacking cell wall α-glucan. Possible roles for these enzymes in the synthesis and/or maintenance of the fungal cell wall are discussed.
Aspergillus niger is a filamentous ascomycete fungus with a worldwide distribution. As a saprophyte, the fungus produces and secretes a large variety of extracellular enzymes, especially proteases and polysaccharide hydrolases to convert plant cell walls and storage compounds into growth substrates (see, e.g., references 19 and 44). This quality is exploited for the production of enzymes for the food and feed industry on a large scale. Recently, the full genome sequence of A. niger CBS 513.88 was determined and annotated (55). A high level of synteny was observed between A. niger and other sequenced aspergilli, although more extracellular hydrolytic enzymes were annotated for A. niger. A detailed full genome search showed the presence of a considerable number of previously unknown, predicted enzymes belonging to the α-amylase superfamily.
The α-amylase superfamily (38) comprises a large variety of enzymes that are active towards polysaccharides with α-glycosidic linkages, such as starch and glycogen (42). Most members of this family are involved in either production of storage compounds, such as glycogen or starch, or degradation of these compounds as extracellular carbon and energy sources. The tertiary structure of these enzymes is characterized by a (β/α)8 barrel containing four highly conserved amino acid regions that form the catalytic site (42) (see also the CAZy website at http://afmb.cnrs-mrs.fr/CAZY/). Based on sequence similarity, members of the α-amylase superfamily are divided over glycoside hydrolase (GH) families 13, 70, and 77 (GH13, GH70, and GH77, respectively). Here, we focus on family GH13, which mostly contains enzymes that perform a hydrolytic reaction, i.e., they cleave an α-glycosidic linkage using water as an acceptor molecule. The best known hydrolytic enzyme of GH13 is α-amylase (EC 3.2.1.1), which hydrolyzes the internal α-(1,4)-glycosidic bonds in starch, glycogen, and maltooligosaccharides [α-(1,4)-linked glucose oligomers], producing shorter maltooligosaccharides and maltose [O-α-d-Glc-(1,4)-α-d-Glc]. Other GH13 members perform a transglycosylation (or glucanotransferase) reaction in which they cleave an α-(1,4) glycosidic bond in a donor substrate and subsequently do not use water but instead use another oligosaccharide as the acceptor substrate to form a new α-glycosidic linkage.
A. niger produces a number of extracellular enzymes classified as members of GH13, which are involved in the degradation of starch. These include acid amylase, which is well-known for its stability at low pH (10) and two almost identical α-amylase enzymes (AmyA/B) (34). We have identified several additional members of GH13 in the A. niger genome sequence, three of which clustered together in the phylogenetic tree of the GH13 members and showed a typical glycosylphosphatidylinositol (GPI) anchoring signal at the protein C terminus. A GPI anchor serves as a targeting signal to the cell membrane and/or the cell wall.
The cell walls of aspergilli have been shown to contain four major classes of polysaccharides: chitin, α-glucan, β-(1,3)-glucan, and galactomannan (1, 8, 27, 64). In addition, it contains covalently attached cell wall proteins (12). The α-glucan fraction is composed of an α-(1,3)-glucan with 3 to 10% α-(1,4)-glycosidic linkages (24, 27), and nigeran, a glucan with alternating α-(1,3) and α-(1,4) glycosidic bonds (9). α-(1,3)-Glucan synthases have been identified and functionally studied in a number of different fungi (3, 16, 23, 62). These enzymes are large proteins (∼2,400 amino acids long) consisting of three conserved domains which are predicted to be involved in the synthesis, transport, and cross-linking of the α-(1,3)-glucan (23). Detailed structural studies in Schizosaccharomyces pombe have revealed that the α-glucan is a linear glucose polymer of 260 residues in length consisting of two α-(1,3)-glucan chains that are interconnected via α-(1,4)-linked glucose residues (22). A mutation in the N-terminal part of the α-(1,3)-glucan synthases (the proposed cross-linking domain) abolished the linkage between the two α-(1,3)-glucan chains, indicating that this part of the protein acts as a glucanotransferase, connecting the glucan chains.
Recently, two types of putative GH13 enzymes have been shown to play a role in fungal cell wall α-(1,3)-glucan formation. Marion et al. (43) showed the involvement of a putative α-amylase (Amy1p) in the formation of α-(1,3)-glucan in the cell wall of Histoplasma capsulatum. In this pathogenic fungus, α-(1,3)-glucan is known to play an important role in virulence (61). A functional knockout of AMY1 resulted in a lack of α-(1,3)-glucan formation and decreased virulence. The second α-amylase homologue, Aah3p, was studied in S. pombe (47). A knockout strain of this GPI-anchored protein was hypersensitive towards cell wall-degrading enzymes and showed aberrant cell shape. The enzymatic activities of Amy1p and Aah3p have not been studied.
In this paper, we report the first biochemical characterization of two GH13 enzymes putatively involved in α-(1,3)-glucan formation. We expressed and purified two GPI-anchored enzymes from A. niger, both homologues of Aah3p from S. pombe. The biochemical characterization showed that the two A. niger enzymes are GH13 α-glucanotransferases, making them the first of their kind to be described for fungi. A gene knockout of one of the enzymes in A. niger resulted in increased sensitivity towards calcofluor white (CFW), a cell wall-disrupting compound.
MATERIALS AND METHODS
Bioinformatics tools.
The full genome sequence of A. niger strain CBS 513.88 was provided by DSM (a biotechnology company based in The Netherlands) (55). A Hidden Markov model (HMM) profile was built using the HMMR package (20) based on the amino acid sequences of described α-amylases, which were retrieved from the CAZy website (http://afmb.cnrs-mrs.fr/CAZY/) (14). The obtained profile was used to screen the A. niger CBS 513.88 genomic database using the WISE 2 package (7). The presence of a signal peptidase cleavage site and a GPI attachment site were predicted by web-based search tools (http://www.cbs.dtu.dk/services/SignalP/ [4] and http://mendel.imp.univie.ac.at/sat/gpi/gpi_server.html [21], respectively). The GPI attachment prediction was confirmed by a manual comparison of the protein sequences with the consensus sequence for yeast GPI proteins as described by De Groot et al. (17). Amino acid sequence alignments and phylogenetic analysis were performed using MEGA3.1 (37) and adjusted manually if necessary. Sequences from other fungal genomes were retrieved via the option genomic BLAST at NCBI (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=fungi).
Aspergillus niger strains, growth conditions, and transformation.
A. niger strain N402 (cspA1 derivative of ATCC 9029 [11]) mRNA was used for the construction of a cDNA library. Genomic DNA from A. niger NRRL3122 and N402 strains was isolated and used as a template in PCRs. A. niger strain MGG029-ΔaamA (68) was used as a host for protein overexpression. This strain, derived from strain MGG029 (prtT glaA::fleor pyrG), is deficient in the expression of several extracellular proteases, and it has no glucoamylase gene (glaA) and acid amylase gene (aamA), resulting in very poor growth on starch (68). Strain MA70.15 (pyrG− ku70::amdS) (46) was used for disruption of the agtA gene.
Aspergillus strains were grown in Aspergillus minimal medium (MM) or complete medium (CM) which is MM with the addition of 0.1% Casamino Acids and 0.5% yeast extract (Oxoid, Basingstoke, United Kingdom) (6). Cultures for protein production were grown in CMS (CM supplemented with 2% [wt/vol] sucrose and 1% [wt/vol] glucose). Spores were obtained by growing A. niger on CM with 2% (wt/vol) agar for 4 days and scraping off the spores in 0.9% (wt/vol) NaCl. Liquid cultures were inoculated with 106 spores liter−1 medium and subsequently grown at 30°C while shaking at 280 rpm. Transformation of A. niger was performed as described previously (58) using lysing enzymes (Sigma, Zwijndrecht, The Netherlands). Selection of positive clones was performed on the basis of their ability to grow successively on MM containing 15 mM CsCl and 10 mM aceetamide or acrylamide as the sole nitrogen source, brought about by the expression of the amdS gene (32).
Cloning procedures.
All basic molecular techniques were performed according to standard procedures (63). Escherichia coli TOP10 (Invitrogen, Carlsbad, CA) or DH5α (Stratagene, La Jolla, CA) was used for transformation and amplification of recombinant DNA. The primers used were obtained from Eurogentec (Seraing, Belgium) or Biolegio (Nijmegen, The Netherlands). All steps during the construction of the overexpression vectors were checked by restriction analysis, and the final constructs were checked by sequencing (GATC Biotech AG, Konstanz, Germany). Genomic DNA was isolated from A. niger N402 and NRRL3122 as described by Kolar et al. (33). All PCRs were performed with 2.5 units of Pwo DNA polymerase (Roche, Indianapolis, IN), 1× buffer, and 1 mM of each deoxynucleoside triphosphate in a total volume of 25 μl. A cDNA library was produced from A. niger N402 grown on MM with the addition of starch as the sole carbon source. Primers used are indicated in Table 1.
TABLE 1.
Primers used for the production of plasmids for overexpression and deletion in A. niger
| Primer | Primer sequencea | Restriction enzyme(s) |
|---|---|---|
| Primers used for construction of pΔagtA | ||
| AgtAP1for | ATAAGAATGCGGCCGCTGTCCTGTGTGTTCCAGCCT | NotI |
| AgtAP2rev | GCTCTAGAAATGATCAAGGGTTGCGTACA | XbaI |
| AgtAP3for | GCTCTAGATATGCTGATAGCTACAGATGG | XbaI |
| AgtAP4rev | CGGGATCCGGAGTGGATAGCTGGTAAGGC | BamHI |
| Primers used for construction of Ppki-agtA-SBD-amdS | ||
| SBD-fw | GGCCCTATGCATGGCCCTGGGCCCACCTGTGCGGCCACATCTGC | NsiI, ApaI |
| SBD-rev | CCCGCTGCGGCCGCCTACCGCCAGGTGTCAGTCAC | NotI |
| AgtA-SBD-fw | GGCCCTATGCATATGGTCTCAATGTCGGCCCTGC | NsiI |
| AgtA-SBD-rev | CCGGGAGGGCCCTCCGCACAGCCCACTGCC | ApaI |
| Primers used for construction of Ppki-agtA-amdS and Ppki-agtB-amdS | ||
| AgtA-fw | GGCCCTATGCATGTCTCAATGTCGGCCCTGC | NsiI |
| AgtA-rev | CCCGCTGCGGCCGCTTACCACATCCCCACAATCA | NotI |
| AgtB-for | GGCCCTATGCATTTTCGAAAATCCGCTTCCCTC | NsiI |
| AgtB-rev | CCCGCTGCGGCCGCTTATATCCGGAATGCCAAAAAT | NotI |
The restriction sites used for cloning are underlined.
The overexpression vector for the transformation of A. niger was provided by J. Benen (Wageningen University, Wageningen, The Netherlands) and was produced as follows. Gene pgaII (encoding polygalacturonase II from A. niger) was cloned into pPROM-S (5) using NsiI and KpnI restriction sites. A NotI site was generated immediately downstream of the stop codon of the pgaII gene by site-directed mutagenesis. The gene encoding acetamidase (amdS [33]) was amplified by PCR with specific primers from plasmid p3SR2 (69) and cloned in front of the pki promoter region (54) using XbaI restriction sites, resulting in vector Ppki-pgaII-amdS. The construct for the overexpression of the chimeric protein AgtA-SBD (a fusion between AgtA and the starch binding domain [SBD] of A. niger glucoamylase GlaA) was made as follows. Genomic DNA from A. niger N402 was used as a template in a PCR with specific primers to generate the DNA fragment encoding the SBD, including the linker region. The primers were designed to amplify nucleotides 3643 to 4149 (numbering according to glaA coding sequence EMBL accession no. AY250996). An ApaI restriction site was built into the forward primer to allow the subsequent cloning of the agtA gene fragment in frame with the SBD. The SBD-encoding fragment was cloned into vector Ppki-pgaII-amdS using NsiI and NotI restriction sites, thereby replacing the pgaII gene. The cDNA fragment encoding agtA was generated by PCR with specific primers on the cDNA library. The primers were designed to amplify the gene up to nucleotide 1766 (in gene sequence), which does not include the C-terminal GPI-anchoring part. The agtA cDNA was cloned in frame N-terminally of the SBD using NsiI and ApaI, resulting in the expression vector Ppki-agtA-SBD-amdS. Sequencing of the construct revealed one point mutation compared to the original gene sequence in the genomic database. Nucleotide 1420 (in the coding sequence of agtA) was changed from A to G, resulting in Ala474 instead of Thr474 in the derived amino acid sequence. This mutation was consistently found in several independent clones and was therefore considered to represent a difference between strains N402 and CBS 513.88. The same mutation was found in the equivalent protein sequence published by the Department of Energy Joint Genome Institute (http://genome.jgi-psf.org/Aspni1/Aspni1.home.html).
The constructs for overexpression of agtA and agtB were produced as follows. The complete gene sequences of genes agtA and agtB were amplified with specific primers from genomic DNA isolated from A. niger NRRL3122 (67). The primers contained restriction sites for NsiI and NotI, which were used to clone the gene fragments into vector Ppki-pgaII-amdS, thereby replacing the pgaII gene. This resulted in the vectors Ppki-agtA-amdS and Ppki-agtB-amdS.
Protein production, purification, and detection procedures. (i) Production of AgtA-SBD.
Several stable transformants were checked for their level of extracellular production of the chimeric protein AgtA-SBD by Western blotting with polyclonal antiserum raised against purified SBD (antiserum kindly provided by D. Archer and D. McKensie [University of Nottingham & IFR Norwich, United Kingdom]) (41). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed according to standard protocols. Immobilon-P (Millipore, Billerica, MA) was used as the blotting membrane. The untransformed strain was included as a negative control, while purified SBD (M. J. van der Maarel et al., unpublished data) served as a positive control for immunodetection. The A. niger transformant producing the highest levels of AgtA-SBD was grown in a 5-liter batch fermentor (New Brunswick Scientific, Edison, NJ) inoculated with 100 ml culture pregrown on potato dextrose broth (Difco). The medium used for batch fermentation consisted of the following components: 3% (wt/vol) glucose, 117 mM NaNO3, 3.25 mM MgSO4·7H2O, 14.7 mM KH2PO4, 0.69 mM CaCl2·H2O, 0.5% yeast extract, and spore elements. Fermentor conditions were pH 4.5, a temperature of 30°C, and aeration at 1.2 liter min−1. The feed contained 20% (wt/vol) glucose, 74 mM KH2PO4, 350 mM NaNO3, 1% yeast extract, and 1% tryptone and was added at a rate of 5 ml h−1. Three days after the feed was started, the growth medium was collected by filtration over miracloth (Calbiochem, EMD Biosciences, La Jolla, CA). The pH of the medium was subsequently adjusted to pH 6 with 1 M NaOH, and AgtA-SBD was extracted from the medium via binding of the SBD to starch granules based on the procedure described by Paldi et al. (53). One liter of medium was added to 13 g waxy maize starch that was prewashed with elution buffer (10 mM sodium acetate [NaAc], pH 6) and incubated for 2 h at 4°C while shaking gently. The starch with bound proteins was collected by centrifugation (15 min at 5,000 × g) and washed once with ice-cold elution buffer followed by another centrifugation step. AgtA-SBD was eluted from the starch granules by the addition of 25 ml of elution buffer per batch of 13 g starch and subsequent incubation at 40°C for 1 h while shaking gently. After the starch granules were removed by centrifugation, the protein was collected from the supernatant. A second round of binding was performed to remove residual proteins and components of the medium. The protein was concentrated, and the buffer was changed to 20 mM Tris-HCl, pH 8, using a Centriprep YM50 column (Millipore, Bedford, MA). The sample was applied to an anion-exchange column (ResourceQ [1 ml]; Amersham Biosciences, Piscataway, NJ) equilibrated with 20 mM Tris-HCl, pH 8. Proteins were eluted with a NaCl gradient (0 to 1 M NaCl) at a flow rate of 1 ml min−1. AgtA-SBD was eluted as a single activity peak at 280 mM NaCl. Purification of AgtA-SBD was confirmed by Western blot analysis. Deglycosylation was performed with 850 U of endoglycosidase H (Endo H) (New England Biolabs, Ipswich, MA) on 1.5 μg of purified protein for 20 h in a total volume of 15 μl according to the manufacturer's instructions.
(ii) Production of AgtA and AgtB.
After transformation of each of the plasmids Ppki-agtA-amdS and Ppki-agtB-amdS into A. niger MGG029-ΔaamA, nine transformants were selected which showed the best growth on selective medium. The best transformant for overexpression of each protein was selected by growth in liquid CMS and visual inspection of protein production on SDS-polyacrylamide gels. The selected transformants were grown in CMS for 3 days at 30°C and 200 rpm. Mycelium was removed from the culture medium by filtration over miracloth. The medium was concentrated over a Centriprep YM-50 membrane filter, and the concentrated protein was taken up in 20 mM Tris-HCl buffer, pH 8. The proteins were purified via anion-exchange chromatography as described above. Both AgtA and AgtB were eluted as a single activity peak at a concentration of 150 mM NaCl. At each stage of the protein purification, the protein amount was measured using the Bradford method with reagent from Bio-Rad (Hercules, CA), and purity was checked using SDS-PAGE analysis (39) and staining with Biosafe Coomassie (Bio-Rad).
(iii) Enzymatic assays.
All oligosaccharides used were obtained from Sigma, except for nigerotriose [O-α-d-Glc-(1,3)-α-d-Glc-(1,3)-d-Glc], which was purchased from Dextra Laboratories (Reading, United Kingdom), and nigerose [O-α-d-Glc-(1,3)-α-d-Glc], which was a kind gift from Nihon Shokuhin Kako Co. Ltd. (Shizuoka, Japan). Lactobacillus reuteri polysaccharide was a gift from S. Kralj (University of Groningen, The Netherlands), and α-(1,3)-glucan isolated from A. nidulans was a kind gift from B. J. Zonneveld (Leiden University, Leiden, The Netherlands). As soluble starch, Paselli SA2 with an average degree of polymerization (DP) of 50 (AVEBE, Foxhol, The Netherlands), was used. All reactions were performed at 37°C. To determine the optimum pH for activity of AgtA and AgtB, 0.5 μg purified enzyme was incubated with 20 mM maltopentaose (a maltooligosaccharide with a DP of 5) in a 20-μl reaction volume for 30 min at 11 different pH values. The reaction was buffered by either 30 mM NaAc buffer at pH 4.2 to 7.0 or K2HPO4/KH2PO4 buffer at pH 6.3 to 8.0. Subsequently, 2 μl of the reaction mixture was spotted onto a thin-layer chromatography (TLC) plate (silica gel 60 F254; Merck, Darmstadt, Germany), and after the plate was dried, it was run for 6 h in 75 ml running buffer (butanol-ethanol-MilliQ, 5/5/3 [vol/vol/vol]) in a container of 22 by 6 by 22 cm. After the plate was run, it was dried and sprayed with 50% sulfuric acid in methanol and left to develop for 10 min at 110°C.
Hydrolyzing activity on potato starch was determined by the incubation of 1 μg of purified enzyme with 600 μl of 0.02%, 0.2%, or 2% (wt/vol) dissolved potato starch in 50 mM NaAc buffer containing 1 mM CaCl2. Reactions were buffered at pH 5.5 (AgtA) or pH 4.8 (AgtB) and performed in duplicate. Samples of 50 μl were taken after several time intervals up to 4 h and used for the determination of reducing ends and glucose. The formation of reducing ends was measured with the bicinchoninic acid method (45), and glucose formation was measured with the glucose GOD-PAP assay (Roche, Mannheim, Germany). Appropriate calibration curves and negative controls were included for all assays and reactions.
Standard assay conditions for all further enzymatic reactions were as follows: 0.4 μg of purified enzyme was incubated in 20 μl of 25 mM NaAc buffer (pH 5.5) containing 1 mM CaCl2 and 0.01% sodium azide in the presence of 20 mM oligosaccharide substrate and/or 4% (wt/vol) soluble starch or other polysaccharide, except for nigeran and α-(1,3)-glucan. Nigeran and α-(1,3)-glucan were dissolved in 1 M NaOH, after which the pH of a 1% solution was adjusted to pH 5.5 with hydrogen acetate. The final concentration used in reaction mixtures was 0.5% (wt/vol). Reaction products were detected either by TLC (as described above) or high-performance liquid chromatography (HPLC) (Dionex) analysis. For HPLC analysis, 5 μl of the reaction mixture was diluted in 1.5 ml 90% dimethyl sulfoxide. Separation of oligosaccharides was achieved by the method of Kralj et al. (36).
Disruption of the agtA gene.
The plasmid used to disrupt the agtA gene was constructed as follows. The DNA fragments flanking the agtA gene were amplified by PCR using strain N402 genomic DNA as the template: 1.5 kb of 5′ flanking DNA and 1.0 kb of 3′ flanking DNA were amplified by PCR using primers AgtAP1for and AgtAP2rev and primers AgtAP3for and AgtAP4rev (Table 1), respectively. Each primer was adapted with a restriction site for further cloning. The amplified PCR fragments were digested with NotI and XbaI or XbaI and BamHI, respectively, and cloned in a three-way ligation into NotI- and BamHI-digested pBluescript II SK to obtain plasmid pAgtAF53. Subsequently, pAgtAF53 was digested with XbaI and ligated with the 2.7-kb XbaI fragment encoding the Aspergillus oryzae pyrG gene, obtained from plasmid pAO4-13 (18), which resulted in the agtA disruption plasmid pΔagtA. Before transformation into strain MA70.15, pΔagtA was linearized with NotI. Uridine prototrophic transformants were selected by incubating protoplasts on agar plates containing MM without uridine. Transformants were purified, and genomic DNA was isolated and analyzed by Southern blot analysis to identify possible ΔagtA strains. Deletion of the agtA locus by homologous recombination was expected to result in the appearance of a 4.8-kb fragment after digestion of the genomic DNA with KpnI and the loss of a 2.0-kb fragment which was expected in the wild type. For the hybridization, the NotI-XbaI fragment (containing the 5′ flanking region of the agtA gene) was used. Independently obtained transformants were purified, and strains with the expected hybridization pattern (MA71.1, MA71.3, MA71.4, and MA71.7) were used for phenotypic analysis.
Phenotypic characterization of the ΔagtA strain and AgtA/AgtB overexpression strains.
Sensitivity towards CFW was assayed as described previously (59). Conidiospores from the control strains (strain MA70.15 transformed with pAO4-13 containing A. oryzae pyrG and strain MGG029-ΔaamA), the ΔagtA strains (MA71.1, MA71.3, MA71.4, and MA71.7), and the agtA and agtB overexpression strains were spotted on CFW plates. All strains were also checked for their ability to grow on starch as the sole carbon source by inoculating 3 μl of spore solution containing 106, 105, 104, or 103 spores on plates containing MM with either 1% (wt/vol) potato starch or 1% (wt/vol) glucose as the carbon source. Colony growth was monitored daily.
Nucleotide sequence accession numbers.
The full genome sequence of A. niger strain CBS 513.88 has been deposited at the EMBL database with accession numbers AM270980 to AM270998 (55). The locus tags of the genes studied here are An09g03100 (AgtA), An12g02460 (AgtB), and An15g07800 (AgtC).
RESULTS
Sequence analysis.
The full genome sequence of A. niger CBS 513.88 was searched for genes encoding proteins belonging to GH13 using a HMM profile based on known α-amylases. Apart from three genes encoding previously described extracellular α-amylases, eight genes coding for as-yet undescribed GH13 proteins were identified, as well as five genes predicted to encode membrane-bound α-(1,3)-glucan synthases (55). Within the group of eight α-amylase-type proteins, three were characterized by the presence of an N-terminal signal sequence for secretion and a hydrophobic C-terminal sequence predicted to act as an attachment site for a GPI anchor (4, 17). These proteins, called AgtA, AgtB, and AgtC, form the topic of this study. The predicted sites for the attachment of a GPI anchor (the ω-sites) were largely in accordance with the consensus sequence for fungal GPI proteins as described by De Groot et al. (17) (Fig. 1). An exception to this consensus sequence was residue Thr543 at the ω+2 position in AgtC: an alternative ω-site could be residue Ser538 instead of Ser541 as predicted by the online GPI prediction tool (21). Homologous enzymes with predicted GPI-anchoring sites were also identified in the available genome sequences of other aspergilli as well as in Neurospora crassa, Magnaporthe grisea, and S. pombe. In many cases, the genes encoding the Agt homologues are located next to genes encoding predicted α-(1,3) glucan synthases. No homologous GH13 proteins containing GPI anchoring sites were found in the genome sequences of members of the subphylum Saccharomycotina, such as Candida albicans, Kluyveromyces lactis, and Saccharomyces cerevisiae.
FIG. 1.
GPI anchor-specific amino acid features in fungal proteins as described previously compared to the C-terminal sequences of AgtA, AgtB, and AgtC. (A) Consensus sequence for GPI attachment according to De Groot et al. (17), with X representing any amino acid. The ω-site is indicated in bold type. (B) C-terminal ends of AgtA, AgtB, and AgtC indicating the potential GPI modification site. The ω-site as predicted by the online tool for the prediction of a GPI modification site (21) is indicated in bold type, and an alternative ω-site is indicated by bold underlined type. The hydrophobic tail is underlined.
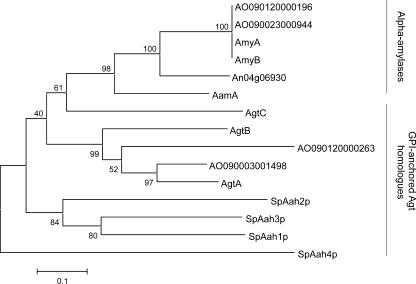
The amino acid sequences of AgtA, AgtB, and AgtC show 54 to 56% similarity to that of A. niger acid amylase (10). A phylogenetic analysis of the Agt proteins compared to the A. niger α-amylases, the homologous proteins in A. oryzae, and the Aah proteins in S. pombe is given in Fig. 2. The Agt proteins contain most of the amino acids generally conserved in GH13 (Table 2). However, in all three protein sequences, the highly conserved His143, which is part of conserved region I, is absent (numbering used is according to acid α-amylase of A. niger, unless indicated otherwise). In AgtB and AgtC, a second conserved histidine in conserved region IV is replaced by glutamate (Table 2).
FIG. 2.
Bootstrapped phylogenetic tree of the A. niger extracellular α-amylases (acid amylase AamA, α-amylase A and B, and putative α-amylase An04g06930) and α-glucanotransferases (AgtA/B/C), the homologous putative proteins identified in the genome of A. oryzae, and the S. pombe Aah proteins (SpAah1p to SpAah4p). The alignment and phylogenetic analysis were performed with MEGA version 3.1 using default settings. A bootstrapped tree was constructed with the neighbor-joining method using 500 replicates.
TABLE 2.
Alignment of the generally conserved regions of the α-amylase familya
| Enzyme | Sequenceb in:
|
|||
|---|---|---|---|---|
| Region I | Region II | Region III | Region IV | |
| Acid amylase | LMVDVVPNH | DGLRIDSVLE | YCVGEVDN | NFIENHD |
| AmyA | LMVDVVANH | DGLRIDTVKH | YCIGEVLD | TFVENHD |
| AgtA | LMMDTVINN | DGLRIDAAKH | FMTGEVLQ | SFSENHD |
| AgtB | LLLDVVINN | DGLRIDAAKS | FMTGEVMD | NFIEDQD |
| AgtC | LMLDIVVGD | DGLRIDSVLN | FTVGEGAT | TFTANQD |
| Aah3p (S. pombe) | VMLDSIVNS | DGLRIDAVKM | YSVGEVFS | TFIENHD |
| Amino acid numberingc | 135-143 | 222-231 | 247-254 | 312-318 |
Alignment of the generally conserved regions of the α-amylase family as present in A. niger acid amylase and α-amylase AmyA compared to homologous regions in AgtA, AgtB, and AgtC from A. niger and the homologous protein Aah3p from S. pombe.
Catalytic residues are underlined, and generally conserved residues are indicated in bold type.
Amino acid numbering according to A. niger acid amylase.
Purification and enzyme activity of AgtA fused to an SBD.
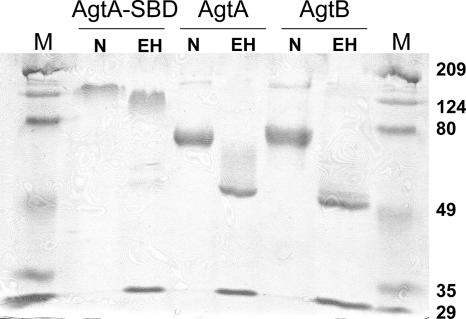
The agtA gene, lacking its C-terminal anchoring domain, was fused to the SBD of the A. niger glucoamylase gene serving as a protein tag, and the fusion construct was transformed into A. niger MGG029-ΔaamA, resulting in 25 stable transformants. The transformant with the highest expression level of the AgtA-SBD fusion protein in the culture medium as determined by Western blotting with polyclonal antiserum raised against purified SBD (41) was selected for larger-scale protein production. AgtA-SBD was isolated from the culture medium by binding it to waxy maize starch granules, and after further purification by anion-exchange chromatography, a single protein band with an apparent molecular mass of about 130 kDa was obtained (Fig. 3). This band was recognized by anti-SBD antibodies, a strong indication that this band contained the AgtA-SBD fusion protein (calculated molecular mass of 73.6 kDa). Treatment with Endo H to remove N glycosylation resulted in a small decrease in the apparent molecular mass, indicating that the protein was N glycosylated (Fig. 3). The high molecular mass of the fusion protein after Endo H treatment could be caused by heavy O glycosylation of the linker region of the SBD (70).
FIG. 3.
SDS-PAGE analysis of purified AgtA-SBD, AgtA and AgtB in native form (N) and after treatment with Endo H (EH). The M lanes contain molecular mass markers. The positions of molecular mass markers (in kilodaltons) are indicated to the right of the gel.
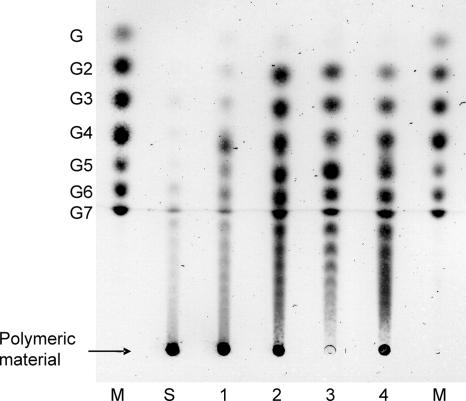
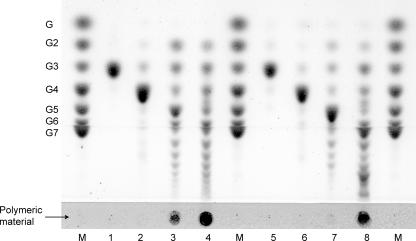
Purified AgtA-SBD was incubated with potato starch to determine its hydrolyzing activity. Hydrolysis of starch may result in the formation of glucose, maltose, or longer maltooligosaccharides, the formation of which can be quantified by the measurement of the reducing ends formed during the reaction. For AgtA-SBD, a low hydrolyzing activity of 0.46 ± 0.02 μmol of reducing ends mg−1 min−1 was detected when incubated with 0.2% starch, and no formation of glucose was observed. TLC analysis of the reaction mixture showed that short oligosaccharides were formed in small amounts from soluble starch (Fig. 4, lane 1). Incubation with both maltose and soluble starch resulted in the formation of more oligosaccharides than made from starch alone (Fig. 4, lane 2). No products were formed from maltose alone (result not shown). When AgtA-SBD was incubated with maltopentaose or maltoheptaose (maltooligosaccharide with a DP of 7), a variety of oligosaccharides ranging from maltose to oligosaccharides with a DP of at least 13 to 18, respectively, were formed (Fig. 4, lanes 3 and 4). This result indicated that AgtA-SBD hydrolyzed starch to some extent but mainly acted as a glucanotransferase, transferring parts of a donor oligosaccharide, which might be starch, to an acceptor substrate, e.g., maltose, thereby producing a variety of oligosaccharides of different lengths.
FIG. 4.
TLC analysis of reaction products of AgtA-SBD from different substrates. Enzyme (0.4 μg) was incubated with 20 mM oligosaccharide and/or 4% soluble starch for 1 h at 37°C. The arrow indicates where the following samples were loaded: molecular size markers (lanes M) containing a mix of maltooligosaccharides ranging from glucose (G1) to maltoheptaose (G7), unmodified soluble starch (lane S), and reaction products of AgtA-SBD incubated with soluble starch (lane 1), maltose and soluble starch (lane 2), maltopentaose (lane 3), or maltoheptaose (lane 4).
Production and purification of AgtA and AgtB.
To rule out any effect of the incorporated SBD on the enzymatic activity of AgtA, both AgtA and AgtB were overexpressed in A. niger in their native form for further biochemical analysis. All selected transformants overproduced a protein with an estimated molecular mass of 85 kDa not observed in the untransformed strain. In a similar procedure of cloning and transformation, we also attempted to produce the AgtC protein. Although insertion of the overexpression construct in selected transformants was confirmed by Southern blot analysis, none of the transformants overproduced the AgtC protein. The characterization of AgtC was therefore not included in this study.
Proteins expressed in the culture medium of A. niger MGG029-ΔaamA-AgtA and of A. niger MGG029-ΔaamA-AgtB were concentrated and subsequently submitted to anion-exchange chromatography, resulting in the purification of AgtA and AgtB. Both proteins had an apparent molecular mass of approximately 70 kDa, but after removal of N glycosylation, the apparent protein masses decreased to approximately 55 kDa, close to their theoretical masses (58.8 kDa for AgtA and 57.7 kDa for AgtB, after removal of the C-terminal end for GPI anchoring) (Fig. 3).
Glucanotransferase activity of AgtA and AgtB on maltooligosaccharides.
Both AgtA and AgtB were incubated with maltooligosaccharides ranging in size from maltose to maltohexaose (a maltooligosaccharide with a DP of 6), and the products were analyzed by TLC. With maltopentaose or maltohexaose as substrates, both enzymes produced a range of oligosaccharides with a DP of 15 or larger, similar to what was observed previously for AgtA-SBD (Fig. 5, lanes 3, 4, 7, and 8). Incubation with maltopentaose at lower concentrations (2 or 10 mM) also resulted in the formation of products with a DP of 6 and larger (results not shown). Activity of both enzymes on the smaller maltooligosaccharides was limited (Fig. 5, lanes 1 and 2 and lanes 5 and 6). Neither of the enzymes produced glucose in detectable amounts. When incubated with dissolved potato starch, both enzymes produced small amounts of reducing ends (hydrolysis on 2% starch, 0.55 ± 0.19 μmol mg−1 min−1 for AgtA and 0.26 ± 0.16 μmol mg−1 min−1 for AgtB; the hydrolysis rate was approximately four times lower when measured on 0.2% starch). TLC analysis of the products produced from maltoheptaose at different pH values indicated that AgtA was active between pH 4.5 and pH 6, and AgtB showed activity between pH 4 and pH 5.5 (data not shown). Glucose polymers with other types of glycosidic linkages, like dextran [a glucan polymer with α-(1,6)-glycosidic bonds], nigeran, A. nidulans α-(1,3) glucan, an L. reuteri polysaccharide containing α-(1,3) and α-(1,6) glycosidic bonds (35), and cellulose [β-(1,4) glycosidic bonds] were tested alone or in combination with maltose as an acceptor substrate. None of these polysaccharides acted as a substrate for AgtA or AgtB (data not shown).
FIG. 5.
TLC analysis of the reaction products of AgtA (left side) and AgtB (right side) incubated with different substrates. Purified enzyme (0.4 μg) was incubated with 20 mM substrate for 1 h at 37°C. The spots where the samples are applied are indicated by the arrow. The molecular size marker (M) lanes contain a mix of maltooligosaccharides ranging from glucose (G1) to maltoheptaose (G7). The figure shows reaction products of AgtA (lanes 1 to 4) or AgtB (lanes 5 to 8) incubated with maltotriose (lanes 1 and 5), maltotetraose (lanes 2 and 6), maltopentaose (lanes 3 and 7), or maltohexaose (lanes 4 and 8).
Identification of the reaction products of AgtA and AgtB.
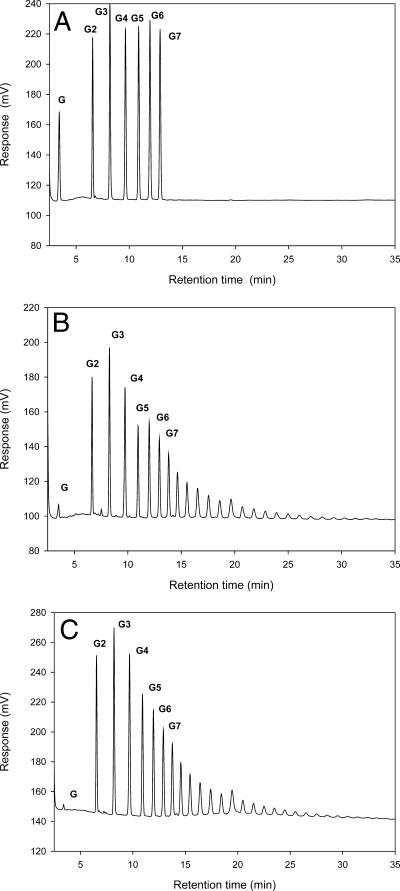
To study the nature of the glycosidic linkages formed and the maximum length of the oligosaccharide products, reaction products of AgtA and AgtB were analyzed qualitatively by HPLC. AgtA and AgtB produced a similar mixture of oligosaccharides with various lengths from maltoheptaose as a substrate (Fig. 6B and C). The maximum detectable product length had a DP of approximately 28 after 1 hour of incubation for both enzymes. A similar mixture of products was produced from maltopentaose as a substrate (results not shown). The retention time of the products was the same as the retention time of standard maltooligosaccharides (Fig. 6A), except for one small peak among the products of AgtA, which was identified as panose (4-α-isomaltosylglucose).
FIG. 6.
HPLC analysis of the reaction products formed by incubation of AgtA and AgtB on maltoheptaose. (A) Elution profile of a standard mixture of maltooligosaccharides containing glucose (G1) to maltoheptaose (G7). (B and C) Reaction products of AgtA (B) and AgtB (C) after incubation of 0.4 μg purified enzyme with 20 mM maltoheptaose at 37°C for 1 h.
AgtA and AgtB can use α-(1,3)-glucooligosaccharides as acceptor.
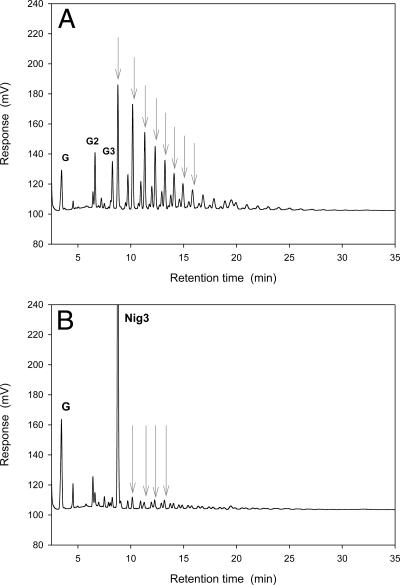
The presence of the putative GPI-anchoring sequence in both AgtA and AgtB indicate that both enzymes are possibly present in the cell wall or cell membrane. The α-glucans present in the cell wall of A. niger are α-(1,3)-glucan (24) and nigeran (9). In the previous paragraph we showed that neither nigeran nor α-(1,3)-glucan was used as a (donor) substrate by AgtA and AgtB. To investigate whether small substrates with α-(1,3) glycosidic bonds could be used as an acceptor substrate, both enzymes were incubated with soluble starch as a donor substrate, combined with glucose, maltose, nigerose, or nigerotriose as an acceptor substrate. Analysis of the reaction products by TLC and HPLC revealed that glucose was not used as an acceptor substrate by either of the enzymes; maltose was an efficient acceptor as shown previously for AgtA-SBD (results not shown). AgtA also formed a series of oligosaccharides using either nigerose or nigerotriose as an acceptor substrate, although the amount of products formed was smaller than when maltose acted as an acceptor substrate (Fig. 7A). AgtB did not use nigerose or nigerotriose efficiently as an acceptor substrate (Fig. 7B). No activity of AgtA or AgtB was observed on nigerose or nigerotriose as the sole substrate (result not shown). These results indicated that small α-(1,3)-linked oligosaccharides can be used as acceptor substrates by AgtA, and to a very limited extent by AgtB, but only in combination with an α-(1,4)-linked donor molecule.
FIG. 7.
HPLC analysis of the reaction products formed by AgtA (A) and AgtB (B) upon incubation with soluble starch and nigerotriose (Nig3). G, G2, and G3 indicate peaks representing glucose, maltose, and maltotriose, respectively. Peaks representing products most likely containing α-(1,3) glycosidic bonds, resulting from the use of nigerotriose as an acceptor substrate, are indicated with gray arrows. Enzyme (0.4 μg) was incubated with 20 mM nigerotriose and 4% soluble starch at 37°C for 18 h.
ΔagtA and agtA/agtB overexpression strains are CFW hypersensitive.
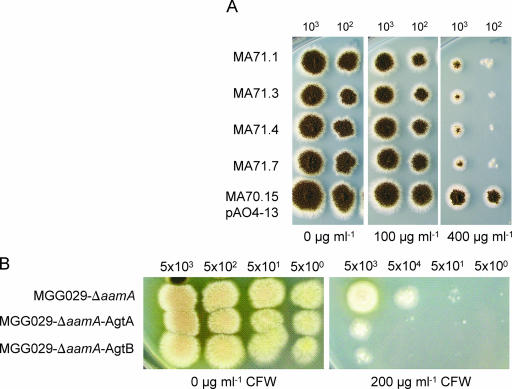
To examine the consequence of the loss of the agtA gene in A. niger and to analyze the physiological role of this enzyme, a deletion mutant of the agtA gene was constructed. Ten randomly chosen pΔagtA transformants were subjected to Southern blot analysis which revealed that in 8 out of the 10 transformants, the agtA gene was properly deleted (data not shown). Phenotypic analysis of several ΔagtA strains revealed that their growth rate on solid media was slightly reduced, but no changes in the morphology of the hyphae or conidia were observed. We analyzed the sensitivity of the ΔagtA strain to the cell wall-disturbing compound CFW. Hypersensitivity towards CFW has been shown to be indicative of mutants with impaired cell wall strength (15, 60). As shown in Fig. 8A, the ΔagtA strains showed an increased sensitivity towards CFW. The observed sensitivity is not as strong as for the deletion of other A. niger cell wall-related proteins, such as α-(1,3)-glucan synthase A or the cell wall protein A (15, 16). The overexpression of agtA and agtB also resulted in an increased sensitivity towards CFW (Fig. 8B). All deletion and overexpression strains were also tested for their ability to grow on starch as the sole carbon source. Deletion of agtA and overexpression of AgtA or AgtB had no significant effect on the ability of A. niger to grow on starch compared to the untransformed strains.
FIG. 8.
Effect of agtA deletion (A) or agtA and agtB overexpression (B) on the susceptibility of the resulting strains towards CFW-induced cell wall stress. (A) A. niger ΔagtA strains (MA71.1, MA71.3, MA71.4, and MA71.7) and the control strain (MA70.15 transformed with pAO4-13), grown on complete medium containing 0, 100, or 400 μg ml−1 CFW for 96 h. (B) A. niger strains overexpressing AgtA or AgtB and the parental strain (MGG029-ΔaamA) grown on complete medium containing 0 or 200 μg ml−1 CFW for 72 h. The number of spores applied per spot is indicated above the panels.
DISCUSSION
All putative GPI-anchored GH13 enzymes identified in the genome sequences of four aspergilli, as well as N. crassa, M. grisea, and S. pombe, were originally annotated as α-amylases, because of their high similarity to known extracellular fungal α-amylases. However, most of the protein sequences missed the commonly conserved His143 in region I (26). Mutation of this residue, which is located in the active site (66), may result in a strongly reduced activity in α-amylases (13, 50) or in altered reaction specificities in other GH13 enzymes (see, e.g., references 40 and 50). Despite the missing His143 residue, the A. niger Agt enzymes and their homologues are clearly members of GH13, based on their high similarity with known proteins in this family. The second generally conserved His residue, His317 in conserved region IV, is replaced by Gln in AgtB and AgtC. Although His317 is overall highly conserved in the α-amylase family, this residue appears to be less important for the determination of the catalytic activity because some α-amylases are known which also do not possess a His in this position (25, 28). In conclusion, the Agt enzymes in A. niger and homologues in other fungi are highly similar to the well described fungal α-amylases, but aberrant conserved regions combined with the presence of a GPI-anchoring signal make them clearly distinguishable. We produced two of the A. niger Agt enzymes and determined their biochemical activities.
As the enzymatic activity of the Agt proteins was not known at the start of this research, it was decided to start with the production of the enzyme encoded by gene An09g03100 (AgtA) fused with a clearly recognizable tag, the SBD. This allowed isolation and purification of the protein from media via binding to starch granules, as well as recognition of the purified protein using antibodies. Subsequently, both AgtA and AgtB (encoded by gene An12g02460) were produced in their native form for further biochemical characterization. A host A. niger strain with very low α-amylase activity was chosen to strongly reduce interference by native activities when searching for the biochemical activity of the investigated enzymes. AgtA-SBD as well as native AgtA and AgtB had a very low hydrolyzing activity on starch but clearly showed α-glucanotransferase activity on maltooligosaccharides alone and on maltooligosaccharides plus starch. We therefore propose to name the A. niger GPI-anchored GH13 enzymes AgtA and B (Agt for α-glucanotransferase). Since the predicted protein encoded by gene An15g07800 has the same sequence characteristics, we propose naming it AgtC.
AgtA and AgtB were overproduced from their entire predicted coding sequences, which included the predicted GPI-anchoring signal. The presence of both AgtA and AgtB in the medium suggested that these proteins were not fully retained at the plasma membrane or cell wall but that at least part of the proteins was released into the medium. This might be an indication that the proteins are not GPI anchored, although their C-terminal signal sequence as well as the investigation of the homologous Aah3p from S. pombe (47) suggest otherwise. Another explanation is that the enzymes had been released by endogenous phospholipase C activity, as was previously shown to occur with GPI-anchored proteins in A. niger and S. cerevisiae (12).
AgtA and AgtB produced similar ranges of products consisting of maltooligosaccharides, indicating that both enzymes formed α-(1,4) glycosidic bonds and can therefore be classified as 4-α-glucanotransferases (EC 2.4.1.25). Also, a small amount of panose was produced by AgtA, indicating an ability to synthesize α-(1,6) linkages. Alternatively, panose may have been produced by a minor contamination of α-glucosidase, which is known to produce α-(1,6) linkages (30). The enzymatic activities of AgtA and AgtB are unique among the α-glucanotransferases from bacteria as well as eukarya. The α-glucanotransferases that have been described until now usually release one glucose molecule for every transfer event (65), but AgtA and AgtB did not produce glucose in significant amounts. Additionally, bacterial amylomaltases use very small donor and acceptor molecules (maltotriose and glucose, respectively) (29), while AgtA and AgtB prefer longer donor molecules with a minimum length of five glucose residues and maltose as the smallest possible acceptor substrate. The use of the α-(1,3)-linked oligosaccharides nigerose and nigerotriose as acceptor substrates by GH13 α-glucanotransferases has not been reported before. We conclude that the A. niger α-glucanotransferases represent a new subgroup of GH13 in view of their atypical donor and acceptor profiles and their C-terminal GPI-anchoring sequences. Based on their common putative cell wall-associated location and amino acid sequences, it is expected that the closely related GPI-anchored GH13 proteins in other fungi will show similar glucanotransferase activities, although their precise substrate and product profiles remain to be determined.
Most extracellular members of GH13 are involved in the degradation of starch to supply energy and carbon to the cells. There are strong indications that this is not the case for AgtA and AgtB. This study describes that neither a knockout of agtA nor overexpression of AgtA or AgtB had an effect on the ability of A. niger strains to grow on starch, even if the parental strain was severely hampered in this trait. In another study, we have also shown that expression of the agtA and agtB genes is not regulated by AmyR, the general regulator for starch-processing enzymes in aspergilli (X.-L. Yuan et al., unpublished data) (56), and similar results were found for the homologous genes in Aspergillus nidulans (51). Taken together, these data indicate that the Agt proteins most likely are not involved in starch catabolism. An alternative function could be the production or modification of α-glucans in the fungal cell wall, which was suggested for one of the GPI-anchored Agt homologues in the fission yeast S. pombe, Aah3p, in a functional study (47). Deletion of aah3 resulted in a morphological defect and hypersensitivity towards cell wall-degrading enzymes. The knockout could not be rescued by transformation with the aah3 gene in which the catalytic residues had been mutated, showing the importance of the enzymatic activity rather than the structural properties of the protein. Our finding of a clear enzymatic activity for AgtA and AgtB confirms the importance of the catalytic residues for their physiological role. The proposed role for AgtA and its homologues in cell wall α-glucan production or maintenance is strengthened by the analysis of agtA knockout strains, which showed increased sensitivity towards CFW. The overexpression of AgtA and AgtB in A. niger caused a similar effect, which might be an indication that the unnaturally large amounts of these enzymes have a negative effect on cell wall strength.
The function of the Agt proteins in the fungal cell wall might be analogous to the function of GPI-anchored β-glucanosyltransferases. These enzymes, identified in several yeasts and fungi, including aspergilli, play a role in the cross-linking of cell wall β-glucan (48, 57). In Aspergillus fumigatus, the β-glucan component of the cell wall is used as a target for antifungal drugs (2). A knockout of one of its GPI-anchored β-glucanosyltransferases, Gel2p, resulted in an altered cell wall composition, increased sensitivity for CFW, and reduced virulence (49). We tested the abilities of AgtA and AgtB to process the two α-glucan cell wall components, nigeran and α-(1,3) glucan, but no activity was detected. Because of their poor solubility at low pH, these substrates were offered at a relatively low concentration and partly in crystalline form, which might prevent the enzymes from acting on these cell wall components in vitro. It was shown, however, that AgtA performed a transglycosylation reaction involving an α-(1,4)-linked donor substrate and an α-(1,3)-linked acceptor substrate. A similar reaction was thought to occur in S. pombe cell walls, where two linear polysaccharide chains of α-(1,3)-glucan with several α-(1,4) linkages at the reducing end were interconnected by a transglycosylation reaction (22). Although this process was suggested to be performed by the transferase domain of α-(1,3)-glucan synthase Ags1p, a similar cross-linking reaction could be performed by AgtA. This would also explain the clustering of agt and ags (α-glucan synthase) genes conserved in many ascomycetes. Clustering of genes involved in the same metabolic pathway is well described in fungi (31).
To conclude, we have studied two novel putatively GPI-anchored GH13 enzymes of A. niger, with homologues in many other fungi. The AgtA and AgtB enzymes both showed a unique type of α-(1,4) glucanotransferase activity, and our functional characterization indicated that their involvement in the facilitation of growth on starch is unlikely. The characterization of a knockout of agtA suggested that this enzyme could be involved in cell wall α-glucan synthesis, which is in line with the results on a knockout of a homologous protein from S. pombe (47). More study is needed to confirm the proposed physiological role for these α-glucanotransferases and to identify their exact in vivo reaction.
Acknowledgments
We are very grateful to Jacques Benen and Harrie Kools of Wageningen University for providing the pki-pgaII plasmid and help with the HMMR searches. We also acknowledge the contribution of Peter Sanders (TNO Quality of Life, The Netherlands) for the HPLC analysis and Mark Arentshorst (Leiden University) for technical assistance with the knockout. We thank DSM for providing the A. niger genome sequence.
This work was supported by SenterNovem in the framework of the IOP-Genomics program (project IGE1021).
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Bardalaye, P. C., and J. H. Nordin. 1977. Chemical structure of the galactomannan from the cell wall of Aspergillus niger. J. Biol. Chem. 252:2584-2591. [PubMed] [Google Scholar]
- 2.Beauvais, A., and J. P. Latgé. 2001. Membrane and cell wall targets in Aspergillus fumigatus. Drug Resist. Updates 4:38-49. [DOI] [PubMed] [Google Scholar]
- 3.Beauvais, A., D. Maubon, S. Park, W. Morelle, M. Tanguy, M. Huerre, D. S. Perlin, and J. P. Latgé. 2005. Two α(1-3) glucan synthases with different functions in Aspergillus fumigatus. Appl. Environ. Microbiol. 71:1531-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. Von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Benen, J. A., H. C. Kester, and J. Visser. 1999. Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II and C. Eur. J. Biochem. 259:577-585. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, J. E., and L. L. Lasure. 1991. More gene manipulations in fungi. Academic Press, San Diego, CA.
- 7.Birney, E., M. Clamp, and R. Durbin. 2004. GeneWise and Genomewise. Genome Res. 14:988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumenthal, H. J., and S. Roseman. 1957. Quantitative estimation of chitin in fungi. J. Bacteriol. 74:222-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bobbitt, T. F., J. H. Nordin, M. Roux, J. F. Revol, and R. H. Marchessault. 1977. Distribution and conformation of crystalline nigeran in hyphal walls of Aspergillus niger and Aspergillus awamori. J. Bacteriol. 132:691-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boel, E., L. Brady, A. M. Brzozowski, Z. Derewenda, G. G. Dodson, V. J. Jensen, S. B. Petersen, H. Swift, L. Thim, and H. F. Woldike. 1990. Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus. Biochemistry 29:6244-6249. [DOI] [PubMed] [Google Scholar]
- 11.Bos, C. J., A. J. Debets, K. Swart, A. Huybers, G. Kobus, and S. M. Slakhorst. 1988. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14:437-443. [DOI] [PubMed] [Google Scholar]
- 12.Brul, S., A. King, J. M. van der Vaart, J. Chapman, F. Klis, and C. T. Verrips. 1997. The incorporation of mannoproteins in the cell wall of S. cerevisiae and filamentous Ascomycetes. Antonie Leeuwenhoek 72:229-237. [DOI] [PubMed] [Google Scholar]
- 13.Chang, C. T., H. F. Lo, M. C. Chi, C. Y. Yao, W. H. Hsu, and L. L. Lin. 2003. Identification of essential histidine residues in a recombinant alpha-amylase of thermophilic and alkaliphilic Bacillus sp. strain TS-23. Extremophiles 7:505-509. [DOI] [PubMed] [Google Scholar]
- 14.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. J. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 15.Damveld, R. A., M. Arentshorst, P. A. Vankuyk, F. M. Klis, C. A. van den Hondel, and A. F. Ram. 2005. Characterisation of CwpA, a putative glycosylphosphatidylinositol-anchored cell wall mannoprotein in the filamentous fungus Aspergillus niger. Fungal Genet. Biol. 42:873-885. [DOI] [PubMed] [Google Scholar]
- 16.Damveld, R. A., P. A. Vankuyk, M. Arentshorst, F. M. Klis, C. A. van den Hondel, and A. F. Ram. 2005. Expression of agsA, one of five 1,3-alpha-D-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet. Biol. 42:165-177. [DOI] [PubMed] [Google Scholar]
- 17.De Groot, P. W., K. J. Hellingwerf, and F. M. Klis. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781-796. [DOI] [PubMed] [Google Scholar]
- 18.de Ruiter-Jacobs, Y. M., M. Broekhuijsen, S. E. Unkles, E. I. Campbell, J. R. Kinghorn, R. Contreras, P. H. Pouwels, and C. A. van den Hondel. 1989. A gene transfer system based on the homologous pyrG gene and efficient expression of bacterial genes in Aspergillus oryzae. Curr. Genet. 16:159-163. [DOI] [PubMed] [Google Scholar]
- 19.Duarte, J. C., and M. Costa-Ferreira. 1994. Aspergilli and lignocellulosics: enzymology and biotechnological applications. FEMS Microbiol. Rev. 13:377-386. [DOI] [PubMed] [Google Scholar]
- 20.Durbin, R., and S. Eddy. 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids. A tutorial introduction to hidden Markov models and other probabilistic modelling approaches in computational sequence analysis. Cambridge University Press, Cambridge, United Kingdom.
- 21.Eisenhaber, B., G. Schneider, M. Wildpaner, and F. Eisenhaber. 2004. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 337:243-253. [DOI] [PubMed] [Google Scholar]
- 22.Grün, C. H., F. Hochstenbach, B. M. Humbel, A. J. Verkleij, J. H. Sietsma, F. M. Klis, J. P. Kamerling, and J. F. Vliegenthart. 2005. The structure of cell wall alpha-glucan from fission yeast. Glycobiology 15:245-257. [DOI] [PubMed] [Google Scholar]
- 23.Hochstenbach, F., F. M. Klis, H. van den Ende, E. van Donselaar, P. J. Peters, and R. D. Klausner. 1998. Identification of a putative alpha-glucan synthase essential for cell wall construction and morphogenesis in fission yeast. Proc. Natl. Acad. Sci. USA 95:9161-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horisberger, M., B. A. Lewis, and F. Smith. 1972. Structure of a (1,3)-α-D-glucan (pseudonigeran) of Aspergillus niger NNRL 326 cell wall. Carbohydr. Res. 23:183-188. [DOI] [PubMed] [Google Scholar]
- 25.Hoshiko, S., O. Makabe, C. Nojiri, K. Katsumata, E. Satoh, and K. Nagaoka. 1987. Molecular cloning and characterization of the Streptomyces hygroscopicus α-amylase gene. J. Bacteriol. 169:1029-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jespersen, H. M., E. A. MacGregor, M. R. Sierks, and B. Svensson. 1991. Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem. J. 280:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston, I. R. 1965. The composition of the cell wall of Aspergillus niger. Biochem. J. 96:651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang, H. K., J. H. Lee, D. Kim, D. F. Day, J. F. Robyt, K. H. Park, and T. W. Moon. 2004. Cloning and expression of Lipomyces starkeyi alpha-amylase in Escherichia coli and determination of some of its properties. FEMS Microbiol. Lett. 233:53-64. [DOI] [PubMed] [Google Scholar]
- 29.Kaper, T., M. J. van der Maarel, G. J. Euverink, and L. Dijkhuizen. 2004. Exploring and exploiting starch-modifying amylomaltases from thermophiles. Biochem. Soc. Trans. 32:279-282. [DOI] [PubMed] [Google Scholar]
- 30.Kato, N., S. Suyama, M. Shirokane, M. Kato, T. Kobayashi, and N. Tsukagoshi. 2002. Novel α-glucosidase from Aspergillus nidulans with strong transglycosylation activity. Appl. Environ. Microbiol. 68:1250-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 32.Kelly, J. M., and M. J. Hynes. 1985. Transformation of Aspergillus niger by the amdS gene of Aspergillus nidulans. EMBO J. 4:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolar, M., P. J. Punt, C. A. van den Hondel, and H. Schwab. 1988. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene 62:127-134. [DOI] [PubMed] [Google Scholar]
- 34.Korman, D. R., F. T. Bayliss, C. C. Barnett, C. L. Carmona, K. H. Kodama, T. J. Royer, S. A. Thompson, M. Ward, L. J. Wilson, and R. M. Berka. 1990. Cloning, characterization, and expression of two alpha-amylase genes from Aspergillus niger var. awamori. Curr. Genet. 17:203-212. [DOI] [PubMed] [Google Scholar]
- 35.Kralj, S., G. H. van Geel-Schutten, M. M. Dondorff, S. Kirsanovs, M. J. van der Maarel, and L. Dijkhuizen. 2004. Glucan synthesis in the genus Lactobacillus: isolation and characterization of glucansucrase genes, enzymes and glucan products from six different strains. Microbiology 150:3681-3690. [DOI] [PubMed] [Google Scholar]
- 36.Kralj, S., G. H. van Geel-Schutten, M. J. van der Maarel, and L. Dijkhuizen. 2004. Biochemical and molecular characterization of Lactobacillus reuteri 121 reuteransucrase. Microbiology 150:2099-2112. [DOI] [PubMed] [Google Scholar]
- 37.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: Molecular Evolutionary Analysis, version 1.01. The Pennsylvania State University, University Park, PA.
- 38.Kuriki, T., and T. Imanaka. 1999. The concept of the alpha-amylase family: structural similarity and common catalytic mechanism. J. Biosci. Bioeng. 87:557-565. [DOI] [PubMed] [Google Scholar]
- 39.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 40.Leemhuis, H., U. F. Wehmeier, and L. Dijkhuizen. 2004. Single amino acid mutations interchange the reaction specificities of cyclodextrin glycosyltransferase and the acarbose-modifying enzyme acarviosyl transferase. Biochemistry 43:13204-13213. [DOI] [PubMed] [Google Scholar]
- 41.Le Gal-Coeffet, M. F., A. J. Jacks, K. Sorimachi, M. P. Williamson, G. Williamson, and D. B. Archer. 1995. Expression in Aspergillus niger of the starch-binding domain of glucoamylase. Comparison with the proteolytically produced starch-binding domain. Eur. J. Biochem. 233:561-567. [DOI] [PubMed] [Google Scholar]
- 42.MacGregor, E. A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the alpha-amylase family of enzymes. Biochim. Biophys. Acta 1546:1-20. [DOI] [PubMed] [Google Scholar]
- 43.Marion, C. L., C. A. Rappleye, J. T. Engle, and W. E. Goldman. 2006. An alpha-(1,4)-amylase is essential for alpha-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol. Microbiol. 62:970-983. [DOI] [PubMed] [Google Scholar]
- 44.Martens-Uzunova, E. S., J. S. Zandleven, J. A. Benen, H. Awad, H. J. Kools, G. Beldman, A. G. Voragen, J. A. van den Berg, and P. J. Schaap. 2006. A new group of exo-acting family 28 glycoside hydrolases of Aspergillus niger that are involved in pectin degradation. Biochem. J. 400:43-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meeuwsen, P. J., J. P. Vincken, G. Beldman, and A. G. Voragen. 2000. A universal assay for screening expression libraries for carbohydrases. J. Biosci. Bioeng. 89:107-109. [DOI] [PubMed] [Google Scholar]
- 46.Meyer, V., M. Arentshorst, A. El Ghezal, A. C. Drews, R. Kooistra, C. A. van den Hondel, and A. F. Ram. 2007. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 128:770-775. [DOI] [PubMed] [Google Scholar]
- 47.Morita, T., N. Tanaka, A. Hosomi, Y. Giga-Hama, and K. Takegawa. 2006. An alpha-amylase homologue, aah3, encodes a GPI-anchored membrane protein required for cell wall integrity and morphogenesis in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 70:1454-1463. [DOI] [PubMed] [Google Scholar]
- 48.Mouyna, I., T. Fontaine, M. Vai, M. Monod, W. A. Fonzi, M. Diaquin, L. Popolo, R. P. Hartland, and J. P. Latgé. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275:14882-14889. [DOI] [PubMed] [Google Scholar]
- 49.Mouyna, I., W. Morelle, M. Vai, M. Monod, B. Lechenne, T. Fontaine, A. Beauvais, J. Sarfati, M. C. Prevost, C. Henry, and J. P. Latgé. 2005. Deletion of GEL2 encoding for a beta(1-3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus. Mol. Microbiol. 56:1675-1688. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura, A., K. Haga, and K. Yamane. 1993. Three histidine residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of the replacement on pH dependence and transition-state stabilization. Biochemistry 32:6624-6631. [DOI] [PubMed] [Google Scholar]
- 51.Nakamura, T., Y. Maeda, N. Tanoue, T. Makita, M. Kato, and T. Kobayashi. 2006. Expression profile of amylolytic genes in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 70:2363-2370. [DOI] [PubMed] [Google Scholar]
- 52.Reference deleted.
- 53.Paldi, T., I. Levy, and O. Shoseyov. 2003. Glucoamylase starch-binding domain of Aspergillus niger B1: molecular cloning and functional characterization. Biochem. J. 372:905-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parenicova, L., J. A. Benen, H. C. Kester, and J. Visser. 1998. pgaE encodes a fourth member of the endopolygalacturonase gene family from Aspergillus niger. Eur. J. Biochem. 251:72-80. [DOI] [PubMed] [Google Scholar]
- 55.Pel, H. J., J. H. de Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. De Vries, R. Albang, K. Albermann, M. R. Andersen, J. D. Bendtsen, J. A. Benen, M. van den Berg, S. Breestraat, M. X. Caddick, R. Contreras, M. Cornell, P. M. Coutinho, E. G. Danchin, A. J. Debets, P. Dekker, P. W. van Dijck, A. van Dijk, L. Dijkhuizen, A. J. Driessen, C. d'Enfert, S. Geysens, C. Goosen, G. S. Groot, P. W. De Groot, T. Guillemette, B. Henrissat, M. Herweijer, J. P. van den Hombergh, C. A. van den Hondel, R. T. van der Heijden, R. M. van der Kaaij, F. M. Klis, H. J. Kools, C. P. Kubicek, P. A. van Kuyk, J. Lauber, X. Lu, M. J. van der Maarel, R. Meulenberg, H. Menke, M. A. Mortimer, J. Nielsen, S. G. Oliver, M. Olsthoorn, K. Pal, N. N. van Peij, A. F. Ram, U. Rinas, J. A. Roubos, C. M. Sagt, M. Schmoll, J. Sun, D. Ussery, J. Varga, W. Vervecken, P. J. van de Vondervoort, H. Wedler, H. A. Wosten, A. P. Zeng, A. J. van Ooyen, J. Visser, and H. Stam. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221-231. [DOI] [PubMed] [Google Scholar]
- 56.Petersen, K. L., J. Lehmbeck, and T. Christensen. 1999. A new transcriptional activator for amylase genes in Aspergillus. Mol. Gen. Genet. 262:668-676. [DOI] [PubMed] [Google Scholar]
- 57.Popolo, L., M. Vai, E. Gatti, S. Porello, P. Bonfante, R. Balestrini, and L. Alberghina. 1993. Physiological analysis of mutants indicates involvement of the Saccharomyces cerevisiae GPI-anchored protein gp115 in morphogenesis and cell separation. J. Bacteriol. 175:1879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Punt, P. J., and C. A. van den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216:447-457. [DOI] [PubMed] [Google Scholar]
- 59.Ram, A. F., and F. M. Klis. 2006. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor White and Congo Red. Nature Protocols 1:2253-2256. [DOI] [PubMed] [Google Scholar]
- 60.Ram, A. F., A. Wolters, H. R. Ten, and F. M. Klis. 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 10:1019-1030. [DOI] [PubMed] [Google Scholar]
- 61.Rappleye, C. A., J. T. Engle, and W. E. Goldman. 2004. RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol. Microbiol. 53:153-165. [DOI] [PubMed] [Google Scholar]
- 62.Reese, A. J., and T. L. Doering. 2003. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 50:1401-1409. [DOI] [PubMed] [Google Scholar]
- 63.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 64.Stagg, C. M., and M. S. Feather. 1973. The characterization of a chitin-associated D-glucan from the cell walls of Aspergillus niger. Biochim. Biophys. Acta 320:64-72. [DOI] [PubMed] [Google Scholar]
- 65.Takaha, T., M. Yanase, S. Okada, and S. M. Smith. 1993. Disproportionating enzyme (4-alpha-glucanotransferase; EC 2.4.1.25) of potato. Purification, molecular cloning, and potential role in starch metabolism. J. Biol. Chem. 268:1391-1396. [PubMed] [Google Scholar]
- 66.Uitdehaag, J. C., R. Mosi, K. H. Kalk, B. A. van der Veen, L. Dijkhuizen, S. G. Withers, and B. W. Dijkstra. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the alpha-amylase family. Nat. Struct. Biol. 6:432-436. [DOI] [PubMed] [Google Scholar]
- 67.van Dijck, P. W., G. C. Selten, and R. A. Hempenius. 2003. On the safety of a new generation of DSM Aspergillus niger enzyme production strains. Regul. Toxicol. Pharmacol. 38:27-35. [DOI] [PubMed] [Google Scholar]
- 68.Weenink, X. O., P. J. Punt, C. A. van den Hondel, and A. F. Ram. 2006. A new method for screening and isolation of hypersecretion mutants in Aspergillus niger. Appl. Microbiol. Biotechnol. 69:711-717. [DOI] [PubMed] [Google Scholar]
- 69.Wernars, K., T. Goosen, L. M. Wennekes, J. Visser, C. J. Bos, H. W. van den Broek, R. F. van Gorcom, C. A. van den Hondel, and P. H. Pouwels. 1985. Gene amplification in Aspergillus nidulans by transformation with vectors containing the amdS gene. Curr. Genet. 9:361-368. [DOI] [PubMed] [Google Scholar]
- 70.Williamson, G., N. J. Belshaw, and M. P. Williamson. 1992. O-glycosylation in Aspergillus glucoamylase. Conformation and role in binding. Biochem. J. 282:423-428. [DOI] [PMC free article] [PubMed] [Google Scholar]