Abstract
The study of eukaryotic membrane proteins has been hampered by a paucity of systems that achieve consistent high-level functional protein expression. We report the use of a modified membrane protein hyperexpression system to characterize three classes of fungal membrane proteins (ABC transporters Pdr5p, CaCdr1p, CaCdr2p, CgCdr1p, CgPdh1p, CkAbc1p, and CneMdr1p, the major facilitator superfamily transporter CaMdr1p, and the cytochrome P450 enzyme CaErg11p) that contribute to the drug resistance phenotypes of five pathogenic fungi and to express human P glycoprotein (HsAbcb1p). The hyperexpression system consists of a set of plasmids that direct the stable integration of a single copy of the expression cassette at the chromosomal PDR5 locus of a modified host Saccharomyces cerevisiae strain, ADΔ. Overexpression of heterologous proteins at levels of up to 29% of plasma membrane protein was achieved. Membrane proteins were expressed with or without green fluorescent protein (GFP), monomeric red fluorescent protein, His, FLAG/His, Cys, or His/Cys tags. Most GFP-tagged proteins tested were correctly trafficked within the cell, and His-tagged proteins could be affinity purified. Kinetic analysis of ABC transporters indicated that the apparent Km value and the Vmax value of ATPase activities were not significantly affected by the addition of His tags. The efflux properties of seven fungal drug pumps were characterized by their substrate specificities and their unique patterns of inhibition by eight xenobiotics that chemosensitized S. cerevisiae strains overexpressing ABC drug pumps to fluconazole. The modified hyperexpression system has wide application for the study of eukaryotic membrane proteins and could also be used in the pharmaceutical industry for drug screening.
The resolution and exploitation of protein structure and function are among the greatest biological challenges in the postgenomic era. These challenges, and their potential dividends, are greatest for membrane proteins, which are notoriously difficult to functionally express and purify in the quantities and forms needed for drug discovery or for high-resolution X-ray crystallography (1, 16). About a quarter of the cellular proteome consists of membrane proteins (5), which often play vital physiological roles: from environmental sensing to energy transduction, from nutrient uptake to drug efflux, and from cellular proliferation to programmed cell death. Membrane proteins are involved in many prominent diseases, including cystic fibrosis (48), type 2 diabetes (49), heart disease (52), and the drug resistance of numerous cancers (57). Hence, they are the targets for many therapies and constitute up to 70% of the drug targets used in medicine today. Membrane proteins also play key roles in drug modification, detoxification, and resistance in a wide variety of prokaryotic and eukaryotic systems (7). A fundamental understanding of cell biology, cell physiology, and cell-drug interactions therefore requires a detailed analysis of membrane protein function. Furthermore, the development of new drugs needs both the presentation of membrane protein targets in suitable screening systems and the high-resolution structures of membrane proteins for the implementation of structure-directed drug design and virtual screening regimes. While thousands of unique high-resolution protein structures are deposited in databases, less than 0.3% of these are for membrane proteins (http://www.mpibp-frankfurt.mpg.de/michel/public/memprotstruct.html and http://blanco.biomol.uci.edu/membrane_proteins_xtal.html). In addition to requiring specialist techniques for the solubilization and purification of membrane proteins, most of the membrane protein structures resolved thus far have been obtained for proteins that are naturally highly expressed. In contrast, the vast majority of membrane proteins are expressed at low levels. Of the ∼80 unique membrane protein structures obtained to date, about 75% are from bacteria and only 3 of the ∼20 structures obtained for eukaryotic membrane proteins have been obtained through heterologous expression. Overcoming this well-recognized structural analysis bottleneck requires heterologous expression systems that can deliver 10- to 100-mg quantities of functional, monodisperse eukaryotic membrane proteins with minimized microheterogeneity that are suitable for crystallography (16).
Resistance of the pathogenic fungus Candida albicans to the azole antifungals has been shown to involve three classes of membrane protein: ATP-binding cassette (ABC) pumps, major facilitator superfamily (MFS) pumps, and overexpression and mutation of the cytochrome P450 drug target Erg11p (2, 47, 51, 63). The development of strategies to combat the azole resistance of fungal pathogens would be facilitated greatly by studying the structures and functions of these membrane proteins. We have further developed a system for expressing membrane proteins in the yeast Saccharomyces cerevisiae (36, 38) and used it to study membrane proteins involved in fungal azole resistance. S. cerevisiae has many advantages over other expression systems as a robust eukaryotic host for membrane protein expression (5, 46). It is the best-characterized model eukaryotic organism. It can be cultured on both a micro and industrial scale, including formats suitable for protein production and drug screening, at a fraction of the cost of insect or mammalian cells, and it is readily genetically manipulated to incorporate desirable features into individual proteins or expression pathways (5, 20, 55). We previously described an S. cerevisiae expression system (36, 38) that utilizes a host strain deleted in several ABC transporters (13). This system has been used by us and others to heterologously express and study fungal proteins involved in drug efflux (26, 36, 38, 41, 43, 50, 54). We report here development of the system to facilitate the cloning of membrane protein genes with and without affinity, epitope, or reporter tags.
The modified S. cerevisiae membrane protein hyperexpression system resulted in reproducible, constitutive functional expression of fungal membrane proteins. We demonstrate that the system is well suited for the production of large quantities of membrane proteins for structural and functional studies and that yeast expressing heterologous membrane proteins can be used to screen for compounds that overcome fungal drug resistance.
MATERIALS AND METHODS
Strains and culture conditions.
The fungal isolates and yeast strains used in this study are listed in Table 1. All S. cerevisiae strains created in this study were based on AD1-8u− (13, 38). Strain ADΔ is identical to strain AD1-8u− except that the entire chromosomal URA3 locus, which corresponds to the URA3 marker of plasmid pABC3, was deleted by replacing the ura3 gene of strain AD1-8u− with the 422-bp repeat region of the CaURA3 blaster cassette (Table 1) (64). All fungal strains were grown in yeast extract, peptone, and glucose (YPD) medium: 1% (wt/vol) Bacto-yeast extract (Difco Laboratories, Detroit, MI), 2% (wt/vol) Bacto-peptone (Difco), and 2% (wt/vol) glucose. Yeast transformants were selected on plates containing complete supplement medium without uracil (CSM−ura) (Bio 101, Vista, CA) or histidine (CSM−his; Bio 101), 0.67% (wt/vol) yeast nitrogen base without amino acids (Difco), 2% (wt/vol) glucose, and 2% (wt/vol) agar (Difco). Plasmids were maintained in Escherichia coli strain DH5α. E. coli cells were grown in Luria-Bertani medium to which ampicillin was added (100 μg/ml) as required.
TABLE 1.
Fungal and yeast strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| Candida albicans strain ATCC 10261 | Wild-type isolate | ATCC collection |
| Candida glabrata strain CBS138 | Database strain | 15 |
| Candida krusei strain B2399 | Wild-type isolate | CDC |
| Cryptococcus neoformans strain CDC551 | Serotype A, sexual type α | Chiba University, Japan |
| Saccharomyces cerevisiae strain AD124567 | MATα PDR1-3 ura3 his1 Δyor1::hisG Δsnq2::hisG Δpdr10::hisG Δpdr11::hisG Δycf1::hisG Δpdr3::hisG | 13 |
| AD1-8u− | AD124567 Δpdr5::hisG Δpdr15::hisG | 13 |
| ADΔ | AD1-8u, Δura3 | This study |
| AD/ScPDR5 | AD1-8u− Δpdr5::pABC3-ScPDR5 | This study |
| AD/ScPDR5-HIS | AD1-8u−, Δpdr5::pABC5′-ScPDR5-HIS | This study |
| AD/ScPDR5-GFP | AD1-8u−, Δpdr5::pABC3-ScPDR5-GFP | This study |
| ADΔ/CaCDR1A | ADΔ, Δpdr5::pABC3-CaCDR1A | This study |
| AD/CaCDR1B | AD1-8u−, Δpdr5::pABC3-CaCDR1B | 26 |
| ADΔ/CaCDR1A-HIS | ADΔ, Δpdr5::pABC3-CaCDR1A-HIS | This study |
| ADΔ/CaCDR1A-CYS | ADΔ, Δpdr5::pABC3-CaCDR1A-CYS | This study |
| ADΔ/CaCDR1A-HIS-CYS | ADΔ, Δpdr5::pABC3-CaCDR1A-HIS-CYS | This study |
| ADΔ/CaCDR1A-FLAG-HIS | ADΔ, Δpdr5::pABC3-CaCDR1A-FLAG-HIS | This study |
| ADΔ/CaCDR1A-GFP | ADΔ, Δpdr5::pABC3-CaCDR1A-GFP | This study |
| ADΔ/CaCDR1A-mRFP | ADΔ, Δpdr5::pABC3-CaCDR1A-mRFP | This study |
| AD/CaCDR2A | AD1-8u−, Δpdr5::pABC3-CaCDR2A | 26 |
| AD/CaCDR2A-HIS | AD1-8u−, Δpdr5::pABC5′-CaCDR2A-HIS | This study |
| AD/CaCDR2A-GFP | AD1-8u−, Δpdr5::pABC3-CaCDR2A-GFP | This study |
| AD/CaMDR1A | AD1-8u−, Δpdr5::pABC3-CaMDR1A | This study |
| AD/CaMDR1A-GFP | AD1-8u−, Δpdr5::pABC5′-CaMDR1A-GFP | This study |
| AD/CaERG11A | AD1-8u−, Δpdr5::pABC3-CaERG11A | This study |
| AD/CaERG11A-GFP | AD1-8u−, Δpdr5::pABC5′-CaERG11A-GFP | This study |
| AD/CgCDR1 | AD1-8u−, Δpdr5::pSK-PDR5-PPUS-CgCDR1 | 61 |
| AD/CgCDR1-HIS | AD1-8u−, Δpdr5::3′pABC5′-CgCDR1-HIS | This study |
| AD/CgCDR1-GFP | AD1-8u−, Δpdr5::3′pABC3-CgCDR1-GFP | This study |
| AD/CgPDH1 | AD1-8u−, Δpdr5::pSK-PDR5-PPUS-CgPDH1 | 61 |
| AD/CgPDH1-HIS | AD1-8u−, Δpdr5::3′pABC5′-CgPDH1-HIS | This study |
| AD/CgPDH1-GFP | AD1-8u−, Δpdr5::3′pABC3-CgPDH1-GFP | This study |
| ADΔ/CkABC1g | ADΔ, Δpdr5::pABC3-CkABC1g | This study |
| ADΔ/CkABC1g-HIS | ADΔ, Δpdr5::pABC5′-CkABC1g-HIS | This study |
| ADΔ/CkABC1g-GFP | ADΔ, Δpdr5::pABC3-CkABC1g-GFP | This study |
| AD/CneMDR1 | AD1-8u−, Δpdr5::pABC3-CneMDR1 | This study |
| AD/CneMDR1-HIS | AD1-8u−, Δpdr5::pABC5′-CneMDR1-HIS | This study |
| AD/CneMDR1-GFP | AD1-8u−, Δpdr5::pABC3-CneMDR1-GFP | This study |
| AD/HsABCB1 | AD1-8u−, Δpdr5::pABC3-HsABCB1 | This study |
| AD/HsABCB1-GFP | AD1-8u−, Δpdr5::pABC5′-HsABCB1-GFP | This study |
Materials.
Molecular biology reagents and restriction and modifying enzymes were from New England Biolabs (Beverly, MA) or from Roche Diagnostics N.Z. Ltd. (Auckland, New Zealand). High-performance liquid chromatography-purified DNA oligonucleotides were purchased from Hermann GbR Synthetische Biomoleküle (Denzlingen, Germany). PCR and DNA fragments were purified using kits from Qiagen Pty. Ltd. (Clifton Hill, Victoria, Australia). Genomic DNA (gDNA) was isolated from yeast using the Y-DER yeast DNA extraction reagent kit from Pierce (Rockford, IL) by downscaling 50-fold. Yeast were transformed using the alkali cation yeast transformation kit from Bio 101. A modified protocol took into account the sensitivity of strains AD1-8u− and ADΔ to high concentrations of Li+ cations. A log-phase YPD culture of 250 ml (instead of 50 ml) was harvested at an optical density at 600 nm (OD600) of 0.6 and the washed cells incubated with the Li+ cation solution for only 25 min at 30°C. All plasmids and DNA fragments were verified by DNA sequencing using the DYEnamic ET Terminator cycle sequencing kit, version 3.1 (Amersham Pharmacia Biotech), and analyzed at the Micromon DNA Sequencing Facility (Monash University, Melbourne, Australia). PCRs used the high-fidelity KOD+ DNA polymerase (Toyobo, Osaka, Japan, or Novagen, San Diego, CA). Site-directed mutagenesis was carried out using the Chameleon site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Compounds.
Fluconazole (FLC) (Diflucan; aqueous solution) was purchased from Pfizer Laboratories Ltd. (Auckland, New Zealand), itraconazole (ITC) and ketoconazole (KTZ) were purchased from Janssen-Kyowa (Tokyo, Japan), aureobasidin A was purchased from Takara Bio Inc. (Shiga, Japan), miconazole (MCZ), rhodamine 6G (R6G), nystatin (NYS), cycloheximide, cerulenin, enniatin, vanadate, and oligomycin were purchased from Sigma-Aldrich New Zealand Ltd. (Auckland, New Zealand), FK506 was a gift from Astellas Pharma Inc. (Tokyo, Japan), and the milbemycins α11, α20, α25, β9 and β11 were a gift from Sankyo Co. Ltd. (Tokyo, Japan).
Construction of plasmids.
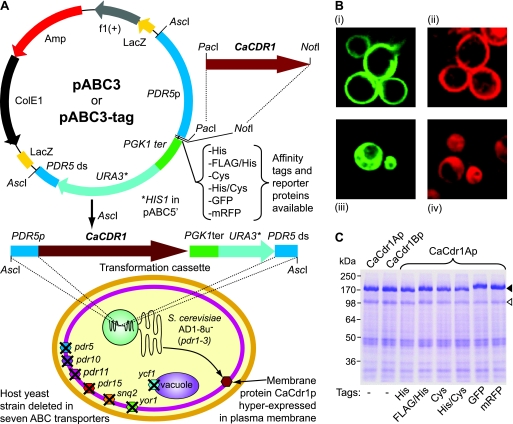
Plasmids used in this study are listed in Table 2. pABC3 was constructed from the pBluescript SK(+) (Stratagene, La Jolla, CA) derivative pSK-PDR5PPUS (38). To ensure the efficient termination of highly expressed genes, the S. cerevisiae PGK1 transcription terminator was PCR amplified as a HindIII/BamHI fragment from gDNA and used to replace the HindIII/BamHI fragment of pSK-PDR5PPUS at a site immediately 3′ of the PDR5 promoter sequence. PacI and NotI sites were introduced 5′ to the terminator and AscI sites introduced at each end of the transformation cassette by site-directed mutagenesis (Fig. 1A). DNA encoding a modified version of the green fluorescent protein (GFP) from Aequorea victoria (yEGFP3; a gift of B. P. Cormack [11]) or the monomeric red fluorescent protein (mRFP) from Discosoma coral (mRFP1; a gift of R. Y. Tsien [9]) was PCR amplified using 5′ primers containing NotI sites just upstream of the ATG start codon and 3′ primers containing EagI sites downstream of the reporter gene stop sequence TAA AT and directionally cloned into the pABC3 NotI site. Sequences encoding hexahistidine (His) or tetracysteine (Cys) or the double-affinity tags FLAG/His and His/Cys were ligated into the NotI site of pABC3 as double-stranded molecules obtained by annealing complementary synthetic primers which contained the tag sequence followed by the same TAA AT sequence and giving 5′ NotI- and 3′ EagI-compatible overhangs. Plasmid constructs containing the NotI site immediately upstream of the coding sequence of GFP, mRFP, or the tag sequences were selected and confirmed by DNA sequencing.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pABC3 | Empty expression vector | This study |
| pABC3-HIS | Vector pABC3 containing a C-term. hexahistidine affinity tag | This study |
| pABC3-CYS | pABC3 containing a C-terminal tetracysteine tag | This study |
| pABC3-HIS-CYS | pABC3 containing a C-terminal tandem tag (hexa-histidine and tetra-cysteine) | This study |
| pABC3-FLAG-HIS | pABC3 containing a C-terminal tandem tag (FLAG and hexa-histidine tag) | This study |
| pABC3-GFP | pABC3 containing C-terminal YEGFP3 | This study |
| pABC3-mRFP | pABC3 containing C-terminal mRFP1 | This study |
| pABC5' | Identical to pABC3 but contains HIS1 instead of URA3 | This study |
| pABC5′-HIS | Identical to pABC3-HIS but contains HIS1 instead of URA3 | This study |
| pABC5′-CYS | Identical to pABC3-CYS but contains HIS1 instead of URA3 | This study |
| pABC5′-HIS-CYS | Identical to pABC3-HIS-CYS but contains HIS1 instead of URA3 | This study |
| pABC5′-FLAG-HIS | Identical to pABC3-FLAG-HIS but contains HIS1 instead of URA3 | This study |
| pABC5′-GFP | Identical to pABC3-GFP but contais HIS1 instead of URA3 | This study |
| pABC5′-mRFP | Identical to pABC3-mRFP but contains HIS1 instead of URA3 | This study |
| pABC5′-ScPDR5 | pABC3 with PDR5 ORF from Saccharomyces cerevisiae AD1-8u− | This study |
| pABC3-CaCDR1A | pABC3 with A allele of CDR1 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaCDR1A-HIS | pABC3-HIS with A allele of CDR1 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaCDR1A-CYS | pABC3-CYS with A allele of CDR1 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaCDR1A-HIS-CYS | pABC3-HIS-CYS with A allele of CDR1 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaCDR1A-FLAG-HIS | pABC3-FLAG-HIS with A allele of CDR1 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaCDR1A-GFP | pABC3-GFP with A allele of CDR1 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaCDR1A-mRFP | pABC3-mRFP with A allele of CDR1 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaCDR1B | pABC3 with B allele of CDR1 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaCDR2A | pABC3 with A allele of CDR2 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaMDR1A | pABC3 with A allele of MDR1 ORF from Candida albicans strain 10261 | This study |
| pABC3-CaERG11A | pABC3 with A allele of ERG11 ORF from Candida albicans strain 10261 | This study |
| pABC3-CneMDR1 | pABC3 with MDR1 ORF from Cryptococcus neoformans strain CDC551 | This study |
| pABC3-HsABCB1 | pABC3 with HsABCB1 ORF from Homo sapiens | This study |
FIG. 1.
(A) Schematic diagram of the yeast membrane protein hyperexpression system. The system comprises a set of vectors based on pBluescript SK(+), containing a transformation cassette that consists of the PDR5 promoter (dark blue), the PGK1 terminator (green), a selection marker (light blue) (URA3 [pABC3 series of vectors] or HIS1 [pABC5′ series of vectors]), and a small part of the 3′ end of the PDR5 gene (dark blue). Unique 8-bp PacI and NotI cloning sites are located between the PDR5 promoter and the PGK1 terminator for the directional cloning of any ORF. Plasmids pABC3 and pABC5′ provided templates for derivative plasmids that contain a choice of six different C-terminal tags located 3′ to the NotI cloning site (pABC3-tag and pABC5′-tag). The transformation cassette containing an ORF cloned into the PacI and NotI sites can be excised as an AscI fragment and used to transform the host strain AD1-8u−. (B) Confocal microscopy images of AD1-8u− cells overexpressing either the azole drug pump CaCdr1p (i and ii) or the cytosolic protein CaUra3p (iii and iv) tagged with either green (GFP) (i and iii) or red (mRFP) (ii and iv) fluorescent protein. (C) PM proteins (30 μg per lane) of AD1-8u− cells hyperexpressing CaCdr1Ap, CaCdr1Bp, or CaCdr1Ap C-terminally tagged with each of the six tags were separated by SDS-PAGE and visualized with Coomassie blue.
An identical set of plasmids was engineered to contain the HIS1 auxotrophic marker (including 973 bp upstream and 378 bp downstream of the open reading frame [ORF]) instead of the URA3 marker of vector pABC3. This series of vectors was named pABC5′ and pABC5′-tag by analogy to the pABC3 series of vectors (Fig. 1A). The parental pABC5 plasmid included a second PacI site within the HIS1 promoter that was destroyed to facilitate directional cloning of ORFs as PacI/NotI fragments. Vector pABC5 was partially digested with PacI, and the full-length fragment was blunt ended with T4 DNA polymerase, gel purified, and religated to form pABC5′. The deletion of the PacI site in the HIS1 promoter of plasmid pABC5′ was confirmed by DNA sequence analysis and did not affect its function.
Construction of yeast strains overexpressing heterologous membrane proteins.
Recombinant pABC3, pABC5′, or tagged plasmids containing heterologous ORFs (2 μg) were digested with AscI and the gel-purified transformation cassette used to transform the AD1-8u− or ADΔ strain (Fig. 1A). Transformants were selected on CSM−ura or CSM−his agar plates after incubation at 30°C for 48 h to 72 h. About 100 transformants were usually obtained per μg of DNA. In each experiment, 10 transformants were selected and tested by colony PCR for the proper integration of the complete transformation cassette at the chromosomal PDR5 locus. PCRs designed to amplify products across both the predicted 5′ and 3′ integration sites were used to verify the expected integration of the transformation cassette at the PDR5 locus. For a clone to be selected as a positive transformant, both PCRs were required to amplify PCR fragments with sizes and sequences expected for integration via homologous double crossover as shown in Fig. 1A. More than 90% of transformants showed correct integration of the cassette. Three positive transformants for each construct were selected for further analysis. HsABCB1 (MDR1) was cloned from cDNA contained on plasmid pMDRA1, kindly provided by K. Ueda, Kyoto University, Kyoto, Japan.
Alternative strategies for obtaining strains overexpressing tagged membrane proteins.
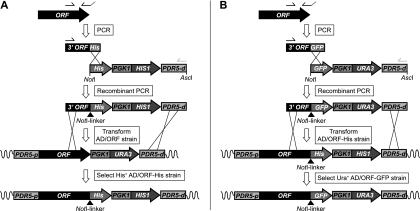
A set of strains overexpressing C-terminally tagged membrane proteins in AD1-8u− or ADΔ were obtained by two separate protocols using recombinant PCR (Fig. 2). Strains overexpressing ScPdr5p, CaCdr2Ap, CgCdr1p, CgPdh1p, CkAbc1p, and CneMdr1p with a C-terminal His tag were obtained by modifying yeast strains overexpressing the nontagged proteins (Fig. 2A). The overexpressing yeast strains were directly transformed with a DNA fragment that contained approximately 500 bp of the 3′ end of the ORF fused to the hexahistidine tag coding sequence (His), followed by the PGK1 terminator, the HIS1 marker, and the PDR5 downstream region of plasmid pABC5′. The required PCR products were obtained in a two-step recombinant PCR amplification. First, the 3′ end of the ORF was amplified using ORF-containing DNA as a template (either plasmid or gDNA), an ORF-specific forward primer (25-mer) binding about 500 bp upstream of the ORF stop codon, and a reverse primer containing 16 bp of the His tag sequence (reverse complement), the NotI site, an extra C, and the last 25 bp of the ORF without the stop codon (Fig. 2A, PCR). The His tag-containing fragment from vector pABC5′-His was isolated as a NotI/AscI fragment and gel purified. Equal amounts (10 ng) of the two DNA fragments were combined and amplified by 15 cycles of recombinant PCR, using the ORF-specific forward primer and a reverse primer that binds to the 3′ end of the NotI/AscI fragment of plasmid pABC5′-His (Fig. 2A, recombinant PCR). The recombinant PCR product was used to transform the Ura+ His− ORF-expressing S. cerevisiae strain to His+ Ura−. Yeast transformants with the URA3 marker downstream of the ORF replaced by HIS1 were selected on CSM−his plates.
FIG. 2.
Strategies for adding His or GFP tags to ORFs. (A) His tags were added by recombinant PCR, followed by transformation of S. cerevisiae strains already containing the heterologous ORF to His+. (B) GFP tags were added by recombinant PCR, followed by transformation of S. cerevisiae strains already containing His tags to Ura+.
Yeast strains overexpressing proteins with a C-terminal GFP tag were obtained by transforming yeast strains overexpressing the above C-terminal His-tagged proteins with a recombinant DNA fragment that contained 500 bp of the 3′ end of the ORF fused to the GFP tag, the PGK1 terminator, the URA3 marker, and the PDR5 downstream region of plasmid pABC3-GFP (Fig. 2B, PCR). The reverse primers used to amplify the small 500-bp ORF-specific fragments contained (in reverse complement order) the first 16 bp of the GFP tag sequence, the NotI site, an extra C, and the last 25 bp of the ORF minus the stop codon. The GFP tag-containing fragment was isolated from vector pABC3-GFP as a NotI/AscI fragment and gel purified. The two fragments were fused and amplified by recombinant PCR (Fig. 2B, recombinant PCR). Yeast transformants with the HIS1 marker downstream of the ORF replaced by the URA3-containing transformation PCR fragment were selected on CSM−ura plates.
Yeast strains overexpressing C-terminally GFP-tagged CaMdr1Ap, CaErg11Ap, and HsAbcb1p were obtained by transformation of yeast cells overexpressing nontagged protein using a recombinant PCR strategy similar to that shown in Fig. 2A. In the recombinant PCR, however, the AscI/NotI fragment of plasmid pABC5′-GFP containing the HIS1 marker was used, and positive transformants were selected on CSM−his plates. All transformants were confirmed by colony PCR across the whole region of integration and sequencing of the resulting PCR fragments.
Cloning of CkABC1 for expression in yeast.
Because plasmid pABC3-CkABC1 could not be amplified in E. coli, a cloning strategy using PCR of a ligation mixture was used to clone CkABC1 in S. cerevisiae AD1-8u−. Plasmid pABC3 (10 ng) cut with PacI/NotI and 50 ng CkABC1 ORF, obtained by 35 PCR cycles from genomic DNA and cut to completion with PacI/NotI, were ligated for 1 h at room temperature in a 20-μl volume using a rapid ligation protocol (Promega, Madison, WI). A 1-μl portion of the ligation mixture was then amplified in a standard 35-cycle PCR using primers that bind to the AscI regions of pABC3-CkABC1 (Fig. 1A). The resultant 7.3-kb transformation cassette was gel purified and used to transform ADΔ to Ura+.
Analysis, visualization, and purification of PM proteins.
Plasma membrane (PM) fractions of S. cerevisiae cells were prepared as described previously (40). Protein samples (30 μg) were separated on 8% acrylamide gels and stained with Coomassie blue R250. The relative amounts of Coomassie-stained protein bands in images of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE)-separated membrane preparations were calculated using NIH Image software. S. cerevisiae strains expressing GFP- or mRFP-fusion proteins were examined using a Zeiss 510 Axiovert 200 M inverted confocal laser scanning microscope (Otago Centre for Confocal Microscopy). His-tagged CaCdr1Ap was purified from PM preparations using the protocol of Ferreira-Pereira et al. (18).
Functional analysis of membrane transporters.
The susceptibility of yeast to antifungal agents, the energy-dependent efflux of R6G from yeast cells, and the ATPase activities of PM preparations were measured as described previously (26, 40). For R6G efflux experiments, yeast cells were glucose deprived, and preloaded with 15 μM R6G, and washed. Efflux was initiated by the addition of glucose (22 mM). Agarose diffusion assays were performed to test the susceptibility of a panel of strains overexpressing drug pumps. In brief, a 10-ml YPD overnight culture of each test strain was diluted 1:20 into 3 ml CSM medium and grown at 30°C for another 4 h to mid-logarithmic growth phase (OD600, ∼1; ∼107 cells/ml). The cells were diluted to an OD600 of 0.008 in 5 ml of melted CSM containing 0.6% agarose (50°C) and plated on 20-ml CSM agarose plates. Whatman 3MM paper disks (5-mm diameter) containing xenobiotics were placed on the solidified top agarose, and the plates were incubated at 30°C for 48 h. The amounts of xenobiotics used on the disks are described in the figure legends.
Screening for inhibitors of fungal drug pumps (chemosensitization assay).
The chemosensitization of a range of yeast strains overexpressing fungal drug pumps to FLC was carried out as described previously (40). In brief, a 10-ml YPD overnight culture of cells was diluted 1:20 into CSM and incubated at 30°C for a further 4 h. Each test strain (OD600, ∼1) was diluted to an OD600 of 0.008 in 5 ml of melted CSM containing 0.6% agarose (50°C) with either no FLC (to determine the toxicity of each drug; control) or FLC at 0.25 × the MIC of FLC (MICFLC). The chemosensitizing effect of each drug was visualized at the following concentrations of FLC for each strain: AD/ScPDR5, 62.5 μg/ml; ADΔ/CaCDR1A, 50 μg/ml; AD/CaCDR2A, 15.6 μg/ml; AD/CgCDR1, 62.5 μg/ml; AD/CgPDH1, 4.0 μg/ml; ADΔ/CkABC1, 15.6 μg/ml; AD/CaMDR1A, 12.5 μg/ml. The cell suspension was poured into a rectangular Omnitray plate (126 by 86 by 19 mm; Nunc, Roskilde, Denmark) that contained 20 ml of CSM solidified with 0.6% agarose either without (control) or with 0.25× MICFLC. Whatman 3MM paper disks containing potential drug pump inhibitors (5 μg of milbemycins α11, α20, α25, β9, and β11, 5 μg enniatin, 25 μg FK506, or 50 nmol oligomycin) were placed on the solidified top agarose, and the plates were incubated at 30°C for 48 h.
Nucleotide sequence accession numbers.
The DNA sequences of all 14 pABC3- and pABC5′-derived vectors shown in Fig. 1A were confirmed by DNA sequence analysis and submitted to the GenBank database (accession numbers DQ903883 to DQ903896).
RESULTS
Versatile yeast membrane protein hyperexpression system.
The membrane protein hyperexpression system consists of plasmids pABC3 and pABC5′, and their derivative plasmids, and the host S. cerevisiae strains AD1-8u− and its close relative ADΔ (see Materials and Methods) (Fig. 1A; Tables 1 and 2) (36). Both strains are deleted in seven genes encoding ABC transporters and the gene for the Pdr3p transcription factor but contain the gain-of-function mutant transcription factor Pdr1-3p (38). ABC transporters constitute one of the largest protein families known and are involved in many cellular processes, including lipid translocation and the transport of a variety of compounds across biological membranes (8, 35, 66). The lack of the seven ABC transporters makes strains AD1-8u− and ADΔ exquisitely sensitive to a range of xenobiotics (38). The AD1-8u− strain contains ura3 and his1 null alleles that permit transformation using either the URA3 and/or HIS1 selectable markers. The ADΔ strain had the URA3 gene completely deleted to minimize ectopic integration of the PDR5-flanked cassette at the URA3 locus and to facilitate future gene disruptions using the URA3 blaster strategy. Additional refinements to the system included the development of pABC3 and pABC5′ variants containing affinity tags and reporter proteins (Fig. 1A). Plasmids with a hexahistidine affinity tag (His), tetracysteine affinity tag (Cys) (21, 59), or double-affinity FLAG/His (28) or His/Cys tags allow the affinity purification and chemical detection or immunodetection of heterologously hyperexpressed membrane proteins. Plasmids containing the ORFs of GFP (yEGFP3) (11) or mRFP (mRFP1) (9) provide transformation cassettes that allow the visualization of heterologous proteins in S. cerevisiae (Fig. 1B).
Cloning heterologous ORFs into pABC plasmids.
ORFs to be cloned in the expression system were amplified from gDNA or first-strand cDNA by PCR using a 5′ primer containing the PacI site and a 3′ reverse primer containing the NotI site just downstream of the ORF stop codon. To achieve high levels of expression, all 5′ primers contained the PacI site 11 bp upstream of the ATG start codon, followed by AAA and then the ORF ATG start codon. This maintained the size of the 5′ untranslated region (UTR) of the PDR5 mRNA leader sequence and, most importantly, preserved the Kozak consensus sequence for highly expressed S. cerevisiae genes, particularly the A at position −3. The 3′ reverse primers were designed so that the ORF sequence was followed by the stop codon sequence TAA AT to ensure efficient termination of highly expressed genes (6), followed by the NotI site. Three extra nucleotides (suitable random sequences) were added at the 5′ ends of each primer to ensure efficient cutting of the PCR-amplified ORF fragments with the PacI and NotI restriction enzymes. The general design of the 5′ and 3′ primers used to clone an ORF into pABC3 and pABC5′ comprised the 5′ primer 5′-NNN TTAATTAA AAA ATG plus the first 20 bp of the ORF and the 3′ reverse primer 5′-NNN GCGGCCGC AT TTA plus the reverse complement sequence of the last 20 bp of the ORF. Most ORFs could be amplified as PacI/NotI fragments, cut with PacI and NotI restriction enzymes, gel purified, and cloned into pABC vectors. In rare cases where the ORF contained an extra internal PacI or NotI site, the ORF fragment was first partially digested with the enzyme that cut within the ORF (e.g., PacI for PDR5) and then the full-length partially digested ORF fragment was gel purified and fully digested with the second enzyme before cloning into pABC3 or pABC5′ was done.
Cloning heterologous ORFs into pABC3-tag or pABC5′-tag plasmids.
Directional cloning of ORFs as PacI/NotI fragments into any of the 12 tagged vectors required a slight modification of the 3′ reverse primer described above. The modifications included the removal of the AT TTA reverse stop codon sequence and the addition of one extra C nucleotide 3′ to the NotI site to ensure an in-frame fusion of the ORF with the chosen tag. Thus, the 3′ reverse primer sequence to PCR amplify any ORF for cloning into a tagged vector comprised 5′-NNN GCGGCCGCC plus the reverse complement sequence of the last 20 bp of the ORF (the extra C nucleotide is in bold). Cloning ORFs into pABC3-tag or pABC5′-tag vectors with these primers introduced a three-amino-acid C-terminal linker (GGC GGC CGC, encoding Gly-Gly-Arg) between the ORF and the tag.
Cloning heterologous ORFs in S. cerevisiae.
ORFs encoding genes to be hyperexpressed in S. cerevisiae AD1-8u− or ADΔ were PCR amplified and directionally cloned into the PacI and NotI restriction sites of pABC3, pABC5′, or their derivative plasmids (see Materials and Methods) (Fig. 1A). A transformation cassette containing the inserted ORF with or without a C-terminal fusion to any of the six tags, the S. cerevisiae PGK1 terminator, the URA3 or HIS1 selection marker, and flanking PDR5 sequences was excised from the recombinant plasmid with AscI. The toxicity of plasmid pABC3-CkABC1 precluded cloning the CkABC1 gene in E. coli. In this case, the entire transformation cassette was amplified by PCR directly from the ligation mix (see Materials and Methods). The selection of Ura+ or His+ transformants of AD1-8u− directs the integration of the linearized transformation cassette, via homologous recombination of its terminal sequences, at the PDR5 chromosomal locus (Fig. 1A).
Hyperexpression of ABC efflux pump ScPdr5p and CaCdr1p in S. cerevisiae.
The efficiency of the S. cerevisiae membrane protein hyperexpression system was demonstrated by cloning the homologous ScPDR5 gene into strain AD1-8u− to form AD/ScPDR5. The level of ScPdr5p expression in AD/ScPDR5 was compared to the expression of ScPdr5p in the control strain, AD124567 (13). AD124567 is a precursor to strain AD1-8u− that still contains the wild-type PDR5 gene. SDS-PAGE analysis of PM preparations and measurement of the strains' MICFLCs showed that the levels of ScPdr5p expression were identical between the two strains (data not shown). This control experiment confirmed that the modifications to the 5′ and 3′ UTR of PDR5 introduced using our simple cloning strategy (Fig. 1A) maintained the high level of protein expression achieved with the wild-type PDR5 UTR.
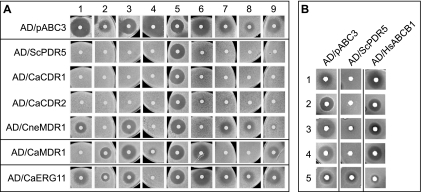
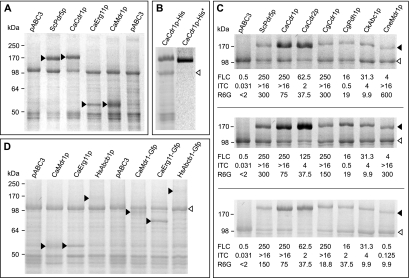
The membrane protein hyperexpression system was used to study proteins involved in the FLC resistance of Candida albicans. This yeast is an opportunistic pathogen of the immunocompromised and can cause a range of infections in the very young, the elderly, cancer patients, organ transplant recipients, and HIV-positive/AIDS patients (45, 63). Triazole antifungals, such as FLC, which inhibit the microsomal cytochrome P450 enzyme 14α-lanosterol demethylase (Erg11p), involved in ergosterol biosynthesis, are widely used to treat candidiasis patients (63). Prolonged courses of the antifungal FLC lead to the emergence of C. albicans strains resistant to FLC. Low-level resistance can be due to overexpression of the drug target Erg11p or mutations in Erg11p. Higher levels of resistance correlate with overexpression of the MFS drug pump Mdr1p or the ABC pump Cdr1p or Cdr2p (2, 47, 51). C. albicans is diploid, and we cloned the A allele of CDR1 of C. albicans strain ATCC 10261 into the PacI and NotI sites of pABC3 and transformed S. cerevisiae AD1-8u− with the AscI fragment of the recombinant plasmid (Fig. 1A). Hyperexpression of CaCdr1Ap in the PM (Fig. 1C) conferred on AD1-8u− resistance to the azole drugs FLC, ITC, and other azoles but not to the polyene NYS (Table 3 and Fig. 3), as expected, since NYS is not a substrate of ABC pumps. Some of the phenotypic effects could, however, be caused by the background mutations in host strains AD1-8u− and ADΔ. These mutations and the expression of certain pumps could potentially affect PM composition and concomitantly alter ABC pump function, thus leading to slightly changed substrate specificities that are not seen in the natural host. Allelic variation caused by single-nucleotide polymorphisms can affect gene function. The CaCDR1A, CaCDR1B, CaCDR2A, and CaCDR2B alleles of C. albicans ATCC 10261 were individually cloned and expressed in AD1-8u− (Fig. 1C and 4C). The expression of each CaCdr1p and CaCdr2p allele to the same extent enabled identification of functional differences between each of these pairs of alleles (26) and further characterization of these proteins' functions.
TABLE 3.
Phenotypes conferred on S. cerevisiae AD1-8u− by hyperexpression of fungal drug efflux pumps and other membrane proteins
| Hyperexpressed protein | MIC of drug (μg/ml)
|
R6G effluxa (pmol/107 cells/min) | ATPase activitya,b (nmol Pi/min/mg protein) | ||
|---|---|---|---|---|---|
| FLC | ITC | NYS | |||
| None (pABC3) | 0.5 | 0.031 | 2 | 0.0 | 7 ± 2 |
| ScPdr5p | 300 | >32 | 2 | 93 ± 5 | 199 ± 12 |
| CaCdr1Ap (allele A) | 200 | >32 | 2 | 75 ± 9 | 185 ± 4 |
| CaCdr1Bp (allele B) | 300 | >32 | 2 | 79 ± 7 | 188 ± 3 |
| CaCdr2Ap (allele A) | 75 | 4 | 2 | 22 ± 5 | 128 ± 9c |
| CaCdr2Bp (allele B) | 150 | >32 | 2 | 33 ± 3 | 164 ± 16c |
| CaMdr1Ap | 60 | 0.031 | 2 | 0.0 | 6 ± 2 |
| CaErg11Ap | 5 | 0.125 | 2 | 0.0 | 5 ± 2 |
| CgCdr1p | 300 | >32 | 4 | 105 ± 1 | 200 ± 14 |
| CgPdh1p | 8 | 0.062 | 4 | 15 ± 3 | 141 ± 19 |
| CkAbc1p | 32 | 8 | 2 | 82 ± 5 | 57 ± 2 |
| CneMdr1p | 5 | >32 | 2 | 92 ± 20 | 45 ± 3 |
Mean ± SD for three experiments carried out in triplicate.
Oligomycin (20 μM)-sensitive ATPase activity, measured at pH 7.5.
Two hundred micromolar oligomycin used in assays.
FIG. 3.
Agarose diffusion assays of AD1-8u− cells overexpressing a selection of fungal ABC drug pumps (ScPdr5p, CaCdr1Ap, CaCdr2Ap, and CneMdr1p), a fungal MFS drug pump, CaMdr1Ap, the azole drug target CaErg11Ap, and the sensitive control strain AD1-8u− containing the empty pABC3 transformation cassette integrated at the chromosomal PDR5 locus (AD/pABC3). Cells were analyzed using agarose diffusion assays as described in Materials and Methods. (A) Susceptibilities to the following xenobiotics were tested: 1, FLC (10 μg); 2, ITC (0.16 μg); 3, MCZ (0.2 μg); 4, KTZ (0.2 μg); 5, NYS (60 μg); 6, R6G (48 μg); 7, cycloheximide (0.2 μg); 8, cerulenin (1 μg); 9, Triton X-100 (1 mg). (B) As in panel A, the substrate specificities of cells overexpressing HsAbcb1p were tested and compared to those of cells overexpressing ScPdr5p using the control strain AD/pABC3. Drugs applied were as follows (top to bottom): 1, FLC (4 μg); 2, ITC (0.5 μg); 3, R6G (80 μg); 4, Triton X-100 (1 mg); 5, aureobasidin A (20 μg).
FIG. 4.
SDS-PAGE analysis of PM proteins (30 μg per lane) from AD1-8u− cells overexpressing different fungal and human membrane proteins. In each panel, filled arrowheads indicate heterologous proteins and open arrowheads indicate the endogenous 100-kDa PM H+ ATPase (Pma1p). (A) AD1-8u− cells overexpressing different classes of fungal membrane protein: ABC transporters ScPdr5p and CaCdr1Ap, MFS transporter CaMdr1Ap, and the azole drug target CaErg11Ap, a cytochrome P450 enzyme. pABC3 refers to the drug-sensitive control strain as described in the legend to Fig. 3. (B) The left lane shows 30 μg of PM protein isolated from AD/CaCDR1A-His-p cells. CaCdr1A-His-p after Ni affinity chromatography is shown in the right lane. (C) Fungal drug pumps hyperexpressed in AD1-8u− cells (top panel). The addition of a C-terminal His tag (middle panel) or GFP tag (bottom panel) does not affect their level of expression. Shown below the gel in each panel are the FLC, ITC, and R6G MICs (μg/ml) for each of the overexpressing strains. (D) SDS-PAGE analysis of PM proteins isolated from AD/CaMDR1A, AD/CaERG11A, and the AD/HsABCB1 cells and their GFP-tagged derivatives.
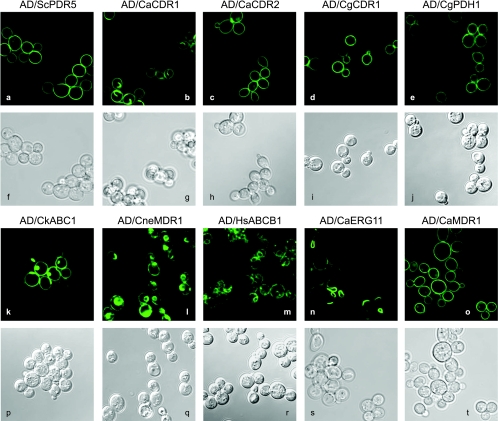
To analyze the trafficking of heterologous CaCdr1Ap to the S. cerevisiae PM, CaCDR1A was cloned in AD1-8u− fused to GFP or mRFP. When S. cerevisiae cells expressing these fusion proteins were grown under standard conditions, confocal microscopy indicated that the proteins were correctly trafficked to the cell surface (Fig. 1B, i and ii), which did not occur with overexpression of plasmid-encoded CaCdr1p (19). Only a minor proportion of the CaCdr1Ap protein was located in the cell interior, seemingly around the nucleus. In contrast, CaUra3p fusion proteins in control strains, created by expressing C. albicans ATCC 10261 URA3 (encoding the cytoplasmic enzyme orotidine-5′-phosphate decarboxylase) in our system, were uniformly distributed throughout the cytoplasm and excluded from organelles (Fig. 1B, iii and iv).
The effect of tags on heterologous gene expression was investigated by cloning CaCDR1A into pABC3 derivatives so that the following tags were fused to the CaCdr1Ap C terminus: His, FLAG/His, Cys, and His/Cys. None of these tags or fusion to GFP or mRFP caused a significant reduction in CaCdr1Ap expression in PM preparations (Fig. 1C). Only the Cys-containing tags (Cys and His/Cys) slightly reduced the expression of CaCdr1Ap (Fig. 1C).
Hyperexpression and characterization of other fungal membrane proteins involved in azole resistance.
The S. cerevisiae membrane protein hyperexpression system was used to functionally express three classes of fungal membrane protein that are involved in resistance of C. albicans to FLC. Individual alleles of C. albicans ATCC 10261 CDR1 (GenBank accession numbers DQ462358 and DQ462359) (26), CDR2 (GenBank accession numbers DQ470007 and DQ470008) (26), ERG11 (GenBank accession number DQ903897), and MDR1 (GenBank accession number DQ903899) were cloned and expressed in S. cerevisiae AD1-8u−. SDS-PAGE analysis of recombinant S. cerevisiae strains indicated that polypeptides of the expected molecular masses were hyperexpressed in PM fractions (Fig. 4). The identity of the hyperexpressed protein could be confirmed for CaCdr1Ap, CaCdr2Ap, and CaMdr1Ap by immunodetection (26, 38) and for CaErg11Ap and ScPdr5p by mass spectrometry of tryptic fingerprints (data not shown). The expression of these proteins, measured by NIH Image analysis of Coomassie blue-stained SDS-PAGE profiles, ranged from 29% (for CaCdr1Ap) to 4.9% (for Erg11Ap) of PM protein. The hyperexpressed membrane proteins were shown to be functional with three assays of whole-cell or in vitro function: antifungal susceptibility, energy-dependent efflux of the fluorescent pump substrate R6G, and ATPase activity. The host strain AD1-8u− is hypersusceptible to the triazole antifungals FLC and ITC (Table 3). Expression of CaCdr1Ap or the S. cerevisiae ortholog ScPdr5p increased the resistance of AD1-8u− to FLC and ITC between 400 and >1,000-fold (Table 3). Expression of the MFS membrane transporter CaMdr1Ap conferred less resistance to FLC and no resistance to ITC (Table 3; Fig. 3), as has been found for clinical C. albicans isolates expressing Mdr1p (47). Hyperexpression of the azole drug target CaErg11Ap conferred only a 4- to 10-fold increase in azole resistance (Table 3). Hyperexpression of fungal transporters did not confer on S. cerevisiae resistance to the polyene antifungal NYS, which is not a substrate for the pumps (Table 3; Fig. 3). S. cerevisiae cells expressing fungal ABC transporters but not the MFS pump CaMdr1Ap demonstrated R6G efflux in a time- and energy-dependent fashion (Table 3; Fig. 5). Finally, the drug efflux pump ATPase activity of cells expressing ABC transporters was measured (Table 3). PM fractions from cells expressing ABC but not MFS pumps had significant ATPase activities. The pump ATPase activity could be distinguished from the PM H+ ATPase activity because it was oligomycin sensitive and active at pH 7.5 (Fig. 6C).
FIG. 5.
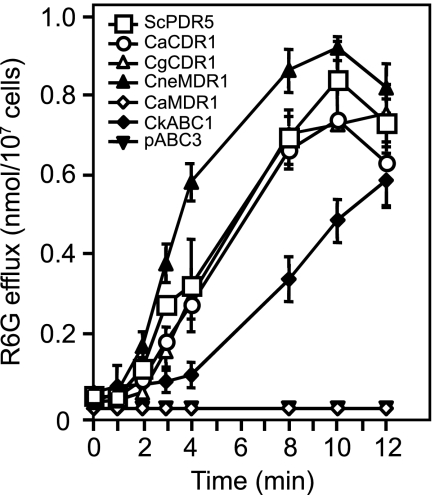
In vivo pumping activities of different fungal drug pumps using the fluorescent pump substrate R6G. AD1-8u− cells overexpressing drug pumps were grown to mid-logarithmic phase, harvested, and loaded with the fluorescent substrate R6G, and the assay was started by the addition of glucose as an energy source as described in Materials and Methods. The fluorescence intensity accumulating in the supernatant after filtration of the cells was measured and the R6G pumping activity per 107 cells determined. The results are the means ± standard deviations for triplicate determinations.
FIG. 6.
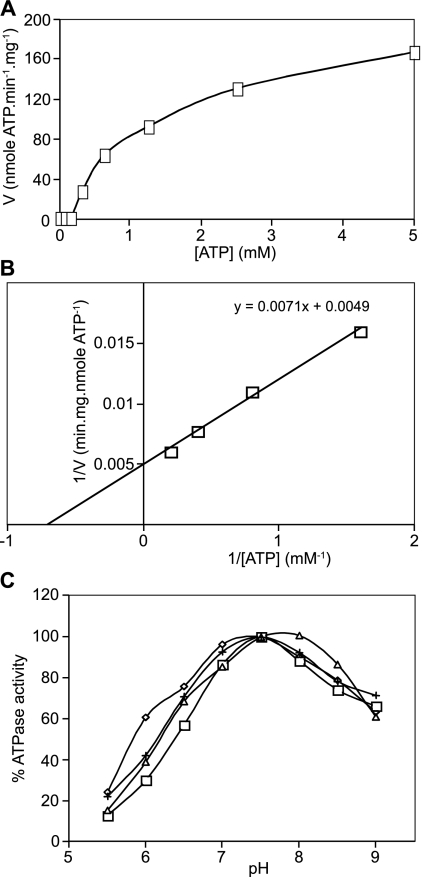
Kinetic parameters for the ATPase activities of fungal ABC drug transporters. (A) Vanadate-sensitive ATPase activity (V) versus substrate concentration plot for Cdr1Ap-His/Cys-tagged drug pump at pH 7.5. (B) Lineweaver Burke plot for the vanadate-sensitive ATPase activity of the CaCdr1Ap-His/Cys-tagged drug pump at pH 7.5. The data were taken from panel A. (C) The pH profile for the ATPase activity of untagged CaCdr1Ap (diamond) or CaCdr1Ap with a His tag (square), Cys tag (triangle), or His/Cys tag (cross) is shown. Results are expressed as percentages of activity at pH 7.5.
A panel of S. cerevisiae strains hyperexpressing fungal ABC transporters.
The hyperexpression system was used to express ABC transporters from other fungal pathogens, including the Candida glabrata database strain CBS138 (CgCDR1 and CgPDH1), Candida krusei strain B2399 (CkABC1), and Cryptococcus neoformans strain CDC551 (CneMDR1) (Tables 1 and 2; Fig. 4C). The AD/CgCDR1 and AD/CgPDH1 strains were described previously (61). The putative azole drug pump encoded by CkABC1, thought to be involved in the innate azole drug resistance of C. krusei, was originally described by Katiyar and Edlind (29). They reported the cloning of a 300-bp fragment of CkABC1 corresponding to a highly conserved consensus region of ABC transporters. We used inverse PCR to clone CkABC1 and discovered that the 4.65-kb CkABC1 gene contains an 88-bp intron at the 5′ end of the ORF (GenBank accession numbers DQ903906 and DQ903907). A strain expressing CkABC1 cDNA (AD/CkABC1-c) was created and was found to have properties similar to those of AD-CkABC1. The PM preparations from AD/CkABC1-c cells contained 50% to 100% more CkAbc1p than PM preparations from intron-containing AD/CkABC1 cells (data not shown). This indicated that although S. cerevisiae recognizes the CkABC1 intron, its processing gave a reduced level of CkAbc1p expression. CneMDR1 was isolated from C. neoformans strain CDC551 and found to be identical to the published sequence of CneMDR1 isolated from a different strain (GenBank accession number DQ903909).
The overexpression of each of these fungal ABC transporters conferred increased azole drug resistance on AD1-8u− (Fig. 4C, top panel; Table 3). As expected, each transporter effluxed R6G in a time- and energy-dependent fashion, albeit at different rates and with different lag periods after the addition of glucose (Table 3; Fig. 5) (13, 26, 61, 62). These data confirmed the hypothesis that all these transporters are energy-dependent azole drug efflux pumps. There was a positive correlation between the R6G efflux activity and the resistance of strains expressing ABC transporters (apart from CneMdr1p) to FLC determined by liquid MIC measurements. All the ABC drug efflux pumps showed ATPase activity, and although activity was generally higher in strains with higher MICFLCs, the relationship was not linear, indicating an indirect relationship between ATPase activity and FLC susceptibility. This may reflect different degrees of coupling between energy utilization in the nucleotide binding domains and drug transport mediated by the membrane sector of the pumps (61).
Effect of affinity and epitope tags on membrane protein function.
The overexpression, purification, and crystallization of membrane proteins constitute a challenging scientific endeavor. Besides achieving high levels of expression, it is paramount for the crystallization of membrane proteins that the overexpressed membrane protein can be solubilized with mild detergent and remain stable in solution as a monodisperse molecule. After successful overexpression of an array of fungal membrane proteins, C-terminally hexahistidine-tagged derivatives of these fungal ABC drug pumps were created to facilitate their affinity purification. The addition of a His tag did not affect the level of overexpression of any of the seven ABC transporters tested (Fig. 4C, middle panel). Initial purification efforts using PMs obtained from ADΔ/CaCDR1A-His were very promising. CaCdr1Ap-His was readily solubilized with the mild detergent dodecylmaltoside, and a one-step nickel affinity purification led to a CaCdr1Ap-His protein fraction with greater than 95% purity (Fig. 4B).
Effects of C-terminal affinity tags on kinetics and inhibition of fungal ABC transporter ATPase activities.
The effects of C-terminal tags on the function of fungal ABC transporters was determined by the measurement of kinetic parameters and responses to known inhibitors of their ATPase activity in PM preparations. All pumps, including those modified with C-terminal His, Cys, and His/Cys tags, showed rate-versus-ATP concentration profiles that approximated Michaelis-Menten kinetics at higher ATP concentrations. Figure 6A and B show the results for PMs containing CaCdr1pA-His/Cys as a typical example. The apparent Km (Km app) values for the unmodified ABC transporters lay within a narrow range of 0.6 mM ATP for CgPdh1p to 2.45 mM ATP for CaCdr2Ap, with most ABC transporters having a Km app value of about 1.5 mM ATP. The addition of a C-terminal His tag had only minor effects on the Km app values (Table 4). At ATP concentrations at least twofold below the Km app value, all pumps showed positive cooperativity with ATP, as expected for ABC transporters with two ATP molecules in the active site (Fig. 6A). The Vmax values of ATPase activity in the PM preparations ranged between 95 nmol/min/mg for CneMdr1p and 270 nmol/min/mg for CgPdh1p among the nontagged pumps. The addition of C-terminal His tags also had only modest effects on Vmax values. The pH optimum for ScPdr5p, CaCdr1Ap, CaCdr2Ap, CgCdr1p, and CkAbc1p was 7.5, with a slight shift to neutral pH for CgPdh1p (7.0) and CneMdr1p (6.5 to 7.0). The addition of a C-terminal His tag to the ABC transporters had no effect on any of the pH profiles. For CaCdr1Ap, the influence of a C-terminal Cys tag or the double-affinity tag His/Cys on its kinetic parameters was tested. Both tags had minor effects on the Km app, Vmax, and pH optimum values. Most notably, a slight alkaline shift in the pH optimum was seen for the C-terminal Cys tag (7.5 to 8.0) (Fig. 6C; Table 4).
TABLE 4.
Effect of tags on pump ATPase activity and response to inhibitors
| Hyperexpressed pump | Kinetics of ATPase activity
|
IC50 of:
|
||||
|---|---|---|---|---|---|---|
| Km app (mM ATP) | Vmax (nmol/min/mg) | pH optimum | Vanadate (μM) | Oligomycin (μM) | FK506 (μg/ml) | |
| None (AD1-8u−) | 2.61 | 30 | 6-8 | |||
| ScPDR5 | 1.42 | 303 | 7.5 | 1.3 | 0.25 | 0.013 |
| ScPDR5-His | 1.30 | 333 | 7.5 | 1.3 | 0.30 | 0.013 |
| CaCDR1A | 1.31 | 135 | 7.5 | 3.1 | 0.30 | 0.63 |
| CaCDR1A-His | 1.20 | 133 | 7.5 | 3.1 | 0.30 | 0.63 |
| CaCDR1A-Cys | 1.82 | 111 | 7.5-8.0 | 12.5 | 0.70 | 0.80 |
| CaCDR1A-His-Cys | 1.45 | 204 | 7.5 | 6.3 | 0.70 | 0.60 |
| CaCDR2A | 2.45 | 250 | 7.5 | 1.3 | >20 | 20 |
| CaCDR2A-His | 1.05 | 278 | 7.5 | 3.0 | >20 | 20 |
| CgCDR1 | 1.95 | 227 | 7.5 | 1.5 | 0.063 | 0.0063 |
| CgCDR1-His | 1.27 | 303 | 7.5 | 1.6 | 0.01 | 0.013 |
| CgPDH1 | 0.60 | 270 | 7.0 | 0.9 | 0.30 | 0.013 |
| CgPDH1-His | 0.85 | 303 | 7.0 | 1.3 | 0.30 | 0.013 |
| CkABC1 | 1.31 | 161 | 7.5 | 0.8 | 0.40 | 1.0 |
| CkABC1-His | 1.45 | 204 | 7.5 | 1.3 | 0.40 | 1.0 |
| CneMDR1 | 1.71 | 95 | 6.5-7.0 | 3.1 | 0.50 | 20 |
| CneMDR1-His | 1.03 | 79 | 6.5-7.0 | 4.0 | 1.00 | 20 |
As independent measures of ABC transporter function, the 50% inhibitory concentrations (IC50s) for known inhibitors of ATPase activity were determined. The vanadate IC50s of all ABC transporters were within a narrow range between 0.8 μM for CkAbc1p and 3.1 μM for CneMdr1p. The addition of a C-terminal His tag hardly affected the IC50s of vanadate for any of the pumps except for CaCdr2Ap (3.0 μM for CaCdr2Ap-His versus 1.3 μM for CaCdr2Ap). The addition of a C-terminal Cys tag led to a fourfold increase in the IC50 for vanadate (12.5 μM for CaCdr1Ap-Cys versus 3.1 μM for CaCdr1Ap). The IC50s for the ATPase activities of the panel of fungal ABC transporters for oligomycin and FK506 were not significantly affected by the presence of any of the tags (Table 4). With the exception of CgCdr1p, which was highly sensitive to inhibition by oligomycin (IC50 = 0.063 μM), and CaCdr2Ap, which was poorly inhibited by oligomycin (IC50 > 20 μM), the oligomycin IC50s for the ABC transporters lay within a very narrow range of 0.25 μM for ScPdr5p to 0.5 μM for CneMdr1p. FK506 was a very effective inhibitor of the ATPase activities of CgCdr1p (IC50 = 0.0063 μM), CgPdh1p (IC50 = 0.013 μM), and ScPdr5p (IC50 = 0.013 μM). Severalfold-higher concentrations were required to inhibit CaCdr1Ap and CkAbc1p (IC50 = 0.63 μM and 1.0 μM, respectively). FK506 was an even less effective inhibitor of the ATPase activities of CaCdr2Ap and CneMdr1p (IC50 > 20 μM), with 200 μM FK506 required to obtain 90% inhibition (data not shown). The cross-resistance of CaCdr2p to FK506 and oligomycin was not found for CneMdr1p, indicating that the FK506 and oligomycin binding sites are not identical. In summary, apart from the Cys and His/Cys double tags, the addition of C-terminal affinity tags did not affect significantly the level of expression or the kinetic characteristics of any of the fungal ABC transporters tested. Furthermore, the addition of a C-terminal His tag also did not affect the overall function of any of these ABC transporters, as measured by their in vivo resistance to the drugs FLC, ITC, and R6G (Fig. 4C).
Cellular location of heterologously overexpressed membrane proteins.
While the addition of a C-terminal GFP tag to any of the seven fungal ABC transporters tested did not affect their level of expression in the PM, the drug sensitivities of these yeast strains and the determination of the cellular location of the transporters by confocal microscopy indicated that in some cases, function and/or cellular localization could be affected (Fig. 4C and 7). The FLC, ITC, and R6G MICs for some strains overexpressing ABC transporters indicated that they are affected in their in vivo transport function with some but not other substrates (Fig. 4C, lower panel). The addition of the GFP tag reduced the MICFLC and MICITC for AD/CneMDR1 but not for the other six strains overexpressing fungal ABC transporters. The GFP tag had more complex effects on R6G transport. It reduced the MICR6G of AD/CneMDR1 128-fold, that AD/ScPDR5 2-fold, and that of AD/CgCDR1 16-fold. The C-terminal GFP tag on CgPdh1p, however, increased the MICITC fourfold and the MICR6G twofold. Confocal microscopy revealed that ScPdr5p, CaCdr2Ap, CgCdr1p, and CgPdh1p were properly and exclusively located in the PM (Fig. 7). However, a small fraction of CaCdr1Ap and CkAbc1p and the largest fraction of CneMdr1p were localized inside the cell in structures reminiscent of the endoplasmic reticulum (56). Since the C-terminal GFP tag did not increase the susceptibility of cells overexpressing CaCdr1Ap or CkAbc1p to FLC, ITC, or R6G, the confocal images of GFP-tagged proteins probably reflect the localization of the native untagged proteins. For CneMdr1p, the addition of the C-terminal GFP tag increased the susceptibilities of AD/CneMDR1 to azoles and R6G (Fig. 4C). This suggests that either the function of CneMdr1p was affected or the localization of CneMdr1p-GFP determined by confocal microscopy did not reflect the localization of nontagged CneMdr1p. CaMdr1Ap and CaErg11Ap were also labeled with a C-terminal GFP tag. CaMdr1Ap localized exclusively to the cell surface. CaErg11Ap localized primarily to the endoplasmic reticulum, as expected, with a small amount present on the cell surface. That result is consistent with CaErg11Ap being detected in the PM preparations (Fig. 4A and D). Upon closer inspection, it appeared that CaErg11Ap was also significantly concentrated in the same internal karmella-like structures (often round and sometimes hook-like structures depending on the plane of observation) that possibly surround the nucleus as described for CaCdr1Ap, CkAbc1p, CneMdr1p, and also HsAbcb1p (see below).
FIG. 7.
Confocal microscopy images of AD1-8u− cells overexpressing different classes of membrane proteins. ABC drug pumps ScPdr5p (a), CaCdr2Ap (c), CgCdr1p (d), and CgPdh1p (e) localized exclusively to the PM, whereas the fungal drug pumps CaCdr1Ap (b) and CkAbc1p (k) also partially localized in internal membrane structures. CneMdr1p (l) and HsAbcb1p (m) were not evenly distributed throughout the PM and were also found in internal structures. The MFS drug pump CaMdr1Ap (o) localized exclusively to the PM, and the azole drug target CaErg11Ap (n) localized to the endoplasmic reticulum, as expected. Light microscopy images of the cells (f to j and p to t) are shown below their confocal images (a to e and k to o).
Substrate specificities of overexpressed drug pumps and affinities of the azole drug target CaErg11Ap to a range of azoles.
The panel of S. cerevisiae strains expressing heterologous membrane proteins enabled the use of agarose diffusion assays to identify drug efflux pump substrates and compare the drug sensitivities of the azole target Erg11p. The specific overexpression of the fungal drug pumps ScPdr5p, CaCdr1Ap, and CaCdr2Ap gave reduced susceptibilities to all nine xenobiotics tested except to the negative control, NYS, indicating that the remaining eight drugs are substrates of these pumps (Fig. 3A). The overexpressed fungal efflux pumps CgCdr1p and CgPdh1p have previously been shown to confer similar phenotypes (61). In contrast, CneMdr1p pumped only ITC, KTZ, R6G, Triton X-100, and, slightly less effectively, FLC. AD/CneMDR1 remained sensitive to MCZ, cycloheximide, and cerulenin, and these drugs are therefore not substrates. Although CaMdr1p is known to confer resistance to benomyl, cycloheximide, and cerulenin (4, 24) and has been reported to pump FLC (65), we found that KTZ is also a CaMdr1p substrate (Fig. 3A). The detection of this activity may be due to the high CaMdr1p expression level achieved, or it may reflect the fact that our genetically modified host strains AD1-8u− and ADΔ are more sensitive to xenobiotics, which allows the detection of pump substrates that could not be identified using their natural hosts due to the masking of these activities by their other endogenous pumps. As expected, the overexpression of the drug target CaErg11Ap gave significantly reduced susceptibilities to different azole drugs but not to the polyene antibiotic NYS or the pump substrates R6G, cycloheximide, cerulenin, or Triton X-100 (Fig. 3A). For comparable amounts of ITC, MCZ, and KTZ, the relative sizes of the growth inhibition zones indicated that KTZ is a significantly less effective inhibitor of CaErg11Ap than either ITC or MCZ.
Expression of human membrane protein HsAbcb1p in S. cerevisiae.
Human P glycoprotein (HsAbcb1p) is an ABC transporter implicated in the development of drug resistance by cancer cells that has been studied by the scientific community for three decades (22, 57). HsABCB1 was successfully cloned and expressed in S. cerevisiae AD1-8u− (Fig. 4D). Although the expression of HsAbcb1p at 3.3% of total PM protein as measured by NIH Image software was significantly lower than that achieved for fungal ABC transporters, expression conferred on S. cerevisiae resistance to the known substrates aureobasidin A and R6G, and to ITC (Fig. 3B). The clarity of the AD/HsABCB1 R6G inhibitory zone, compared to the zone for AD/pABC3 cells, is partly due to efflux of (red) R6G from AD/HsABCB1 cells and retention of the dye by AD/pABC3 cells. The resistance phenotype conferred by HsAbcb1p was confirmed by liquid MIC assays. HsAbcb1p expression increased R6G resistance relative to that of AD/pABC3 16-fold (from 0.25 to 4 μg/ml), daunorubicin resistance 16-fold (from 2 to 32 μg/ml), and ITC resistance 40-fold (from 0.031 to 1.28 μg/ml). The faint Coomassie-stained band indicated in the HsAbcb1p lane of Fig. 4D was confirmed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry of tryptic fragments (42) to be HsAbcb1p (data not shown). The addition of a C-terminal GFP tag did not significantly affect the level of HsAbcb1p expression in the PM (Fig. 4D), but confocal microscopy indicated that most HsAbcb1p-GFP was localized inside the cell in membrane structures similar to those in cells overexpressing CneMdr1p (Fig. 7). Only a small proportion of the protein localized to the cell surface. It is not yet clear whether the apparent “mislocalization” was caused by the addition of the C-terminal GFP tag or whether it reflects the localization of the untagged protein.
Identification of compounds that chemosensitize S. cerevisiae cells hyperexpressing membrane pumps to FLC.
The panel of strains overexpressing fungal drug pumps can be used in screens to identify and/or characterize broad-spectrum drug pump inhibitors that overcome the multidrug-resistant (MDR) phenotype of fungal pathogens. We therefore tested the abilities of eight known or suspected fungal ABC drug pump inhibitors to chemosensitize cells to FLC. The macrolide FK506 is widely used in cancer and organ transplant patients as an immunosuppressant. FK506 was originally discovered as an antifungal, and as an inhibitor of CaCdr1p (19) it reverses multidrug efflux in C. albicans (34, 53). The milbemycins are 16-membered ring macrolides belonging to families that are used as insecticides, acaricides, and anthelminthics (44, 60). Some milbemycins have also been identified as chemosensitizers of fungal multidrug efflux mediated by CaCdr1p (33). Enniatin is another circular molecule that has been found to inhibit antifungal drug efflux (25, 33). Oligomycin is used to distinguish fungal drug efflux pump ATPase activities from those of other PM ATPases, including the PM H+ ATPase.
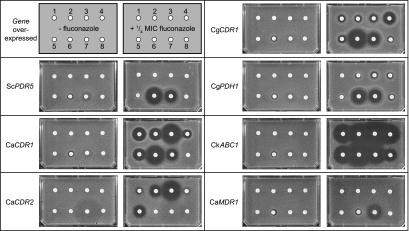
Chemosensitization to FLC of S. cerevisiae strains overexpressing drug efflux pumps was determined using agarose diffusion assays (Fig. 8). Toxic effects of the test compounds on cells were identified using assays carried out in the absence of FLC. A strain overexpressing the structurally unrelated, membrane potential-dependent MFS drug efflux pump CaMdr1Ap provided a counterscreen to identify growth-inhibitory effects of a compound on cells in the presence of FLC that were independent of the ABC transporters. Although the ScPdr5p, CaCdr1Ap, CaCdr2Ap, CgCdr1p, and CkAbc1p transporters have highly similar primary sequences (≥50% similarity) and appear to produce efflux of a comparable range of substrates (Fig. 3), each overexpressing strain showed a unique pattern of responses to the panel of eight compounds (Fig. 8). Of the compounds tested, only enniatin and FK506 gave detectable toxicity with S. cerevisiae cells in the absence of FLC (Fig. 8). Enniatin weakly inhibited the growth of all the overexpressing strains tested except AD/CaCDR2A. In the presence of FLC, enniatin dramatically enhanced the growth inhibition of all the overexpressing strains except AD/CaCDR2A and AD/CaMDR1A. This indicated specific inhibition by enniatin of the in vivo activities of all the ABC transporters tested except CaCdr2Ap. In the absence of FLC, FK506 caused barely detectable toxicity with all the strains except AD/CaMDR1A. In the presence of FLC, FK506 appeared to chemosensitize cell growth in all strains except AD/CaCDR2A. The lack of inhibition of AD/CaMDR1A in the absence of FLC and its growth inhibition in the presence of FLC indicate that the effects of FK506 are more complex and are consistent with known effects of FK506 (34, 53), including its interaction with the calcineurin stress response pathway (31), its known inhibitory effects on CaCdr1p (19) and ScPdr5p (17), and its poor inhibition of CaCdr2p in vitro (Table 4). In the absence of FLC, the five milbemycins tested were not toxic to the overexpressing cells, and none of them chemosensitized AD/CaMDR1A to FLC. In the presence of FLC, each strain overexpressing an ABC transporter responded differently to the milbemycins. Both α and β milbemycins significantly inhibited the ABC transporters CaCdr1Ap, CaCdr2Ap, CgCdr1p, CgPdh1p, and CkAbc1p but not ScPdr5p. Of the affected ABC transporters, CkAbc1p was most strongly inhibited by all milbemycins tested. CaCdr1Ap was strongly inhibited by only milbemycin α25 and was more modestly inhibited by the remaining milbemycins, with β9 showing the weakest effect. CaCdr1Ap and CaCdr2Ap showed comparable inhibition profiles for the α25, α20, and β11 milbemycins, but α11 or β9 was ineffective against CaCdr2Ap. In contrast, the FLC efflux activity of CgCdr1p was modestly inhibited by milbemycin β9 and weakly by the remaining milbemycins. Its homolog, CgPdh1p, was weakly chemosensitized by only three (α20, α25, and β9) of the five milbemycins tested. Oligomycin affected the in vivo pump function of only three of the seven ABC transporters. It strongly inhibited FLC efflux by AD/CkABC1 and, as expected, weakly inhibited AD/CgCDR1 and AD/CgPDH1 (61). With the exception of CaCdr2Ap, which is resistant to oligomycin, this result is in contrast with the in vitro inhibition at submicromolar concentrations of all the efflux pumps, including ScPdr5p and CaCdr1Ap (Table 4).
FIG. 8.
Effects of ABC drug pump inhibitors. AD1-8u− cells overexpressing fungal ABC drug pumps ScPdr5p, CaCdr1Ap, CaCdr2Ap, CgCdr1p, CgPdh1p or CkAbc1p, or the control strain overexpressing the MFS drug pump CaMdr1Ap, were analyzed by agarose diffusion assays using CSM solidified with 0.6% agarose and containing either no FLC (left hand plates; control) or FLC at 0.25× MICFLC for each test strain, as described in Materials and Methods. Whatman 3MM paper disks containing the indicated amounts of drug pump inhibitors (1, 5 μg milbemycin α11; 2, 5 μg milbemycin α20; 3, 5 μg milbemycin α25; 4, 5 μg milbemycin β9; 5, 5 μg milbemycin β11; 6, 5 μg enniatin; 7, 25 μg FK506; 8, 50 nmol oligomycin) were placed on each plate.
DISCUSSION
This study investigated the functions of membrane proteins involved in fungal drug resistance. It used a membrane protein hyperexpression system that, unlike plasmid-based overexpression systems, drives the constitutive overexpression of heterologous membrane proteins from the chromosomal PDR5 gene promoter with the help of the gain-of-function mutant transcription factor Pdr1-3p. Pdr1p and Pdr3p are two transcription factors regulating the expression of a set of genes involved in membrane protein biosynthesis and play a key role in the pleiotropic drug resistance (PDR) network (3, 14, 23, 32, 66). The pdr1-3 mutation in AD1-8u−, together with the disruption of pdr3, lead to the constitutive hyperexpression of PDR5 and the coordinated overexpression of other members of the PDR gene network (10, 14). The S. cerevisiae host strain AD1-8u− has been deleted in seven of its major ABC transporters. This makes it exquisitely sensitive to a wide range of xenobiotics and reduces the background of large proteins in the PM fractions that might interfere with the biochemical analysis and purification of the similarly sized overexpressed heterologous ABC drug pumps (13, 36, 38, 40). These features facilitated our study of proteins conferring drug resistance on S. cerevisiae. It is possible that the genetic backgrounds of AD1-8u− and ADΔ or the overexpression of certain efflux pumps affects PM composition, and this may in turn affect membrane protein function, but this still permits comparison of the properties of heterologous membrane proteins. In addition, our data are consistent with reports of known drug pump substrates, including those for the human MDR pump HsAbcb1p (22). The fact that we found additional pump substrates, for example, KTZ for CaMdr1p, is likely due to the increased sensitivity of the expression system.
In applying this hyperexpression system to the analysis of fungal drug resistance, a panel of strains that overexpress ABC- and MFS-type transporters and the azole drug target Erg11p from a range of fungal pathogens has been created. This panel of strains was used to search for broad-spectrum chemosensitizers and will be an invaluable tool for a variety of other purposes. The ability to determine the substrate specificities of multidrug efflux pumps involved in resistance to azole drugs (Fig. 3 and 8) (61) and to characterize these pumps in vitro (Fig. 6; Table 4) (62) will help elucidate structure-function relationships of proteins that are normally difficult to distinguish from a background of related molecules. The ability to rapidly clone and express individual alleles at equivalent levels enables the study of the effects of single-nucleotide polymorphisms in ABC transporters, such as CaCdr1p, CaCdr2p, or HsAbcb1p, on pump function and substrate specificity (26, 27). The panel of pump-expressing strains provides a powerful counterscreen to identify drug candidates that are potential pump substrates and would quickly lead to the development of MDR (41). These drug candidates can then be eliminated from the drug discovery pipeline. The same panel of pump-expressing strains can be used to identify and characterize new classes of antifungal chemosensitizers that specifically target the pumps responsible for the MDR phenotype of fungal pathogens (Fig. 8) (37, 40). The panel of overexpressing strains is currently being extended to include drug pumps and azole targets from other human fungal pathogens, such as Aspergillus fumigatus.
Stable functional overexpression of membrane proteins can be used to determine the effectiveness of new drugs against targets such as the azole drug target Erg11p. For example, the overexpression of CaErg11Ap gave moderate resistance to ITC and MCZ but a high level of resistance to KTZ. This result indicates MCZ and ITC are much more effective inhibitors of CaErg11Ap than KTZ. Finally, the ability to express functionally active His-tagged membrane proteins and their ease of affinity purification will facilitate their structural analysis. A similar expression and affinity purification strategy was recently combined with anion exchange chromatography to isolate enzymatically active CaCdr1p-His from a crude membrane fraction (54).
Application of the yeast expression system to the analysis of different classes of membrane protein involved in drug resistance has highlighted features that allow consistent expression. Unlike the case with other expression systems, such as that of Pichia pastoris, nearly all the transformants we obtained with the same construct showed identical phenotypes. This made the cloning process much more rapid and robust. The integration of a single copy of the expression cassette into the chromosomal PDR5 locus was sufficient to obtain high levels of protein expression, probably because the induction of multiple genes by the Pdr1-3 transcription factor conditions the system at the transcriptional, translational and posttranslational levels (14, 39) for efficient delivery of membrane proteins to the PM. Other hyperexpression systems based on a yeast multicopy plasmid (19) may overload the posttranslational secretory machinery and lead to protein turnover or sequestration of proteins in internal membranes, analogous to the inclusion bodies found with the overexpression of certain proteins in E. coli. Often expression of membrane proteins encoded on multicopy plasmids can result in relatively low levels of functional protein expression in the PM that confer only modest phenotypes (27). Expression of CaCdr1p in our system increased ITC resistance 1,000-fold. In some cases we see GFP-tagged material in structures reminiscent of stacks of smooth PM or karmellae that are induced when membrane proteins are overexpressed. While incorrect trafficking appeared to be enhanced for the ABC transporters that are most phylogenetically distant from S. cerevisiae, it has not yet been possible to determine whether this reflects the normal distribution of the untagged recombinant protein or is an adventitious consequence of the GFP tag (56).
In an initial attempt to extend our analysis to membrane proteins from higher eukaryotes involved in drug pumping, we functionally overexpressed human HsAbcb1p at 3.3% of PM protein. This made the AD1-8u− host strain 40 times more resistant to ITC and 16-fold more resistant to the anticancer drug daunorubicin, a more significant change in phenotype than the 2-fold increase seen with another S. cerevisiae expression system (27). If genetic distance does reduce the yield of heterologously expressed membrane protein because of differences in biosynthetic, quality control, and trafficking machineries, codon optimization strategies could be used to overcome limitations for transcription and translation that may also affect protein function (30). In addition, the genetic tractability of S. cerevisiae opens many avenues for the improved expression and correct delivery of functional membrane proteins. For example, hosts selected for the expression of high levels of an endogenous membrane protein from a particular functional class may provide optimal vehicles for the heterologous expression of related membrane proteins. Improved hosts could be generated by directed modification of genes, such as those involved in the unfolded protein response and protein turnover pathways, or by selecting extragenic mutations that enhance the relevant phenotypes of targets currently expressed at modest levels. Our system for constitutive protein overexpression in a background depleted of efflux pumps provides extra dimensions for chemogenomic screens that aim to identify both the on- and off-target effects of drugs (12). If the system was further modified to allow regulated expression of the heterologous protein, then it should be possible to overexpress lethal targets and proteins involved in cell cycle check points (58). Finally, the expression of regulated membrane proteins within defined physiological windows could be used to minimize protein microheterogeneity and make them more amenable to structural analysis by X-ray crystallography. We expect this expression system, therefore, to be a valuable addition to current technologies that will enable a more systematic study of a variety of fungal, plant, and human membrane proteins, including targets for drug discovery.
Acknowledgments
This study was supported by the Japan Health Sciences Foundation, the National Institutes of Health (grant numbers R21DE015075-RDC and R01DE016885-01-RDC), and the Japan Society for the Promotion of Sciences (grant numbers L02705-EL and S04718-EL).
We thank A. Goffeau and A. Decottignies for providing strains AD1-7 and AD1-8u−, R. B. Wilson for plasmid pDDB57, which we used for the URA3 blaster cassette strategy, B. P. Cormack for yEGFP3, R. Tsien for mRFP1, K. Ueda for the HsABCB1 cDNA, Sankyo Co. Ltd., for the milbemycins, Astellas Pharma, Inc., for FK506, K. Nakamura for help in cloning CneMDR1, A. Kaneko for identification of overexpressed proteins by mass spectrometry of tryptic fingerprints, and A. McNaughton at the Otago Centre for Confocal Microscopy for his expert assistance.
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Abbott, A. 2000. Structures by numbers. Nature 408:130-132. [DOI] [PubMed] [Google Scholar]
- 2.Albertson, G. D., M. Niimi, R. D. Cannon, and H. F. Jenkinson. 1996. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 40:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balzi, E., W. Chen, S. Ulaszewski, E. Capieaux, and A. Goffeau. 1987. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem. 262:16871-16879. [PubMed] [Google Scholar]
- 4.Ben-Yaacov, R., S. Knoller, G. A. Caldwell, J. M. Becker, and Y. Koltin. 1994. Candida albicans gene encoding resistance to benomyl and methotrexate is a multidrug resistance gene. Antimicrob. Agents Chemother. 38:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bill, R. M. 2001. Yeast—a panacea for the structure-function analysis of membrane proteins? Curr. Genet. 40:157-171. [DOI] [PubMed] [Google Scholar]
- 6.Bonetti, B., L. Fu, J. Moon, and D. M. Bedwell. 1995. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 251:334-345. [DOI] [PubMed] [Google Scholar]
- 7.Borges-Walmsley, M. I., K. S. McKeegan, and A. R. Walmsley. 2003. Structure and function of efflux pumps that confer resistance to drugs. Biochem. J. 376:313-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borst, P., and R. O. Elferink. 2002. Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 71:537-592. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvajal, E., H. B. van den Hazel, A. Cybularz-Kolaczkowska, E. Balzi, and A. Goffeau. 1997. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 256:406-415. [DOI] [PubMed] [Google Scholar]
- 11.Cormack, B. P., G. Bertram, M. Egerton, N. A. Gow, S. Falkow, and A. J. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 12.De Backer, M. D., and P. Van Dijck. 2003. Progress in functional genomics approaches to antifungal drug target discovery. Trends Microbiol. 11:470-478. [DOI] [PubMed] [Google Scholar]
- 13.Decottignies, A., A. M. Grant, J. W. Nichols, H. de Wet, D. B. McIntosh, and A. Goffeau. 1998. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273:12612-12622. [DOI] [PubMed] [Google Scholar]
- 14.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 15.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 16.Edwards, A. M., C. H. Arrowsmith, D. Christendat, A. Dharamsi, J. D. Friesen, J. F. Greenblatt, and M. Vedadi. 2000. Protein production: feeding the crystallographers and NMR spectroscopists. Nat. Struct. Biol. 7(Suppl.):970-972. [DOI] [PubMed] [Google Scholar]
- 17.Egner, R., B. E. Bauer, and K. Kuchler. 2000. The transmembrane domain 10 of the yeast Pdr5p ABC antifungal efflux pump determines both substrate specificity and inhibitor susceptibility. Mol. Microbiol. 35:1255-1263. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira-Pereira, A., S. Marco, A. Decottignies, J. Nader, A. Goffeau, and J. L. Rigaud. 2003. Three-dimensional reconstruction of the Saccharomyces cerevisiae multidrug resistance protein Pdr5p. J. Biol. Chem. 278:11995-11999. [DOI] [PubMed] [Google Scholar]
- 19.Gauthier, C., S. Weber, A. M. Alarco, O. Alqawi, R. Daoud, E. Georges, and M. Raymond. 2003. Functional similarities and differences between Candida albicans Cdr1p and Cdr2p transporters. Antimicrob. Agents Chemother. 47:1543-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerngross, T. U. 2004. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 22:1409-1414. [DOI] [PubMed] [Google Scholar]
- 21.Griffin, B. A., S. R. Adams, and R. Y. Tsien. 1998. Specific covalent labeling of recombinant protein molecules inside live cells. Science 281:269-272. [DOI] [PubMed] [Google Scholar]
- 22.Higgins, C. F. 2007. Multiple molecular mechanisms for multidrug resistance transporters. Nature 446:749-757. [DOI] [PubMed] [Google Scholar]
- 23.Hikkel, I., A. Lucau-Danila, T. Delaveau, P. Marc, F. Devaux, and C. Jacq. 2003. A general strategy to uncover transcription factor properties identifies a new regulator of drug resistance in yeast. J. Biol. Chem. 278:11427-11432. [DOI] [PubMed] [Google Scholar]
- 24.Hiller, D., D. Sanglard, and J. Morschhauser. 2006. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob. Agents Chemother. 50:1365-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiraga, K., S. Yamamoto, H. Fukuda, N. Hamanaka, and K. Oda. 2005. Enniatin has a new function as an inhibitor of Pdr5p, one of the ABC transporters in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 328:1119-1125. [DOI] [PubMed] [Google Scholar]
- 26.Holmes, A. R., S. Tsao, S. W. Ong, E. Lamping, K. Niimi, B. C. Monk, M. Niimi, A. Kaneko, B. R. Holland, J. Schmid, and R. D. Cannon. 2006. Heterozygosity and functional allelic variation in the Candida albicans efflux pump genes CDR1 and CDR2. Mol. Microbiol. 62:170-186. [DOI] [PubMed] [Google Scholar]
- 27.Jeong, H., I. Herskowitz, D. L. Kroetz, and J. Rine. 2007. Function-altering SNPs in the human multidrug transporter gene ABCB1 identified using a Saccharomyces-based assay. PLoS Genet. 3:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko, A., T. Umeyama, N. Hanaoka, B. C. Monk, Y. Uehara, and M. Niimi. 2004. Tandem affinity purification of the Candida albicans septin protein complex. Yeast 21:1025-1033. [DOI] [PubMed] [Google Scholar]
- 29.Katiyar, S. K., and T. D. Edlind. 2001. Identification and expression of multidrug resistance-related ABC transporter genes in Candida krusei. Med. Mycol. 39:109-116. [DOI] [PubMed] [Google Scholar]
- 30.Kimchi-Sarfaty, C., J. M. Oh, I. W. Kim, Z. E. Sauna, A. M. Calcagno, S. V. Ambudkar, and M. M. Gottesman. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525-528. [DOI] [PubMed] [Google Scholar]
- 31.Kraus, P. R., D. S. Fox, G. M. Cox, and J. Heitman. 2003. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48:1377-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Crom, S., F. Devaux, P. Marc, X. Zhang, W. S. Moye-Rowley, and C. Jacq. 2002. New insights into the pleiotropic drug resistance network from genome-wide characterization of the YRR1 transcription factor regulation system. Mol. Cell. Biol. 22:2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, M. D., J. L. Galazzo, A. L. Staley, J. C. Lee, M. S. Warren, H. Fuernkranz, S. Chamberland, O. Lomovskaya, and G. H. Miller. 2001. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco 56:81-85. [DOI] [PubMed] [Google Scholar]
- 34.Maesaki, S., P. Marichal, M. A. Hossain, D. Sanglard, H. Vanden Bossche, and S. Kohno. 1998. Synergic effects of tactolimus and azole antifungal agents against azole-resistant Candida albicans strains. J. Antimicrob. Chemother. 42:747-753. [DOI] [PubMed] [Google Scholar]
- 35.Martinoia, E., M. Klein, M. Geisler, L. Bovet, C. Forestier, U. Kolukisaoglu, B. Muller-Rober, and B. Schulz. 2002. Multifunctionality of plant ABC transporters—more than just detoxifiers. Planta 214:345-355. [DOI] [PubMed] [Google Scholar]
- 36.Monk, B. C., R. D. Cannon, K. Nakamura, M. Niimi, K. Niimi, D. R. K. Harding, A. R. Holmes, E. Lamping, A. Goffeau, and A. Decottignies. August 2002. Membrane protein expression system and its application. International patent PCT/NZ02/00163.
- 37.Monk, B. C., K. Niimi, S. Lin, A. Knight, T. B. Kardos, R. D. Cannon, R. Parshot, A. King, D. Lun, and D. R. Harding. 2005. Surface-active fungicidal d-peptide inhibitors of the plasma membrane proton pump that block azole resistance. Antimicrob. Agents Chemother. 49:57-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura, K., M. Niimi, K. Niimi, A. R. Holmes, J. E. Yates, A. Decottignies, B. C. Monk, A. Goffeau, and R. D. Cannon. 2001. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob. Agents Chemother. 45:3366-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nawrocki, A., S. J. Fey, A. Goffeau, P. Roepstorff, and P. M. Larsen. 2001. The effects of transcription regulating genes PDR1, pdr1-3 and PDR3 in pleiotropic drug resistance. Proteomics 1:1022-1032. [DOI] [PubMed] [Google Scholar]
- 40.Niimi, K., D. R. Harding, R. Parshot, A. King, D. J. Lun, A. Decottignies, M. Niimi, S. Lin, R. D. Cannon, A. Goffeau, and B. C. Monk. 2004. Chemosensitization of fluconazole resistance in Saccharomyces cerevisiae and pathogenic fungi by a d-octapeptide derivative. Antimicrob. Agents Chemother. 48:1256-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niimi, K., K. Maki, F. Ikeda, A. R. Holmes, E. Lamping, M. Niimi, B. C. Monk, and R. D. Cannon. 2006. Overexpression of Candida albicans CDR1, CDR2, or MDR1 does not produce significant changes in echinocandin susceptibility. Antimicrob. Agents Chemother. 50:1148-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niimi, M., Y. Nagai, K. Niimi, S. Wada, R. D. Cannon, Y. Uehara, and B. C. Monk. 2002. Identification of two proteins induced by exposure of the pathogenic fungus Candida glabrata to fluconazole. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 782:245-252. [DOI] [PubMed] [Google Scholar]
- 43.Niimi, M., S. Wada, K. Tanabe, A. Kaneko, Y. Takano, T. Umeyama, N. Hanaoka, Y. Uehara, E. Lamping, K. Niimi, S. Tsao, A. R. Holmes, B. C. Monk, and R. D. Cannon. 2005. Functional analysis of fungal drug efflux transporters by heterologous expression in Saccharomyces cerevisiae. Jpn. J. Infect. Dis. 58:1-7. [PubMed] [Google Scholar]
- 44.Nonaka, K., T. Tsukiyama, Y. Okamoto, K. Sato, C. Kumasaka, T. Yammoto, F. Maruyama, and H. Yoshikawa. 2000. New milbemycins from Streptomyces hygroscopicus subsp. aureolacrimosus: fermentation, isolation and structure elucidation. J. Antibiot. (Tokyo) 53:694-704. [DOI] [PubMed] [Google Scholar]
- 45.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Baillière Tindall, London, United Kingdom.
- 46.Osterberg, M., H. Kim, J. Warringer, K. Melen, A. Blomberg, and G. von Heijne. 2006. Phenotypic effects of membrane protein overexpression in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103:11148-11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ratjen, F., and G. Doring. 2003. Cystic fibrosis. Lancet 361:681-689. [DOI] [PubMed] [Google Scholar]
- 49.Reis, A. F., and G. Velho. 2002. Sulfonylurea receptor-1 (SUR1): genetic and metabolic evidences for a role in the susceptibility to type 2 diabetes mellitus. Diabetes Metab. 28:14-19. [PubMed] [Google Scholar]
- 50.Saini, P., T. Prasad, N. A. Gaur, S. Shukla, S. Jha, S. S. Komath, L. A. Khan, Q. M. Haq, and R. Prasad. 2005. Alanine scanning of transmembrane helix 11 of Cdr1p ABC antifungal efflux pump of Candida albicans: identification of amino acid residues critical for drug efflux. J. Antimicrob. Chemother. 56:77-86. [DOI] [PubMed] [Google Scholar]
- 51.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitz, G., and W. E. Kaminski. 2002. ATP-binding cassette (ABC) transporters in atherosclerosis. Curr. Atheroscler. Rep. 4:243-251. [DOI] [PubMed] [Google Scholar]
- 53.Schuetzer-Muehlbauer, M., B. Willinger, R. Egner, G. Ecker, and K. Kuchler. 2003. Reversal of antifungal resistance mediated by ABC efflux pumps from Candida albicans functionally expressed in yeast. Int. J. Antimicrob. Agents 22:291-300. [DOI] [PubMed] [Google Scholar]
- 54.Shukla, S., V. Rai, D. Banerjee, and R. Prasad. 2006. Characterization of Cdr1p, a major multidrug efflux protein of Candida albicans: purified protein is amenable to intrinsic fluorescence analysis. Biochemistry 45:2425-2435. [DOI] [PubMed] [Google Scholar]
- 55.Simon, J. A., and A. Bedalov. 2004. Yeast as a model system for anticancer drug discovery. Nat. Rev. Cancer 4:481-492. [DOI] [PubMed] [Google Scholar]
- 56.Snapp, E. L., R. S. Hegde, M. Francolini, F. Lombardo, S. Colombo, E. Pedrazzini, N. Borgese, and J. Lippincott-Schwartz. 2003. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stege, A., A. Priebsch, C. Nieth, and H. Lage. 2004. Stable and complete overcoming of MDR1/P-glycoprotein-mediated multidrug resistance in human gastric carcinoma cells by RNA interference. CancerGene Ther. 11:699-706. [DOI] [PubMed] [Google Scholar]
- 58.Stevenson, L. F., B. K. Kennedy, and E. Harlow. 2001. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl. Acad. Sci. USA 98:3946-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorn, K. S., N. Naber, M. Matuska, R. D. Vale, and R. Cooke. 2000. A novel method of affinity-purifying proteins using a bis-arsenical fluorescein. Protein Sci. 9:213-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsukiyama, T., H. Kajino, F. Kajino, S. Furuta, Y. Tsukamoto, K. Sato, A. Kinoshita, R. Ichinose, and K. Tanaka. 2002. Synthesis of milbemycins alpha9, alpha10, alpha11, alpha12, alpha14, alpha15, alpha20, alpha21, alpha22, alpha23, alpha26, alpha27, delta(2,3), delta(4,26)-milbemycins A3, A4 from milbemycins A3, A4, and their acaricidal activities. J. Antibiot. (Tokyo) 55:993-1003. [DOI] [PubMed] [Google Scholar]
- 61.Wada, S., M. Niimi, K. Niimi, A. R. Holmes, B. C. Monk, R. D. Cannon, and Y. Uehara. 2002. Candida glabrata ATP-binding cassette transporters Cdr1p and Pdh1p expressed in a Saccharomyces cerevisiae strain deficient in membrane transporters show phosphorylation-dependent pumping properties. J. Biol. Chem. 277:46809-46821. [DOI] [PubMed] [Google Scholar]
- 62.Wada, S., K. Tanabe, A. Yamazaki, M. Niimi, Y. Uehara, K. Niimi, E. Lamping, R. D. Cannon, and B. C. Monk. 2005. Phosphorylation of Candida glabrata ATP-binding cassette transporter Cdr1p regulates drug efflux activity and ATPase stability. J. Biol. Chem. 280:94-103. [DOI] [PubMed] [Google Scholar]
- 63.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 65.Wirsching, S., S. Michel, and J. Morschhauser. 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36:856-865. [DOI] [PubMed] [Google Scholar]
- 66.Wolfger, H., Y. M. Mamnun, and K. Kuchler. 2001. Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 152:375-389. [DOI] [PubMed] [Google Scholar]