Abstract
A new family of site-specific repeated elements identified in Trypanosoma cruzi, which we named TcTREZO, is described here. TcTREZO appears to be a composite repeated element, since three subregions may be defined within it on the basis of sequence similarities with other T. cruzi sequences. Analysis of the distribution of TcTREZO in the genome clearly indicates that it displays site specificity for insertion. Most TcTREZO elements are flanked by conserved sequences. There is a highly conserved 68-bp sequence at the 5′ end of the element and a sequence domain of ∼500 bp without a well-defined borderline at the 3′ end. Northern blot hybridization and reverse transcriptase PCR analyses showed that TcTREZO transcripts are expressed as oligo(A)-terminated transcripts whose length corresponds to the unit size of the element (1.6 kb). Transcripts of ∼0.2 kb derived from a small part of TcTREZO are also detected in steady-state RNA. TcTREZO transcripts are unspliced and not translated. The copy number of TcTREZO sequences was estimated to be ∼173 copies per haploid genome. TcTREZO appears to have been assembled by insertions of sequences into a progenitor element. Once associated with each other, these subunits were amplified as a new transposable element. TcTREZO shows site specificity for insertion, suggesting that a sequence-specific endonuclease could be responsible for its insertion at a unique site.
Reassociation kinetic studies of the genomic DNA of the protozoan parasite Trypanosoma cruzi, the causative agent of Chagas’ disease, showed that repetitive DNA sequences account for a substantial portion of the nuclear genome of this parasite (12, 30). The complete sequencing of the T. cruzi genome confirmed that at least 50% of the genome is repetitive sequence, consisting mostly of micro- and minisatellites, retrotransposon-like elements, large gene families of surface proteins, and subtelomeric repeats (15). The current T. cruzi genome assembly shows that 57% of the contigs have repeats at one or both ends, reflecting the highly repetitive nature of the genome (15). Furthermore, the numerous repetitive regions of the genome were collapsed and/or misassembled.
Long terminal repeats (LTR) and non-LTR retrotransposons constitute the most abundant mobile elements described in the T. cruzi genome (accounting for ∼5% of the nuclear genome), while no element that transposes through a DNA-based pathway has been identified to date in this parasite (15, 16, 38, 43, 47). The most abundant non-LTR elements are L1Tc and NARTc (5, 8, 31), with 320 (L1Tc) and 133 (NARTc) copies per haploid genome (15). The autonomous L1Tc (4.9-kb) and nonautonomous NARTc (0.25-kb) elements share the first 78 residues and other conserved blocks, suggesting that NARTc was derived from L1Tc by a 3′ deletion (5, 8, 15). The autonomous T. cruzi L1Tc non-LTR retroelement encodes a protein containing four domains (endonuclease, reverse transcriptase, RNase H, and DNA-binding motifs) presumably involved in its own retrotransposition (15, 23, 34, 35). On the other hand, the nonautonomous NARTc elements presumably use the L1Tc-encoded enzymatic activities for their own transposition (5, 8).
Retrotransposons can also be divided into site-specific and non-site-specific elements. The T. cruzi CZAR was the only site-specific retroelement (“siteposon”) identified prior to this study and is found in the spliced-leader (SL) RNA genes (45). The CZAR siteposon encodes a sequence-specific endonuclease that could be responsible for its insertion at a unique site in the SL gene locus. L1Tc and NARTc have been reported as randomly distributed; however, analysis of their genomic context in the complete T. cruzi genome indicates that they are preferentially inserted into conserved motifs (5, 8, 15).
Repeats represent an extraordinary source of information about biological processes. They constitute molecular paleontological records, giving clues about evolutionary events and forces. It has been suggested that repetitive DNA elements may be involved in genetic rearrangements and control of gene expression in trypanosomes (4, 38, 44, 47). Most repeated sequences are species specific and have been shown to be useful tools for diagnostic and taxonomic purposes (24, 38, 47). In the course of studying repetitive DNA in the T. cruzi genome, we identified a new site-specific repeated element, which we named TcTREZO (Trypanosoma cruzi tandem repetitive element ZO). Some unusual structural features clearly differentiate TcTREZO from other retrotransposons in trypanosomes described to date. We describe the characterization of TcTREZO and establish its relationship with other repetitive DNA families.
MATERIALS AND METHODS
Parasites.
T. cruzi clone CL Brener (49) was used in this study. Parasites were maintained by cyclic passage in mice and in axenic cultures in liver infusion tryptose medium containing 10% fetal calf serum at 28°C. Trypanosoma rangeli, Trypanosoma brucei, and Leishmania (Leishmania) amazonensis were kindly provided by Marta M. Teixeira (Department of Parasitology, ICB-USP), Maria L. C. Almeida (UNIFESP), and Clara L. Barbieri (UNIFESP), respectively.
Separation of T. cruzi chromosomal DNA by PFGE.
Epimastigotes from T. cruzi clone CL Brener (49) were grown to late logarithmic phase. Cells were collected in phosphate-buffered saline and mixed with an equal volume of 1% low-melting-point agarose. Approximately 1 × 108 cells (100 μl) were used for each gel plug; these were incubated in a solution containing 0.5 M EDTA (pH 8.0), 1% sodium lauroylsarcosinate, and 1 mg/ml proteinase K at 50°C for 48 h and stored at 4°C in 0.5 M EDTA (pH 8.0). Chromosomal bands were separated on agarose gels in a Gene Navigator apparatus (Amersham Pharmacia Biotech, NJ) using a hexagonal electrode array. Pulsed-field gel electrophoresis (PFGE) was carried out in 1.2% agarose gels in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.3) at 13°C for 132 h as previously described (10). Gels were stained with ethidium bromide (0.5 μg/ml), photographed, transferred to nylon filters, and hybridized as described below.
Two-dimensional PFGE.
Blocks containing chromosomal-size T. cruzi DNA were placed in 1% low-gelling agarose (Invitrogen) gels as described previously (13). Samples were first run as described above, and then a 5-mm agarose strip containing the resolved chromosomal bands was washed with TE (10 mM Tris-HCl, pH 7.6, 1 mM EDTA) buffer overnight at 4°C. The strip was equilibrated in BamHI or EcoRI restriction buffer for 3 h at 4°C. Next, we added 3,000 U of BamHI or EcoRI enzymes and incubated the digest at 37°C for 6 h. The second dimension was carried out in a contour-clamped homogeneous electric field apparatus (Bio-Rad, CA). The strip was placed in a well made in a 1% agarose-0.5× TBE gel and run in a linear gradient of switching times from 30 to 70 s in a voltage gradient of 6 V cm−1 for 18 h at 14°C. DNA fragments were transferred to nylon filters as described below and then hybridized with the TcTREZO probe.
DNA isolation, Southern blotting, and hybridization.
DNA samples isolated from epimastigotes of T. cruzi strains as previously described (4) were digested with restriction enzymes, separated by electrophoresis on agarose gels (0.8%), and stained with ethidium bromide (0.5 μg/ml). They were then incubated with 0.25 M HCl for 45 min, denatured with 0.5 M NaOH-1 M NaCl for 20 min, neutralized with 1 M Tris-base-0.5 M NaCl for 20 min, and transferred to nylon membranes in 20× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate). The membranes were prehybridized in a solution containing 50% formamide-5× SSC-5× Denhardt's solution (Invitrogen)-0.1 mg/ml salmon sperm DNA-0.1 mg/ml tRNA at 42°C for 1 h and then hybridized overnight at the same temperature with the 32P-labeled probes. Following hybridization, the membranes were subjected to two washes (30 min each at 42°C) in 2× SSC containing 0.1% sodium dodecyl sulfate (SDS) and 0.1% sodium pyrophosphate and to two additional washes at 56°C in 0.1× SSC containing 0.1% SDS and 0.1% sodium pyrophosphate. They were then exposed to X-ray film.
Solutions containing genomic DNA from Trypanosoma rangeli, Trypanosoma brucei, L. (L.) amazonensis, and T. cruzi were used in a dot blot experiment as described below. DNA samples were denatured with NaOH (0.4 M) for 10 min, chilled on ice, and diluted with an equal volume of 2 M ammonium acetate. Samples were applied to nylon membranes (Amersham) using a dot blot apparatus (Bio-Rad). DNA was fixed by exposure to 150 mJ of UV light in a GS Gene Linker UV chamber (Bio-Rad). Filters were hybridized exactly as were Southern blots.
RNA isolation, Northern blotting, and hybridization.
Total RNA was extracted from epimastigotes with TRIzol. The poly(A)+ and poly(A)− RNA fractions were isolated by chromatography on oligo(dT) cellulose columns and subjected to electrophoresis on agarose-formaldehyde gel. Five or 10 μg of RNA was subjected to electrophoresis on agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized with the different probes. The following probes were amplified by PCR using the TcTREZO clone (GenBank accession no. AF508945) as the template (primers and primer sequences are in parentheses): ISPB (intergenic spacer of the pyrimidine-biosynthetic gene cluster) probe plus spacer (ISPB forward, 5′ CCA TCT GTT TCG TGG TTT GTG CGG C 3′; spacer reverse, 5′ GCA GCG GCA ACC CCG GCA GTA AGG 3′) and mucin-like probe (M13 forward and MUC reverse, 5′ CGC ACA AAC CAC GAA ACA GAT GG 3′). The 55-bp repeat was amplified from clone F4.12 (GenBank accession no. AF503285) using M13 forward and reverse primers.
Cloning of TcTREZO transcripts by RT-PCR.
First- and second-strand cDNA was prepared using SuperScript One-Step reverse transcriptase PCR (RT-PCR) with Platinum Taq according to the manufacturer's instructions (Life Technologies). Specific forward or reverse primers based on the nucleotide sequences of TcTREZO were used with oligo(dT) to amplify sequences by PCR. The T. cruzi tubulin gene sequences (used as an internal control) were amplified with appropriated primers in a final volume of 50 μl at a denaturing temperature of 95°C for 30 s, an annealing temperature of 55°C for 30 s, and an elongation temperature of 72°C for 30 s. The amplified PCR products were then cloned into plasmid pGEM-T Easy vector (Promega) and transformed into the DH5α Escherichia coli strain. Nucleotide sequences of cDNA clones were determined by the dideoxynucleotide chain termination method using BigDye Terminator cycle sequencing chemistry (Applied Biosystems) in an ABI PRISM 377 DNA sequencer.
Sequence similarity searches.
The shotgun sequencing readings obtained from five clones of the TcTREZO element were analyzed using PHRED/PHRAP/Consed (17, 18, 25) to read sequencer trace data, write the base calls and quality values, assemble shotgun DNA sequence data, and visually inspect, edit, and finish the alignments.
The T. cruzi clone CL Brener (49) genome sequence used in this study was obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/GenBank). A locally compiled database (DB) of T. cruzi sequences was built by parsing sequences from GenBank, GeneDB (http://www.genedb.org), and The Institute for Genomic Research and used for sequence similarity searches. Similarity searches against this locally compiled DB were carried out using the BLAST and FASTA program package algorithms (4, 36). The search for and identification and extraction of TcTREZO full copies in this locally compiled sequence DB were performed by two different approaches: (i) using a PERL script specifically developed for this study and loaded with a regular expression specifically describing the element presented in Fig. 1 and (ii) using RepeatMasker (A. F. A. Smit, R. Hubley, and P. Green, RepeatMasker Open 3.0, 1996 to 2004, http://www.repeatmasker.org) loaded with Tcruzi DB and the TcTREZO element as a custom library. The copy numbers of TcTREZO sequences were estimated according to the equation proposed by Agüero et al. (1). For this calculation, the haploid T. cruzi genome size used was 55 Mb (16), the total number of sequences was 32,746, and the size of TcTREZO was 1,573 bp. Global multiple sequence alignments were performed using Clustal W (40) and were followed by visual inspection and manual adjustment with SeaView (21) (http://pbil.univ-lyon1.fr/software/seaview.html) and GeneDoc (http://www.psc.edu/biomed/genedoc). The annotation and graphical output of the TcTREZO element and flanking regions were obtained using ARTEMIS (39) (http://www.sanger.ac.uk/Software/Artemis) and in-house-developed PERL scripts to analyze and format the results of the similarity searches. Prediction of RNA secondary structure was performed with the Vienna RNA Package (http://www.tbi.univie.ac.at/∼ivo/RNA/) (27).
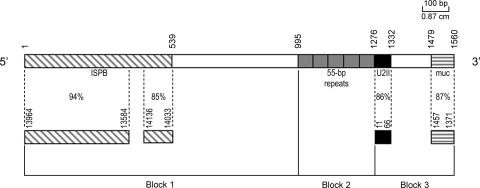
FIG. 1.
Schematic representation of TcTREZO element showing its composite nature. Block 1, sequence with similarity to the intergenic spacer of the pyrimidine-biosynthetic (ISPB) gene cluster (boxed diagonal lines) and a sequence without homology to previously reported T. cruzi sequences (open box); block 2, tandem arrangement of 55-bp repeats (gray box); block 3, sequence with similarity to the U2II RNA gene (black box), sequence without homology to previously reported T. cruzi sequences (open box), and sequence with similarity to the untranslated region of mucin-like protein pseudogene (boxed horizontal lines). The length (in bp) of the TcTREZO consensus sequence is indicated. The regions and percentages of identity of sequences that share homology to TcTREZO are indicated. The regions are as follows (GenBank accession numbers are in parentheses): ISPB gene cluster (AB017765), U2II RNA gene (X56457), and muc2 gene for mucin-like glycoprotein (AJ239063).
Nucleotide sequence accession numbers.
The TcTREZO sequences have been submitted to GenBank and assigned accession numbers EF577449, EF577450, EF577451, EF577452, EF577443, EF577444, EF577445, EF577446, EF577447, and EF577448.
RESULTS
Isolation and sequence analysis of the TcTREZO element.
In the course of studying the presence of repetitive sequences in the T. cruzi genome, we isolated a 220-bp fragment composed of tandemly arranged 55-bp repeats (GenBank accession no. AF503285). The fragment strongly hybridized with a single ∼1.6-kb band in EcoRI digests of total T. cruzi DNA (see Fig. 5).
FIG. 5.
Genomic organization of TcTREZO. Southern (B to D) and dot blot (E) hybridizations of genomic DNA of different T. cruzi strains and trypanosomatids with 32P-labeled TcTREZO. (A) Schematic representation of TcTREZO elements in the contig with GenBank accession no. AAHK01000548. The DraI, PstI, and EcoRI restriction sites are indicated. (B) Southern blot containing genomic DNA of T. cruzi (clone CL Brener) digested to completion with HindIII (H), EcoRI (E), or BamHI (B) or with two restriction enzymes (H/B, HindIII plus BamHI; H/E, HindIII plus EcoRI; B/E, BamHI plus EcoRI). (C) DNA of T. cruzi (clone CL Brener) digested partially (Ep) or to completion (Et) with EcoRI. (D) Southern blot of genomic DNA of different isolates of T. cruzi (clone CL Brener [CLB], strain Y, clone Sylvio [S], and strain G) digested with EcoRI. (E) Demonstration of species specificity of TcTREZO by dot blot hybridization using genomic DNA from Trypanosoma rangeli (Tr), Trypanosoma brucei (Tb), L. (L.) amazonensis (LLa), and T. cruzi (Tc). Samples were hybridized with randomly primed 32P-labeled TcTREZO. After exposure, the membranes were incubated at 100°C for 30 min in 0.1% SDS and rehybridized with the tubulin gene probe.
Two different approaches were adopted in order to isolate the full-length repetitive element. First, T. cruzi genomic DNA was digested with EcoRI and the 1.6-kb fragment was cloned into the pUC18 plasmid. With the 55-bp repeat as a probe, 27 recombinants were selected and named Z1 to Z27. End sequencing of all the clones was analyzed for similarity against public-domain DB sequences, and five of them were fully sequenced (GenBank accession no. AF508945, EF577449, EF577450, EF577451, and EF577452). Alignment of these clones showed that they were very similar, indeed almost identical. TcTREZO corresponds to the sequence consensus obtained from the alignment of the five completely sequenced clones. The most striking structural feature of the ∼1.6-kb repetitive element TcTREZO is the presence of a central core composed of the 55-bp repeat arranged in tandem (Fig. 1, block 2) and flanked by different ∼995-bp and 254-bp domains (Fig. 1, blocks 1 and 3, respectively).
Figure 1 shows the physical map and a comparison of the element with other T. cruzi repetitive sequences reported in the literature. TcTREZO's block 1, from nucleotide (nt) 1 to 539, has 85 to 94% identity with sequences in the intergenic spacer of the pyrimidine biosynthetic gene cluster previously described (22) and with a small T. cruzi repetitive sequence called TcIRE (1). This subregion is followed by a 455-bp sequence (nt 540 to 995) that has no predicted function associated with it. TcTREZO's block 3, from nt 1277 to 1560, exhibits a high sequence identity (86%) with the T. cruzi U2II RNA gene (nt 1277 to 1332) and the untranslated region of the mucin-like protein pseudogene (nt 1479 to 1560).
The central core of the element (TcTREZO's block 2) is composed of repetitive 55-bp units in head-to-tail tandem arrays. The number of copies varies from one to six, with 84% sequence identity among them. According to our experimental results and in silico analysis against the latest data from the T. cruzi genome DB (TcGeneDB; http://www.genedb.org/), these 55-bp repeats are specific to this element. On the other hand, our similarity searches indicate that TcTREZO's blocks 1 and 3 have domains that could be found not only in locations associated with the TcTREZO element but also in several other genomic locations. These results show the composite nature of TcTREZO and could reflect the origin of this element in the T. cruzi genome.
The second approach was a detailed scan over the entire locally compiled copy of TcGeneDB in order to corroborate our experimental results and to find other complete copies of the TcTREZO element in the genome. Using regular expression implemented in a PERL script, we scanned the T. cruzi genome and found 49 full-length copies of this element. The average degree of conservation between these full copies is 94%, and the schematic representation of the 1,560-bp consensus sequence is shown in Fig. 1. Such a high intrafamily identity suggests that the mobilization of TcTREZO in the T. cruzi genome is an ongoing process. Our comparative in silico analyses also revealed the conservation pattern in TcTREZO's blocks 1 and 3 and showed that these blocks are extremely conserved (96% sequence identity in both of them).
A BLAST search with the 55-bp repeats identified 1,906 contigs (with lengths ranging from 183 to 93,048 bp). Because TcTREZO is ∼1.6 kb, 56 contigs that were longer than 1.9 kb were retained for this study. We identified 73 complete copies of TcTREZO, which include the 49 full copies described above, 7 truncated elements, and 41 incomplete elements, the last of which were located at one extremity of the contigs. TcTREZO elements were distributed as follows: 12 contigs carried a single copy of the element, 37 contigs carried tandemly arranged copies, and 7 contigs contained truncated forms (six elements had lost mucin and U2II RNA sequences, and one copy had lost part of the central region of the element) (Fig. 2).
FIG. 2.
TcTREZO is located in the nonsyntenic “islands” of the T. cruzi genome. (A) Detailed representation of T. cruzi contigs containing TcTREZO (gray boxes), each line representing the entire contig. The length (in bp) is indicated to the right of each contig. Locus names are shown above each black rectangle. The symbol ϕ indicates a pseudogene. The shaded area contains the TcTREZO elements and is shown in panel B. The contigs have the following GenBank accession numbers: 1, AAHK01000217; 2, AAHK01000548; 3, AAHK01001486; 4, AAHK01001836; 5, AAHK01001811; 6, AAHK01000143; 7, AAHK01000451; 8, AAHK01000836; 9, AAHK01000647. Loci are as follows: pref, prefoldin subunit 2; cyc, cyclin 1; hel, ATP-dependent DNA helicase; leuc, leucine-rich repeat protein; sat, 196-bp DNA satellite; kin, kinesin; mlh1, mismatch repair protein MLH1; if-2, translation initiation factor IF-2; syn, syntaxin 7-like protein; masp, MASP; ts, surface glycoprotein gene from trans-sialidase superfamily; muc, mucin; L1Tc, non-LTR retrotransposon encoding APE, reverse transcriptase (rt), and gag (RNase H); rhs, retrotransposon hot spot protein; gp63, surface metalloproteinase; dgf-1, dispersed gene family 1 (putative surface protein); sire, short interspersed repetitive element; dire, degenerate ingi/L1Tc-related element; viper, vestigial interposed retroelement. (B) Schematic representation of TcTREZO. The upstream and downstream regions flanking the element are indicated by hatched boxes.
TcTREZO elements are found in large and nonsyntenic regions of the T. cruzi genome, consisting mostly of interspersed arrays of surface antigen (pseudo)genes (mucin-associated surface protein [MASP], TS, GP63, mucin, and DGF-1 genes), the RHS (retrotransposon hot spot protein) gene, and retrotransposon-like elements (VIPER, L1Tc, SIRE, and DIRE) (Fig. 2; derivations of the designations are in the legend). These regions often appear to lie between internal chromosome synteny blocks (15, 16), suggesting that they could be genetically unstable.
Analysis of TcTREZO adjacent sequences in T. cruzi genome.
To study the insertion site of TcTREZO, we compared the 5′ and 3′ flanking regions of the element with sequences in the TcGeneDB. We selected 100 bp of the 5′ region and 600 bp of the 3′ region flanking TcTREZO and rigorously analyzed them using the fasta3 algorithm of the FASTA suite (36, 37) with a ktup parameter value of 1 for highest selectivity. After careful inspection of alignments, we found that TcTREZO elements lack obvious target site duplications (TSDs). At the 5′ and 3′ regions of TcTREZO, we found several inverted repeats and small repetitions that resemble microsatellites. We did not detect any direct repeats only a few base pairs in length (∼10 bp) flanking TcTREZO that could represent a TSD. To study the insertion site of TcTREZO further, we analyzed the conservation of nucleotides 1,000 bp upstream and downstream of each element. For the analysis of the upstream and downstream conservation patterns, we considered only contigs greater than 1.9 kb.
In 27 (79.4%) of the 34 elements analyzed in the upstream region, we identified a 68-bp highly conserved sequence flanking the 5′ region of the TcTREZO element. The average percentage of identity among the copies of the 68-bp conserved domain is 89% (Fig. 3A). In our analysis of the downstream region of the TcTREZO element we found a 500-bp conserved sequence domain without a well-defined borderline. Among 25 elements analyzed, 18 (72%) are flanked at the 3′ end by this conserved sequence, which is related to the ISPB subregion of TcTREZO's block 1 (Fig. 3B). This analysis showed that some TcTREZO elements are framed by two conserved sequences, indicating that they are inserted at specific genomic sites.
FIG. 3.
TcTREZO insertion site. Comparison of the 5′ and 3′adjacent sequences flanking TcTREZO in the contigs of the TcGeneDB. The accession numbers of the corresponding contigs are indicated at the left. Alignments were done by Clustal W. The first block (A) shows a conserved region of 68 nt upstream of TcTREZO, and the second block (B) shows the 3′ region of this element. Sequences corresponding to the conserved residues are shaded in black (>80% conservation) and gray (60 to 80% conservation); no shading denotes residues with <60% conservation. A schematic representation of TcTREZO is shown at the top. The upstream and downstream regions are shown by hatched boxes.
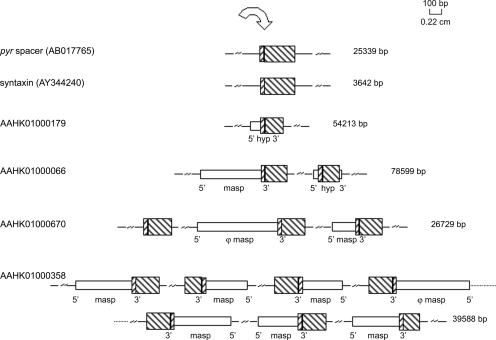
The finding of the 68-bp upstream motif and the downstream flanking sequence framing each TcTREZO raised the question of whether they could be found as isolated units in the T. cruzi genome. Using regular expressions implemented in a PERL script, we scanned the T. cruzi genome and identified 213 sequences harboring the 68-bp motif associated with the first 250 nt of the downstream flanking region, 486 sequences carrying only the 68-bp motif, and 1,481 sequences containing only the downstream flanking region. The length of the downstream flanking region is variable (Fig. 4). Remarkably, many of the 68-bp motifs are located within the 3′ end of the coding sequence of the MASP gene. Using a BLASTN search against the TcGeneDB DB, we identified 852 copies of the MASP gene carrying the 68-bp motif plus approximately 40 nt from the downstream region. This indicates that most TcTREZO elements are inserted into MASP (pseudo)genes, suggesting that there is one homologous recombination hot spot in this gene (Fig. 2). We have also identified several TcTREZO copies (AAHK01001139, AAHK01000629, AAHK01001119, AAHK01002727, and AAHK01000985) located in regions of the genome other than those of the MASP genes. Taken together, these results confirm that TcTREZO is not randomly distributed in the T. cruzi genome.
FIG. 4.
Schematic representation of T. cruzi sequences containing the insertion site for TcTREZO. The upstream (68-bp motif) and downstream (ISPB) sequences flanking TcTREZO are shown by rising and falling diagonal lines, respectively. The accession numbers of the corresponding sequences are indicated at the left, and the lengths are indicated at the right. Loci are as follows: hyp, hypothetical protein gene; masp, MASP gene. The orientations (5′ to 3′) of (pseudo)genes are indicated. The symbol ϕ indicates a pseudogene.
Genomic organization of the TcTREZO element.
When a Southern blot of T. cruzi genomic DNA digested with several restriction enzymes was hybridized with TcTREZO, a very simple pattern of hybridization was observed (Fig. 5). The probe hybridized with a single ∼1.6-kb band in EcoRI digests and with a large, broad, ∼23-kb band obtained by digestion with HindIII or BamHI (Fig. 5B). Double digestion with EcoRI (EcoRI plus BamHI or EcoRI plus HindIII) produced a single 1.6-kb band, suggesting the presence of several copies of the EcoRI 1.6-kb fragment in the 23-kb band (Fig. 5B). This hybridization profile was confirmed by using other restriction enzymes. For instance, a 23-kb hybridizing fragment was obtained by single digestion with BclI, KpnI, SmaI, XhoI, or BglII (data not shown), which do not have restriction sites within the TcTREZO element, while digestion with DraI or PstI resulted in a 1.6-kb fragment (see the restriction map of TcTREZO in Fig. 5A). The intensity of the hybridization signal indicated that the ∼23-kb band must contain fragments of different sizes. Because of the low resolution in this region of the agarose gel, the exact sizes of the restriction fragments contained in the band cannot be determined exactly. Partial digestion of the genomic DNA with EcoRI produced a ladder of hybridizing fragments, suggesting that the TcTREZO loci carry tandemly arranged and well-conserved 1.6-kb units (Fig. 5C).
Sequences homologous to TcTREZO were detected by hybridization in all T. cruzi isolates tested: strains G and Y and clones Sylvio and CL Brener (Fig. 5D). Southern blots of endonuclease-restricted T. cruzi DNA also showed the existence of both qualitative and quantitative differences in the hybridization patterns of TcTREZO in different T. cruzi strains (Fig. 5D). No hybridization signal was found with other Trypanosoma species (Trypanosoma rangeli and Trypanosoma brucei) or with a related trypanosomatid, Leishmania (Leishmania) amazonensis (Fig. 5E). These results indicate that TcTREZO is species specific for T. cruzi.
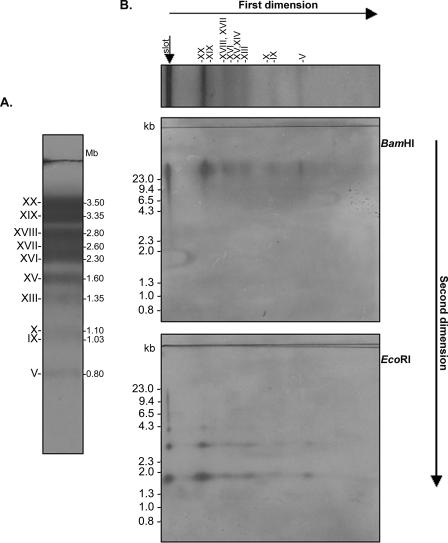
Hybridization of T. cruzi (clone CL Brener) chromosomes separated by PFGE using the central region of TcTREZO as a probe showed that this element is located in 10 chromosomal bands (3.5 to 0.8 Mb) (Fig. 6A). To determine how TcTREZO elements are organized in the different chromosomal bands, we performed Southern analysis of T. cruzi two-dimensional pulsed-field gels. Southern blot hybridization with the labeled 55-bp repeat as the probe revealed a common BamHI or EcoRI pattern of DNA fragments for every T. cruzi chromosome. A large BamHI fragment (>23 kb) and two EcoRI fragments of 1.6 and 3.2 kb can be seen for all chromosomes that hybridized in the first dimension (compare panels A and B). The hybridization signal for the 3.2-kb EcoRI fragment was weaker than that observed with the 1.6-kb EcoRI fragment, suggesting that the 3.2-kb fragment could be due to partial digestion with EcoRI. A ladder of hybridizing fragments can be visualized in the region corresponding to the gel slot (Fig. 6B). Taken together, these results confirmed that TcTREZO elements are distributed in tandemly arranged and well-conserved 1.6-kb units.
FIG. 6.
Mapping of TcTREZO in the chromosomal bands. (A) Chromosomal bands of clone CL Brener were separated by PFGE and hybridized with 32P-labeled TcTREZO. (B) Two-dimensional PFGE. The chromosomal bands were separated in the first dimension by PFGE in a Gene Navigator apparatus. Then a 5-mm agarose strip containing the resolved chromosomal bands was incubated with either the BamHI or EcoRI restriction enzyme. In the second dimension, the strip was placed in a well made in a 1% agarose-0.5× TBE gel and subjected to a linear gradient of switching times from 30 to 70 s in a voltage gradient of 6 V cm−1 for 18 h at 14°C in a contour-clamped homogeneous electric field apparatus. DNA fragments were hybridized with 32P-labeled TcTREZO. Vertical arrow, ladder of hybridizing fragments in the region corresponding to the gel slot.
We used the RepeatMasker script to estimate the number of copies of TcTREZO in the T. cruzi DB. Using the 1,573-bp sequence of TcTREZO (GenBank accession no. AF508945) formatted as a custom library, we found 173 copies of this element per haploid genome.
Transcription of TcTREZO.
To investigate whether or not TcTREZO is transcribed, Northern blots carrying total and poly(A)+ RNAs from epimastigotes were hybridized with the complete TcTREZO element, which hybridized with transcripts of 1.6 kb and ∼0.2 kb in the total and poly(A)+ RNA fractions (Fig. 7A). The 1.60-kb RNA has the size expected for a TcTREZO transcript. The transcripts of 0.2-kb were also detected in the poly(A)− RNA fraction (Fig. 7A). Figure 7B shows the hybridization of different subregions of TcTREZO with total epimastigote RNA. TcTREZO's block 1 hybridized only with the 1.60-kb transcript, while block 2, which carries the 55-bp repeats, strongly hybridized with the 0.2-kb transcript and to a lesser degree with the 1.60-kb transcript (Fig. 7B).
FIG. 7.
Northern blot of T. cruzi epimastigotes (CL Brener clone). (A) Total RNA (T), poly(A)− (−), and poly(A)+ (+) RNA fractions were separated by gel electrophoresis (left), transferred to a nylon membrane, and hybridized with the TcTREZO probe (right). (B) Equivalent amounts of total RNA were separated by electrophoresis (left) and transferred to a nylon membrane. The material was hybridized with the following probes: block 1, ISPB plus spacer; block 2, 55-bp repeats, mucin-like protein probe, and the entire TcTREZO. The schematic representation of each probe is shown. α-Tubulin was used as an internal positive hybridization control.
To isolate TcTREZO transcripts, cDNAs from epimastigotes were synthesized by RT-PCR using a primer from block 1 and an antisense primer from an internal sequence found exclusively in TcTREZO. TcTREZO transcripts were amplified by one-step RT-PCR and also from a single-stranded cDNA strand synthesized using oligo(dT) as a primer followed by PCR amplification. Both reactions generated amplicons with the expected sizes, and their identities were confirmed by sequencing. There was a high degree of similarity among these sequences, and six were deposited in the GenBank DB (accession no. EF577443 to EF577448). A nucleotide sequence identity search of the GenBank DB revealed that TcTREZO exhibits a high degree of identity with seven expressed tag sequences, confirming that this repeated element is transcribed. Sense transcripts (n = 5) are preferentially synthesized compared to antisense transcripts (n = 2). In conclusion, TcTREZO is not silent in the genome. Transcripts from TcTREZO are relatively abundant in epimastigotes and expressed as oligo(A)-terminated transcripts with a length corresponding to the unit size of the element.
To determine whether some TcTREZO transcripts were linked to potential protein-coding sequences, we checked for the presence of an SL sequence that is found at the 5′ ends of all protein-coding transcripts in trypanosomes. Single-stranded cDNA generated by reverse transcription of total RNA was PCR amplified using an SL-specific primer and TcTREZO-specific primers. Two kinds of RT-PCRs were used: (i) one-step RT-PCR and (ii) synthesis of a cDNA strand using oligo(dT) as a primer followed by PCR amplification. As a control, α-tubulin and SL-specific primers were used to amplify full-length transcripts from tubulin cDNA. A Southern blot of the PCR products was probed at high stringency with TcTREZO and tubulin. No hybridization was obtained after an overnight exposure of the blot to X-ray film (data not shown). Reprobing the blot with an α-tubulin sequence confirmed that a full-length α-tubulin sequence had been successfully amplified from T. cruzi cDNA using one-step RT-PCR or oligo(dT) and SL-specific primers. The tubulin amplicon was cloned and its identity confirmed by sequencing.
In another set of experiments, RT-PCR was carried out using a combination of sense and antisense primers derived from different regions of the TcTREZO element. All these reactions produced fragments of the expected sizes, whose identity was confirmed by sequencing. These results suggest that the majority if not all TcTREZO-containing transcripts lack the SL sequence indicative of trypanosome mRNA transcripts targeted for translation. Taken together, these results suggest that TcTREZO transcripts are unspliced. Ours results are in agreement with those from other authors (26, 42) showing that transcripts of RIME/ingi (T. brucei) and L1Tc (T. cruzi) are unspliced.
DISCUSSION
We have described here a new site-specific repetitive element named TcTREZO, which appears to display a high degree of integration site specificity. TcTREZO is also an exceptional element in terms of its distinct substructure, which consists of sequences derived from different regions of the genome. We view TcTREZO as a composite element that could have been generated by insertion of an ancient sequence formed by the three cassettes into the insertion site, which was then amplified as a transpositional unit. Two lines of evidence suggest that TcTREZO could be a nonautonomous non-LTR retrotransposon that migrates via an RNA intermediary. First, TcTREZO is transcribed as oligo(A)-terminated sequences that are not derived from read-through transcripts from putative promoters far upstream. Although transcribed and processed, these sequences do not code for proteins, indicating that these transcripts are not translated. Second, analysis using the mfold program (50) (http://www.bioinfo.rpi.edu/applications/mfold/rna/form1.cgi) to predict secondary structures indicated the presence of stack and hairpin loop structures in TcTREZO RNA (R. T. Souza, J. C. Ruiz, and J. Franco da Silveira, unpublished data). The 3′ terminus of the TcTREZO transcript could be used as a self primer for reverse transcriptase-like activity, generating a cDNA copy of TcTREZO that is then inserted into the genome.
TcTREZO elements are not flanked by TSDs. At the 5′ and 3′ regions of TcTREZO, we found several inverted repeats and small repetitions that resemble microsatellites. TSD is the (retro)transposition signature of many mobile elements. However, there are some retrotransposons that do not generate TSDs, suggesting that other mechanisms of insertion could generate distinctive signatures. In trypanosomes, copies of retroelements that lack TSDs can be found (5, 7). Among the 220 full-length T. cruzi L1Tc elements analyzed (5), 103 (46.8%) are not flanked by a TSD. It has been suggested that the loss of TSD could be the consequence of homologous recombination between retroelements, which would result in chimeric retrotransponsons flanked by unrelated sequences (7). Another possible explanation would be the existence of an endonuclease-independent transposition that could generate retroelements lacking the flanking TSDs. Morrish et al. (32, 33) have described a pathway for LINE-1 retrotransposition in Chinese hamster ovary cells that acts independently of endonuclease but is dependent upon reverse transcriptase. In this case the retrotransposition is not associated with the duplication of the target DNA. It was shown that approximately 30% of endonuclease-independent LINE-1 retrotransposition events in Chinese hamster ovary cells occur in an orientation-specific manner adjacent to a perfect telomere repeat (5′-TTAGGG-3′) (32).
Retroelements lacking TSDs in organisms other than mammalians and trypanosomes have been also described. For instance, neither CR1-like elements nor SINE3 retroelements found in the zebrafish genome are flanked by TSDs; rather, their 3′ termini are composed of 3-bp to 5-bp microsatellites (28, 29). It was suggested that the 3′ microsatellites have been inserted into the genome together with CR1-like elements and that they can be considered to be distinctive hallmarks or signatures of different families.
TcTREZO is specifically inserted between two conserved sequences: a 68-bp motif at the 5′ end and a variable-length domain related to the subregion ISPB of TcTREZO's block 1 at the 3′ end. Computational analysis of sequences from TcGeneDB revealed the presence of the 68-bp motif plus approximately 40 nt from the ISPB in TcTREZO's block 1 at the 3′ ends of the coding sequences of many MASP genes. The insertion of TcTREZO elements at exactly the same relative position in the MASP gene suggests that there is one homologous recombination hot spot in this gene.
MASP genes comprise a large T. cruzi-specific gene family (771 genes, 433 pseudogenes), whose members are characterized by the presence of conserved 5′- and 3′-terminal domains that encode a signal peptide and a glycosylphosphatidylinositol anchor addition site, respectively, suggesting a surface location in the parasite (15). The site specificity of TcTREZO insertion into MASP genes could lead to the observed tandem arrays of this element. The relationship of TcTREZO with the MASP gene family resembles that previously described for T. brucei between non-LTR retrotransposons ingi/RIME and the RHS (retrotransposon hot spot) gene (6). RIME/ingi elements can be inserted in frame with the RHS genes.
Site-specific retrotransposons, or siteposons, have been described in trypanosomatids (T. cruzi, Trypanosoma brucei gambiense, Trypanosoma brucei, Crithidia fasciculata, and Herpetomonas samuelpessoai) (2, 3, 11, 19, 20, 45) and have identical site specificity of integration in the SL RNA gene. The determinants of site-specific integration of TcTREZO are not known but could be multifactorial. One factor could be a retrotransposon-encoded nuclease capable of cleaving the insertion site within the 68-bp and ISPB sequences. Most non-LTR retrotransposons encode an apurinic endonuclease-like endonuclease (APE) upstream of a common reverse transcriptase domain. Although the APE domain has not been found in the site-specific retrotransposons, these retrotransposons encode an integrase-like domain that has the characteristics of a nonpalindromic sequence-specific endonuclease (46, 48). In Bombyx mori, the siteposon R2Bm encodes a sequence-specific endonuclease that is responsible for its insertion at a unique site in the 28S rRNA genes (9). It is possible that in T. cruzi a similar enzyme encoded by the CZAR siteposon (45, 48) is responsible for TcTREZO insertion.
TcTREZO is a species-specific sequence, suggesting that its appearance occurred after the separation of the T. cruzi ancestor from other trypanosomatid lineages. Our results indicate that the composite nature of TcTREZO may reflect the T. cruzi genome plasticity and capacity for sequence rearrangement. The relative abundance of TcTREZO in T. cruzi isolates from the groups T. cruzi I and T. cruzi II was examined by Southern (Fig. 5C) and dot blot (not shown) hybridizations. The TcTREZO element was two- to threefold more abundant in T. cruzi II isolates. Previous studies showed that the genome sizes of T. cruzi II strains are higher than that of T. cruzi I (41) and that some repeated elements (satellite DNA, L1Tc, C6, and E12) are more abundant in T. cruzi II stocks (14, 41). T. cruzi I strains circulate in the sylvatic cycle of parasite transmission, whereas T. cruzi II strains are related to the domestic cycle and predominate where Chagas’ disease is more severe. The abundance of repetitive sequences could be related to the chromosome and genome size variations among T. cruzi isolates. It has been suggested that the increase in the numbers and sizes of chromosomes in T. cruzi II strains may reflect a speciation event and supports the distant relationship between T. cruzi I and T. cruzi II groups (41). The increase of the genome size of T. cruzi II and hybrid genotypes could reflect an adaptive characteristic related to the epidemiological attributes of these strains (41).
Acknowledgments
This work was supported by grants from FAPESP and CNPq (Brazil) to J.F.S. R.T.S. and F.M.L. were awarded doctoral fellowships by FAPESP.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Agüero, F., R. E. Verdun, A. C. Frasch, and D. O. Sanchez. 2000. A random sequencing approach for the analysis of the Trypanosoma cruzi genome: general structure, large gene and repetitive DNA families, and gene discovery. Genome Res. 10:1996-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aksoy, S., T. M. Lalor, J. Martin, L. H. Van der Ploeg, and F. F. Richards. 1987. Multiple copies of a retroposon interrupt spliced leader RNA genes in the African trypanosome, Trypanosoma gambiense. EMBO J. 6:3819-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aksoy, S., S. Williams, S. Chang, and F. F. Richards. 1990. SLACS retrotransposon from Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res. 18:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araya, J., M. I. Cano, H. B. Gomes, E. M. Novak, J. M. Requena, C. Alonso, M. J. Levin, P. Guevara, J. L. Ramirez, and J. F. Da Silveira. 1997. Characterization of an interspersed repetitive DNA element in the genome of Trypanosoma cruzi. Parasitology 115(Pt. 6):563-570. [DOI] [PubMed] [Google Scholar]
- 5.Bringaud, F., D. C. Bartholomeu, G. Blandin, A. Delcher, T. Baltz, N. M. El-Sayed, and E. Ghedin. 2006. The Trypanosoma cruzi L1Tc and NARTc non-LTR retrotransposons show relative site specificity for insertion. Mol. Biol. Evol. 23:411-420. [DOI] [PubMed] [Google Scholar]
- 6.Bringaud, F., N. Biteau, S. E. Melville, S. Hez, N. M. El-Sayed, V. Leech, M. Berriman, N. Hall, J. E. Donelson, and T. Baltz. 2002. A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryot. Cell 1:137-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bringaud, F., N. Biteau, E. Zuiderwijk, M. Berriman, N. M. El-Sayed, E. Ghedin, S. E. Melville, N. Hall, and T. Baltz. 2004. The ingi and RIME non-LTR retrotransposons are not randomly distributed in the genome of Trypanosoma brucei. Mol. Biol. Evol. 21:520-528. [DOI] [PubMed] [Google Scholar]
- 8.Bringaud, F., J. L. Garcia-Perez, S. R. Heras, E. Ghedin, N. M. El-Sayed, B. Andersson, T. Baltz, and M. C. Lopez. 2002. Identification of non-autonomous non-LTR retrotransposons in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 124:73-78. [DOI] [PubMed] [Google Scholar]
- 9.Burke, W. D., C. C. Calalang, and T. H. Eickbush. 1987. The site-specific ribosomal insertion element type II of Bombyx mori (R2Bm) contains the coding sequence for a reverse transcriptase-like enzyme. Mol. Cell. Biol. 7:2221-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cano, M. I., A. Gruber, M. Vazquez, A. Cortes, M. J. Levin, A. Gonzalez, W. Degrave, E. Rondinelli, B. Zingales, J. L. Ramirez, et al. 1995. Molecular karyotype of clone CL Brener chosen for the Trypanosoma cruzi genome project. Mol. Biochem. Parasitol. 71:273-278. [DOI] [PubMed] [Google Scholar]
- 11.Carrington, M., I. Roditi, and R. O. Williams. 1987. The structure and transcription of an element interspersed between tandem arrays of mini-exon donor RNA genes in Trypanosoma brucei. Nucleic Acids Res. 15:10179-10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castro, C., S. P. Craig, and M. Castaneda. 1981. Genome organization and ploidy number in Trypanosoma cruzi. Mol. Biochem. Parasitol. 4:273-282. [DOI] [PubMed] [Google Scholar]
- 13.Chiurillo, M. A., I. Cano, J. F. Da Silveira, and J. L. Ramirez. 1999. Organization of telomeric and sub-telomeric regions of chromosomes from the protozoan parasite Trypanosoma cruzi. Mol. Biochem. Parasitol. 100:173-183. [DOI] [PubMed] [Google Scholar]
- 14.Elias, M. C., N. S. Vargas, B. Zingales, and S. Schenkman. 2003. Organization of satellite DNA in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 129:1-9. [DOI] [PubMed] [Google Scholar]
- 15.El-Sayed, N. M., P. J. Myler, D. C. Bartholomeu, D. Nilsson, G. Aggarwal, A. N. Tran, E. Ghedin, E. A. Worthey, A. L. Delcher, G. Blandin, S. J. Westenberger, E. Caler, G. C. Cerqueira, C. Branche, B. Haas, A. Anupama, E. Arner, L. Aslund, P. Attipoe, E. Bontempi, F. Bringaud, P. Burton, E. Cadag, D. A. Campbell, M. Carrington, J. Crabtree, H. Darban, J. F. da Silveira, P. de Jong, K. Edwards, P. T. Englund, G. Fazelina, T. Feldblyum, M. Ferella, A. C. Frasch, K. Gull, D. Horn, L. Hou, Y. Huang, E. Kindlund, M. Klingbeil, S. Kluge, H. Koo, D. Lacerda, M. J. Levin, H. Lorenzi, T. Louie, C. R. Machado, R. McCulloch, A. McKenna, Y. Mizuno, J. C. Mottram, S. Nelson, S. Ochaya, K. Osoegawa, G. Pai, M. Parsons, M. Pentony, U. Pettersson, M. Pop, J. L. Ramirez, J. Rinta, L. Robertson, S. L. Salzberg, D. O. Sanchez, A. Seyler, R. Sharma, J. Shetty, A. J. Simpson, E. Sisk, M. T. Tammi, R. Tarleton, S. Teixeira, S. Van Aken, C. Vogt, P. N. Ward, B. Wickstead, J. Wortman, O. White, C. M. Fraser, K. D. Stuart, and B. Andersson. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409-415. [DOI] [PubMed] [Google Scholar]
- 16.El-Sayed, N. M., P. J. Myler, G. Blandin, M. Berriman, J. Crabtree, G. Aggarwal, E. Caler, H. Renauld, E. A. Worthey, C. Hertz-Fowler, E. Ghedin, C. Peacock, D. C. Bartholomeu, B. J. Haas, A. N. Tran, J. R. Wortman, U. C. Alsmark, S. Angiuoli, A. Anupama, J. Badger, F. Bringaud, E. Cadag, J. M. Carlton, G. C. Cerqueira, T. Creasy, A. L. Delcher, A. Djikeng, T. M. Embley, C. Hauser, A. C. Ivens, S. K. Kummerfeld, J. B. Pereira-Leal, D. Nilsson, J. Peterson, S. L. Salzberg, J. Shallom, J. C. Silva, J. Sundaram, S. Westenberger, O. White, S. E. Melville, J. E. Donelson, B. Andersson, K. D. Stuart, and N. Hall. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404-409. [DOI] [PubMed] [Google Scholar]
- 17.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 18.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel, A., and J. D. Boeke. 1991. Reverse transcriptase encoded by a retrotransposon from the trypanosomatid Crithidia fasciculata. Proc. Natl. Acad. Sci. USA 88:9794-9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabriel, A., T. J. Yen, D. C. Schwartz, C. L. Smith, J. D. Boeke, B. Sollner-Webb, and D. W. Cleveland. 1990. A rapidly rearranging retrotransposon within the miniexon gene locus of Crithidia fasciculata. Mol. Cell. Biol. 10:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 22.Gao, G., T. Nara, J. Nakajima-Shimada, and T. Aoki. 1999. Novel organization and sequences of five genes encoding all six enzymes for de novo pyrimidine biosynthesis in Trypanosoma cruzi. J. Mol. Biol. 285:149-161. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Perez, J. L., C. I. Gonzalez, M. C. Thomas, M. Olivares, and M. C. Lopez. 2003. Characterization of reverse transcriptase activity of the L1Tc retroelement from Trypanosoma cruzi. Cell. Mol. Life Sci. 60:2692-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez, A., E. Prediger, M. E. Huecas, N. Nogueira, and P. M. Lizardi. 1984. Minichromosomal repetitive DNA in Trypanosoma cruzi: its use in a high-sensitivity parasite detection assay. Proc. Natl. Acad. Sci. USA 81:3356-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 26.Heras, S. R., M. C. Lopez, M. Olivares, and M. C. Thomas. 2007. The L1Tc non-LTR retrotransposon of Trypanosoma cruzi contains an internal RNA-pol II-dependent promoter that strongly activates gene transcription and generates unspliced transcripts. Nucleic Acids Res. 35:2199-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofacker, I. L. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 31:3429-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapitonov, V. V., and J. Jurka. 2003. The esterase and PHD domains in CR1-like non-LTR retrotransposons. Mol. Biol. Evol. 20:38-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapitonov, V. V., and J. Jurka. 2003. A novel class of SINE elements derived from 5S rRNA. Mol. Biol. Evol. 20:694-702. [DOI] [PubMed] [Google Scholar]
- 30.Lanar, D. E., L. S. Levy, and J. E. Manning. 1981. Complexity and content of the DNA and RNA in Trypanosoma cruzi. Mol. Biochem. Parasitol. 3:327-341. [DOI] [PubMed] [Google Scholar]
- 31.Martin, F., C. Maranon, M. Olivares, C. Alonso, and M. C. Lopez. 1995. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J. Mol. Biol. 247:49-59. [DOI] [PubMed] [Google Scholar]
- 32.Morrish, T. A., J. L. Garcia-Perez, T. D. Stamato, G. E. Taccioli, J. Sekiguchi, and J. V. Moran. 2007. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 446:208-212. [DOI] [PubMed] [Google Scholar]
- 33.Morrish, T. A., N. Gilbert, J. S. Myers, B. J. Vincent, T. D. Stamato, G. E. Taccioli, M. A. Batzer, and J. V. Moran. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31:159-165. [DOI] [PubMed] [Google Scholar]
- 34.Olivares, M., C. Alonso, and M. C. Lopez. 1997. The open reading frame 1 of the L1Tc retrotransposon of Trypanosoma cruzi codes for a protein with apurinic-apyrimidinic nuclease activity. J. Biol. Chem. 272:25224-25228. [DOI] [PubMed] [Google Scholar]
- 35.Olivares, M., J. L. Garcia-Perez, M. C. Thomas, S. R. Heras, and M. C. Lopez. 2002. The non-LTR (long terminal repeat) retrotransposon L1Tc from Trypanosoma cruzi codes for a protein with RNase H activity. J. Biol. Chem. 277:28025-28030. [DOI] [PubMed] [Google Scholar]
- 36.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson, W. R., and W. Miller. 1992. Dynamic programming algorithms for biological sequence comparison. Methods Enzymol. 210:575-601. [DOI] [PubMed] [Google Scholar]
- 38.Requena, J. M., M. C. Lopez, and C. Alonso. 1996. Genomic repetitive DNA elements of Trypanosoma cruzi. Parasitol. Today 12:279-283. [DOI] [PubMed] [Google Scholar]
- 39.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vargas, N., A. Pedroso, and B. Zingales. 2004. Chromosomal polymorphism, gene synteny and genome size in T. cruzi I and T. cruzi II groups. Mol. Biochem. Parasitol. 138:131-141. [DOI] [PubMed] [Google Scholar]
- 42.Vassella, E., I. Roditi, and R. Braun. 1996. Heterogeneous transcripts of RIME/ingi retroposons in Trypanosoma brucei are unspliced. Mol. Biochem. Parasitol. 82:131-135. [DOI] [PubMed] [Google Scholar]
- 43.Vazquez, M., C. Ben-Dov, H. Lorenzi, T. Moore, A. Schijman, and M. J. Levin. 2000. The short interspersed repetitive element of Trypanosoma cruzi, SIRE, is part of VIPER, an unusual retroelement related to long terminal repeat retrotransposons. Proc. Natl. Acad. Sci. USA 97:2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vazquez, M. P., A. G. Schijman, and M. J. Levin. 1994. A short interspersed repetitive element provides a new 3′ acceptor site for trans-splicing in certain ribosomal P2 beta protein genes of Trypanosoma cruzi. Mol. Biochem. Parasitol. 64:327-336. [DOI] [PubMed] [Google Scholar]
- 45.Villanueva, M. S., S. P. Williams, C. B. Beard, F. F. Richards, and S. Aksoy. 1991. A new member of a family of site-specific retrotransposons is present in the spliced leader RNA genes of Trypanosoma cruzi. Mol. Cell. Biol. 11:6139-6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volff, J. N., C. Korting, A. Froschauer, K. Sweeney, and M. Schartl. 2001. Non-LTR retrotransposons encoding a restriction enzyme-like endonuclease in vertebrates. J. Mol. Evol. 52:351-360. [DOI] [PubMed] [Google Scholar]
- 47.Wickstead, B., K. Ersfeld, and K. Gull. 2003. Repetitive elements in genomes of parasitic protozoa. Microbiol. Mol. Biol. Rev. 67:360-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, J., H. S. Malik, and T. H. Eickbush. 1999. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. USA 96:7847-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zingales, B., M. E. Pereira, K. A. Almeida, E. S. Umezawa, N. S. Nehme, R. P. Oliveira, A. Macedo, and R. P. Souto. 1997. Biological parameters and molecular markers of clone CL Brener—the reference organism of the Trypanosoma cruzi genome project. Mem. Inst. Oswaldo Cruz 92:811-814. [DOI] [PubMed] [Google Scholar]
- 50.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]