Abstract
Cryptococcus neoformans is a human pathogenic fungus with a capsule composed primarily of glucuronoxylomannan (GXM) that is important for virulence. Current views of GXM structure postulate a polymer composed of repeating mannose trisaccharide motifs bearing a single β(1,2) glucuronic acid with variable xylose and O-acetyl substitutions to form six triads. GXM from different strains is notoriously variable in triad composition, but it is not known if the polymer consists of one or more motif-repeating units. We investigated the polymeric organization of GXM by using mass spectrometry to determine if its compositional motif arrangement was similar to that of bacterial capsular polysaccharides, namely, a polymer of a single repeating unit. The results were consistent with, and confirmatory for, the current view that the basic unit of GXM is a repeating mannose trisaccharide motif, but we also found evidence for the copolymerization of different GXM repeating units in one polysaccharide molecule. Analysis of GXM from isogenic phenotypic switch variants suggested structural differences caused by glucuronic acid positional effects, which implied flexibility in the synthetic pathway. Our results suggest that cryptococcal capsule synthesis is fundamentally different from that observed in prokaryotes and employs a unique eukaryotic approach, which theoretically could synthesize an infinite number of structural combinations. The biological significance of this capsule construction scheme is that it is likely to confer a powerful avoidance strategy for interactions with the immune system and phagocytic environmental predators. Consistent with this premise, the antigenic variation of a capsular epitope recognized by a nonprotective antibody was observed under different growth conditions.
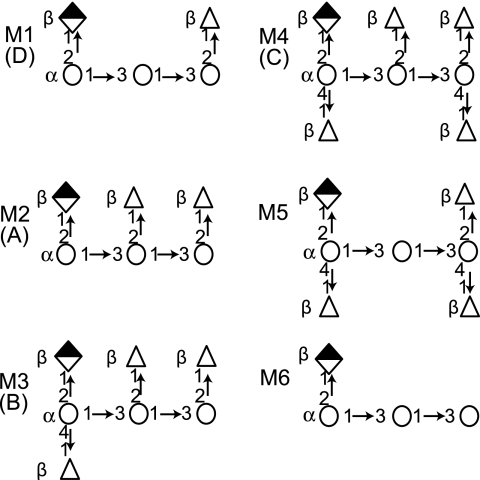
Polysaccharide (PS) capsules are important for microbial survival in the environment and in hosts. Although many details of capsular synthesis in prokaryotes are known, there are few comparable data for the fungi. Gram-negative bacteria synthesize the repeating units for group I capsular PSs at their inner membrane for eventual translocation to the surface (25), while the human fungal pathogen Cryptococcus neoformans employs vesicular transport to export capsule components (7, 12, 24, 27). Bacteria polymerize a single oligosaccharide-repeating unit. In contrast, at least six different oligosaccharide repeating units have been identified for glucuronoxylomannan (GXM), the major capsular PS of C. neoformans (Fig. 1) (5).
FIG. 1.
Diagram of identified structural repeating units for GXM (based on data from reference 5). Repeating units designated M1 to M4 are highly abundant in specific serotypes, listed in parentheses. M5 and M6 do not confer serotype specificity. Mannose, open circles; glucuronic acid, half-filled diamond; xylose, open triangles.
GXM is an acetylated linear PS (4). The six identified repeating units have in common a mannose (Man) trisaccharide bearing a single β(1,2) glucuronic acid side group. The six repeats differ in their molar ratios of β(1,2) and β(1,4)-xylose (Xyl) attached to the Man trisaccharide (4, 5). GXM is composed of a single repeating unit in some strains, while most strains produce GXM that contains multiple units (5). Furthermore, strains utilizing the same set of repeat units often differ from one another by the ratio of those units within their respective GXM molecules (5). Consequently, strain-to-strain comparisons of GXM demonstrate a compositional similarity but antigenic differences. This is unlike bacteria, such as Streptococcus pneumoniae and Neisseria meningitidis, where different serotypes synthesize unique oligosaccharide units, and PS antigenic differences can be explained by compositional differences.
Solving the structure of GXM has posed a formidable problem because it is a large and polydispersed polysaccharide. Structural studies of GXM have relied heavily on the nuclear magnetic resonance (NMR) analysis of de-O-acetylated GXM and the comparison of chemically modified GXM (4, 5). Given that GXM molecules have a mass on the order of 106 daltons (20), NMR analysis is limited in its ability to discern inter- and intramolecular relationships between different repeating units. Further complicating NMR analysis is evidence that purified GXM preparations are not homogeneous. Analysis of GXM purified from C. neoformans cultures by electrophoresis and immunoblotting has demonstrated significant heterogeneity in electrophoretic migration consistent with a heterogeneous composition (20). Likewise, antibodies to GXM produce different staining patterns on cells within a single culture (8, 11). The finding that some C. neoformans switch variants manifest changes in their GXM structure also suggests that GXM preparations cannot be homogeneous (8). Hence, we currently do not know whether the structural diversity found in GXM preparations is the result of mixtures of homopolymers or whether GXM is itself a heteropolymer.
The GXM basic unit is composed of a single hexose, uronic acid, and pentose residues and is modified only by O-acetyl groups on the mannose sugar. Hence, unlike many PSs, each component of GXM has a unique mass, permitting the use of mass spectrometry (MS) to solve the composition of GXM-derived oligosaccharides. The negative charge of GXM, imparted by glucuronic acid, and physical laws of repulsion provide a secondary means with which to examine structural aspects of GXM and to test assumptions imposed by hypothesized structural models of GXM. MS analysis of GXM-derived oligosaccharides suggested that GXM molecules are composed of combinations of different repeating units. Viscosity studies of GXM from isogenic phenotypic switch variants further supported copolymerization, as salt-dependent differences were suggestive of altered GlcA substitution patterns. This unique approach toward capsular PS synthesis may affect C. neoformans pathogenesis and antigenicity by providing a mechanism with which to alter the expression of immunologically relevant PS epitopes.
MATERIALS AND METHODS
Strains.
The C. neoformans strains used were representative of the four major serotypes: A, H99; B, I23; C, 106.97; and D, B-3501, 24067, and RC-2. Strains H99, B-3501, and 24067 are common laboratory strains. Strains 106.97 and I23 are more recent clinical isolates. RC-2 is a variant of 24067 that was identified during microevolution studies. Table 1 summarizes the analyses performed with the isolated GXM from each of these strains.
TABLE 1.
Methods of analysis performed with GXM from different strains used in this study
| Method | Strain
|
|||||
|---|---|---|---|---|---|---|
| H99 serotype A | 106.97 serotype B | I23 serotype C | B-3501 serotype D | 24067 serotype D | RC-2 serotype D | |
| FACE | x | |||||
| HPAEC | x | |||||
| MS | x | x | x | x | x | |
| Elemental | x | x | x | x | x | |
| NMR; light scattering; viscosity | x | |||||
| FACS | x | x | ||||
PS isolation.
GXM was isolated from 2-week-old cultures grown in Sabouraud dextrose (SD) broth at 30°C with shaking at 150 rpm and purified as described in reference 5, with minor modifications (20). The sugar composition of GXM isolated by this methodology has repeatedly shown the presence of either GXM-containing sugars exclusively or the additional presence of glucose. Isolated GXM samples were screened for protein and DNA contamination by using Coomassie blue and ethidium bromide staining, respectively, and found to be negative. Elemental analysis of isolated GXM for carbon, hydrogen, nitrogen, and phosphorus was performed by Quantitative Technologies, Inc. (Whitehouse, NJ).
GXM hydrolysis.
GXM (2 mg) was hydrolyzed in 100 μl of 0.5 M trifluoroacetic acid (TFA) at 95°C for 1 h, unless otherwise stated. The sample was vacuum dried and then dissolved in 20 mM ammonium bicarbonate. Partial hydrolysis was confirmed by the presence of multiple products visible by fluorophore-assisted carbohydrate electrophoresis (FACE) (14) and by thin-layer chromatography (15). For FACE, samples were run on a 30% polyacrylamide gel. The results were recorded as an inverted image using UV transillumination and image documentation software.
Mass spectrometry.
Masses were determined by flow injection analysis with an LTQ quadruple linear ion trap mass spectrometer (ThermoFinnigan, San Jose, CA). An HP model 1100 high-performance liquid chromatography unit was used to pump 50% methanol containing 0.5% ammonium hydroxide at a flow rate of 50 μl min−1 and deliver it to the mass spectrometer. A sample (2 μl) was injected for analysis. The MS, equipped with an electrospray ionization source, was operated in negative mode to detect ions in the m/z range of 300 to 2,000. A tandem mass spectrometry (MS-MS) spectrum was acquired at the mass isolation window of 3 mass units and the relative collision energy of 30%.
Oligosaccharide composition determination.
To determine the composition of an oligosaccharide from individual MS peaks, masses of the individual sugars were added in various compositions. GlcA has a mass of 194.04264 g/mol, Xyl has a mass of 150.05285 g/mol, and Man has a mass of 180.06335 g/mol. However, during bond formation, the mass of water (18.01055 g/mol) is lost from the reducing end (C1) of the sugar. Therefore, in calculations, the following masses were used: GlcA, 176.03209 g/mol (bond formed with Man); Xyl, 132.0423 g/mol (bond formed with Man); Man, 162.05280 g/mol (if C1 has bond with adjacent Man); Man, 180.06335 g/mol (if Man is at the terminal end of the oligosaccharide). As an example, the M1 repeat has a mass of 1 GlcA, 176.03209 g/mol + 1 Xyl, 132.04230 g/mol + 2 Man, 162.05280 g/mol (with bonds to adjacent Man) + 1 Man, 180.06335 g/mol (terminal) = 812.24334 g/mol. The mass of the oligosaccharide is 1 mass unit larger than its m/z peak, as 1 proton was lost during the ionization process. Thus, the M1 triad would ionize at an m/z of approximately 811.2. Larger mass oligosaccharides were occasionally double ionized, which results in the m/z appearing as one-half of the expected m/z value. Due to the imprecision of the hydrolysis procedure, oligosaccharide structures observed for mass spectral analysis varied between experiments, necessitating the use of several MS scans to determine the structures present in a given GXM sample.
Molecular mass and radius of gyration.
The molecular mass and radius of gyration (Rg) of GXM were determined by multiangle laser light scattering, as previously described. The weight-averaged mass (Mw) was calculated by the Zimm equation as described in reference 20 and is the inverse of the y intercept.
Viscosity.
Viscosity was measured using a modified Ostwald-type capillary glass viscometer (Cannon-Manning Semi-Micro, Technical Glass Co., Dover, NJ) at 25°C with 2.25 ml of each sample. GXM (10 mg ml−1), in ultrapure water, from the phenotypic switch variants of the RC-2 strain was solubilized for 4 days at room temperature. Samples were diluted and equilibrated to 25°C prior to testing. Flow time was measured in triplicate and averaged. The actual flow times for a given sample typically did not differ by more than 1 s from the mean. The relative viscosity increment (ηi [also known as specific viscosity, ηsp]) is defined as the change in the ratio of the flow time of the sample to the solvent that is specifically due to the sample particles.
Time course of antigenic variation expression.
Cells from an individual colony grown on SD agar were patched onto a new SD agar plate. Plates were incubated at 30°C. After 3 days, half of the cells were scraped from the plate and washed in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8.5 mM Na2HPO4). A loopful of cells was used to initiate a 50-ml SD broth culture, which was incubated at 30°C with gentle shaking (150 rpm). Cells were scraped from the agar plate again at day 5. Aliquots of cells were removed from the broth culture after growth for 1, 2, and 4 days. Cells from each time point were immediately prepared for analysis by fluorescence-activated cell sorting (FACS): 1 × 107 to 2 × 107 cells in 100 to 200 μl of PBS were labeled overnight at 4°C with 10 μg ml−1 of either monoclonal antibody (MAb) 12A1 or 21D2. The cells were washed twice in PBS and then incubated with 0.1 mg ml−1 of fluorescein isothiocyanate-coupled goat anti-mouse immunoglobulin M antibody (Southern Biotechnologies, Birmingham, AL) for 1 h at room temperature. Following this incubation, the cells were washed twice in PBS, resuspended in 500 μl of PBS, and kept on ice until analyzed. Data were collected and analyzed using CellQuest software from DakoCytomation. Statistical significance was determined by analysis of variance (ANOVA).
RESULTS
Partial acid hydrolysis of GXM.
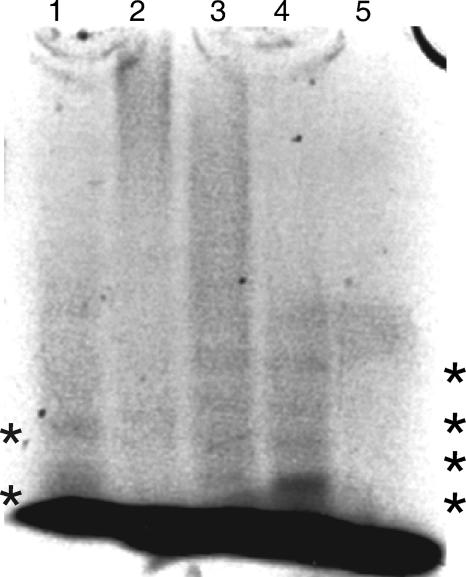
To analyze GXM by MS, it is necessary to break the macromolecule into oligosaccharides suitable for mass analysis. Alternative means to degrade GXM, such as enzymes, do not exist, and physical techniques, such as sonication, do not produce oligosaccharides suitable for MS. Attempts to design alternative protocols using cold alkaline treatment, hydrochloric acid, and exposure to massive doses of ionizing radiation were not successful (unpublished data). Hence, we relied on acid hydrolysis to generate oligosaccharides suitable for MS, since it is the only method currently available for cleaving GXM into small parts. We established that GXM was susceptible to partial acid hydrolysis using conditions developed for the hydrolysis of polysaccharides from other cryptococcal species (0.3 M TFA at 95°C for 5 h) (Fig. 2) (15). However, MS analysis of the hydrolyzed GXM using those conditions revealed only ions with m/z values that corresponded to oligosaccharides composed only of Man and GlcA. To obtain oligosaccharides that reflected the predicted GXM composition, different acid concentrations and hydrolysis times were evaluated for the cleavage of GXM (Fig. 2), and we selected a higher concentration of trifluoroacetic acid (0.5 M) and a shorter hydrolysis time (1 h) than used previously. To determine the extent of sample degradation (i.e., monosaccharide release) under this condition, 2 mg of GXM from strain B-3501 was hydrolyzed using these conditions and analyzed for monosaccharide composition by high-pH anion-exchange chromatography (HPAEC) and electrochemical detection (data not shown). The hydrolysate contained 45.6 μg of Man monosaccharides, whereas the amount of Xyl monosaccharides was below the limit of detection. The GlcA analysis was inconclusive. Since Man comprises 56% of the mass in an average repeating unit from this strain, only 4% of the total Man present in the sample was released by acid hydrolysis as monosaccharides. Thus, the hydrolysis conditions produced only minimal degradation of the sample into free sugars, and the preponderance of Man in the monosaccharide analysis suggested that hydrolysis was limited largely to the mannose backbone.
FIG. 2.
Fluorophore-assisted carbohydrate electrophoresis of GXM from strain B-3501 under different hydrolysis conditions at 95°C. Lane 1, 0.3 M TFA for 5 h; lane 2, 0.1 M TFA for 1 h; lane 3, 0.5 M TFA for 1 h; lane 4, 0.5 M TFA for 5 h; lane 5, no TFA for 5 h. The dark band at the lower edge of the figure is unreacted fluorophore. *, visible oligosaccharide bands in lane 1, 3, and 4.
MS analysis of GXM-derived oligosaccharides.
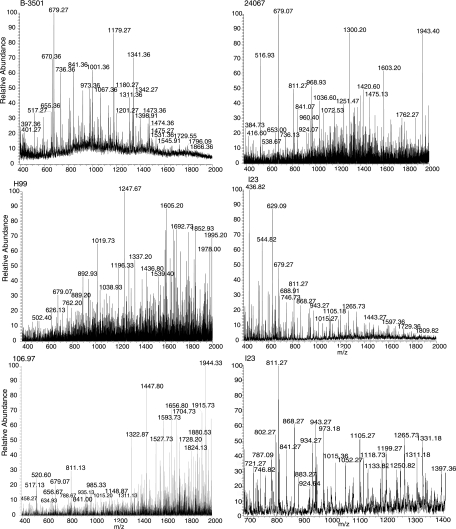
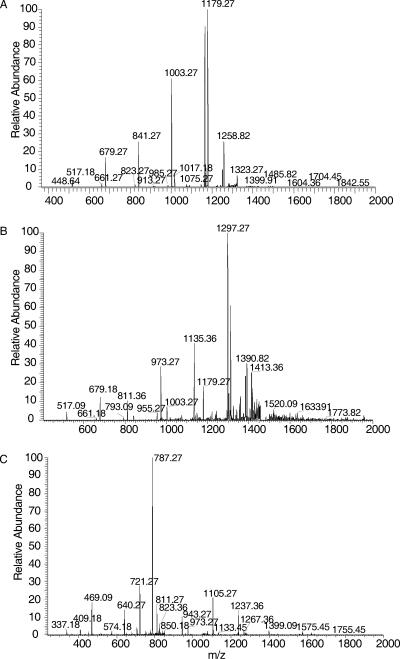
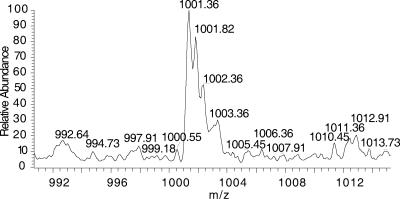
Many of the masses identified by MS were assignable to sugar compositions that agreed with the repeating units described previously (Fig. 1, Fig. 3, and Table 2). This confirmed prior structural inferences about GXM from NMR data and provided confidence in the feasibility of this approach. The analysis was restricted to acidic oligosaccharides, since neither negative nor positive mode ionization detected neutral oligosaccharides. As expected, the 3 Man-to-1 GlcA ratio predicted for the six proposed repeating units (5) was frequently observed in the composition of the oligosaccharides from GXM, confirming earlier NMR findings (Fig. 1 and Table 2). Oligosaccharides that were detected ranged from 2 to 12 sugars, and several retained an acetyl group. Given that the classical GXM triads are composed of three Man residues, oligosaccharides with at least six Man units must represent more than one repeating unit. These oligosaccharides were critical in determining if a structural model of GXM based on the polymerization of identical repeating units was consistent with MS data, since their masses could be compared to those expected from combinations of the published repeating units (Table 3). The six repeating units form 21 possible paired combinations with nine different masses. These nine masses represented groups consisting of only a single repeating unit (groups 1 and 9), combinations of different repeating units exclusively (groups 2, 4, and 8), and combinations inclusive of either a single repeating unit or different repeating units (groups 3, 5, 6, and 7). Masses congruent with groups 1 to 5 were identified in the mass spectra of the hydrolyzed GXM from the different strains. Groups 6 to 9 were not identified. Most importantly, the identification of masses representing groups 1 to 5 suggested that GXM-produced synthesis involved the combination of identical units as well as nonidentical units and that both comprise the GXM from a single strain, as shown by the serotype B and D strains (Table 3). Although secondary analysis of every MS peak by collision-induced dissociation was precluded by the quantity of peaks, analysis of select peaks did confirm the loss of expected sugar masses. Fragmentation of the group-1 and group-2 ions from strain B-3501 GXM and the group-4 ion from I23 displayed m/z values which differed by the mass of GlcA, Xyl, or Man (Fig. 4). The presence of an m/z value of 1,001 in B-3501 GXM further confirmed that the M6 repeating unit is present in GXM as a homopolymeric span of three M6 units (Fig. 5).
FIG. 3.
Representative mass spectra of the GXM hydrolysis products from strains B-3501 (serotype D), 24067 (serotype D), H99 (serotype A), I23 (serotype B), and 106.97 (serotype C). The bottom right panel shows a close-up of the I23 spectrum between a m/z of 700 and 1,400.
TABLE 2.
m/z peaks observed with GXM from different strains of C. neoformans and proposed composition for the oligosaccharide represented by the peaka
| m/z valueb | Strain | Serotype | Proposed composition |
|---|---|---|---|
| 397.4 | B-3501 | D | Man1GlcA1 + 1 acetyl |
| 516.8 | 106.97 | C | Man2GlcA1 |
| 516.9 | 24067 | D | |
| 517.3 | B-3501 | D | |
| 517.2 | I23 | B | |
| 670.4* | B-3501 | D | Man6GlcA2 |
| 679.1 | 24067 | D | Man3GlcA1 |
| 679.1 | H99 | A | |
| 679.3 | B-3501 | D | |
| 679.3 | I23 | B | |
| 679.3 | 106.97 | C | |
| 736.3* | 24067 | D | Man6GlcA2Xyl1 |
| 736.4* | B-3501 | D | |
| 802.4* | B-3501 | D | Man6GlcA2Xyl2 |
| 802.4* | 24067 | D | |
| 802.4* | I23 | B | |
| 811.1 | 106.97 | C | Man3GlcA1Xyl1 |
| 811.3 | B-3501 | D | |
| 811.3 | 24067 | D | |
| 811.3 | I23 | B | |
| 841.1 | 24067 | D | Man4GlcA1 |
| 841.4 | B-3501 | D | |
| 868.4* | I23 | B | Man6GlcA2Xyl3 |
| 883.4* | B-3501 | D | Man7GlcA2Xyl2 |
| 934.3* | I23 | B | Man6GlcA2Xyl4 |
| 935.1*c | 106.97 | C | |
| 943.3 | I23 | B | Man3GlcA1Xyl2 |
| 973.4 | 24067 | D | Man4GlcA1Xyl1 |
| 973.5 | B-3501 | D | |
| 1,001.4* | B-3501 | D | Man9GlcA3 |
| 1,038.9 | H99 | A | Man4GlcA1Xyl1 + 1 acetyl + 1 Na+ |
| 1,067.9 | B-3501 | D | Man9GlcA3Xyl1 |
| 1,148.9 | 106.97 | C | Man4GlcA2Xyl1 |
| 1,179.3 | B-3501 | D | Man5GlcA2 |
| 1,199.8 | I23 | B | Man5GlcA1Xyl1 + 1 acetyl + 1 Na+ |
| 1,199.9 | 106.97 | C | |
| 1,201.3 | B-3501 | D | Man5GlcA2 + 1 Na+ |
| 1,341.4 | B-3501 | D | Man6GlcA2 |
| 1,473.4 | B-3501 | D | Man6GlcA2Xyl1 |
| 1,484.8 | 24067 | D | Man5GlcA2Xyl2 + 1 acetyl |
| 1,605.2 | H99 | A | Man6GlcA2Xyl2 |
| 1,605.3 | I23 | B | |
| 1,605.4 | B-3501 | D | |
| 1,636.1 | 24067 | D | Man7GlcA2Xyl1 |
| 1,664.5 | B-3501 | D | Man8GlcA1Xyl1 + 1 acetyl |
| 1,692.7 | H99 | A | Man6GlcA1Xyl4 |
| 1,944.3 | 106.97 | C | Man7GlcA3Xyl2c |
Mass spectra were obtained in the negative mode in 50% MeOH/0.5% NH4OH. Peaks from different strains representing the same oligosaccharide are listed below the first entry.
*, values for doubly charged ions.
Tentative identification.
TABLE 3.
Masses of oligosaccharides from GXM demonstrate different combinations of repeating units
| Group | Expected m/z value(s)a | Mass (g/mol) | Possible repeating unit combinations | Strain(s) (serotype) containing the oligosaccharideb |
|---|---|---|---|---|
| 1 | 670.2,* 1,341.4 | 1,342.4 | M6 + M6 | B-3501 (D); 24067 (D) |
| 2 | 736.2,* 1,473.4 | 1,474.4 | M1 + M6 | B-3501 (D); 24067 (D) |
| 3 | 802.3,* 1,605.5 | 1,606.5 | M1 + M1, M2 + M6 | B-3501 (D); 24067 (D); H99 (A); I23 (B) |
| 4 | 868.3,* 1,737.5 | 1,738.5 | M1 + M2, M3 + M6, M5 + M6 | I23 (B) |
| 5 | 934.3,* 1,869.6 | 1,870.6 | M1 + M5, M1 + M3, M2 + M2, M4 + M6 | I23 (B); 106.97 (C)c |
| 6 | 1,000.3* | 2,002.6 | M1 + M4, M2 + M3, M2 + M5 | NI |
| 7 | 1,066.3* | 2,134.6 | M2 + M4, M3 + M3, M3 + M5, M5 + M5 | NI |
| 8 | 1,132.4* | 2,266.7 | M3 + M4, M4 + M5 | NI |
| 9 | 1,198.4* | 2,398.7 | M4 + M4 | NI |
*, values for doubly charged ions.
NI, not identified.
Tentative identification.
FIG. 4.
The MS-MS spectra of m/z of 1,341 and 1,473 from B-3501 GXM and a m/z of 868 from I23 GXM. The fragmentation patterns show the loss of Man, GlcA, and Xyl from the ions, confirming the oligosaccharide composition. (A) A m/z of 1,341, singly charged ion; (B) m/z of 1,473, singly charged ion; (C) m/z of 868, doubly charged ion.
FIG. 5.
Mass spectrum m/z of 1,001 from B-3501 GXM. This ion represents an oligosaccharide of 2,004 g/mol mass, owing to its double ionization. The double ionization is observable by the difference of a m/z of 0.5 between ionized peaks, instead of the anticipated m/z of 1 for singly ionized peaks. The predicted composition is Man9GlcA3, equivalent to three M6 repeating units.
Possible concerns and limitations of MS analysis.
The spectra exhibited more intense baseline noise than control samples not containing GXM, and some assignable peaks were of lower abundance than peaks of unknown compositions. To control for the quality of the GXM samples and to identify the source of the noise within the spectra, GXM was analyzed for possible contaminants. Neither protein nor DNA contamination was detectable by Coomassie blue or ethidium bromide detection, respectively. Carbon and hydrogen elemental analysis demonstrated a molar ratio of 1:1.84 to 1:1.98, consistent with the samples being composed of almost pure carbohydrates. The nitrogen content of the samples ranged from 0.006 to 0.05 mol per mole carbon, essentially ruling out significant protein or DNA contamination. Two independent preparations of GXM from strain H99, which contained the highest amount of nitrogen, were evaluated for phosphorus content. The nitrogen-to-phosphorus molar ratio in these samples was approximately 7:1, much lower than the expected ratio for DNA (∼1.25:1). However, contamination during the charge-based precipitation step of the isolation procedure by small amounts of DNA, peptide, or non-GXM acidic polysaccharide material cannot be completely eliminated by the established protocol and would not have been detected by less sensitive methods. However, the greatest potential source of contamination comes from the nitrogen-containing chemical hexadecyltrimethylammonium bromide (CTAB) used during the GXM precipitation step. CTAB is notoriously difficult to remove from GXM during dialysis. Consequently, some of the noise in the MS spectra may have been from CTAB adducts of GXM-derived oligosaccharides in the sample. Despite these difficulties, the MS results produced interpretable results.
GXM from phenotypic switch variants manifests differences in the GXM repeat unit not detected by NMR.
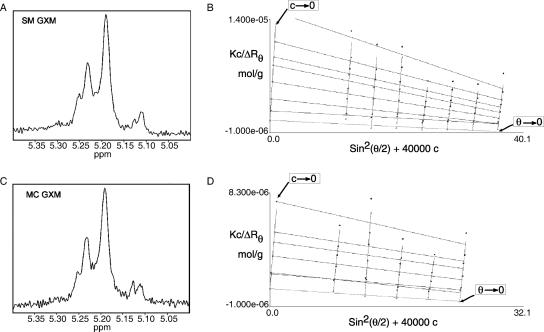
The heterogeneity in GXM repeat units in the above-described results received additional support in studies of GXM from isogenic switch variants of C. neoformans strain RC-2. This strain switches from a parent smooth (SM) to a more virulent mucoid (MC) colony phenotype during chronic infection (9, 10). 1H-NMR spectroscopy of SM and MC GXM revealed that both of the PSs were composed of the M1 repeat (Fig. 6A and C). Identical structures were unexpected as the biophysical and immunomodulatory abilities of SM and MC GXM differed greatly and NMR-detectable changes in GXM repeat were reported in other switch variants with increased virulence (8, 9, 23).
FIG. 6.
1H-NMR and light scattering analyses of GXM from smooth and mucoid phenotypic switch variants, strain RC-2, do not suggest differences in the repeating units or molecular masses. (A) 1H-NMR of GXM from the smooth phenotypic variant. (B) Zimm plot of light scattering data obtained from the GXM from the smooth phenotypic variant. (C) 1H-NMR of GXM from the mucoid phenotypic variant. (D) Zimm plot of light scattering data obtained from the GXM from the mucoid phenotypic variant.
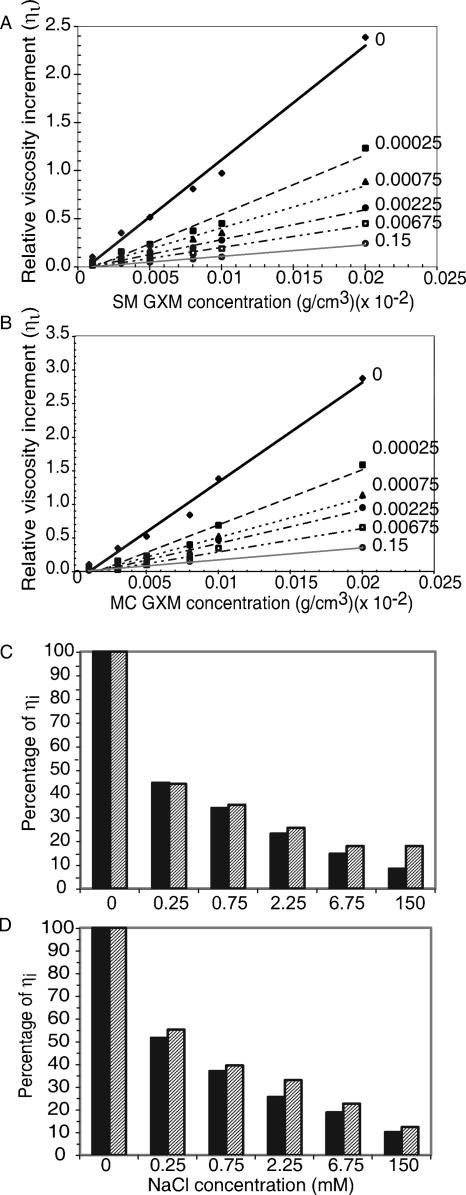
Despite identical NMR patterns, biophysical properties of the PS differed, and size-exclusion chromatography revealed that SM and MC GXM eluted at different times, suggesting a molecular mass difference (10). However, extrapolation to mass standards can be inaccurate as multiple hydrodynamic properties affect particle flow. Consequently, their masses were measured by light scattering as described previously (20), which makes no hydrodynamic assumptions nor requires a mass standard. The molecular masses of the SM and MC GXM were (1.9 ± 0.2) × 106 and (2.3 ± 0.2) × 106 g/mol, respectively, which were within the error of the method. This implied that the differences observed by column chromatography originated from different hydrodynamic characteristics. Consistent with this notion, differences were discovered in the Rg for SM and MC GXMs, 369 nm and 298 nm, respectively. The Rg is the average distance from the center point to the outer edge of the molecule in solution and was calculated from the light scattering data. A difference in Rg despite comparable mass suggested a difference in secondary structure. We measured Rg differences in a nonionic solvent where repulsion between the negatively charged GlcA sugars would be maximized. Hence, it was reasonable to hypothesize that differences in electrostatic repulsion existed and were caused by difference in the spacing of negatively charged GlcA. Consequently, we predicted that viscosities of GXM from SM and MC strains would be different if measured in solutions of different ionic strength, as cations neutralized the negative charges. The relative viscosity increment (ηi) was expected to decrease for both SM and MC GXM (Fig. 7A and B). The SM and MC measurements were compared directly by arbitrarily assigning the ηi at an ionic strength of 0 to 100% and determining the percentage of change of ηi at different ionic strengths (Fig. 7C and D). SM GXM had a larger decrease in ηi than the MC GXM in response to the changing salt concentrations. From this result, we inferred that the larger Rg of SM GXM was due to greater electrostatic repulsion within the PS. The elementary compositions of both SM and MC GXM were identical (data not shown), and therefore, repulsion can only be caused by the proximity of GlcA sugars. From the viscosity data, we conclude that the spacing of GlcA along the Man backbone must differ between the GXM of the SM and that of the MC strain. Because overall sugar ratios and elementary composition do not differ for SM and MC GXM and because NMR measures an average repeat unit over the entire Man backbone, these important structural difference are not revealed by standard 1H-NMR.
FIG. 7.
The viscosity of the smooth GXM solution decreased more rapidly than the mucoid GXM solution in the presence of salt. (A and B) The relative viscosity increment was determined for various concentrations of GXM from the smooth (A) or mucoid (B) variants. GXM was dissolved in NaCl solutions of different ionic strengths. Ionic strength is listed to the right of the linear regression line for each concentration series. (C and D) Comparison of the relative viscosity increment for GXM from the smooth (solid bars) and mucoid (gray hatched bars) variants at densities of 5 × 10−5 g/cm3 (C) or 2 × 10−4 g/cm3 (D). The accurate reproducibility of sample flow times results in minimal standard error for ηi, which is not visible when plotted.
Antigenic variation within the capsule.
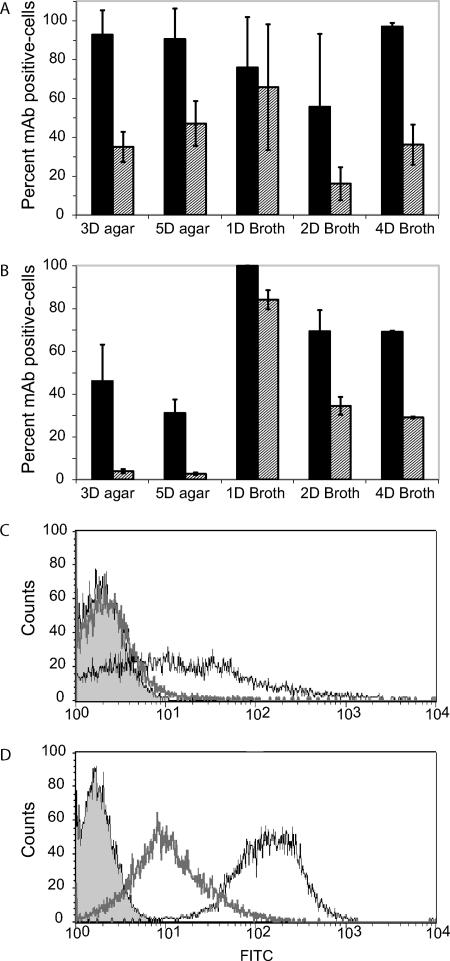
To obtain additional evidence for diversity in the capsular polysaccharide of C. neoformans, we examined the expression of PS epitopes in serotype D and serotype C strains during different growth phases as revealed by MAb binding. MAb 12A1 is a protective antibody that recognizes more than one epitope and binds to both native and de-O-acetylated GXM (19). MAb 21D2 is a nonprotective antibody that binds preferentially to de-O-acetylated GXM (2). Although capsular heterogeneity exists within a given population of cells, these studies determined a great variability of antibody binding during different growth phases. In strain B-3501, the Bonferroni correction of an ANOVA indicated that MAb 21D2 binding increased significantly during logarithmic growth in broth compared to growth on agar. Binding decreased once the yeast cells transitioned into stationary growth, where it eventually increased again to the baseline level, as seen on agar growth. In contrast, there was significantly less variation in MAb 12A1 binding to this strain as a function of culture growth. Antibody binding to serotype C strain 106.97 also varied significantly. Cells from strain 106.97 colonies bound very little MAb 21D2 relative to that of cells in liquid broth, where binding decreased when cells entered stationary phase. Similar staining patterns were also observed for MAb 12A1, although binding was also detected in cells isolated from agar colonies (Fig. 8). In combination, these data suggest that the epitope for MAb 21D2 is variably expressed in the capsule of the serotype C strain and that of the D strain and is most abundant during logarithmic growth in broth cultures.
FIG. 8.
Expression of epitopes in GXM for MAbs 12A1 and 21D2 under different growth conditions. (A and B) Percentage of cells stained positive for either MAb 12A1 (solid bars) or 21D2 (gray hatched bars) as determined by FACS analysis. Error bars represent the standard errors. (A) Serotype D strain B-3501; (B) serotype C strain 106.97. The growth conditions are listed by the number of days (D) of incubation and the medium used (agar or broth). Data are averages of two to three experiments. (C and D) Histograms from a representative experiment of the FACS data obtained for MAb 21D2 (thick gray line, unfilled) and MAb 12A1 (thin black line, unfilled) after 3 days of growth on agar (C) and 1 day of growth in broth (D) for strain 106.97. The unstained population is represented by the filled area.
DISCUSSION
GXM accounts for almost 90% of the PS capsule mass of C. neoformans (3). The capsule is an essential virulence factor that protects the fungal cells from phagocytosis and immune responses (6, 26). In recent years, several studies have shed considerable light on the architecture of C. neoformans polysaccharide capsule (13, 22, 29). Although much work has been done to elucidate pathways of sugar addition and to identify genes responsible for capsule synthesis (16, 17), the biochemistry of GXM polymer synthesis is largely unknown. In this regard, knowledge of the final GXM structure is essential for interpreting biochemical data and clarifying the synthetic pathways. Based upon what is known about glycan and polysaccharide synthesis in other systems, three broad models of GXM synthesis in C. neoformans can be hypothesized: (i) the polymerization of presynthesized oligosaccharide units, (ii) the processive extension of an oligosaccharide through glycosyl transferase reactions, or a combination of presynthesized oligosaccharide units and (iii) processive extension of the oligosaccharide. Capsules are common among prokaryotes, and the synthesis of bacterial capsular PSs involves polymerization of a single repeating unit. In contrast, C. neoformans GXM has at least six repeating units (5). This raises the question of whether six different types of homogenous GXM are synthesized or whether a single PS molecule can contain more than one repeat type. Unfortunately, NMR cannot easily resolve this question because the analysis reflects average repeats of a given PS. Fortunately, the fact that each constituent of GXM has a different mass allows the application of MS to this structure problem. Due to the accuracy of mass measurements, the probability that such peaks are not GXM derived is extremely small.
MS revealed that the Man/GlcA ratio of GXM-derived oligosaccharides was consistent with the averaged 3:1 ratio predicted from the repeat structures by NMR. The identification here of numerous oligosaccharides with the expected masses provided additional sequence-level information and the first independent confirmation that the triad scheme for GXM structure is fundamentally correct. Our analysis also provided evidence for intramolecular structural heterogeneity. Since there is no evidence for microbial capsule synthesis via the sequential joining of monosaccharide units, a conservative assumption is that the repeating units are first synthesized and then joined together to form the C. neoformans capsule. The copolymerization of different repeating units was deduced from the mass analysis of oligosaccharides derived from GXM originating from one serotype B strain and two serotype D strains. The most straightforward interpretation for the unique oligosaccharide masses detected by MS is that these arise from the presence of different repeating units in one molecule. The doubly charged ion at an m/z of 868, discovered in strain I23, corresponded to the mass expected for an oligosaccharide with the composition of distinct repeating units. Thus, even without knowing the precise structural detail of this strain, the MS analysis of GXM provided evidence for a synthetic process that was dissimilar to that of prokaryotes. MS analysis of oligosaccharides derived after partial hydrolysis from the serotype D strains demonstrated that homopolymeric tracts of M6 exist within the M1 copolymer (e.g., m/z of 1,341 and 1,001). Homopolymeric tracts of other types of repeating units may also occur but were not distinguishable by this method. It should be noted, however, that the three mass groups identified in the serotype D strains are distinguishable by the mass of one or two Xyl.
HPAEC studies of monosaccharide release from hydrolyzed GXM did not detect Xyl, even though a wide range of hydrolysis products were detected by FACE. Thus, we conclude that the serotype D oligosaccharides are representative of the original structure. The reduced sensitivity of the Xyl linkage to hydrolysis was surprising since our initial conditions had not produced oligosaccharides with this critical sugar. However, recent evidence that GXM exists as a fiber containing several molecules of GXM (20) raises the possibility that Xyl is located in the interior spaces of the fibers, a location where it may be relatively less susceptible to acid hydrolysis. If this is the case, a rapid hydrolysis protocol may permit preferential attack of backbone mannose linkages without immediate cleavage of the Xyl linkage. Acid hydrolysis is currently the only available means with which to obtain oligosaccharides from GXM of a size suitable for MS. Interestingly, charge analysis of different regions of the capsule provided evidence that the negative charge of the capsule was not evenly distributed (18). The uneven distribution of charge in the capsule could be the result of homopolymeric tracts of M6, which were identified in the MS analysis of GXM oligosaccharides, a finding that is also consistent with a GXM synthetic model of nonuniform copolymerization of repeating units.
Physical studies of GXM from phenotypic switch variants provided strong support for the conclusions drawn from the MS analysis and, at the same time, demonstrated a structural difference to support the clear association between the variant GXMs and virulence (10, 23). GXM from MC and SM switch variants has indistinguishable NMR profiles, similar sugar and glycosyl linkage compositions (data not shown), and similar masses. However, their Rg values differed, suggesting different secondary structures in nonionic solvents. The differences in viscosity as a function of electrolyte concentration implied a disparity in charge neutralization and repulsion, caused by the interaction of salt and GlcA, and confirmed that secondary structural dissimilarities existed. Since GlcA is the only charged residue in GXM, these differences imply positional differences for GlcA in SM and in MC GXM. The previously defined repeating units state that GlcA is linked to every third Man in the backbone. The viscosity data suggest that at least one GXM does not contain GlcA at the expected interval. The identification of oligosaccharides from other strains that had Man/GlcA ratios of 4:1, 4:2, or 5:2 could also be consistent with deviations from the 3:1 ratio existing. This structural difference between the variants was undetectable by 1H-NMR but is only explainable by copolymerization. The structural studies that deduced the repeating units did not determine the relationship of GlcA to the reducing end of the units. Therefore, if, for example, the M1 repeat exists with GlcA linked either to the nonreducing Man or to the reducing Man [-Manα1,3(GlcAβ1,2)Man α1,3Man (Xylβ1,2) versus -Manα1,3(Xylβ1,2)Man α1,3Man (GlcAβ1,2)-], then copolymerization of these similar units could lead to secondary structural differences. Such a difference would also not be identifiable either by MS of hydrolyzed GXM or by linkage analysis, as neither method could provide details of sequence order that would be necessary. Hence, viscosity measurements provided invaluable structural insight.
By combining repeating units to synthesize a fiber of GXM, the PS capsule of C. neoformans has the potential for enormous structural complexity. For example, the GXM from strain 3501 has a mass of 2.6 × 106 g/mol (20) and contains four of the six identified repeating units (M1, M2, M5, and M6), according to NMR studies (5). Assuming that any repeating unit can occur in any position in the PS and that all Man units have an equal chance of being acetylated, GXM could consist of 162,894 or 5.2 × 103,484 combinations. By expanding the molecular diversity to the capsule (1), there are potentially 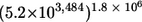 or
or 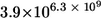 PS combinations on the surface of C. neoformans. These enormous combinatorial estimates do not include unknown structural repeats or the polydispersed size of PSs, either of which could add more complexity. Although the inherent assumption here is that GXM synthesis occurs by polymerized units, a large number of combinations would also occur if synthesis occurred by sequential addition of sugars to a growing oligosaccharide. This could be the case if C. neoformans was utilizing its glycan synthesis machinery to manufacture GXM. Either way, the number of PS structural combinations that could exist in the capsule of C. neoformans approaches infinity, and biological relevance to these calculations is supported by the MS data and the differences in the GXM from phenotypic switch variant.
PS combinations on the surface of C. neoformans. These enormous combinatorial estimates do not include unknown structural repeats or the polydispersed size of PSs, either of which could add more complexity. Although the inherent assumption here is that GXM synthesis occurs by polymerized units, a large number of combinations would also occur if synthesis occurred by sequential addition of sugars to a growing oligosaccharide. This could be the case if C. neoformans was utilizing its glycan synthesis machinery to manufacture GXM. Either way, the number of PS structural combinations that could exist in the capsule of C. neoformans approaches infinity, and biological relevance to these calculations is supported by the MS data and the differences in the GXM from phenotypic switch variant.
By having the flexibility to alter the composition of an individual GXM molecule, C. neoformans demonstrates a unique approach to capsule diversification and construction that distinguishes it from the capsulated bacteria. Combinatorial diversity may involve the synthesis of different repeating units at different intracellular locations, with some potentially present in vesicles (7, 12, 24, 27). Hence, the eukaryotic secretion apparatus may provide the means for generating significantly more complex capsules in fungi than in bacteria and could include a distinct mechanism.
Immunofluorescence studies have shown tremendous variability in capsular staining within a C. neoformans culture (11). Such variability appears to be a dynamic process that is inducible and reversible. Nutrient accessibility does not influence epitope expression since nutrients are readily available from agar or during logarithmic growth in broth. The change in capsule structure, as inferred from MAbs 21D2 and 12A1, during logarithmic growth implies that metabolism, unrelated to nutrient availability, influences GXM synthesis. These changes could affect the outcome of infection if they occurred in vivo. For example, increased expression of specific epitopes during periods of active replication, such as the epitope for MAb 21D2, could elicit antibodies early during human infection that would be ineffective later when the epitopes have changed. Consistent with this, the initial polyclonal response of mice to cryptococcal infection is biased toward antibodies that share a characteristic of nonprotective MAbs (28). The structural changes responsible for the altered expression of these epitopes are not known. However, the acetyl modification of GXM is important for antibody recognition. Acetylation reduces MAb 21D2 binding, and MAb 12A1 can recognize acetyl-dependent and non-acetyl-dependent epitopes (21). Therefore, metabolic changes due to growth conditions may affect the acetyl modification of GXM. A specific motif for acetylation has not been determined, and the extent of acetylation would be predicted to change if the altering of structural combinations resulted in the creation or loss of the acetylation motif.
In summary, the combinatorial capability within the GXM structure provides a new mechanism for C. neoformans to change the physical and antigenic properties of its capsule. Variation in the capsule structure could provide a survival advantage for a fungal population subjected to predation by amoebae in the environment and contribute to their survival in humans.
Acknowledgments
We thank Robert Cherniak for sharing unpublished data and for the 1H-NMR data. Strains I23 and 106.97 were kindly provided by Uma Banerjee, All India Institute of Medical Sciences, and Thomas Mitchell, Duke University, respectively. We thank the Complex Carbohydrate Research Center (CCRC) at The University of Georgia for HPAEC analysis. The CCRC is supported by the DOE Center for Plant and Microbial Complex Carbohydrates, DE-FG09-93ER-20097.
This work was supported by NIH grants AI33774, AI33142, and HL59842 to A.C. and by AI59681 to B.C.F. D.C.M. was supported by a Burroughs Wellcome Fund fellow award through the Life Science Research Foundation.
Footnotes
Published ahead of print on 29 June 2007.
REFERENCES
- 1.Bryan, R. A., O. Zaragoza, T., Zhang, G. Ortiz, A. Casadevall, and E. Dadachova. 2005. Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot. Cell 4:465-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadevall, A., J. Mukherjee, S. J. Devi, R. Schneerson, J. B. Robbins, and M. D. Scharff. 1992. Antibodies elicited by a Cryptococcus neoformans glucuronoxylomannan-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. J. Infect. Dis. 65:1086-1093. [DOI] [PubMed] [Google Scholar]
- 3.Cherniak, R., E. Reiss, and S. Turner. 1982. A galactoxylomannan antigen of Cryptococcus neoformans serotype A. Carbohydr. Res. 103:239-250. [Google Scholar]
- 4.Cherniak, R., and J. B. Sundstrom. 1994. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 62:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherniak, R., H. Valafar, L. C. Morris, and F. Valafar. 1998. Cryptococcus neoformans chemotyping by quantitative analysis of 1H NMR spectra of glucuronoxylomannans using a computer simulated artificial neural network. Clin. Diagn. Lab. Immunol. 5:146-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Poeta, M. 2004. Role of phagocytosis in the virulence of Cryptococcus neoformans. Eukaryot. Cell 3:1067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldmesser, M., Y. Kress, and A. Casadevall. 2001. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 147:2355-2365. [DOI] [PubMed] [Google Scholar]
- 8.Fries, B. C., D. L. Goldman, R. Cherniak, R. Ju, and A. Casadevall. 1999. Phenotypic switching in Cryptococcus neoformans strain 24067A associated with changes in virulence, polysaccharide structure, and cellular morphology. Infect. Immun. 67:6076-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fries, B. C., S. C. Lee, R. Kennan, W. Zhao, A. Casadevall, and D. L. Goldman. 2005. Phenotypic switching of Cryptococcus neoformans can produce variants that elicit increased intracranial pressure in a rat model of cryptococcal meningoencephalitis. Infect. Immun. 73:1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fries, B. C., C. P. Taborda, E. Serfass, and A. Casadevall. 2001. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J. Clin. Investig. 108:1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Hermoso, D., F. Dromer, and G. Janbon. 2004. Cryptococcus neoformans capsule structure evolution in vitro and during murine infection. Infect. Immun. 72:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Rivera, J., Y. C. Chang, K. J. Kwon-Chung, and A. Casadevall. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gates, M. A., P. Thorkildson, and T. R. Kozel. 2004. Molecular architecture of the Cryptococcus neoformans capsule. Mol. Microbiol. 52:13-24. [DOI] [PubMed] [Google Scholar]
- 14.Goins, T. L., and J. E. Cutler. 2000. Relative abundance of oligosaccharides in Candida species as determined by fluorophore-assisted carbohydrate electrophoresis. J. Clin. Microbiol. 38:2862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda, R., and T. Maeda. 2004. Structural studies of the capsular polysaccharide of a non-neoformans Cryptococcus species identified as C. laurentii, which was reclassified as Cryptococcus flavescens, from a patient with AIDS. Carbohydr. Res. 339:503-509. [DOI] [PubMed] [Google Scholar]
- 16.Janbon, G. 2004. Cryptococcus neoformans capsule biosynthesis and regulation. FEMS Yeast Res. 4:765-771. [DOI] [PubMed] [Google Scholar]
- 17.Klutts, J. S., A. Yoneda, M. C. Reilly, I. Bose, and T. L. Doering. 2006. Glycosyltransferases and their products: cryptococcal variations on fungal themes. FEMS Yeast Res. 6:499-512. [DOI] [PubMed] [Google Scholar]
- 18.Maxson, M. E., E. Dadachova, A. Casadevall, and O. Zaragoza. 2007. Radial mass density, charge, and epitope distribution in the Cryptococcus neoformans capsule. Eukaryot. Cell 6:95-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFadden, D. C., and A. Casadevall. 2004. Unexpected diversity in the fine specificity of monoclonal antibodies that use the same V region gene to glucuronoxylomannan of Cryptococcus neoformans. J. Immunol. 172:3670-3677. [DOI] [PubMed] [Google Scholar]
- 20.McFadden, D. C., M. De Jesus, and A. Casadevall. 2005. The physical properties of the capsular polysaccharides from Cryptococcus neoformans suggest features for capsule construction. J. Biol. Chem. 281:1868-1875. [DOI] [PubMed] [Google Scholar]
- 21.McFadden, D. C., O. Zaragoza, and A. Casadevall. 2004. Immunoreactivity of cryptococcal antigen is not stable under prolonged incubations in human serum. J. Clin. Microbiol. 42:2786-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierini, L. M., and T. L. Doering. 2001. Spatial and temporal sequence of capsule construction in Cryptococcus neoformans. Mol. Microbiol. 41:105-115. [DOI] [PubMed] [Google Scholar]
- 23.Pietrella, D., B. Fries, P. Lupo, F. Bistoni, A. Casadevall, and A. Vecchiarelli. 2003. Phenotypic switching of Cryptococcus neoformans can influence the outcome of the human immune response. Cell. Microbiol. 5:513-522. [DOI] [PubMed] [Google Scholar]
- 24.Rodrigues, M. L., L. Nimrichter, D. L. Oliveira, S. Frases, K. Miranda, O. Zaragoza, M. Alvarez, A. Nakouzi, M. Feldmesser, and A. Casadevall. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 6:48-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitfield, C., and A. Paiment. 2003. Biosynthesis and assembly of Group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr. Res. 338:2491-2502. [DOI] [PubMed] [Google Scholar]
- 26.Yauch, L. E., J. S. Lam, and S. M. Levitz. 2006. Direct inhibition of T-cell responses by the Cryptococcus capsular polysaccharide glucuronoxylomannan. PLoS Pathog. 2:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoneda, A., and T. L. Doering. 2006. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 17:5131-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaragoza, O., and A. Casadevall. 2004. Antibodies produced in response to Cryptococcus neoformans pulmonary infection in mice have characteristics of nonprotective antibodies. Infect. Immun. 72:4271-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaragoza, O., A. Telzak, R. A. Bryan, F. Dadachova, and A. Casadevall. 2006. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol. Microbiol. 59:67-83. [DOI] [PubMed] [Google Scholar]