Abstract
Infection of the chestnut blight fungus, Cryphonectria parasitica, by hypovirus CHV1-EP713 or by reovirus MyRV1-Cp9B21 or MyRV2-CpC18 results in reduced fungal virulence (hypovirulence). However, additional phenotypic changes caused by the two groups of mycoviruses are quite different. We now report that the loss of female fertility and the resulting absence of virus transmission through sexual spores observed after hypovirus infection was not observed for reovirus-infected C. parasitica. Consistent with this result, expression of two genes involved in sexual reproduction, the pheromone precursor gene, Mf2/1, and the yeast STE12-like transcriptional factor gene, cpst12, was less reduced in reovirus-infected strains than in the hypovirus CHV1-EP713-infected strain. Analysis with a custom microarray cDNA chip containing expressed sequence tag clones representing approximately 2,200 unique C. parasitica genes identified 140 and 128 host genes that were responsive to MyRV1-Cp9B21 or MyRV2-CpC18 infection, respectively. Comparison of these virus-responsive genes revealed an overlap of 85 genes, even though the nucleotide sequence identity for the two reoviruses is less than 50%. Significantly, 84 of the 85 genes were altered in the same direction. Further comparison revealed that 51% and 48% of the MyRV1-Cp9B21- and MyRV2-CpC18-responsive genes were also responsive to CHV1-EP713 infection. Finally, similar to results reported for CHV1-EP713 infection, a high percentage (59% and 66%) of the reovirus-responsive genes were also differentially expressed following disruption of the cellular G-protein signal transduction pathway. These data support the hypothesis that hypovirus and reovirus infections perturb common and specific C. parasitica regulatory pathways to cause hypovirulence and distinct sets of phenotypic changes.
Mycovirus infections are widespread throughout the kingdom Fungi and can cause phenotypic changes in their fungal hosts that are fundamentally interesting and of potential practical value. For example, the Curvularia thermal tolerance virus was recently reported to be required for the endophytic fungus Curvularia protuberata to confer heat tolerance on a range of plant hosts (23). Other mycoviruses have been shown to interfere with fungus-plant host pathogenic interactions by attenuating fungal virulence (hypovirulence), thereby providing applications for biological control (recently reviewed in reference 26).
Progress in advancing the molecular biology of mycovirus-mediated alterations of fungus-host interactions has come primarily from studies on members of the mycovirus family Hypoviridae (hypoviruses), responsible for hypovirulence of the chestnut blight fungus Cryphonectria parasitica. This is due to the development of a reverse-genetics system for several hypoviruses (7, 8, 22) and robust DNA transformation protocols for C. parasitica (9). The recent construction of an expressed sequence tag (EST)-based microarray representing approximately 2,200 C. parasitica genes has allowed the monitoring of host transcriptional responses to hypovirus infections and to genetic disruptions of host signaling pathways (1, 11). Approximately 13% of the C. parasitica transcriptome was observed to change in accumulation by at least twofold in response to persistent hypovirus CHV1-EP713 infection (1). Comparison of the changes in transcriptional profiles resulting from CHV1-EP713 infection and from disruption of genes encoding heterotrimeric G-protein α and β subunits confirmed previous reports that hypovirus infection alters host G-protein signal transduction (11). An extension of these studies recently led to the identification of a C. parasitica G-protein-regulated transcription factor, CpST12, which is down-regulated by hypovirus CHV1-EP713 infection and is required for fungal virulence, female fertility, and the regulated expression of a subset of hypovirus-responsive genes (12).
C. parasitica has been shown to support the replication of members of five RNA virus families (19), providing additional utility as a system for examining mycovirus-fungal host interactions. In addition to the family Hypoviridae, these include the Partitiviridae, Chrysoviridae, Narnaviridae, and Reoviridae. The C. parasitica-infecting members of the Reoviridae are particularly interesting because the phenotypic changes that result from their infection are quite distinct from hypovirus-mediated phenotypic changes and because reovirus particles are infectious when introduced into fungal spheroplasts (18). Additionally, the complete genome sequences of reoviruses MyRV1-Cp9B21 (32) and MyRV2-CpC18 (R. Festa, M. Stout, and B. Hillman, unpublished) have been determined.
Hypovirus CHV1-EP713 causes a number of phenotypic changes in addition to reduced virulence (8). These include reduced orange pigmentation, reduced asexual sporulation, and loss of female fertility. While infection by reovirus MyRV1-Cp9B21 or MyRV2-CpC18 also causes a severe reduction in virulence, these viruses do not significantly affect fungal pigment production or conidiation (14, 19). Here we report additional differences in the phenotypic changes resulting from hypovirus and reovirus infections of the common fungal host and examine the effects of these unrelated viruses on C. parasitica gene expression. The results strengthen an emerging view of the relationship between mycovirus-mediated changes in host gene expression and phenotype. They also support the hypothesis that hypovirus and reovirus infections perturb common and specific C. parasitica regulatory pathways to cause hypovirulence and distinct sets of phenotypic changes.
MATERIALS AND METHODS
Fungal strains and growth conditions.
C. parasitica virus-free strain EP155 (ATCC 38755), isogenic hypovirus CHV1-EP713-infected strain EP713 (ATCC 52571), and reovirus-infected EP155 strains MyRV1-Cp9B21/EP155 and MyRV2-CpC18/EP155 were maintained on potato dextrose agar (PDA) (Difco, Detroit, MI) at 22 to 24°C with a 12-h light-dark cycle at 1,300 to 1,600 lx. C. parasitica cultures used for RNA preparations were grown under similar conditions on PDA overlaid with a cellophane membrane (PDA/cellophane). The effect of reovirus and hypovirus infection on hyphal growth was measured under three different growth conditions as described by Kasahara and Nuss (20). Colony diameter was measured during growth on PDA (70 ml per 140-mm petri dish) following inoculation of the center of the plate with a plug of freshly grown mycelia (0.7 cm in diameter) and incubation at 22 to 24°C with a 12-h light-dark cycle at 1,300 to 1,600 lx. Mass was determined for colonies grown on PDA/cellophane and in liquid potato dextrose broth (PDB) medium. Mycelia were stripped from the cellophane membranes and directly weighed without further processing (fresh weight). Liquid PDB cultures (50 ml) were inoculated with a single mycelial plug (0.7 cm in diameter) and incubated without stirring under the same conditions as indicated for PDA plates. Hyphae were collected by filtering through Miracloth (Calbiochem, San Diego, CA), washed with distilled water, dried in a desiccator under vacuum for 2 to 3 days, and weighed (dry weight).
Mating tests.
Mating was performed on autoclaved twigs of American chestnut embedded in 2% water agar as described by Anagnostakis (3). Reovirus-infected strains MyRV1-Cp9B21/EP155 and MyRV2-CpC18/EP155 were tested as both male and female in crosses with EP146. Each cross was performed in triplicate, with EP155 (mating type Mat-2, orange pigmentation) × EP146 (mating type Mat-1, brown pigmentation) crosses serving as controls. The twigs were mechanically scored multiple times with a scalpel prior to inoculation to ensure effective colonization by the strains serving as the female parent in the crosses. These twigs were incubated at 20 to 22°C until perithecia could be observed in the stroma.
To determine if reoviruses were transmitted through the sexual cycle into ascospores, perithecia were detached from the stroma using a dissecting needle. Each perithecium was rolled over 4% water agar to remove stromatal debris. Perithecia were then transferred to autoclaved microscopic slides, washed with sterile, distilled water, and squashed with a blunt glass rod. Ascospores were carefully transferred into test tubes, serially diluted, and plated on PDA plates. Single ascospore cultures were isolated after incubation for 36 h, and the phenotype was observed after incubation at 22°C for 1 week under a 12-h light photoperiod. Reovirus infection was further confirmed by extraction of viral double-stranded RNAs (dsRNAs) as described elsewhere (18).
Nucleic acid preparation and analysis.
Cultures grown on PDA/cellophane for a week were harvested, frozen in liquid nitrogen, and ground into a fine powder using a mortar and pestle. RNA isolation was performed as described by Allen et al. (1). Real-time PCR analysis of transcript accumulation was performed by using TaqMan reagents and a 7300 real-time PCR system (Applied Biosystems, Foster City, CA) as described previously (27). Probes and oligonucleotides for the particular clones to be tested were obtained from Integrated DNA Technologies (Coralville, IA). Real-time reverse transcription-PCR (RT-PCR) was performed at least twice, in triplicate for each transcript, with at least two independent RNA preparations, with primers and probes specific for 18S rRNA and the target genes. Calculations of transcript accumulation values in the mutants relative to those in strain EP155 were performed by using the comparative threshold cycle method as described previously (27), using the 18S rRNA values to normalize for variations in template concentration.
Microarray fluorescent probe generation, hybridization, and scanning.
Fluorescence-labeled cDNA probes were prepared from total RNA (25 μg per probe) by the direct incorporation of Cy3- or Cy5-dUTP using a CyScribe first-strand cDNA labeling kit (Amersham Biosciences, Piscataway, NJ) primed with oligo(dT) according to the manufacturer's instructions. Unincorporated nucleotides were removed with a Microcon-30 spin column, and probes were processed according to the method of Allen et al. (1). Prehybridization, hybridization, and posthybridization wash steps were performed as suggested by the manufacturer of the GAPS II slides (Corning, Lowell, MA). Each hybridized chip was scanned in both the Cy3 and Cy5 channels with an Affymetrix 418 scanner as described by Allen et al. (1).
Microarray data analysis and management.
Scanned images were interpreted with Spotfinder (version 1.0) (http://www.tigr.org). Further data processing and analyses were performed as described by Deng et al. (12). Briefly, normalization of raw fluorescent signal was achieved by utilizing algorithms written in Mathematica 5.1 (Wolfram Research, Chicago IL). Dye bias effects were corrected utilizing LOWESS, a local linear regression algorithm, while differences between repeated hybridizations were managed by standard deviation regularization, i.e., by rescaling each spot's Cy3 and Cy5 measurement using the standard deviation of Cy3 and Cy5 values as calculated across all experiments. A gene was designated differentially expressed if its representative spot on the microarray chip produced a log2 ratio of ≥1 standard deviations from the average log2 ratio of each data set in a minimum of three of four hybridizations. Comparisons between multiple profiling experiments and subsequent creation of differentially expressed clone lists were performed as previously described (12).
Microarray accession number.
The microarray data determined in this study have been submitted to the Gene Expression Omnibus at the National Center for Biotechnology Information under series accession number GSE 7555.
RESULTS
Reovirus-infected C. parasitica strains are fertile and transmit virus to ascospore progeny.
C. parasitica strains infected with hypoviruses CHV1, -2, -3, and -4 are unable to serve as females in mating experiments and fail to transmit hypoviruses to ascospore progeny (4, 6, 17, 30). In contrast to the mating deficiency described for hypovirus-infected C. parasitica strains, reovirus-infected strains MyRV1-Cp9B21/EP155 and MyRV2-CpC18/EP155 produced mature perithecia when serving as either the male or the female parent in a mating experiment (data not shown). The control crosses between EP155 and EP146 and the crosses in which reovirus infected-isolates served as the male produced abundant perithecia after 4 to 5 weeks of incubation. Crosses in which the reovirus-infected isolates served as the female partner took longer to produce perithecia, 7 to 8 weeks, and produced only ∼20% as many mature perithecia as were produced in control crosses. However, the ascospores were viable, and a significant number of ascospore-derived colonies were reovirus infected (Table 1).
TABLE 1.
Transmission of reoviruses to ascospore progeny of Cryphonectria parasitica in crosses in which the reovirus-infected strain served as the female parent
| Reovirus | Total no. of ascospores | No. of infected colonies | % Colonies infected |
|---|---|---|---|
| MyRV1-CpC18 | 124 | 71 | 57.30 |
| MyRV2-Cp9B21 | 108 | 66 | 61.10 |
C. parasitica strain EP146 produces a distinctive brown pigment as a monogenic nuclear trait rather than the normal orange pigment produced by EP155 colonies (6). As a result, the ascospore progeny derived from perithecia resulting from crosses in which a reovirus-infected strain served as the female produced brown and orange colonies at a ratio of 1:1. Reovirus-infected phenotypes were observed for both orange and brown ascospore-derived cultures. The presence of reovirus in the progeny was confirmed by extraction of the 11 viral dsRNAs characteristic of MyRV1 or MyRV2 from selected cultures (data not shown). However, transmission of reoviruses to ascospore progeny was not 100% as shown in Table 1 but ranged from 57.3% to 61.1% (Table 1). Thus, reoviruses MyRV1-Cp9B21 and MyRV2-CpC18, unlike hypoviruses, do not cause a complete loss of female fertility and are transmitted in significant numbers to ascospore progeny.
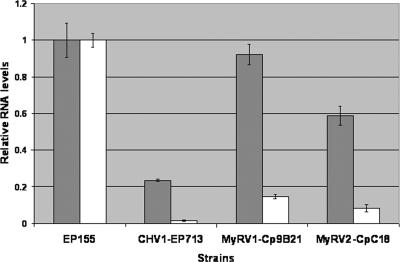
The sex pheromone precursor gene Mf2/1 and the yeast Ste12-like transcription factor gene cpst12 are both required for the mating process in C. parasitica (12, 33), and the expression of both genes has been reported to be down-regulated following hypovirus infection (12, 33). Thus, it was of interest to examine the effect of reovirus infection on Mf2/1 and cpst12 transcript levels. Real-time RT-PCR results revealed that Mf2/1 transcript levels were reduced to 2% of the levels present in the virus-free isolate EP155 as a result of hypovirus CHV1-EP713 infection (Fig. 1). Mf2/1 transcript levels were four- and eightfold higher in EP155 infected with the reoviruses MyRV1-Cp9B21 and MyRV2-CpC18, respectively, than in CHV1-EP713-infected EP155 (Fig. 1). Similarly, hypovirus CHV1-EP713 infection was found to cause a greater reduction in cpst12 transcript levels than did infection by either of the reoviruses (Fig. 1). These results are consistent with the observations that the reoviruses MyRV1-Cp9B21 and MyRV2-CpC18 delay perithecial formation rather than causing female infertility as does the hypovirus CHV1-EP713, providing the means for reovirus transmission to ascospore progeny.
FIG. 1.
Real-time semiquantitative RT-PCR analysis of transcript levels for the sex pheromone precursor gene Mf2/1 (open bars) and the transcription factor gene cpst12 (solid bars) in hypovirus- and reovirus-infected C. parasitica. Transcript accumulation levels are shown for virus-infected strains, normalized to the transcript levels in the virus-free strain EP155. The reference value of 1.0 for transcript levels in EP155 was calculated as described in Materials and Methods. Bars represent standard errors for each data set.
Comparative effects of hypovirus and reovirus infections on C. parasitica growth and colony morphology.
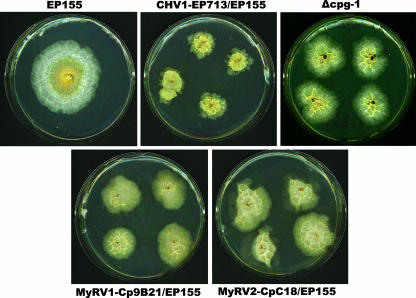
Hillman and colleagues (18) previously compared the phenotypic changes caused by the hypovirus CHV1-EP713 and the reovirus MyRV1-Cp9B21. The reovirus MyRV1-Cp9B21 was found to reduce C. parasitica virulence even more than the hypovirus CHV1-EP713 but to have little effect on fungal asexual sporulation or orange pigment production, two traits that are significantly reduced following CHV1-EP713 infection. Hillman et al. (18) also reported that MyRV1-Cp9B21, like CHV1-EP713, reduces the hyphal growth rate on solid synthetic media. It has been shown previously that culture conditions can influence the magnitude and spectrum of hypovirus-induced symptoms and virus-mediated alterations of C. parasitica gene expression (discussed in reference 27). Thus, it was of interest to examine the effect of culture conditions on the growth and colony morphology of reovirus-infected C. parasitica prior to the analysis of reovirus-mediated effects on C. parasitica gene expression. Growth of the reovirus-infected strains was examined under three different conditions and compared with those of isogenic, virus-free, and CHV1-EP713-infected EP155. As previously reported, hypovirus CHV1-EP713-infected colonies grown on PDA expanded at a rate approximately 20% slower than that for isogenic virus-free strain EP155 (17, 20). Strains infected with the two reoviruses MyRV1-Cp9B21 and MYRV2-CpC18 also grew slower than EP155, with reductions of 28.3% and 15% compared to growth of virus-free strain EP155 (Table 2). Similar results were observed for growth in liquid medium (PDB); reduction in dry weight ranged from 18.0% to 24.0% for hypovirus- and reovirus-infected strains. In contrast, colony masses on PDA/cellophane were significantly different for virus-free strain EP155 and the three virus-infected strains (63.1% reduction for CHV1-EP713, 85.7% for MyRV1-Cp9B21, and 58.1% for MyRV1-CpC18) (Table 2). The slow growth of virus-infected strains on cellophane was accompanied by the formation of ridges on the colony face, giving a “crinkled” appearance. Interestingly, this distinctive colony morphology is exhibited by both reovirus-infected strains and the CHV1-EP713-infected strain and closely resembled the morphology of colonies of a C. parasitica strain in which the gene encoding the G-protein α subunit CPG-1 has been disrupted (Fig. 2).
TABLE 2.
Effect of hypovirus CHV1-EP713 and reoviruses MYRV1-Cp9B21 and MyRV2-CpC18 infections on hyphal growth under three different conditionsa
| Strainb | Diameter of PDA plates (mm) | Growth of strain in:
|
|
|---|---|---|---|
| PDA/cellophane plates (mg [fresh wt]) | Liquid PD broth (mg [dry wt]) | ||
| EP155 | 63.5 | 320.0 | 63.8 |
| CHV1-EP713/EP155 | 49.8 | 114.8 | 48.5 |
| MyRV1-Cp9B21/EP155 | 45.5 | 45.8 | 48.8 |
| MyRV2-CpC18/EP155 | 54.0 | 134.5 | 52.3 |
Measurements were taken at days 3, 5, and 7 after inoculation; only data for day 7 are presented. Values represent a minimum of four replicates. Growth conditions and statistical analysis were as described in Material and Methods. For diameter of PDA plates, standard error is 1.61 mm; least significant difference is 5.08 mm. For growth in PDA/cellophane plates, standard error is 2.6 mg (fresh weight); least significant difference is 8.2 mg. For growth in liquid PD broth, standard error is 3.2 mg (dry weight); least significant difference is 1.0 mg. Two means that differ by an amount greater than the least significant difference are declared to be significantly different with a probability of type I error (α) of 0.05.
EP155 is a virus-free strain.
FIG. 2.
Distinctive crinkled colony morphology exhibited by C. parasitica infected with hypovirus CHV1-EP713 or reovirus MyRV1-Cp9B21 or MyRV2-CpC18 when grown on PDA overlaid with cellophane. The same colony morphology is exhibited by the G-protein α-subunit Δcpg-1 deletion mutant in the absence of virus infection.
Similar transcriptional profiles for strains infected by MyRV1-Cp9B21 and MyRV2-CpC18.
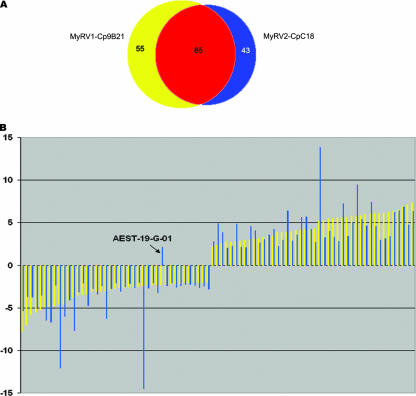
A custom microarray chip (1) containing EST clones representing ca. 2,200 unique C. parasitica genes was used to monitor host transcriptional responses to infection by the two reoviruses. Of the 2,200 genes, 140 were scored as being differentially expressed in response to MyRV1-Cp9B21 infection (72 down-regulated and 68 up-regulated, available at (http://www.umbi.umd.edu/∼cbr/deng/MyRV1Cp9B21.pdf). Similarly, microarray analysis revealed that 128 genes were differentially expressed in the reovirus MyRV2-CpC18-infected isolate (65 down-regulated and 63 up-regulated, available at (http://www.umbi.umd.edu/∼cbr/deng/MyRV2CpC18.pdf). Comparison of the genes responsive to MyRV1-Cp9B21 and MyRV2-CpC18 infections revealed an overlap of 85 genes, representing 60.7% of the total for MyRV1-Cp9B21 and 66.4% of the total for MyRV2CpC18 (Fig. 3A). The list of these genes is presented in Table 3 under headings of putative biological processes as assigned by Dawe et al. (10) according to the classification guidelines outlined by the Gene Ontology Consortium (http://www.geneontology.org) and as previously reported for CHV1-EP713-responsive genes by Allen et al. (1).
FIG. 3.
(A) Venn diagram showing overlap of expression profiles for the two reoviruses MyRV1-Cp9B21 and MyRV2-CpC18. A total of 85 genes were found on both lists of differentially expressed clones. (B) Graphical representation of the relative magnitude of microarray-predicted changes in transcript accumulation for the 85 identified common reovirus-responsive C. parasitica genes (from Table 2). Yellow-shaded bars indicate the magnitude of transcript accumulation change following MyRV1-Cp9B21 infection (n-fold change, y axis), whereas the magnitude of change for the same genes after MyRV2-CpC18 infection is indicated by the blue bars.
TABLE 3.
C. parasitica genes responsive to infection by both MyRV1-Cp9B21 and MyRV2-CpC18a
| Function and AEST | Fold change in expression with infection by:
|
E value | Description (organism) | |
|---|---|---|---|---|
| MyRV1-Cp9B21 | MyRV2-CpC18 | |||
| Alcohol metabolism | ||||
| AEST-34-H-08 | 6.10 | 2.96 | 1.7E−1 | FAD binding family protein (Francisella tularensis subsp. tularensis) |
| Carbohydrate metabolism | ||||
| AEST-08-F-03 | −2.01 | −2.63 | 1.1E−104 | Hypothetical protein (Neurospora crassa) |
| 6.0E−92 | Phosphoglucomutase (Aspergillus oryzae) | |||
| AEST-11-B-12 | −2.01 | −2.48 | 6.7E−82 | Conserved hypothetical protein (Gibberella zeae PH-1) |
| 2.0E−70 | UDP-glucose pyrophosphorylase (Emericella nidulans) | |||
| AEST-26-B-12 | −2.21 | −2.06 | 1.5E−34 | d-Mannonate oxidoreductase (Aspergillus fumigatus Af293) |
| AEST-37-F-03 | −2.31 | −2.07 | 1.6E−37 | Beta-glucosidase homolog (Cochliobolus heterostrophus) |
| Cell cycle | ||||
| AEST-06-F-09 | 2.72 | 2.03 | 3.0E−68 | Hypothetical protein (Neurospora crassa) |
| 5.0E−65 | Cell division control protein Cdc48 (Aspergillus fumigatus Af293) | |||
| AEST-07-B-12 | 3.97 | 2.83 | 2.7E−89 | GTP-binding nuclear protein RAN/TC4 (Gibberella zeae PH-1) |
| Cytoplasm organization and biogenesis | ||||
| AEST-39-A-08 | 3.91 | 2.97 | 2.1E−96 | Dynamin-related protein |
| Electron transport | ||||
| AEST-18-F-06 | −2.14 | −2.40 | 5.9E−62 | Related to flavin-containing monooxygenase (Neurospora crassa) |
| AEST-34-F-11 | −4.56 | −6.03 | 4.2E−94 | Hypothetical protein MG03155.4 (Magnaporthe grisea 70-15) |
| 2.0E−93 | NUAM_NEUCR NADH-ubiquinone oxidoreductase 78-kDa subunit (Gibberella zeae PH-1) | |||
| External protective structure organization and biogenesis | ||||
| AEST-03-D-12 | −4.86 | −6.68 | 4.5E−32 | Cell wall protein (PhiA) |
| AEST-09-D-12 | −4.91 | −6.44 | 1.6E−19 | 22-kDa glycoprotein (Ophiostoma novo-ulmi) |
| AEST-14-D-02 | −3.06 | −3.41 | 1.2E−64 | Hypothetical protein MG10400.4 (Magnaporthe grisea 70-15) |
| 8.0E−52 | Cell wall glucanase (Aspergillus fumigatus Af293) | |||
| Glutathione metabolism | ||||
| AEST-12-G-04 | 3.07 | 4.10 | 1.3E−63 | Theta class glutathione S-transferase (Aspergillus fumigatus Af293) |
| Lipid metabolism | ||||
| AEST-09-B-11 | 4.12 | 3.55 | 4.9E−48 | Hypothetical protein MG01790.4 (Magnaporthe grisea 70-15) |
| 1.0E−33 | Acyl-coenzyme A desaturase (Coccidioides immitis RS) | |||
| Metabolism/nucleobase | ||||
| AEST-05-F-11 | −5.51 | −4.99 | 2.3E−83 | NUP3_PENSQ nuclease PA3 (endonuclease PA3) (DNase PA3) |
| AEST-24-C-10 | −5.24 | −3.53 | 2.9E−30 | JX0127 RNase M (EC 3.1.27.-) (Aspergillus phoenicis) |
| One carbon compound metabolism | ||||
| AEST-01-H-09 | 2.76 | 2.24 | 6.2E−78 | Hypothetical protein MG05155.4 (Magnaporthe grisea 70-15) |
| 3.0E−74 | Adenosyl homocysteinase (Chaetomium globosum CBS 148.51) | |||
| Oxygen and radical metabolism | ||||
| AEST-11-E-09 | 6.03 | 7.43 | 6.4E−38 | Cytochrome P450 (Aspergillus fumigatus Af293) |
| Phosphate metabolism | ||||
| AEST-40-B-12 | −3.02 | −2.43 | 3.1E−33 | Hypothetical protein MG05692.4 (Magnaporthe grisea 70-15) |
| Protein metabolism | ||||
| AEST-01-D-05 | −2.72 | −2.08 | 1.1E−62 | Aspartyl proteinase (Trichoderma asperellum) |
| AEST-03-G-06 | −4.08 | −4.05 | 1.0E−68 | Acid proteinase (Cryphonectria parasitica) |
| AEST-04-D-09 | −5.78 | 3.75 | 1.3E−38 | Acid proteinase (Cryphonectria parasitica) |
| AEST-05-A-09 | −6.99 | −3.67 | 6.0E−33 | Aspergillopepsin |
| AEST-07-G-02 | −3.09 | −2.74 | 1.2E−102 | Microbial aspartic proteinases; precursor endothiapepsin (Cryphonectria parasitica) |
| AEST-29-E-12 | −4.75 | −2.38 | 2.0E−56 | Chain A |
| Secondary metabolism | ||||
| AEST-22-E-05 | 6.00 | 4.61 | 2.2E−18 | Hypothetical protein AN8910.2 (Aspergillus nidulans FGSC A4) |
| 3.0E−14 | Polyketide synthase (Cochliobolus heterostrophus) | |||
| Transport | ||||
| AEST-01-F-12 | 6.25 | 3.39 | 6.6E−18 | Polyamine transporter |
| AEST-23-D-02 | −2.05 | −2.24 | 3.0E−89 | Hypothetical protein (Neurospora crassa) |
| 6.0E−69 | Secretory pathway gdp dissociation inhibitor (Aspergillus fumigatus) | |||
| AEST-31-G-06 | −2.57 | −2.13 | 4.1E−38 | Hypothetical protein FG06562.1 (Gibberella zeae PH-1) |
| 6.0E−32 | Oligopeptide transporter (Candida albicans) | |||
| Biological process unknown | ||||
| AEST-40-G-02 | 6.05 | 4.57 | No hit | |
| AEST-40-G-11 | 2.85 | 2.12 | No hit | |
| AEST-01-C-10 | 5.82 | 9.47 | 2.4E−20 | Hypothetical protein MG09257.4 (Magnaporthe grisea 70-15) |
| 1.0E−08 | Parasitic phase-specific protein PSP-1 (Coccidioides posadasii) | |||
| AEST-01-E-01 | 3.42 | 3.58 | 1.2E−18 | Hypothetical protein AN4568.2 (Aspergillus nidulans FGSC A4) |
| AEST-15-E-03 | 3.92 | 6.39 | 2.8E−16 | Predicted protein (Neurospora crassa) |
| 3.1E−01 | Protein kinase domain-containing protein (Tetrahymena thermophila) | |||
| AEST-05-B-03 | 5.56 | 2.78 | 9.5E−44 | Hypothetical protein FG03413.1 (Gibberella zeae PH-1) |
| 2.0E−24 | Oxidoreductase, zinc-binding (Aspergillus fumigatus Af293) | |||
| AEST-08-B-09 | −2.35 | −2.68 | 2.4E−40 | Hypothetical protein FG01974.1 (Gibberella zeae PH-1) |
| 5.0E−39 | het-c (Podospora anserina), involved in vegetative incompatibility | |||
| AEST-10-F-05 | −2.42 | −14.52 | 1.9E−09 | MUS26 (Neurospora crassa) |
| AEST-37-H-03 | 3.13 | 2.61 | 4.6E−01 | Putative l-aspartate oxidase (Nocardia farcinica IFM 10152) |
| AEST-21-H-03 | −2.57 | −2.53 | 1.6E−56 | Hypothetical protein MG06174.4 (Magnaporthe grisea 70-15) |
| AEST-05-F-09 | −3.40 | −4.73 | 3.3E−01 | Hypothetical protein MG01242.4 (Magnaporthe grisea 70-15) |
| AEST-26-G-12 | 6.52 | 6.38 | 1.2E−44 | Hypothetical protein AN3524.2 (Aspergillus nidulans FGSC A4) |
| 3.0E−09 | NAD-binding Rossmann fold oxidoreductase (Aspergillus fumigatus) | |||
| AEST-01-G-02 | −2.26 | −2.39 | 4.2E−18 | Hypothetical protein MG04873.4 (Magnaporthe grisea 70-15) |
| 2.0E−05 | CipC protein (Emericella nidulans) | |||
| AEST-02-D-07 | −2.54 | −2.64 | 8.1E−03 | Hypothetical protein FG11199.1 (Gibberella zeae PH-1) |
| 4.2E−01 | Cation exchanger (Aspergillus fumigatus Af293) | |||
| AEST-03-F-02 | 3.02 | 4.57 | 2.0E−69 | Hypothetical protein AN3524.2 (Aspergillus nidulans FGSC A4) |
| AEST-04-E-07 | −3.45 | −2.09 | No hit | |
| AEST-08-A-05 | −2.86 | −2.76 | 1.2E−28 | Conserved hypothetical protein (Aspergillus fumigatus Af293) |
| 8.0E−16 | Endoglucanase (Aspergillus fumigatus Af293) | |||
| AEST-08-A-08 | 5.64 | 3.43 | 3.4E−33 | Hypothetical protein AN7268.2 (Aspergillus nidulans FGSC A4) |
| 1.0E−15 | Short-chain dehydrogenase (Aedes aegypti) | |||
| AEST-08-F-11 | 5.78 | 5.05 | 1.8E−31 | Hypothetical protein AN7231.2 (Aspergillus nidulans FGSC A4) |
| 4.0E−28 | Serine peptidase, family S28 (Aspergillus fumigatus Af293) | |||
| AEST-11-E-06 | 2.18 | 2.82 | 6.8E−29 | Hypothetical protein (Neurospora crassa) |
| 2.0E−22 | Transcriptional regulator family protein (Musa acuminata) | |||
| AEST-12-F-07 | −2.51 | −2.00 | 3.6E−23 | Hypothetical protein FG11205.1 (Gibberella zeae PH-1) |
| 1.0E−19 | Immunomodulatory protein (Antrodia camphorata) | |||
| AEST-26-D-02 | 4.31 | 2.71 | 3.3E−48 | Hypothetical protein FG07557.1 (Gibberella zeae PH-1) |
| 9.0E−19 | Putative oxidoreductase (Burkholderia xenovorans LB400) | |||
| AEST-36-D-02 | −2.03 | −2.36 | 4.3E−42 | hypothetical protein FG10724.1 (Gibberella zeae PH-1) |
| 3.0E−34 | Related to SWI/SNF complex 60-kDa subunit (Neurospora crassa) | |||
| AEST-38-E-03 | −4.59 | −12.06 | 3.0E−31 | Hypothetical protein FG02692.1 (Gibberella zeae PH-1) |
| 5.0E−25 | Alpha-ketoglutarate-dependent xanthine dioxygenase (Emericella nidulans) | |||
| AEST-01-G-08 | 4.20 | 4.24 | 1.1E−12 | Hypothetical protein MG02664.4 (Magnaporthe grisea 70-15) |
| 2.9E−01 | Predicted: similar to proto-oncogene Frat1 (Rattus norvegicus) | |||
| AEST-01-H-10 | −2.05 | −2.23 | 1.6E−40 | Hypothetical protein AN0493.2 (Aspergillus nidulans FGSC A4) |
| 5.2E−01 | Predicted: similar to Down syndrome cell adhesion molecule (Mus musculus) | |||
| AEST-02-B-03 | 5.49 | 4.03 | 1.5E−03 | Hypothetical protein MG10137.4 (Magnaporthe grisea 70-15) |
| 2.5 | Predicted: similar to WNK lysine-deficient protein kinase 4 (Danio rerio) | |||
| AEST-02-F-06 | 5.84 | 5.41 | 6.4E−23 | Hypothetical protein AN7893.2 (Aspergillus nidulans FGSC A4) |
| 6.0E−15 | Citrinin biosynthesis oxygenase CtnA (Monascus purpureus) | |||
| AEST-02-F-12 | 2.61 | 3.85 | 3.2E−52 | Hypothetical protein MG10327.4 (Magnaporthe grisea 70-15) |
| 6.0E−32 | AAA family ATPase, putative (Aspergillus fumigatus Af293) | |||
| AEST-04-A-05 | −3.01 | −6.26 | 1.2E−12 | Predicted protein (Gibberella zeae PH-1) |
| 3.8 | Nucleotide exchange factor RasGEF K (Dictyostelium discoideum) | |||
| AEST-05-E-12 | 6.11 | 3.13 | 1.4E−31 | Hypothetical protein MG09414.4 (Magnaporthe grisea 70-15) |
| 5.9 | Rex protein (Human T-cell lymphotropic virus type 1) | |||
| AEST-06-A-07 | 4.19 | 5.69 | 1.1E−27 | Hypothetical protein AN4567.2 (Aspergillus nidulans FGSC A4) |
| 3.6E−01 | LIN-26B protein (Caenorhabditis briggsae) | |||
| AEST-06-D-03 | −2.61 | −3.06 | 1.8E−05 | Hypothetical protein MG01242.4 (Magnaporthe grisea 70-15) |
| 2.1 | Putative mitogen-activated protein kinase (Oryza sativa (japonica cultivar group)) | |||
| AEST-07-A-09 | 5.57 | 7.23 | 3.4E−24 | Predicted protein (Neurospora crassa) |
| 2.0E−06 | Putative monooxygenase (Bordetella pertussis Tohama I) | |||
| AEST-07-D-09 | −3.51 | −3.14 | 5.0E−01 | LD01372p (Drosophila melanogaster) |
| AEST-07-G-09 | −2.00 | −2.78 | 2.7E−29 | Hypothetical protein MG07850.4 (Magnaporthe grisea 70-15) |
| AEST-08-F-04 | 6.88 | 6.85 | 7.9E−14 | Predicted protein (Magnaporthe grisea 70-15) |
| 9.6E−01 | Predicted: similar to zinc finger protein RIZ (Gallus gallus) | |||
| AEST-09-B-02 | 5.20 | 13.82 | 1.7E−06 | Predicted protein (Magnaporthe grisea 70-15) |
| 1.0E−04 | Predicted: similar to transcription factor RREB-1 (Danio rerio) | |||
| AEST-09-G-10 | 5.25 | 3.23 | No hit | |
| AEST-12-B-01 | 7.35 | 6.37 | 8.5E−47 | Hypothetical protein FG03568.1 (Gibberella zeae PH-1) |
| 8.0E−06 | O-Methyltransferase, family 2: generic methyltransferase (Mycobacterium sp. JLS) | |||
| AEST-15-C-09 | 2.36 | 4.89 | 8.6E−26 | Hypothetical protein MG10327.4 (Magnaporthe grisea 70-15) |
| 8.0E−16 | AAA family ATPase (Aspergillus fumigatus Af293) | |||
| AEST-16-A-09 | −2.31 | −3.26 | 1.7E−21 | Hypothetical protein FG01228.1 (Gibberella zeae PH-1) |
| 5.4E−01 | ROK (Kineococcus radiotolerans SRS30216) | |||
| AEST-16-C-08 | 2.97 | 2.09 | 1.8E−47 | Hypothetical protein FG09400.1 (Gibberella zeae PH-1) |
| 6.0E−13 | Putative arylamine N-acetyltransferase (Streptomyces murayamaensis) | |||
| AEST-19-G-01 | −2.31 | 2.12 | 3.3E−04 | Aspergillopepsin (Aspergillus fumigatus Af293) |
| AEST-21-F-04 | 3.47 | 4.23 | 1.4E−25 | SirC (Leptosphaeria maculans) |
| AEST-28-E-02 | 7.10 | 4.75 | 1.8E−15 | Hypothetical protein AN2403.2 (Aspergillus nidulans FGSC A4) |
| 1.0E−09 | l-Aminoadipate-semialdehyde dehydrogenase, putative (Cryptococcus neoformans) | |||
| AEST-28-G-08 | 5.54 | 3.29 | 8.5E−43 | Hypothetical protein AN4567.2 (Aspergillus nidulans FGSC A4) |
| 3.1E−02 | Transcription factor RegA (Aspergillus fumigatus) | |||
| AEST-31-C-06 | 3.27 | 3.01 | 1.7E−17 | Predicted protein (Neurospora crassa) |
| 4.7E−01 | Predicted: mucin 5, subtype B, tracheobronchial (Macaca mulatta) | |||
| AEST-31-D-08 | −2.21 | −2.60 | 1.4E−01 | Hypothetical protein (Neurospora crassa) |
| 4.4 | Novel zinc finger protein (Danio rerio) | |||
| AEST-34-A-06 | −3.80 | −7.67 | 2.4 | DHX37 protein (Homo sapiens) |
| AEST-34-B-09 | 2.83 | 4.84 | 9.8E−29 | Hypothetical protein FG01048.1 (Gibberella zeae PH-1) |
| 5.0E−16 | Cytochrome P450 monooxygenase (Aspergillus fumigatus Af293) | |||
| AEST-34-E-06 | 4.13 | 5.61 | 4.4E−01 | Hypothetical protein (Oryza sativa (japonica cultivar group)) |
| 1.6 | TPR repeat (Desulfovibrio desulfuricans G20) | |||
| AEST-36-A-02 | 3.89 | 2.27 | 4.1E−39 | Hypothetical protein FG11553.1 (Gibberella zeae PH-1) |
| 1.0E−05 | Acyl homoserine lactone-degrading enzyme (Arthrobacter sp. IBN110) | |||
| AEST-38-C-06 | 6.42 | 6.21 | 1.6E−10 | Hypothetical protein AN3284.2 (Aspergillus nidulans FGSC A4) |
| 9.1 | Amelogenin (Xenopus laevis) | |||
| epn1 | −7.72 | −5.34 | 0.0 | Endothiapepsin (Cryphonectria parasitica) |
Column 1 indicates the arrayed express sequence tag (AEST) library identification. The AEST library used for printing of the C. parasitica microarray chip consisted of a 3,864-clone subset of the EST library depleted of ESTs corresponding to the hypovirus genome and the highly abundant hydrophobin gene cryparin (1). Columns 2 and 3 indicate the magnitude of average n-fold change relative to expression in wild-type EP155. Positive numbers indicate increased transcript abundance in the tested strain, while negative numbers indicate a reduction in abundance. Column 4 indicates the strength of the BLAST value corresponding to the biological process description and source organism of the matched sequence in column 5. In cases in which the strongest hit failed to provide informative description, second-hit information was added.
Further analysis of these 85 genes revealed even greater similarity, since all but 1 (AEST-19-G-01) of these genes had altered transcript accumulation in the same direction (Fig. 3B). Moreover, most of the genes were differentially expressed with similar magnitudes in response to infection by the two reoviruses.
Validation of microarray results by real-time RT-PCR.
To confirm the changes predicted by the microarray analyses, real-time RT-PCR was used to validate the microarray results for 19 genes that were scored as differentially expressed and 3 genes that had no change in transcript accumulation. Each clone was measured by real-time RT-PCR in triplicate for each of two independent RNA isolations. There were very few cases in which real-time RT-PCR analysis failed to support microarray-predicted changes in transcript accumulation. Table 4 shows that only 1 clear false-positive result was obtained from 44 different probe/RNA combinations, a rate of 2.3%. These data provided a high level of confidence in the transcriptional changes that were projected by microarray analysis. It is worth noting that the clone (AEST-19-G-01) that was predicted to be regulated in the opposite direction by the two reoviruses was shown by real-time RT-PCR to be down-regulated in both cases, further supporting the similarities in transcriptional profiles between the two reoviruses.
TABLE 4.
Real-time RT-PCR validation of microarray measurements for transcriptional changes in MyRV1-Cp9B21- and MyRV2-CpC18-infected strainsa
| Clone ID | Fold change in expression with infection by:
|
|||
|---|---|---|---|---|
| MyRV1-Cp9B21
|
MyRV2-CpC18
|
|||
| RT-PCR | Microarray | RT-PCR | Microarray | |
| AEST-02-F-06 | 57.32 | 5.84 | 44.75 | 5.41 |
| AEST-02-F-12 | 7.31 | 2.61 | 13.76 | 3.85 |
| AEST-03-F-05 | −2.26 | −1.64 | −1.52 | −1.39 |
| AEST-03-G-06 | −6.20 | −4.08 | −8.93 | −4.05 |
| AEST-03-H-05 | 1.19 | 1.11 | 1.79 | 1.31 |
| AEST-05-A-09 | −3.89 | −6.99 | −5.52 | −3.67 |
| AEST-05-B-03 | 24.84 | 5.56 | 16.60 | 2.78 |
| AEST-06-B-03 | 3.36 | NA | 16.41 | 2.98 |
| AEST-07-B-12 | 2.64 | 3.97 | 2.52 | 2.83 |
| AEST-07-D-09 | −2.84 | −3.51 | −3.93 | −3.14 |
| AEST-08-F-04 | 75.62 | 6.88 | 46.55 | 6.85 |
| AEST-08-F-10 | 1.69 | 1.86 | 1.78 | 1.12 |
| AEST-09-H-07 | 1.58 | 1.69 | 4.03 | 1.67 |
| AEST-10-A-11 | −1.81 | −1.49 | −1.70 | −1.61 |
| AEST-11-B-12 | −1.67 | −2.48 | −2.54 | −2.01 |
| AEST-11-E-06 | 1.49 | 2.18 | 2.18 | 2.80 |
| AEST-11-E-09 | 86.13 | 7.43 | 44.08 | 6.03 |
| AEST-12-G-04 | 60.06 | 4.10 | 20.61 | 3.07 |
| AEST-13-C-05 | 2.84 | NA | 36.87 | 4.21 |
| AEST-19-G-01b | −5.24 | −2.31 | −3.89 | 2.12 |
| AEST-27-H-09 | −3.19 | NA | −5.22 | −3.35 |
| AEST-29-E-12 | −44.79 | −4.75 | −14.19 | −2.38 |
Real-time RT-PCR measurements were made in triplicate for each clone by using two independent total RNA preparations. n-fold changes and direction of differential expression are relative to results for wild-type EP155. Positive numbers indicate increased transcript accumulation, while negative numbers indicate reduced transcript accumulation in the experimental strain relative to transcript accumulation in control strain EP155.
A false-positive result is reported for this clone for reovirus MyRV2-CpC18; the microarray analysis reports an increased level of transcript accumulation, and real-time PCR analysis indicates a decrease in transcript accumulation.
Comparison of transcriptional profiles between reovirus- and hypovirus-infected isolates and G-protein mutants.
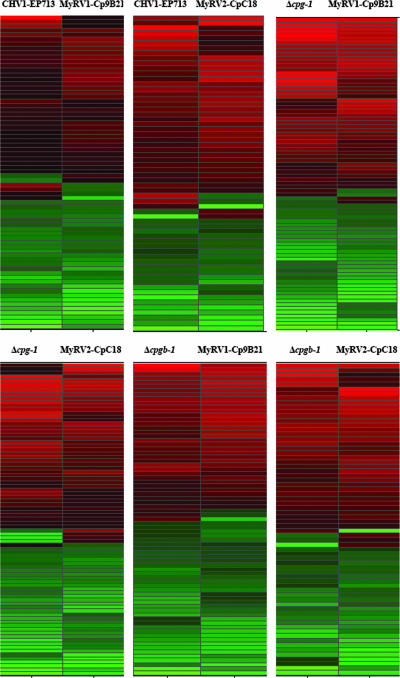
Results of microarry analysis of C. parasitica G-protein deletion mutant strains have provided additional support for previous reports that hypovirus CHV1-EP713 infection disrupts cellular G-protein signal transduction (11). The observation that the reovirus-infected strains produced the distinctive crinkled colony morphology on cellophane overlays that is characteristic of CHV1-EP713-infected and Δcpg-1 strains cultured under the same conditions suggested the possibility that all three mycoviruses alter G-protein signaling. Consequently, lists of genes responsive to reovirus infection, CHV1-EP713 infection, and the disruption of the G-protein α subunit gene cpg-1 and the β subunit gene cpgb-1 were compared. Pairwise comparisons were made between virus infections or between virus infection and G-protein mutants, and lists of commonly differentially expressed genes are presented in hierarchical clustering diagrams in Fig. 4. Since similarly altered transcripts are clustered together in this analysis, the convergence of the coloration in each comparison suggests similarly altered transcriptional profiles. Red lines indicate increases in transcript abundance, while green lines indicate decreases. A comparison of the lists of genes that were differentially expressed in hypovirus CHV1-EP713- and reovirus MyRV1-Cp9B21-infected strains revealed 71 common genes, with 65 of the 71 genes changed in the same direction. When MyRV2-CpC18 infection was similarly compared to CHV1-EP713 infection, 62 genes were in common and 57 of the 62 genes were changed in the same direction (Fig. 4).
FIG. 4.
Pairwise hierarchical clustering diagrams of differentially expressed clones that are in common between virus infected and G-protein mutant strains. Color brightness is proportional to the magnitude of the change, with red indicating genes up-regulated compared to wild-type EP155 and green indicating down-regulation.
The reovirus-responsive genes were also compared with genes differentially expressed in cpg-1 and cpgb-1 deletion mutants (Δcpg-1 and Δcpgb-1) (11). Of the C. parasitica genes that were up- or down-regulated in the Δcpg-1 mutant, 82 were in common with MyRV1-Cp9B21-responsive genes and 85 with MyRV2-CpC18-responsive genes. These represent 58.6% and 66.4% of the totals of MyRV1-Cp9B21- and MyRV2-CpC18-responsive genes, respectively. When the Δcpgb-1 mutant-responsive list was similarly compared, 72 (51.4%) and 69 (53.9%) genes were found to be in common with the MyRV1-Cp9B21- and MyRV2-CpC18-responsive gene lists, respectively (Fig. 4). Moreover, most of the common genes were regulated in the same direction.
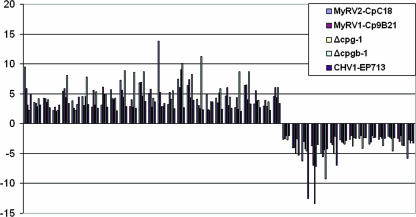
Further examination of the responsive genes for mycoviruses and G-protein mutants revealed that 41 genes were in common across all five samples. These genes are listed in Table 5. Even more remarkably, all the 41 genes were changed in the same direction across all five samples as shown in Fig. 5. These results suggest that hypovirus CHV1-EP713 and reovirus infections have similar impacts on the cellular G-protein signaling pathway.
TABLE 5.
Differentially expressed genes that are in common across infection by three viruses and in two G-protein subunit mutants (Δcpg-1 and Δcpgb-1)
| AEST | Fold change in expression with infection bya:
|
Description of product(s) (organism) | ||||
|---|---|---|---|---|---|---|
| CHV1-EP713 | MyRV1-Cp9B21 | MyRV2-CpC18 | Δcpg-1 mutant | Δcpgb-1 mutant | ||
| AEST-01-C-10 | 5.02 | 5.82 | 9.47 | 3.13 | 2.23 | Hypothetical protein MG09257.4 (Magnaporthe grisea 70-15); Parasitic phase-specific protein PSP-1 (Coccidioides posadasii) |
| AEST-01-E-01 | 3.08 | 3.42 | 3.58 | 2.85 | 4.21 | Hypothetical protein AN4568.2 (Aspergillus nidulans FGSC A4) |
| AEST-01-G-08 | 2.65 | 4.20 | 4.24 | 3.54 | 4.03 | Hypothetical protein MG02664.4 (Magnaporthe grisea 70-15); Predicted: similar to proto-oncogene Frat1 (Rattus norvegicus) |
| AEST-01-H-09 | 3.12 | 2.76 | 2.24 | 2.09 | 2.24 | Hypothetical protein MG05155.4 (Magnaporthe grisea 70-15); adenosyl homocysteinase (Chaetomium globosum CBS 148.51) |
| AEST-02-F-06 | 3.37 | 5.84 | 5.41 | 4.32 | 8.10 | Hypothetical protein AN7893.2 (Aspergillus nidulans FGSC A4); citrinin biosynthesis oxygenase CtnA (Monascus purpureus) |
| AEST-02-F-12 | 4.47 | 2.61 | 3.85 | 2.21 | 3.19 | Hypothetical protein MG10327.4 (Magnaporthe grisea 70-15); AAA family ATPase, putative (Aspergillus fumigatus Af293) |
| AEST-03-F-02 | 3.24 | 3.02 | 4.57 | 4.54 | 7.82 | Hypothetical protein (Microbulbifer degradans 2-40) |
| AEST-05-B-03 | 2.61 | 5.56 | 2.78 | 3.12 | 5.34 | Hypothetical protein FG03413.1 (Gibberella zeae PH-1); oxidoreductase, zinc-binding (Aspergillus fumigatus Af293) |
| AEST-05-E-12 | 2.75 | 6.11 | 3.13 | 5.00 | 4.94 | Hypothetical protein MG09414.4 (Magnaporthe grisea 70-15); rex protein (human T-cell lymphotropic virus type 1) |
| AEST-06-A-07 | 2.14 | 4.19 | 5.69 | 4.01 | 4.27 | Hypothetical protein AN4567.2 (Aspergillus nidulans FGSC A4); LIN-26B protein (Caenorhabditis briggsae) |
| AEST-07-A-09 | 2.86 | 5.57 | 7.23 | 4.41 | 8.88 | Predicted protein (Neurospora crassa); putative monooxygenase (Bordetella pertussis Tohama I) |
| AEST-07-B-12 | 2.63 | 3.97 | 2.83 | 2.73 | 8.62 | GTP-binding nuclear protein RAN/TC4 (Gibberella zeae PH-1); predicted: similar to zinc finger protein RIZ Gallus gallus) |
| AEST-08-F-04 | 3.78 | 6.88 | 6.85 | 4.61 | 8.71 | Predicted protein (Magnaporthe grisea 70-15); serine peptidase, family S28 (Aspergillus fumigatus Af293) |
| AEST-08-F-11 | 3.69 | 5.78 | 5.05 | 2.79 | 4.28 | Hypothetical protein AN7231.2 (Aspergillus nidulans FGSC A4); predicted: similar to transcription factor RREB-1 (Danio rerio) |
| AEST-09-B-02 | 3.39 | 5.20 | 13.82 | 2.84 | 2.87 | Predicted protein (Magnaporthe grisea 70-15) |
| AEST-09-G-10 | 2.46 | 5.25 | 3.23 | 4.05 | 5.53 | transcription factor RegA (Aspergillus fumigatus) |
| AEST-11-E-09 | 2.70 | 6.03 | 7.43 | 9.03 | 10.10 | Cytochrome P450 (Aspergillus fumigatus Af293) |
| AEST-12-B-01 | 3.94 | 7.35 | 6.37 | 4.33 | 8.26 | Hypothetical protein FG03568.1 (Gibberella zeae PH-1); O-methyltransferase, family 2: generic methyltransferase (Mycobacterium sp. JLS) |
| AEST-12-G-04 | 2.34 | 3.07 | 4.10 | 2.51 | 11.24 | Theta class glutathione S-transferase (Aspergillus fumigatus Af293) |
| AEST-15-C-09 | 3.67 | 2.36 | 4.89 | 2.20 | 3.68 | Hypothetical protein MG10327.4 (Magnaporthe grisea 70-15); AAA family ATPase (Aspergillus fumigatus Af293) |
| AEST-21-F-04 | 2.45 | 3.47 | 4.23 | 5.17 | 5.87 | SirC (Leptosphaeria maculans); polyketide synthase (Cochliobolus heterostrophus) |
| AEST-22-E-05 | 2.56 | 6.00 | 4.61 | 3.04 | 4.43 | Hypothetical protein AN8910.2 (Aspergillus nidulans FGSC A4) |
| AEST-26-D-02 | 2.03 | 4.31 | 2.71 | 2.45 | 8.66 | Hypothetical protein FG07557.1 (Gibberella zeae PH-1); putative oxidoreductase (Burkholderia xenovorans LB400) |
| AEST-26-G-12 | 3.31 | 6.52 | 6.38 | 4.01 | 8.68 | Hypothetical protein AN3524.2 (Aspergillus nidulans FGSC A4); NAD-binding Rossmann fold oxidoreductase (Aspergillus fumigatus) |
| AEST-28-G-08 | 2.63 | 5.54 | 3.29 | 3.84 | 3.94 | Hypothetical protein XP_211670 (Homo sapiens); hypothetical protein AN4567.2 (Aspergillus nidulans FGSC A4) |
| AEST-39-A-08 | 2.23 | 3.91 | 2.97 | 2.89 | 3.64 | Dynamin-related protein |
| AEST-40-G-02 | 3.40 | 6.05 | 4.57 | 4.40 | 6.06 | No hit |
| AEST-02-D-07 | −2.00 | −2.54 | −2.64 | −2.04 | −2.70 | Hypothetical protein FG11199.1 (Gibberella zeae PH-1); cation exchanger (Aspergillus fumigatus Af293) |
| AEST-03-G-06 | −5.28 | −4.08 | −4.05 | −5.04 | −2.49 | Acid proteinase (Cryphonectria parasitica) |
| AEST-04-A-05 | −12.52 | −3.01 | −6.26 | −4.21 | −4.60 | Predicted protein (Gibberella zeae PH-1); nucleotide exchange factor RasGEF K (Dictyostelium discoideum) |
| AEST-05-A-09 | −3.53 | −6.99 | −3.67 | −13.45 | −7.11 | Aspergillopepsin |
| AEST-05-F-11 | −4.23 | −5.51 | −4.99 | −4.44 | −9.30 | NUP3_PENSQ nuclease PA3 (endonuclease PA3) (DNase PA3) |
| AEST-07-D-09 | −6.92 | −3.51 | −3.14 | −5.05 | −2.12 | LD01372p (Drosophila melanogaster) |
| AEST-07-G-02 | −2.84 | −3.09 | −2.74 | −3.27 | −3.39 | Microbial aspartic proteinases; precursor endothiapepsin (Cryphonectria parasitica) |
| AEST-08-F-03 | −2.46 | −2.01 | −2.63 | −3.78 | −2.34 | Hypothetical protein (Neurospora crassa); phosphoglucomutase (Aspergillus oryzae) |
| AEST-11-B-12 | −2.34 | −2.01 | −2.48 | −4.11 | −2.34 | Conserved hypothetical protein (Gibberella zeae PH-1); UDP-glucose pyrophosphorylase (Emericella nidulans) |
| AEST-14-D-02 | −2.26 | −3.06 | −3.41 | −2.01 | −2.37 | Hypothetical protein MG10400.4 (Magnaporthe grisea 70-15); cell wall glucanase (Aspergillus fumigatus Af293) |
| AEST-31-D-08 | −2.49 | −2.21 | −2.60 | −2.49 | −3.66 | Hypothetical protein (Neurospora crassa); novel zinc finger protein (Danio rerio) |
| AEST-37-F-03 | −2.38 | −2.31 | −2.07 | −2.55 | −4.57 | Beta-glucosidase homolog (Cochliobolus heterostrophus) |
| AEST-40-B-12 | −3.73 | −3.02 | −2.43 | −2.03 | −3.60 | Hypothetical protein MG05692.4 (Magnaporthe grisea 70-15) |
| epn1 | −3.22 | −2.73 | −5.78 | −3.24 | −2.81 | Endothiapepsin (Cryphonectria parasitica) |
Values are n-fold changes predicted by microarray analysis compared to values for EP155. Positive numbers indicate increased transcript abundance in the tested strain, while negative numbers indicate a reduction in abundance.
FIG. 5.
Similarity in expression profiles for genes responsive to infection by CHV1-EP713, MyRV1-Cp9B21, or MyRV2-CpC18 and deletion of G-protein subunit genes cpg-1 and cpgb-1. The bars represent the relative magnitudes of microarray-predicted changes in transcript accumulation for the 41 common genes (n-fold change, y axis). The order of AEST clones from left to right is as presented in Table 5. That is, values for AEST-01-C-10 are shown at the far left of the figure.
DISCUSSION
The utility of C. parasitica as an experimental system for the study of fungus-mycovirus interactions has benefited from the characterization of associated viruses representing four species within the family Hypoviridae and members of the virus families Reoviridae, Narnaviridae, and Chrysoviridae (reviewed in references 19 and 26). Recent comparative studies showed that viruses as fundamentally different as hypoviruses and reoviruses attenuate C. parasitica virulence but cause quite different effects on other host phenotypic traits, such as conidiation, pigmentation, and colony morphology (31). We were able in this study to show that differences in the responses of C. parasitica to hypovirus and reovirus infection extend to modulation of sexual reproduction. Unlike hypovirus-infected strains, which are unable to form perithecia when serving as the female in a sexual cross, reovirus-infected C. parasitica strains are female fertile and transmit virus to ascospore progeny.
Virus transmission through conidia in filamentous fungi is common and is often an efficient means of vertical transmission, with transmission efficiencies often nearing 100% (19). Although MyRV-1 and -2 are both transmitted through conidia, their rates of transmission are low, often less than 10% in culture (B. I. Hillman, unpublished). Virus transmission through sexual spores in filamentous fungi is generally less common than asexual transmission, and the details are poorly understood. The first C. parasitica virus confirmed to be transmitted through ascospores was the small mitochondrial virus Cryphonectria mitovirus-1/CpNB631 (5, 29). In this instance, once the finding had been made that virus infection did not abolish female fertility, the demonstration that this mitochondrial virus was transmitted vertically but only maternally to sexual progeny was not surprising, since mitochondria had already been demonstrated formally to be inherited maternally in C. parasitica (24). Enebak (13) had previously noted that a C. parasitica isolate infected with reovirus MyRV2-CpC18 was female fertile, but details of fecundity and possible virus transmission were not available. In the current study, we confirmed the finding of female fertility in strain MyRV2-CpC18/EP155 as well as in MyRV1-Cp9B21/EP155 and determined that virus was transmitted maternally in both strains. Interestingly, the efficiency of virus transmission vertically through ascospores, approximately 60%, is about the same as that for the mitochondrial virus (28), although presumably the mechanisms of transmission are quite different.
One of the more interesting findings in this study was the positive correlation between the expression of genes that are known to be associated with female fertility in C. parasitica and other ascomycetes, the sex pheromone precursor gene Mf2/1 and the yeast Ste12-like transcriptional factor gene cpst12, and mating efficiency, further validating the involvement of these genes in the mating process and providing supporting quantitative information. While there remain a number of questions about the mechanism of differential regulation of the mating-associated genes, it is apparent that suppression of mating is not a generalized response to virus infection and does not correlate with fungal virulence, since the two reovirus-infected strains are among the most hypovirulent (14, 18). A detailed understanding of mechanisms underlying hypovirus transmission through sexual spores may provide additional means for modulating virus transmission in ways that enhance biological control potential.
Differential host responses to hypovirus and reovirus infections are not unexpected, considering the fundamental differences between the two viruses and the absence of any conserved nucleotide or amino acid sequences. Reoviruses are “true” dsRNA viruses that have complex capsids containing virtually all of the proteins required for transcription and RNA replication. Although the subcellular replication complexes of fungal reoviruses have not been examined, other similar reoviruses replicate in large viroplasms in the cytoplasm, with no substantial nuclear component in the replication cycle (25). Hypoviruses are most closely related to single-stranded RNA viruses, such as plant-infecting potyviruses and animal-infecting picornaviruses (21), but lack a coat protein. Replication of hypovirus RNA occurs in association with membrane vesicles (15, 16).
In spite of these differences, CHV1-EP713 and reoviruses MyRV1-Cp9B21 and MyRV2-CpC18 significantly reduced C. parasitica virulence and caused a similar change in the pattern of transcript accumulation for a substantial number of host responsive genes (Fig. 4). Remarkably, a significant portion of the host genes that were responsive to both CHV1-EP713 and reovirus infection were also responsive to disruption of the cellular G-protein subunit genes cpg-1 and cpgb-1 (Fig. 5). Moreover, the CHV1-EP713- and reovirus-infected strains exhibited a distinctive “crinkled” colony morphology on PDA/cellophane that is also characteristic of colonies formed by the Gα-subunit cpg-1 deletion strain (Fig. 2). The suggestion raised by this observation that all three mycoviruses modulate the cellular G-protein signaling pathway is strengthened by the fact that the 41 cellular genes that were responsive to the three mycovirus infections and G-protein subunit gene disruption all showed changes in transcript accumulation in the same direction (Fig. 5). This correlation is made even more interesting by previous comparative studies that showed differential modulation of cellular signaling pathways by the mild and severe hypovirus isolates CHV1-Euro7 and CHV1-EP713 (27).
The hypoviruses CHV1-EP713 and CHV1-Euro7 were shown to alter C. parasitica canker formation on chestnut tree tissue to quite different extents (7). Strain EP155 infected by severe hypovirus CHV1-EP713 produces small, superficial cankers on dormant chestnut stems that contain few stromal pustules (asexual spore-forming structures) on the surface (7). In contrast, strains infected with the mild hypovirus CHV1-Euro7 produce cankers that are on the order of two- to threefold larger in area and that contain copious amounts of surface pustules. Using a promoter-reporter system designed to monitor perturbations in C. parasitica G-protein-linked, cAMP-mediated signaling, Parsley et al. (27) showed that infection by the severe hypovirus CHV1-EP713 activated the pathway, while infection by the closely related but mild hypovirus isolate CHV1-Euro7 did not. Significantly for the present study, CHV1-EP713-infected strains were shown to form the crinkled colony morphology on PDA/cellophane, while CHV1-Euro7-infected strains did not (see Fig. 4 of reference 27). Moreover, only 12 of the 41 C. parasitica genes that were found to be coordinately responsive to CHV1-EP713 infection, reovirus infection, and G-protein subunit gene disruption (Fig. 5) were also similarly responsive to CHV1-Euro7 (2; data not shown). Thus, not only does the CHV1-Euro7-infected strain attenuate fungal virulence to a lesser degree than the CHV1-EP713- or reovirus-infected strains, it is also distinguished from the CHV1-EP713- and reovirus-infected strains by the failure both to exhibit the crinkled colony morphology and to similarly modulate the transcript accumulation for two-thirds of the 41 coordinately virus-responsive, G-protein-regulated cellular genes.
The observation that a significant subset of cellular G-protein-regulated genes coordinately respond to two reoviruses that share only 50% nucleotide identity and to a hypovirus, which is in an entirely different virus family, raises the question of whether the transcriptional changes represent a general response to mycovirus infection or are caused by specific viral determinants. The former possibility is not supported by several observations. First, as noted above, only one-third of the 41 coordinately responsive genes respond similarly to infection by the mild hypovirus CHV1-Euro7 (2; F. Deng, T. D. Allen, and D. L. Nuss, unpublished). Second, a number of other mycoviruses found in C. parasitica, e.g., the CHV4 hypoviruses and the OB5 chrysovirus (18), fail to cause any phenotypic changes or symptoms that would be expected to accompany such transcriptional changes. The effects of these mycoviruses on host transcript accumulation remain to be examined, but based on results of the CHV1-EP713 comparison with CHV1-Euro7 (2), we would not expect the dramatic transcriptional responses seen in the current study. The latter possibility, that specific viral determinants target the G-protein signaling pathway, suggests that there may be a selective advantage provided by virus-mediated alterations of cellular G-protein signaling, although what that might be is currently unclear. However, the emerging correlation between virulence attenuation, the distinctive crinkled colony morphology, and coordinate regulation of a subset of virus-responsive, G-protein-regulated cellular genes by unrelated hypoviruses and reoviruses is intriguing and warrants further investigation. In this regard, the anticipated availability of the C. parasitica genome sequence (http://www.jgi.doe.gov/sequencing/cspseqplans2007.html) will provide a greatly expanded view of host transcriptional responses to infection by different mycoviruses, opportunities to analyze regulatory elements of virus-responsive genes, the ability to reconstruct complete signaling pathways, and the means to efficiently explore virus-host protein-protein interactions.
Acknowledgments
This work was supported in part by Public Health Service grant GM55981 to D.L.N.
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Allen, T. D., A. L. Dawe, and D. L. Nuss. 2003. Use of cDNA microarrays to monitor transcriptional responses of the chestnut blight fungus Cryphonectria parasitica to infection by virulence-attenuating hypoviruses. Eukaryot. Cell 2:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. D., and D. L. Nuss. 2004. Specific and common alterations in host gene transcript accumulation following infection of the chestnut blight fungus by mild and severe hypoviruses. J. Virol. 78:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anagnostakis, S. L. 1984. Nuclear gene mutations in Endothia (Cryphonectria) parasitica that affect morphology and virulence. Phytopathology 74:761-765. [Google Scholar]
- 4.Anagnostakis, S. L. 1988. Cryphonectria parasitica, cause of chestnut blight, p. 123-136. In G. S. Sidhu (ed.), Advances in plant pathology. Academic Press Ltd., London, United Kingdom.
- 5.Buck, K. W., R. Esteban, and B. I. Hillman. 2005. Family Narnaviridae, p. 751-756. In C. M. Fauguet, M. A. Mayo, J. Maniloff, U. Desselberger, and A. L. Ball (ed.), Virus taxonomy: eighth report of the International Committee for the Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 6.Chen, B., G. H. Choi, and D. L. Nuss. 1993. Mitotic stability and nuclear inheritance of integrated viral cDNA in engineered hypovirulent strains of the chestnut blight fungus. EMBO J. 12:2991-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, B., and D. L. Nuss. 1999. Infectious cDNA clone of hypovirus CHV1-Euro7: a comparative virology approach to investigate virus-mediated hypovirulence of the chestnut blight fungus Cryphonectria parasitica. J. Virol. 73:985-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, G. H., and D. L. Nuss. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800-803. [DOI] [PubMed] [Google Scholar]
- 9.Churchill, A. C. L., L. M. Ciufetti, D. R. Hansen, H. D. Van Etten, and N. K. Van Alfen. 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17:25-31. [Google Scholar]
- 10.Dawe, A. L., V. C. McMains, M. Panglao, S. Kasahara, B. Chen, and D. L. Nuss. 2003. An ordered collection of expressed sequences from Cryphonectria parasitica and evidence of genomic microsynteny with Neurospora crassa and Magnapothe grisea. Microbiology 149:2273-2284. [DOI] [PubMed] [Google Scholar]
- 11.Dawe, A. L., G. C. Segers, T. D. Allen, C. C. McMains, and D. L. Nuss. 2004. Microarray analysis of Cryphonectria parasitica Gα- and Gβγ-signalling pathways reveals extensive modulation by hypovirus infection. Microbiology 150:4033-4043. [DOI] [PubMed] [Google Scholar]
- 12.Deng, F., T. A. Allen, and D. L. Nuss. 2007. Ste12 transcription factor homologue CpST12 is down-regulated by hypovirus infection and required for virulence and female fertility of the chestnut blight fungus Cryphonectria parasitica. Eukaryot. Cell 6:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enebak, S. A. 1992. Ph.D. dissertation. West Virginia University, Morgantown.
- 14.Enebak, S. A., B. I. Hillman, and W. L. MacDonald. 1994. A hypovirulent isolate of Cryphonectria parasitica with multiple, genetically unique dsRNA segments. Mol. Plant-Microbe Interact. 7:590-595. [Google Scholar]
- 15.Fahima, T., P. Kazmierczak, D. R. Hansen, P. Pfeiffer, and N. K. van Alfen. 1993. Membrane-associated replication of an unencapsidated double-stranded RNA of the fungus, Cryphonectria parasitica. Virology 195:81-89. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, D. R., N. K. van Alfen, K. Gillies, and N. A. Powell. 1985. Naked dsRNA associated with hypovirulence of Endothia parasitica is packaged in fungal vesicles. J. Gen. Virol. 66:2605-2614. [Google Scholar]
- 17.Hillman, B. I., R. Shapira, and D. L. Nuss. 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80:950-956. [Google Scholar]
- 18.Hillman, B. I., S. Supyani, H. Kondo, and N. Suzuki. 2004. A reovirus of the fungus Cryphonectria parasitica is infectious as particles and related to the Coltivirus genus of animal pathogens. J. Virol. 78:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillman, B. I., and N. Suzuki. 2004. Viruses of the chestnut blight fungus. Adv. Virus Res. 63:423-473. [DOI] [PubMed] [Google Scholar]
- 20.Kasahara, S., and D. L. Nuss. 1997. Targeted disruption of a fungal G-protein β subunit gene results in increased vegetative growth but reduced virulence. Mol. Plant-Microbe Interact. 10:984-993. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V., G. H. Choi, D. L. Nuss, R. Shapira, and J. C. Carrington. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-stranded RNA plant viruses. Proc. Natl. Acad. Sci. USA 88:10647-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, H., X. Lan, H. Liao, T. B. Parsley, D. L. Nuss, and B. Chen. 2007. Genome sequence, full-length cDNA clone, and mapping of viral double-stranded RNA accumulation determinant of hypovirus CHV1-EP721. J. Virol. 81:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquez, L. M., R. S. Redman, R. J. Rodriguez, and M. J. Roossinck. 2007. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315:513-515. [DOI] [PubMed] [Google Scholar]
- 24.Milgroom, M. G., and S. E. Lipari. 1993. Maternal inheritance and diversity of mitochondrial DNA in the chestnut blight fungus, Cryphonectria parasitica. Phytopathology 83:563-567. [Google Scholar]
- 25.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 26.Nuss, D. L. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3:632-642. [DOI] [PubMed] [Google Scholar]
- 27.Parsley, T. B., B. Chen, L. M. Geletka, and D. L. Nuss. 2002. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell 1:401-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polashock, J. J., P. J. Bedker, and B. I. Hillman. 1997. Movement of a small mitochondrial double-stranded (ds) RNA element of Cryphonectria parasitica: ascospore inheritance and implications for mitochondrial recombination. Mol. Gen. Genet. 256:566-571. [DOI] [PubMed] [Google Scholar]
- 29.Polashock, J. J., and B. I. Hillman. 1994. A small mitochondrial double-stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc. Natl. Acad. Sci. USA 91:8680-8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smart, C. D., W. Yuan, R. Foglia, D. L. Nuss, D. W. Fulbright, and B. I. Hillman. 1999. Cryphonectria hypovirus 3-GH2, a virus species in the family Hypoviridae with a single open reading frame. Virology 85:491-494. [DOI] [PubMed] [Google Scholar]
- 31.Sun, L., D. L. Nuss, and N. Suzuki. 2006. Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J. Gen. Virol. 87:3703-3714. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, N., S. Supyani, K. Maruyama, and B. I. Hillman. 2004. Complete genome sequence of Mycoreovirus-1/Cp9B21, a member of a novel genus within the family Reoviridae, isolated from the chestnut blight fungus Cryphonectria parasitica. J. Gen. Virol. 85:3437-3448. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, L., R. A. Baasiri, and N. K. van Alfen. 1998. Viral repression of fungal pheromone precursor gene expression. Mol. Cell. Biol. 18:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]