Abstract
The heat shock transcription factor Hsf1 of the yeast Saccharomyces cerevisiae regulates the transcription of a set of genes that contain heat shock elements (HSEs) in their promoters and function in diverse cellular processes, including protein folding. Here, we show that Hsf1 activates the transcription of various target genes when cells are treated with oxidizing reagents, including the superoxide anion generators menadione and KO2 and the thiol oxidants diamide and 1-chloro-2,4-dinitrobenzene (CDNB). Similar to heat shock, the oxidizing reagents are potent inducers of both efficient HSE binding and extensive phosphorylation of Hsf1. The inducible phosphorylation of Hsf1 is regulated by the intramolecular domain-domain interactions and affects HSE structure-specific transcription. Unlike the heat shock, diamide, or CDNB response, menadione or KO2 activation of Hsf1 is inhibited by cyclic-AMP-dependent protein kinase (PKA) activity, which negatively regulates the activator functions of other transcriptional regulators implicated in the oxidative stress response. These results demonstrate that Hsf1 is a member of the oxidative stress-responsive activators and that PKA is a general negative regulator in the superoxide anion response.
Oxidizing reagents can damage critical cellular molecules, including nucleic acids, proteins, and lipids. To protect against oxidant damage, cells contain an array of defense mechanisms. In the yeast Saccharomyces cerevisiae, the transcription factors Yap1 and Skn7 and a pair of related factors, Msn2 and Msn4 (Msn2/4), are implicated in the oxidative stress response (27, 35, 38). Yap1 and Skn7 activate the expression of proteins that intercept and scavenge reactive oxygen species (ROS) (11, 21, 22, 26). The Msn2/4 regulon contains only a small number of antioxidants but also includes heat shock proteins (HSPs), metabolic enzymes, and components of the ubiquitin-proteasome degradation pathway (4, 11, 14, 25, 30). The cyclic-AMP (cAMP)-protein kinase A (PKA) (cAMP-dependent protein kinase) pathway, which plays a role in the control of metabolism, proliferation, and stress resistance, being modulated by the available-nutrient conditions, negatively regulates the activator functions of Yap1, Skn7, and Msn2/4 (6, 9, 12, 14, 32).
The heat shock transcription factor (HSF) is a stress-responsive regulator conserved from yeast to humans. The hydrophobic repeat regions of HSF mediate the formation of a homotrimer, and the DNA-binding domain of each monomer recognizes the sequence nGAAn. Homotrimeric HSF activates the transcription of genes via binding to a conserved DNA sequence motif termed the heat shock element (HSE), which consists of multiple inverted repeats of the nGAAn unit (28, 40). Target genes of S. cerevisiae Hsf1 encode HSPs, chaperones, metabolic enzymes, and cell wall proteins (13, 39, 41). In the heat shock response, the yeast HSF-specific CE2 and CTM domains regulate the hyperphosphorylation of Hsf1 (15). Hyperphosphorylation is dispensable for transcriptional activation by two Hsf1 trimers bound to HSEs with four or more nGAAn units; however, this modification is implicated in a conformational change of a single Hsf1 trimer bound to three-unit HSEs to an active form (16, 17).
Several lines of evidence suggest that Hsf1 is involved in the oxidative stress response. When cells are treated with the superoxide anion generator menadione, Hsf1 activates the transcription of the CUP1 (encoding copper metallothionein) and ERO1 (encoding endoplasmic reticulum oxidoreductin) genes (24, 34). The Hsf1-mediated transcription of ERO1 is also induced by treatment of cells with the thiol-oxidizing reagent diamide (Y. Takemori and H. Sakurai, unpublished data). In the presence of t-butyl hydrogen peroxide (t-bH2O2), Hsf1 cooperates with Skn7 to achieve maximal induction of several HSP genes (29). Partially purified Hsf1 has been shown to change conformation in response to superoxide anion (23). Similar to S. cerevisiae Hsf1, addition of oxidizing reagents to mammalian HSF1 or Drosophila HSF induces a conformational change leading to homotrimer formation and HSE binding (1, 44, 45). In addition, analysis of HSF1 knockout mice has revealed that HSF1 is necessary to maintain redox homeostasis and antioxidative defenses (42).
To gain further insight into the molecular details underlying oxidant activation of Hsf1 function, we investigated the effects of oxidative stress on binding of Hsf1 to the HSE, phosphorylation of the protein, and transcriptional activation of target genes. We show that treatment of cells with menadione, diamide, KO2, or 1-chloro-2,4-dinitrobenzene (CDNB) causes activation of Hsf1 via mechanisms similar to those activated by thermal stress. Unlike the heat shock response, however, menadione- or KO2-induced transcription was negatively regulated by PKA. These results provide important information in understanding both the role of stress-induced modifications of Hsf1 and the organization of the oxidative stress regulon.
MATERIALS AND METHODS
Yeast strains, plasmids, and media.
The yeast strains used in this study are listed in Table 1. Strains containing the hsf1-ba1 or hsf1-CE2Δ-ba1 mutation (see Fig. 5A) and the msn2Δ msn4Δ mutations were described previously (15, 18). Null mutations of SKN7, YAP1, BCY1, and PDE2 were introduced by the one-step gene disruption method. Strain TH2670, in which HSF1 is expressed under the control of the tetO promoter, was obtained from Open Biosystems. Strain ASY62 lacking PKA activity was kindly provided by Yukifumi Uesono (32). The YEp-URA3-HSE4Ptt-CYC1-lacZ reporter gene contains a 4Ptt-type HSE oligonucleotide (TCGACTTCTAGAAGCTTCCAGAAATTAGTGCTACTCGA; GAA repeats are underlined) upstream of the CYC1-lacZ construct (16). The TPK1 gene was cloned into pRS316 to create YCp-URA3-TPK1. Cells were grown in rich glucose (YPD), enriched synthetic glucose (ESD), and synthetic glucose (SD) media (15, 18).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| HS126 | MATα ade2 ura3 leu2 his3 trp1 can1 hsf1::HIS3 YCp-URA3-HSF1 |
| HS170T | YCp-TRP1-HSF1 of HS126 |
| HS171T | YCp-TRP1-hsf1-ba1 of HS126 |
| HS176 | msn2::LEU2 msn4::ADE2 YCp-TRP1-HSF1 of HS126 |
| JU15 | skn7::ADE2 yap1::LEU2 YCp-TRP1-HSF1 of HS126 |
| YN61-l | YCp-TRP1-hsf1-CE2Δ-ba1 of HS126 |
| YAY69 | pde2::ADE2 YCp-TRP1-HSF1 of HS126 |
| YAY70 | bcy1::ADE2 YCp-TRP1-HSF1 of HS126 |
| ASY62 | MATaade8 his3 leu2 trp1 ura3 tpk1::ADE8 tpk2::HIS3 tpk3::TRP1 msn2::HIS3 msn4::LEU2 |
| TH2670 | his3-1 leu2-0 met15-0 URA3::CMV-tTA Kanr-tetO7-TATA-HSF1 |
FIG. 5.
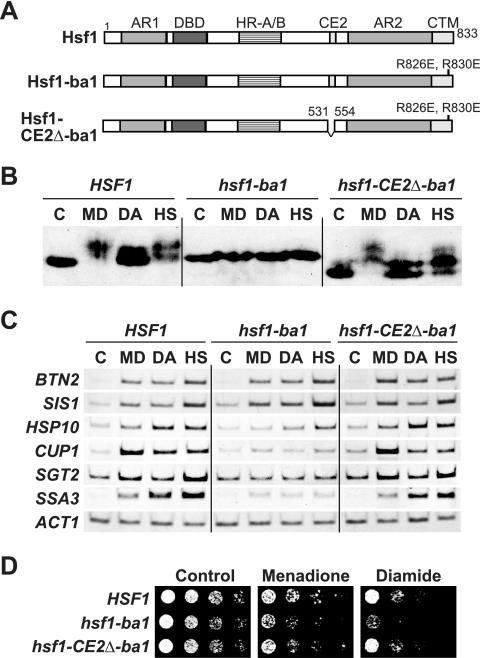
Effects of hsf1 mutations on the menadione or diamide response. (A) Schematic diagram of structural motifs of Hsf1 and Hsf1 mutant constructs. The regions indicated are as follows: AR1 and AR2, activation domains; DBD, DNA-binding domain; HR-A/B, hydrophobic repeat regions A and B; CE2, conserved element 2; CTM, C-terminal modulator. Numbers represent amino acid positions. Hsf1-ba1 contains changes of arginine to glutamate at amino acids 826 and 830 (R826E and R830E), and Hsf1-CE2Δ-ba1 contains the ba1 mutation and lacks the CE2 domain. (B) Phosphorylation of Hsf1 derivatives. Wild-type and HSF1, hsf1-ba1, and hsf1-CE2Δ-ba1 mutant cells were grown in ESD medium at 28°C in the absence (C) or presence of 0.3 mM menadione for 30 min (MD) or 3.0 mM diamide for 30 min (DA) or were grown at 39°C for 15 min (HS). Extracts were prepared from the cells and subjected to immunoblot analysis with an anti-Hsf1 serum. (C) mRNA levels of Hsf1 target genes in hsf1 mutant cells. Cells were grown in ESD medium at 28°C in the absence (C) or presence of 0.3 mM menadione for 45 min (MD) or 3.0 mM diamide for 45 min (DA) or were grown at 39°C for 20 min (HS). Total RNA was prepared from the cells and subjected to RT-PCR analysis. (D) Growth of hsf1 mutant cells on medium containing menadione or diamide. Cells at an A600 of 0.5 were serially diluted fivefold, and 2 μl was spotted onto YPD medium (control) or YPD medium containing 40 μM menadione or 2.0 mM diamide and incubated at 28°C for 2 days.
RNA analysis.
mRNA levels were determined by reverse transcription (RT)-PCR and were compared after normalizing RNA samples with the levels of control ACT1 mRNA (encoding actin), as described previously (15, 16). The experiments were performed at least twice with similar results. The HSE structure of Hsf1 target genes has been previously described (16).
Immunoblot analysis.
Cells were grown under the conditions described in the figure legends. Cell extracts were prepared and subjected to immunoblot analysis with an anti-Hsf1 serum as described previously (15). The experiments were performed at least three times with similar results.
Chromatin immunoprecipitation analysis.
Cells were treated with various stressors as described in the figure legends. Chromatin was immunoprecipitated with an anti-Hsf1 serum and analyzed by PCR as described previously (17). The following regions were amplified by PCR: −386 to −93 of BTN2, −481 to −253 of SIS1, −307 to −133 of HSP10, −315 to +61 of CUP1, −268 to +8 of SGT2, −339 to +36 of SSA3, and −152 to +98 of GAL7.
RESULTS
Menadione and diamide are potent inducers of transcriptional activation of HSE-containing genes.
To examine whether Hsf1 regulates the transcription of genes in response to oxidative stress, we constructed cells harboring the reporter gene HSE4Ptt-CYC1-lacZ, which contains four inverted repeats of the nGAAn sequence upstream of the CYC1 promoter-lacZ fusion. The mRNA levels of lacZ, in parallel with those of the authentic Hsf1 target genes BTN2 and CUP1, were measured by RT-PCR. Oxidative-stress-induced transcription of TRX2 and CTT1, which is regulated by the transcription factors Skn7/Yap1 and Msn2/4, respectively, was analyzed as a positive control (25, 26, 30).
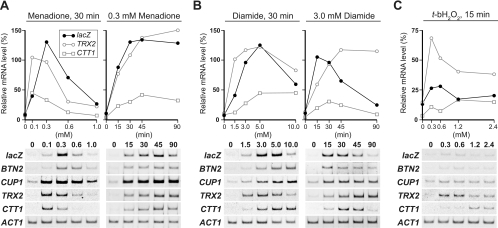
When cells were treated with various concentrations of menadione, the lacZ mRNA levels of the HSE4Ptt-CYC1-lacZ reporter increased more than 10-fold, with a peak response at 0.3 mM menadione (Fig. 1A). The accumulation of lacZ mRNA was apparent within 15 min and remained elevated for 90 min after addition of 0.3 mM menadione. The mRNA levels of BTN2 were also increased by menadione treatment and exhibited a dose response and kinetics similar to those of the lacZ mRNA, whereas the transcription of CUP1 was induced by 0.1 mM menadione. The amounts of TRX2 and CTT1 transcripts reached maximal levels at 0.1 mM menadione, and the kinetics were similar to the HSE-containing genes.
FIG. 1.
Oxidative-stress-induced transcription of various genes. (A) Menadione response. Wild-type cells (strain HS170T) harboring HSE4Ptt-CYC1-lacZ were grown in ESD medium lacking uracil at 28°C and treated with the indicated concentrations of menadione for 30 min (left side) or with 0.3 mM menadione for the indicated times (right side). Total RNA was prepared from the cells and subjected to RT-PCR analysis with primers specific for lacZ, BTN2, CUP1, TRX2, CTT1, and the ACT1 control. The graphs show the mRNA levels normalized to that of ACT1 mRNA, which was 100%. (B) Diamide response. Cells were treated with the indicated concentrations of diamide for 30 min (left side) or with 3.0 mM diamide for the indicated times (right side). RT-PCR analysis was conducted as described above. (C) t-bH2O2 response. Cells were treated with the indicated concentrations of t-bH2O2 for 15 min. RT-PCR analysis was conducted as described above.
Addition of 3 to 10 mM diamide to cultures caused significant increases in transcripts from HSE4Ptt-CYC1-lacZ, BTN2, and CUP1, as well as TRX2 and CTT1 (Fig. 1B). The transcripts of the HSE-containing genes and CTT1 accumulated within 30 min and then gradually decreased after diamide treatment, while TRX2 transcript levels were elevated for 90 min.
The transcription of HSE4Ptt-CYC1-lacZ, BTN2, and CUP1 was induced only twofold by treatment with 0.6 mM t-bH2O2 for 15 min (Fig. 1C). A slight increase (less than twofold) of these transcripts was observed by treatment with 0.3 to 2.4 mM H2O2 for 15 to 90 min (data not shown). In contrast to the low level of activation of the HSE-containing genes, TRX2 transcription was robustly activated by addition of 0.3 mM t-bH2O2 and CTT1 was modestly activated by higher concentrations of t-bH2O2.
These results suggest that Hsf1 is able to activate the transcription of target genes in response to menadione, diamide, and, less efficiently, peroxides. In addition, the concentrations of reagents needed to reach peak induction were different for the different genes. These dose response differences may be associated with distinct regulation of Hsf1, Skn7/Yap1, and Msn2/4 (see Discussion).
Hsf1 activates transcription in response to menadione or diamide.
Cells containing transcription factor mutations were used to ascertain which transcription factors were involved in the stress response of each gene. Because HSF1 is essential for viability, we used tetO-HSF1 cells, in which the promoter region of HSF1 was replaced with the tetracycline-regulated tetO promoter, allowing tetracycline-dependent repression of HSF1 transcription (3). When the tetracycline derivative doxycycline was added to the tetO-HSF1 cell culture, Hsf1 was rapidly depleted and was undetectable by 14 h, as judged by immunoblot analysis with an anti-Hsf1 serum (Fig. 2A). The depletion of Hsf1 caused an inhibition of heat-induced accumulation of the BTN2 and CUP1 transcripts after a temperature shift from 28°C to 39°C (Fig. 2A). The transcriptional activation of these genes by menadione and diamide treatments was not observed in the Hsf1-depleted cells, whereas that of TRX2 and CTT1 occurred normally. In contrast, these oxidizing reagents failed to induce TRX2 transcription in skn7Δ yap1Δ cells and CTT1 transcription in msn2Δ msn4Δ cells; the transcription of HSE4Ptt-CYC1-lacZ, BTN2, and CUP1 was robustly activated in these cells (Fig. 2B). These results demonstrate that menadione- or diamide-induced Hsf1 activates the transcription of target genes independently of Skn7, Yap1, Msn2, or Msn4.
FIG. 2.
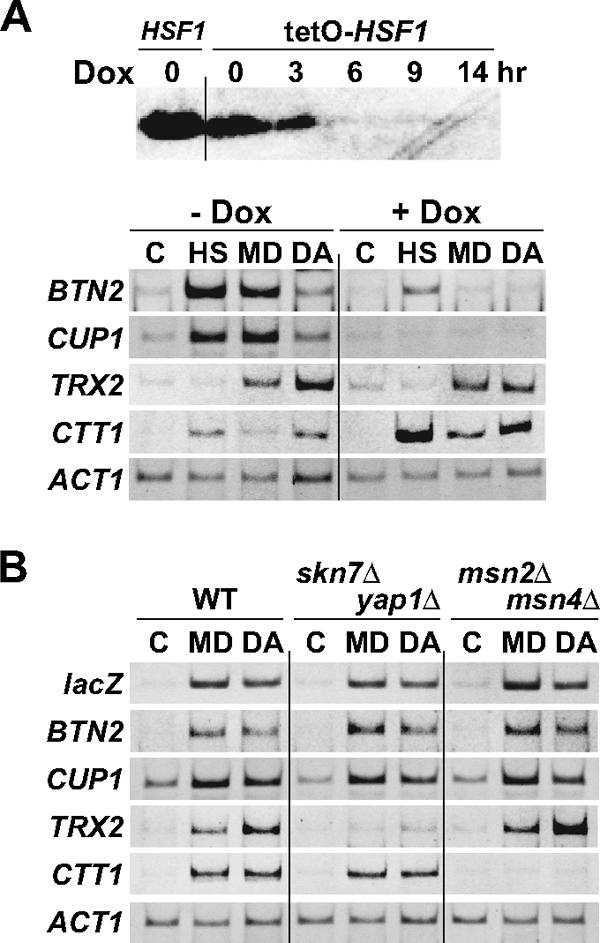
Effects of transcription factor mutations on menadione- or diamide-induced transcription. (A) mRNA levels of various genes in tetO-HSF1 cells. tetO-HSF1 cells were grown at 28°C in SD medium supplemented with 20 μg/ml histidine, 30 μg/ml leucine, and 20 μg/ml methionine. For the upper part of the panel, doxycycline (Dox) was added to a final concentration of 10 μg/ml and cells were grown for the indicated times. Extracts were prepared from the cells and subjected to immunoblot analysis with an anti-Hsf1 serum. The leftmost lane shows the amount of Hsf1 protein in HSF1 wild-type cells. For the lower part of the panel, cells cultured in the absence (− Dox) or presence (+ Dox) of 10 μg/ml doxycycline for 14 h were left untreated (C) or were treated with 0.3 mM menadione for 30 min (MD) or 3.0 mM diamide for 30 min (DA) or were grown at 39°C for 15 min (HS). Total RNA was prepared from the cells and subjected to RT-PCR analysis. (B) mRNA levels of various genes in skn7Δ yap1Δ and msn2Δ msn4Δ mutant cells. Wild-type (WT) and skn7Δ yap1Δ and msn2Δ msn4Δ mutant cells harboring HSE4Ptt-CYC1-lacZ were grown in ESD medium lacking uracil at 28°C in the absence (C) or presence of 0.3 mM menadione for 30 min (MD) or that of 3.0 mM diamide for 30 min (DA). Total RNA was prepared from the cells and subjected to RT-PCR analysis.
PKA activity negatively regulates superoxide anion-induced transcription by Hsf1.
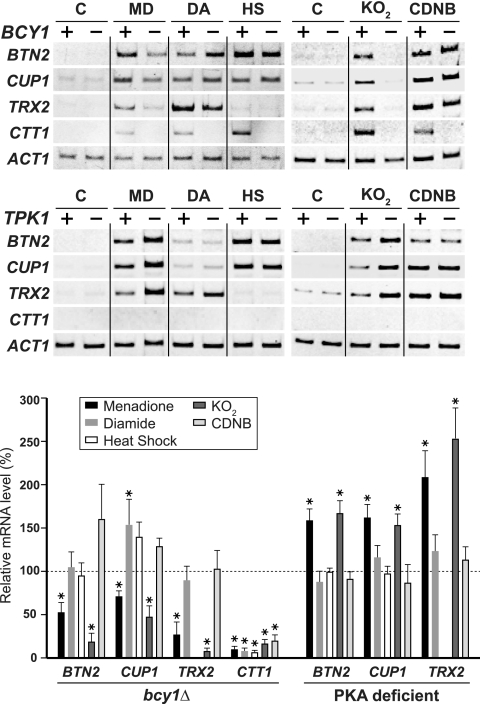
The cAMP-PKA pathway impedes the H2O2 response of Skn7, Yap1, and Msn2/4 (6, 14). We analyzed the effect of PKA activity on the oxidative stress response of Hsf1. Cells lacking BCY1, which encodes the regulatory subunit of PKA, exhibit unrestricted PKA activity (36). When bcy1Δ cells were treated with menadione, the mRNA levels of BTN2 and CUP1 were reproducibly reduced 50 to 70% relative to those in the BCY1 control (Fig. 3). Consistent with this, menadione-induced transcription of these genes was moderately inhibited by a null mutation of PDE2, which encodes the high-affinity cAMP phosphodiesterase and results in constitutive activation of PKA (data not shown). Cells lacking three redundant genes of the PKA catalytic subunit, TPK1, TPK2, and TPK3, are unable to grow, but simultaneous deletion of both MSN2 and MSN4 restores cell viability (32). In the PKA-deficient quintuple-mutant cells, the mRNA levels of Hsf1 target genes were elevated roughly 1.5-fold relative to those in cells harboring a TPK1 expression plasmid (Fig. 3). Transcriptional activation by menadione treatment was inversely proportional to PKA activity, indicating that PKA negatively regulates the menadione response of Hsf1. The effect of PKA was stress specific because BTN2 and CUP1 were activated normally by diamide or heat treatment in bcy1Δ mutant and PKA-deficient cells (Fig. 3).
FIG. 3.
Effects of mutations in cAMP-PKA pathway genes on stress-induced transcription. (Top) wild-type (+) or bcy1Δ mutant (−) cells were grown in ESD medium at 28°C in the absence (C) or presence of 0.3 mM menadione (MD), 3.0 mM diamide (DA), 5.0 mM KO2, or 0.25 mM CDNB for 30 min or were grown at 39°C for 15 min (HS). (Middle) tpk1 tpk2 tpk3 msn2 msn4 mutant cells harboring the empty vector (−) or YCp-TPK1 (+) were grown as described above, except that KO2 was added to a final concentration of 3.0 mM. Total RNA was prepared from the cells and subjected to RT-PCR analysis. (Bottom) The mRNA levels in bcy1Δ mutant and PKA-deficient cells relative to those in cognate wild-type cells are expressed as the mean ± the standard deviation of at least four independent experiments. Asterisks indicate significant differences (P < 0.01) when mutant cells are compared with wild-type cells as determined by the Student t test. Note that the relative TRX2 mRNA levels in heat-shocked cells are not shown because the levels were not substantially induced under the conditions tested.
The above results suggest that activation of Hsf1 function by the two oxidizing reagents menadione and diamide, which are used for a superoxide anion generator and a thiol-oxidizing compound, respectively (11, 38), is mediated by distinct pathways. We then investigated two other oxidizing reagents, potassium superoxide (KO2) and the thiol-specific reagent CDNB (2, 7, 19). In wild-type cells, the BTN2 and CUP1 mRNA levels were increased by KO2 or CDNB treatment (Fig. 3). PKA activity inhibited KO2-induced transcription by Hsf1, as shown by decreased mRNA levels in bcy1Δ cells and by increased mRNA levels in PKA-deficient cells. In contrast, CDNB-induced transcription was not significantly affected by PKA. These results suggest that the negative effect of PKA is involved in the oxidative stress caused by superoxide anion but not in that caused by thiol oxidation. On TRX2 transcription by Skn7/Yap1, PKA specifically inhibited the superoxide anion response, whereas it inhibited Msn2/4-mediated transcription of CTT1 under all of the stress conditions analyzed.
Oxidative stress induces binding of Hsf1 to target promoters.
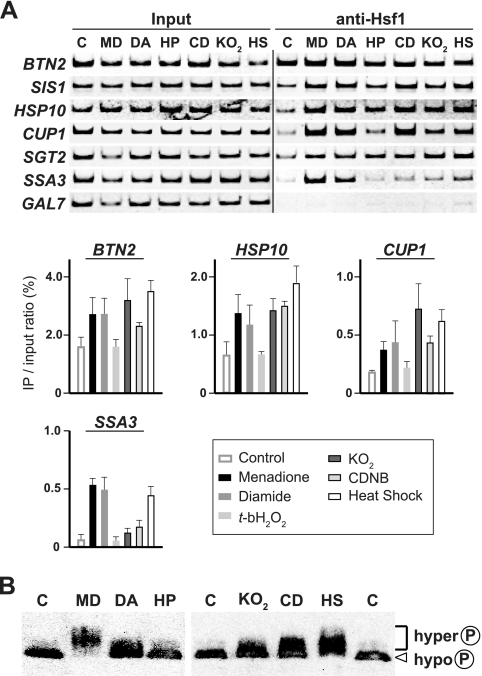
In yeast cells, Hsf1 binds with lower affinity to many target promoters under normal growth conditions and with higher affinity to most of the targets in response to heat or ethanol stress (13, 17). We tested the effect of oxidative stress on Hsf1-promoter interactions by chromatin immunoprecipitation analysis. As shown in Fig. 4A, anti-Hsf1 serum precipitated the promoter fragment of BTN2, but not that of the control nontarget gene GAL7, from extracts of cells grown at 28°C in the absence of oxidizing reagents. Treatment of cells with menadione, diamide, KO2, CDNB, and heat caused a further increase in the amount of the bound BTN2 fragments. The binding of Hsf1 to CUP1 and other target genes SIS1, HSP10, SGT2, and SSA3 was induced under these stress conditions. However, t-bH2O2 treatment had marginal effects on binding of Hsf1 to these genes. Therefore, several oxidizing reagents possess an ability to induce the binding of Hsf1 to target promoters, presumably prior to transcriptional activation.
FIG. 4.
Stress-induced changes in Hsf1 activity. (A) Binding of Hsf1 to target genes under stress conditions. Wild-type cells were grown in ESD medium at 28°C in the absence (C) or presence of 0.3 mM menadione (MD), 3.0 mM diamide (DA), 5.0 mM KO2, or 0.25 mM CDNB (CD) for 30 min or that of 0.6 mM t-bH2O2 for 15 min (HP) or were grown at 39°C for 15 min (HS). Chromatin immunoprecipitation analysis was carried out with an anti-Hsf1 serum. The Input and anti-Hsf1 panels show the PCR products amplified from the extracts before immunoprecipitation (1.0% of each sample used for immunoprecipitation) and from the immunoprecipitates, respectively. The lower part of the panel shows the immunoprecipitation (IP)/input ratios, which are expressed as the mean ± the standard deviation of three independent experiments. (B) Phosphorylation of Hsf1 under stress conditions. Wild-type cells were treated with various stressors as described above. Extracts were prepared from the cells and subjected to immunoblot analysis with an anti-Hsf1 serum.
Oxidative stress induces hyperphosphorylation of Hsf1.
Hsf1 is extensively phosphorylated upon treatment of cells with menadione, as well as in response to heat shock (24). In denaturing gels, hyperphosphorylated Hsf1 migrates more slowly than the hypophosphorylated Hsf1 protein of control cells (15, 24, 33). Addition of diamide, KO2, or CDNB to yeast cultures caused some of the Hsf1 protein to migrate more slowly on gels, as judged by immunoblot analysis, suggesting that these reagents induce the phosphorylation of Hsf1 (Fig. 4B). However, t-bH2O2 treatment did not change the mobility of the protein. These results suggest that hyperphosphorylation of Hsf1 is associated with the acquisition of the activating ability.
hsf1 mutant cells defective in both hyperphosphorylation and transcriptional activation are sensitive to menadione and diamide.
We analyzed the relationships among Hsf1 hyperphosphorylation, transcriptional activation, and oxidative stress protection by the use of hsf1-ba1 cells that contains a loss-of-function mutation in the CTM domain of Hsf1 (Fig. 5A). In the heat shock response, hyperphosphorylation of Hsf1 is inhibited by the hsf1-ba1 mutation whereas the defect is suppressed by simultaneous deletion of the inhibitory CE2 region (15). The Hsf1-ba1 protein from menadione- or diamide-treated cells exhibited the same mobility as control cells; however, the Hsf1-CE2Δ-ba1 protein restored the slower migration (Fig. 5B). When KO2 or CDNB was added to yeast cultures, Hsf1-CE2Δ-ba1, but not Hsf1-ba1, was hyperphosphorylated, judged from the electrophoretic mobility (see supplemental material). These results indicate that hyperphosphorylation of Hsf1 is regulated by the oxidizing reagents in a manner similar to that of heat shock; i.e., phosphorylation is negatively regulated by the CE2 region, and the negative effect is relieved by the CTM domain.
Treatment of HSF1 cells with menadione or diamide caused the activation of various Hsf1 target genes (Fig. 5C). In hsf1-ba1 mutant cells, transcription of genes containing HSEs with four or five nGAAn units (BTN2 and SIS1) was activated normally; however, these reagents failed to induce robust transcription of genes containing three-unit HSEs (HSP10, CUP1, SGT2, and SSA3). The requirement for the CTM domain was circumvented by the CE2 deletion in Hsf1-CE2Δ-ba1. Similar results were obtained in the KO2 and CDNB responses (see supplemental material). These results show that, similar to thermal stress, oxidative-stress-induced hyperphosphorylation of Hsf1 correlates with HSE structure-specific transcriptional activity.
The effect of the hsf1-ba1 mutation on oxidative stress protection was analyzed by a spot dilution assay. As shown in Fig. 5D, hsf1-ba1 mutant cells, but not hsf1-CE2Δ-ba1 mutant cells, were slightly sensitive to menadione and were hypersensitive to diamide. However, hsf1-ba1 mutant cells did not exhibit notable sensitivity to KO2 or CDNB, as well as to t-bH2O2 (see supplemental material). Although the reason why the hsf1-ba1 mutation causes different growth behaviors depending on the oxidizing reagents is unknown, these results suggest that Hsf1-mediated transcription is required for cell growth under oxidizing conditions. Note that the role of Hsf1 in oxidative stress resistance would be underestimated in an analysis of hsf1-ba1 mutant cells because this mutation inhibits the activation of only a subset of Hsf1 target genes. Since Hsf1 is essential for viability even under physiological conditions and activates the transcription of various genes under oxidizing conditions, we suggest that Hsf1 plays an important role in the oxidative stress response.
DISCUSSION
Oxidizing reagents menadione, diamide, KO2, and CDNB stimulate both efficient HSE binding and extensive phosphorylation of Hsf1, thereby leading to the transcriptional activation of all of the Hsf1 target genes analyzed. Target genes of Hsf1 encode chaperones, carbohydrate metabolism enzymes, and cell wall proteins and partially overlap the Msn2/4 targets (13, 41). Although which target gene is involved in oxidative stress protection remains unknown, CUP1 expression is necessary for the menadione tolerance of cells lacking the Cu, Zn superoxide dismutase gene (24). In Hsf1-mediated transcription, higher concentrations of menadione and diamide are necessary to give maximum levels of transcription, relative to Skn7/Yap1-mediated transcription that directs the expression of detoxifying enzymes and antioxidant proteins (Fig. 1A and B). We suggest that the Hsf1 regulon, in concert with the Msn2/4 regulon, is involved in refolding the damaged proteins, resetting the metabolic pathways, and remodeling the cell wall during recovery from severe oxidative stress.
The thiol-oxidizing reagents diamide and CDNB stimulated the activator function of Hsf1. In mammalian HSF1, both heat shock and H2O2 treatment induce disulfide bond formation of cysteine residues located in the DNA-binding domain, which causes binding of HSF1 to the HSE and activation of HSP gene expression (1). However, S. cerevisiae Hsf1 completely lacks cysteine residues, indicating that Hsf1 is not a direct target of thiol oxidation. CDNB preferentially reacts with the antioxidant glutathione and depletes it (19). Diamide leads to a more remarkable decrease in the ratio of reduced to oxidized glutathione in yeast cells than H2O2 or t-bH2O2 (31). In addition, treatment with menadione, a superoxide anion generator, results in a decrease in the intracellular concentration of glutathione, the formation of a menadione-glutathione conjugate, and the export of the conjugate from the cells (43). These findings lead us to infer that thiol oxidation is sensed by glutathione and signaled to Hsf1.
Menadione, through redox cycling, generates superoxide anion, which undergoes a dismutation reaction to generate hydrogen peroxide (37). However, the adaptive response to menadione is different from that to H2O2 in S. cerevisiae (10, 20). Treatment of cells with KO2, but not H2O2 or t-bH2O2, caused the efficient transcription of Hsf1 target genes, suggesting that superoxide anion is an inducer of Hsf1 activity. It should be noted that Hsf1 activation by menadione and KO2 is inhibited by PKA activity. Lee et al. (23) have shown by in vitro DNA-binding assays that Hsf1 undergoes a conformational change to a putative active form in response to superoxide anion and elevated temperatures, but not in response to hydrogen peroxide. Our observations support this idea and further suggest that superoxide anion also stimulates both efficient HSE binding and extensive phosphorylation of Hsf1.
Under normal growth conditions, Hsf1 exhibits a relatively higher binding affinity for some promoters, indicating that the enhanced binding alone does not trigger transcriptional activation. Hyperphosphorylation of Hsf1 is regulated by the CTM-CE2 interaction and required for activation by a single Hsf1 trimer bound to HSEs with three nGAAn units. Although Hsf1 is differentially phosphorylated under distinct stress conditions, as judged by gel electrophoretic (Fig. 4B) and phosphopeptide mapping (24) analyses, some important amino acid residues may be commonly phosphorylated. We suggest that the common phosphorylations have a critical role in the establishment and/or maintenance of the activator function of the single Hsf1 trimer. In contrast, two trimers bound to HSEs with four or more nGAAn units bypass the requirement of hyperphosphorylation for transcriptional activation. Lee et al. (23) have also shown that two trimers, but not a single trimer, adopt the active conformation when stressed. We suggest that the conformational change is involved in hyperphosphorylation-independent activation by Hsf1 trimers.
Transcription factors Hsf1, Msn2/4, Skn7, and Yap1 activated the transcription of the respective target genes when cells were treated with the superoxide anion generators menadione and KO2, the thiol-oxidizing compounds diamide and CDNB, or heat. The activated PKA (bcy1Δ) almost completely inhibited the activator function of Msn2/4 induced by these stressors. In Hsf1- or Skn7/Yap1-mediated transcription, the cAMP-PKA pathway inhibited the stress response induced by superoxide anion but not that induced by thiol oxidation or heat. Although it is unknown why PKA specifically inhibits the superoxide anion response, the negative role of PKA is consistent with the observations that expression of stress-protective genes in the H2O2 response is inversely proportional to PKA activity and that cells with low PKA activity are more H2O2 resistant than cells with high PKA activity (6, 14). In this regard, PKA is a negative regulator in the oxidative stress response caused by ROS but not in that caused by thiol oxidation.
An effect of PKA on Hsf1 activity was recently reported in which PKA inhibits the constitutive phosphorylation and basal-level transcriptional activity of Hsf1 under normal growth condition (8). We found that PKA activity slightly inhibited the menadione-induced slower migration of Hsf1 on gels; however, its significance in the regulation of transcriptional activity remains to be elucidated (data not shown). PKA restrains the binding of Yap1 to target genes and the nuclear localization of Msn2/4 (9, 12). Our preliminary chromatin immunoprecipitation analysis results showed that PKA did not affect promoter occupancy by Hsf1 in menadione-treated cells (data not shown). It is possible that menadione (or superoxide anion) affects the cAMP-PKA pathway regulating the activity of the general RNA polymerase II transcription apparatus (5). Further study is required to address the role of PKA in Hsf1-mediated transcription. Whatever the mechanism, our results demonstrate that PKA is a general negative regulator of the ROS response, especially the superoxide anion response, by its effect on a group of transcriptional activators.
Supplementary Material
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to H.S.
Footnotes
Published ahead of print on 22 June 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Ahn, S. G., and D. J. Thiele. 2003. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 17:516-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso-Monge, R., F. Navarro-García, E. Román, A. I. Negredo, B. Eximan, C. Nombela, and J. Pla. 2003. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belli, G., E. Gari, M. Aldea, and E. Herrero. 1998. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14:1127-1138. [DOI] [PubMed] [Google Scholar]
- 4.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y. W., S. C. Howard, and P. K. Herman. 2004. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell 15:107-116. [DOI] [PubMed] [Google Scholar]
- 6.Charizanis, C., H. Juhnke, B. Krems, and K. D. Entian. 1999. The oxidative stress response mediated via Pos9/Skn7 is negatively regulated by the Ras/PKA pathway in Saccharomyces cerevisiae. Mol. Gen. Genet. 261:740-752. [DOI] [PubMed] [Google Scholar]
- 7.Chauhan, N., D. Inglis, E. Roman, J. Pla, D. Li, J. A. Calera, and R. Calderone. 2003. Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryot. Cell 2:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson, S. B., E. S. Anderson, R. B. Harshaw, T. Thate, N. L. Craig, and H. C. Nelson. 2005. Protein kinase A regulates constitutive expression of small heat-shock genes in an Msn2/4p-independent and Hsf1p-dependent manner in Saccharomyces cerevisiae. Genetics 169:1203-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandes, L., C. Rodrigues-Pousada, and K. Struhl. 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17:6982-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flattery-O'Brien, J., L. P. Collinson, and I. W. Dawes. 1993. Saccharomyces cerevisiae has an inducible response to menadione which differs from that to hydrogen peroxide. J. Gen. Microbiol. 139:501-507. [DOI] [PubMed] [Google Scholar]
- 11.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, J. S., Z. Hu, D. J. Thiele, and V. R. Iyer. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24:5249-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasan, R., C. Leroy, A. D. Isnard, J. Labarre, E. Boy-Marcotte, and M. B. Toledano. 2002. The control of the yeast H2O2 response by the Msn2/4 transcription factors. Mol. Microbiol. 45:233-241. [DOI] [PubMed] [Google Scholar]
- 15.Hashikawa, N., and H. Sakurai. 2004. Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol. Cell. Biol. 24:3648-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashikawa, N., Y. Mizukami, H. Imazu, and H. Sakurai. 2006. Mutated yeast heat shock transcription factor activates transcription independently of hyperphosphorylation. J. Biol. Chem. 281:3936-3942. [DOI] [PubMed] [Google Scholar]
- 17.Hashikawa, N., N. Yamamoto, and H. Sakurai. 2007. Different mechanisms are involved in the transcriptional activation by yeast heat shock transcription factor through two different types of heat shock elements. J. Biol. Chem. 282:10333-10340. [DOI] [PubMed] [Google Scholar]
- 18.Imazu, H., and H. Sakurai. 2005. Saccharomyces cerevisiae heat shock transcription factor regulates cell wall remodeling in response to heat shock. Eukaryot. Cell 4:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izawa, S., Y. Inoue, and A. Kimura. 1995. Oxidative stress response in yeast: effect of glutathione on adaptation to hydrogen peroxide stress in Saccharomyces cerevisiae. FEBS Lett. 368:73-76. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson, D. J. 1992. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J. Bacteriol. 174: 6678-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krems, B., C. Charizanis, and K. D. Entian. 1996. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29:327-334. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040-16046. [DOI] [PubMed] [Google Scholar]
- 23.Lee, S., T. Carlson, N. Christian, K. Lea, J. Kedzie, J. P. Reilly, and J. J. Bonner. 2000. The yeast heat shock transcription factor changes conformation in response to superoxide and temperature. Mol. Biol. Cell 11:1753-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, X. D., and D. J. Thiele. 1996. Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 10:592-603. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227-2235. [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moye-Rowley, W. S. 2003. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot. Cell 2:381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pirkkala, L., P. Nykanen, and L. Sistonen. 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15:1118-1131. [DOI] [PubMed] [Google Scholar]
- 29.Raitt, D. C., A. L. Johnson, A. M. Erkine, K. Makino, B. Morgan, D. S. Gross, and L. H. Johnston. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 11:2335-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shenton, D., G. Perrone, K. A. Quinn, I. W. Dawes, and C. M. Grant. 2002. Regulation of protein S-thiolation by glutaredoxin 5 in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 277:16853-16859. [DOI] [PubMed] [Google Scholar]
- 32.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorger, P. K., and H. R. Pelham. 1988. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54:855-864. [DOI] [PubMed] [Google Scholar]
- 34.Takemori, Y., A. Sakaguchi, S. Matsuda, Y. Mizukami, and H. Sakurai. 2005. Stress-induced transcription of the endoplasmic reticulum oxidoreductin gene ERO1 in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 275:89-96. [DOI] [PubMed] [Google Scholar]
- 35.Temple, M. D., G. G. Perrone, and I. W. Dawes. 2005. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 15:319-326. [DOI] [PubMed] [Google Scholar]
- 36.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 37.Thor, H., M. T. Smith, P. Hartzell, G. Bellomo, S. A. Jewell, and S. Orrenius. 1982. The metabolism of menadione (2-methyl-1,4-naphthoquinone) by isolated hepatocytes: a study of the implications of oxidative stress in intact cells. J. Biol. Chem. 257:12419-12425. [PubMed] [Google Scholar]
- 38.Thorpe, G. W., C. S. Fong, N. Alic, V. J. Higgins, and I. W. Dawes. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. USA 101:6564-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trott, A., and K. A. Morano. 2003. The yeast response to heat shock, p. 71-119. In S. Hohmann and P. W. H. Mager (ed.), Yeast stress responses. Springer-Verlag, Heidelberg, Germany.
- 40.Voellmy, R. 2004. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9:122-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto, A., Y. Mizukami, and H. Sakurai. 2005. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 280:11911-11919. [DOI] [PubMed] [Google Scholar]
- 42.Yan, L. J., E. S. Christians, L. Liu, X. Xiao, R. S. Sohal, and I. J. Benjamin. 2002. Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. EMBO J. 21:5164-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zadzinski, R., A. Fortuniak, T. Bilinski, M. Grey, and G. Bartosz. 1998. Menadione toxicity in Saccharomyces cerevisiae cells: activation by conjugation with glutathione. Biochem. Mol. Biol. Int. 44:747-759. [DOI] [PubMed] [Google Scholar]
- 44.Zhong, M., A. Orosz, and C. Wu. 1998. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol. Cell 2:101-108. [DOI] [PubMed] [Google Scholar]
- 45.Zou, J., W. F. Salminen, S. M. Roberts, and R. Voellmy. 1998. Correlation between glutathione oxidation and trimerization of heat shock factor 1, an early step in stress induction of the Hsp response. Cell Stress Chaperones 3:130-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.