Abstract
Telomerase is an RNA-protein complex responsible for extending one strand of the telomere terminal repeats. Analysis of the telomerase complex in budding yeasts has revealed the presence of one catalytic protein subunit (Est2p/TERT) and at least two noncatalytic components (Est1p and Est3p). The TERT subunit is essential for telomerase catalysis, while the functions of Est1p and Est3p have not been precisely elucidated. In an earlier study, we showed that telomerase derived from a Candida est1-null mutant is defective in primer utilization in vitro; it exhibits reduced initiation and processivity on primers that terminate in two regions of the telomere repeat. Here we show that telomerase derived from a Candida est3-null mutant has nearly identical defects in primer utilization and processivity. Further analysis revealed an unexpected mutual dependence of Est1p and Est3p in their assembly into the full telomerase complex, which accounts for the similarity between the mutant enzymes. We also developed an affinity isolation and an in vitro reconstitution protocol for the telomerase complex that will facilitate future mechanistic studies.
Telomerase is a ribonucleoprotein (RNP) that is responsible for maintaining the terminal repeats of telomeres in most organisms (1, 3, 5, 7, 25). The catalytic core of telomerase consists minimally of two components: an RNA in which the template is embedded (named TER) and a reverse transcriptase (RT)-like protein that mediates catalysis (named TERT in general and Est2p in yeast). In addition, telomerases from different organisms have been shown to possess a number of accessory or regulatory subunits that promote telomerase RNP biogenesis and assembly. However, with few exceptions, these accessory components do not appear to participate directly in the telomere extension function of telomerase. Two notable exceptions to this generalization are Est1p and Est3p in the budding yeast Saccharomyces cerevisiae (12, 24). Both were identified through genetic screens and shown to act in the same pathway as telomerase RNA and TERT and to be subunits of the telomerase complex but not essential for in vitro activity. Est1p has been shown to bind telomerase RNA, telomeric DNA, and Cdc13p (a G-strand telomere-binding protein) (16, 26). Recent studies further implicate Est1p in promoting the recruitment of the telomerase complex to telomere ends in vivo (2, 9, 15). Est1p is also believed to perform a postrecruitment or activation function, but the biochemical nature of this function is obscure (8, 23). The assembly of Est3p into the telomerase complex is known to require both TERT and Est1p, but the mechanisms of this protein in promoting telomere extension are not understood (11, 14).
We recently identified putative orthologues of TERT/EST2, EST1, and EST3 in the genome of the pathogenic fungus Candida albicans. Analysis of knockout strains indicates that the Candida homologues are all required for normal telomere maintenance. C. albicans EST2 was found to be essential for telomerase activity in vitro, consistent with a catalytic role for this protein (21). Unexpectedly, we uncovered a primer-specific impairment of function for the telomerase derived from an est1Δ/est1Δ strain in vitro. The mutant enzyme exhibited reduced abilities to catalyze nucleotide addition at two loci within the Candida telomere repeat (20). This represented the first instance in which a mutation in a noncore component of yeast telomerase was observed to alter the biochemical property of telomerase. In this report, we describe the development of an affinity pulldown protocol for isolating Candida telomerase for in vitro analysis. Using this protocol, we examined the properties of telomerase and found that the est3Δ/est3Δ mutant exhibited defects similar to those of the est1Δ/est1Δ mutant. Further analysis revealed an unexpected mutual dependence of Est1p and Est3p in their assembly into the full telomerase complex, which accounts for the primer specificity of both mutant enzymes. We propose that the previously suggested “activating” function of Est1p is due to its ability to promote Est3p assembly.
MATERIALS AND METHODS
Strains.
C. albicans strain BWP17 (ura3Δ::λimm434/ura3Δ::λimm434 his1:: hisG/his1::hisG arg4::hisG/arg4::hisG) was obtained from A. Mitchell (Columbia University) (27). The Candida est1Δ/est1Δ and est3Δ/est3Δ strains were constructed from BWP17 using URA-Blaster cassettes as previously described (22). For introduction of a FLAG3/TAP-tagged EST2, we transformed Candida strains with the tagging plasmid pBS-CaEST2-TAG-URA linearized by BstEII. This plasmid was constructed as follows. First, a 3.5-kb PCR fragment containing the EST2 open reading frame (ORF) and upstream and downstream sequences was inserted between the KpnI and PstI sites of pBluescript II KS(+). The C terminus of the EST2 ORF was then mutagenized to introduce an NcoI and an AatII site. The TAP (tandem affinity purification) tag from PBS1479 was amplified by PCR and inserted between the NcoI and AatII sites. An oligonucleotide encoding the FLAG3 tag was further inserted into the NcoI site of the plasmid. Finally, a PstI-to-SacII fragment from pGEM-URA3 (containing the URA3 marker; a gift of A. Mitchell, Columbia University) was introduced to yield pBS-CaEST2-TAG-URA. For introduction of protein A-tagged EST1 and EST3, the strains were transformed with pBS-CaEST1-proA-HIS and pBS-CaEST3-proA-HIS linearized by SnaBI. The plasmids were constructed as follows. First, a PCR fragment containing the EST1 (or EST3) ORF and upstream and downstream sequences was inserted between the HindIII and SacI sites of pBluescript II KS(+). The C terminus of the ORF was then mutagenized to introduce an AflII and an NheI site. The protein A tag from pEZZ18 (GE Healthcare Inc.) was amplified by PCR and inserted between the AflII and NheI sites. Finally, a SalI-to-ApaI fragment from pGEM-HIS1 (containing the HIS1 marker; a gift of A. Mitchell, Columbia University) was introduced to yield pBS-CaEST1-proA-HIS (or pBS-CaEST3-proA-HIS). Thus, each tagging plasmid contains the native gene fused at its C terminus to the tag and flanked by native 5′ and 3′ sequences. Proper integration of the tagged genes at the respective native chromosomal loci was confirmed by Southern blot analysis. Each tagged strain contains one copy of the tagged gene.
Telomere length analysis.
Chromosomal DNAs were isolated from 4 to 5 ml of saturated C. albicans culture by the smash-and-grab method, digested with AluI and NlaIII, and fractionated in 0.9% agarose gels, and the telomere restriction fragments were detected by Southern blotting as previously described (21).
Purification of and assay for C. albicans telomerase.
TMG(n) buffer (10 mM Tris-HCl [pH 8.0], 1.2 mM MgCl2, 0.1 mM EDTA, 0.1 mM EGTA, 10% glycerol [n refers to the millimolar concentration of sodium acetate]) was used throughout the study. Whole-cell extracts of C. albicans and DEAE column fractions were prepared as previously described (20, 21). For isolation of TAP-tagged Candida telomerase on immunoglobulin G (IgG)-Sepharose beads, we followed a previously developed protocol for protein A-tagged Saccharomyces telomerase (4). Briefly, 15 μl of IgG-Sepharose was mixed with 4 mg of extracts in TMG(400) plus 0.05% Tween 20 at 4°C for 2 h. The beads were then washed twice in TMG(600) and twice in TMG(0). Telomerase primer extension assays were performed using 12-nucleotide (nt) primers and a single labeled nucleotide triphosphate that can be incorporated at the primer +1 (or primer +1 and primer +2) position (20).
In vitro reconstitution of wild-type telomerase activity.
For in vitro reconstitution, we used extracts derived from est1Δ/est1Δ and est3Δ/est3Δ strains. The est3Δ/est3Δ strain also carried one copy of FLAG3/TAP-tagged EST2 (in addition to two copies of untagged EST2) to facilitate purification. Approximately 3 mg each of the protein extracts was mixed at 22°C for 20 min in 1.2 ml TMG(0). The entire mixture was then subjected to IgG-Sepharose pulldown and a telomerase primer extension assay as described before (20). For in vitro synthesis of Candida Est3p, the EST3 ORF was first amplified by PCR and cloned between the BamHI and EcoRI sites of the pCITE-4a vector (Novagen Inc.) to give pCITE-CaEST3. The Candida EST3 gene encodes two CUG codons (codons 18 and 172), which are abnormally translated in this organism into Ser instead of Leu. These codons in pCITE-CaEST3 were mutated to UCG by the QuikChange protocol (Stratagene Inc.) to allow for expression of the native protein in heterologous systems. The protein was expressed using the plasmid and the TNT-coupled reticulocyte lysate system (Promega Inc.).
Analysis of the association of Est1p and Est3p with the telomerase core complex.
IgG-Sepharose beads (45 μl) were pretreated with 100 μg tRNA in 1 ml TMG(0) at 4°C for 30 min to minimize nonspecific binding. After one wash in TMG(0), the beads were incubated with extracts (4.8 mg) in 1.2 ml TMG(500) and subjected to gentle rotation at 4°C for 2 h. The beads were then washed five times with TMG(800) and twice with TMG(0) and were divided into three equal aliquots. One aliquot each was subjected to Western blot analysis and a telomerase activity assay as previously described (4, 28). The last aliquot was treated with proteinase K and extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and the RNA was isolated by ethanol precipitation. The level of TER (Candida telomerase RNA) was then quantified by semiquantitative RT-PCR (2× RT-PCR master mix; USB Corporation) using primer pairs designed to amplify a 350-bp fragment (forward, 5′-CCCATATTCAATGCTCTTGGAGTGTG-3′; reverse, 5′-CTCCACAAGGTATCATACAAATTATGG-3′). The linearity of the assay was confirmed by titrating the samples and by using different cycle numbers. The pulldown assays for Est1-proA and Est3-proA were each performed three or four times, and the results were highly reproducible.
RESULTS
Development of an affinity isolation procedure for Candida telomerase.
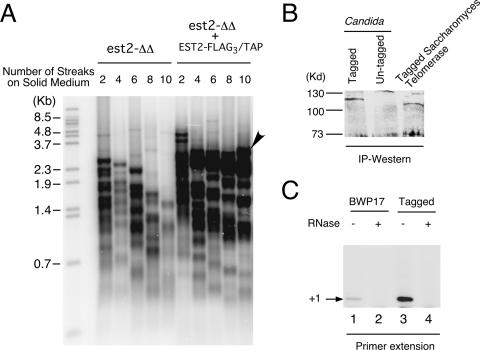
We had earlier developed a partial purification procedure for C. albicans telomerase based on DEAE chromatography (21). However, because the procedure requires elution from the column using a high-salt buffer, it is possible that loosely bound subunits were lost. To develop an alternative purification procedure, we constructed an integrating plasmid with a URA3 marker and an EST2 gene fused at its C terminus to the FLAG3 and TAP tag (17). The functionality of the tagged gene was tested by integrating the plasmid into an est2Δ/est2Δ strain and examining the lengths of telomeres in the resulting strain over many generations. As shown in Fig. 1A, the reconstituted strain exhibited stable telomere hybridization signals over time, in contrast to the progressive loss observed in the est2Δ/est2Δ2 mutant. While some telomeres in the reconstituted strain appear to shorten over time, this loss is apparently compensated by the emergence of long telomeres (Fig. 1A). The telomeres in the reconstituted strain were slightly shorter (lengths, ∼1 to 3.5 kb) than those in the original parental strain, BWP17 (∼2 to 5 kb) (21, 22). This reduction suggests that one copy of Candida EST2 may be unable to maintain telomeres in this strain background. Alternatively, tagging of the C terminus of EST2 might have resulted in partial loss of function. Interestingly, strain BWP17 was recently shown to be monozygous for EST1 due to a large chromosomal deletion (18), yet it maintains substantially longer telomeres than another commonly used laboratory strain, CAI4 (6). Because of widespread aneuploidy in Candida laboratory strains, careful comparison of isogenic strains carrying different copy numbers of telomerase genes is clearly necessary to establish the dosage effect of telomerase genes on telomere length. Notably, the S. cerevisiae telomerase RNA gene was recently demonstrated to be haploinsufficient (13), providing motivation for further analysis of this issue in Candida. In any case, our results clearly show that the tagged EST2 gene is capable of complementing telomerase function in vivo.
FIG. 1.
Functional analysis of FLAG3/TAP-tagged Candida EST2. (A) Telomeres from successive streaks of an est2Δ/est2Δ (est2-ΔΔ) strain and an est2Δ/est2Δ EST2-FLAG3/TAP strain were analyzed by Southern blotting. The streaks from which the DNAs were derived are indicated at the top. Arrowhead indicates the emergence of a long telomere. (B) The expression of FLAG3/TAP-tagged Candida Est2p was analyzed by IgG-Sepharose pulldown and Western blotting. For comparison, an extract derived from strain BWP17 (untagged) and an extract containing protein A-tagged S. cerevisiae Est2p were assayed in parallel. IP, immunoprecipitation. (C) Extracts prepared from BWP17 and a strain with FLAG3/TAP-tagged Est2p were subjected to IgG-Sepharose pulldown, and the telomerase activity in the beads was analyzed by direct primer extension assays.
We then examined the expression of FLAG3/TAP-tagged Est2p in cell extracts by IgG-Sepharose pulldown and Western blotting. As expected, the tagged but not the untagged strain contained an ∼115-kDa protein that reacted with anti-protein A antibodies (Fig. 1B). For comparison, we also analyzed concurrently the expression of a protein A-tagged S. cerevisiae Est2p and identified a protein of slightly greater mobility. The intensities of the Western blot signals suggest that the two telomerase catalytic subunits are expressed at comparable levels in S. cerevisiae and C. albicans. To determine if active Candida telomerase can be isolated on IgG-Sepharose, we subjected the treated beads to primer extension assays. As shown in Fig. 1C, beads incubated with an extract derived from the tagged strain contained much higher levels of RNase-sensitive primer extension products than those incubated with an extract from an untagged strain, consistent with specific binding of telomerase to IgG-Sepharose. Notably, some nonspecific binding of untagged telomerase to the beads can also be observed (Fig. 1C, lanes 1 and 2). This nonspecific binding can be further reduced by higher salt concentrations during the binding reaction (data not shown).
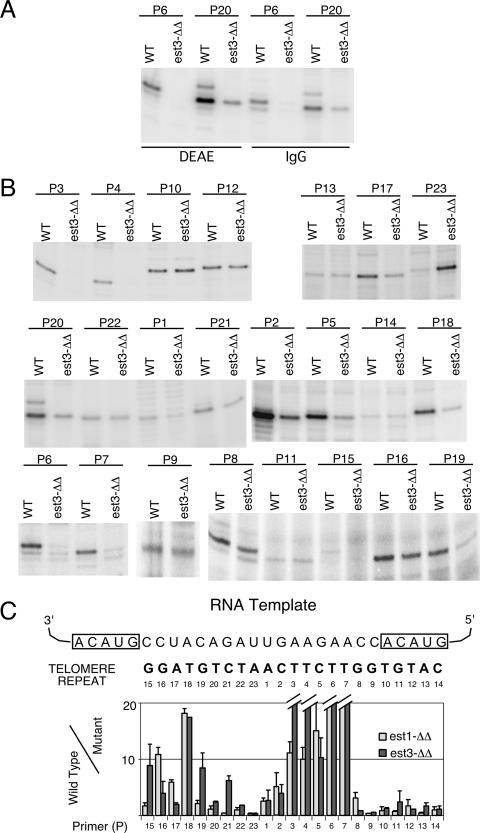
As reported previously, Candida telomerase purified by DEAE chromatography exhibits different activity levels on different primers (20). We compared the relative activities of DEAE- and IgG-Sepharose-derived telomerases using several different primers and found that they exhibited similar activity profiles, suggesting that the methods yielded telomerases of similar compositions and properties (Fig. 2A and data not shown). For the remainder of this study, we used exclusively IgG-Sepharose-bound telomerase.
FIG. 2.
Telomerase from an est3Δ/est3Δ mutant exhibited primer-specific loss of primer extension activity in vitro. (A) Telomerases from the tagged BWP17 (wild-type [WT]) and est3Δ/est3Δ (est3-ΔΔ) strains were isolated by either DEAE chromatography or IgG-Sepharose pulldown and tested for primer extension activity using primers P6 and P20 in the presence of labeled dTTP. (B) Telomerases isolated from the tagged BWP17 and est3Δ/est3Δ strains by IgG-Sepharose pulldown were tested for primer extension activity in vitro using a series of 12-nt primers that correspond to different permutations of the Candida telomere repeat. Pairs of representative assays are shown, and primer names are given at the top. (C) The activities obtained from assays such as those for which results are shown in panels A and B were quantified by a PhosphorImager. The ratios of wild-type to est3Δ/est3Δ telomerase activity on different primers were calculated, and the average ratios (and deviations) from two or three independent experiments were plotted (dark bars). For comparison, the results obtained for est1Δ/est1Δ telomerase from an earlier study were also plotted (light bars).
Candida telomerase derived from the est3Δ/est3Δ mutant is defective in the utilization of specific primers in vitro.
To examine the primer utilization properties of est3Δ/est3Δ telomerase, we adopted the approach that was used earlier for est1Δ/est1Δ telomerase (20). The wild-type and mutant enzymes were isolated from strains with FLAG3/TAP-tagged Est2p by IgG-Sepharose pulldown and were subjected to primer extension assays using a series of 12-mer oligonucleotides (each with a different 3′ end) that correspond to all the different permutations of the Candida repeat (Fig. 2B). Each primer was designated by the position of the first nucleotide added. (Thus, primer P5 has a 3′ end that corresponds to position 4 in Fig. 2C, and the first nucleotide added is dC at position 5.) To simplify the analysis, we assayed wild-type and mutant enzymes using just one labeled nucleotide such that only the primer +1 (or primer +2) product was generated. As shown in Fig. 2B and C, this “single-nucleotide-addition” assay revealed the existence of two regions within the telomere repeat (at positions 3 to 7 and positions 18 to 19) where the deletion of EST3 greatly reduced enzyme activity. At other positions, the loss of Est3p either had no effect or resulted in greater DNA synthesis, especially at position 23. Remarkably, this pattern of primer dependence closely mirrors that observed earlier for the telomerase derived from the est1Δ/est1Δ strain (Fig. 2C) (20).
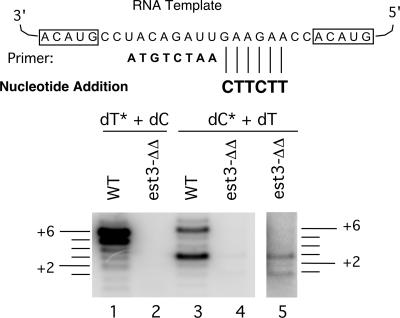
Earlier studies of est1Δ/est1Δ telomerase revealed additionally an elongation or processivity defect in assays that allowed multiple nucleotide addition (20). To determine if this is true for est3Δ/est3Δ telomerase also, we tested the activities of wild-type and mutant enzymes by using an 8-nt primer to which 6 nt can be added in the presence of dCTP and dTTP (Fig. 3). These 6 nt correspond to an EST1- or EST3-responsive region in the single-nucleotide-addition assay (positions 2 to 7). Careful inspection of the banding pattern revealed not only an initiation defect (Fig. 3, compare lanes 1 and 2 with lanes 3 and 4) but also an elongation defect for the est3Δ/est3Δ mutant. Specifically, in the presence of labeled dCTP and unlabeled dTTP, the wild-type enzyme generated predominantly +3 and +6 products (lane 3), while the mutant enzyme generated predominantly +1 and +3 products (lane 5). Thus, both the initiation defect and the elongation defect of the est1Δ/est1Δ telomerase are recapitulated by the est3Δ/est3Δ telomerase.
FIG. 3.
Telomerase from the est3Δ/est3Δ (est3-ΔΔ) mutant exhibited an elongation defect. Telomerases derived from a wild-type (WT) and an est3Δ/est3Δ strain were subjected to primer extension analysis using the indicated 8-nt primer in the presence of dCTP and dTTP, one of which was labeled (*). The labeled and unlabeled nucleotides were included at 0.2 μM and 50 μM, respectively. The sizes of the products relative to the input primer are indicated by horizontal lines to the left and right of the panel. Lane 5 is an overexposure of lane 4.
Reconstitution of full telomerase activity from mutant extracts.
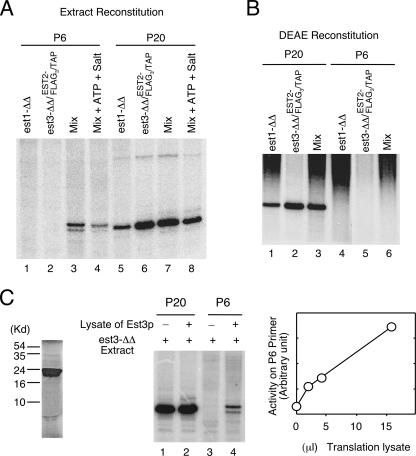
The similarity in the primer extension defects of the est1Δ/est1Δ and est3Δ/est3Δ telomerases raises interesting questions concerning the interaction between the two subunits. One possibility that could explain the similarity is that deletion of one subunit may lead to the destabilization and loss of the other. To address this possibility and to begin to develop an in vitro reconstitution protocol for Candida telomerase, we attempted to recover wild-type telomerase activity by mixing extracts derived from the est1Δ/est1Δ and est3Δ/est3Δ strains. To facilitate the analysis, we used the est3Δ/est3Δ strain that had been modified by the introduction of the EST2-FLAG3/TAP allele. After the extracts were mixed, telomerase was isolated on IgG-Sepharose and subjected to primer extension assays using either an Est1- and Est3-hyperresponsive primer (P6) or a nonresponsive primer (P20) (Fig. 4A). As expected, the mutant telomerase failed to extend P6 but was quite active on P20 (Fig. 4A, lanes 1, 2, 5, and 6). Interestingly, significant activity could be detected on P6 in the mixed sample (Fig. 4A, lanes 3 and 4). This result indicates that the nondeleted component remained present in the extracts and could associate with the core components to reconstitute full telomerase activity. Notably, the inclusion of ATP in the mixing reaction did not enhance reconstitution, suggesting that assembly is not energy dependent (Fig. 4A and data not shown). We also attempted to recover wild-type telomerase by mixing partially purified DEAE fractions but were unable to detect activity on P6 in these assays (Fig. 4B). Together, our results suggest that Est1p and Est3p each remain stable in each other's absence but that each may not associate with the telomerase complex without its partner.
FIG. 4.
Reconstitution of full telomerase activity from mutant extracts in vitro. (A) Extracts derived from an est1Δ/est1Δ (est1-ΔΔ) and an est3Δ/est3Δ (est3-ΔΔ) EST2-FLAG3/TAP strain were either separately subjected to IgG-Sepharose pulldown or subjected to pulldown following mixing. Telomerase activity on the beads was analyzed by primer extension assays using either the P6 or the P20 primer. (B) DEAE fractions (400 mM to 900 mM) were derived from the same strains as those used for panel A. The fractions were assayed separately or after mixing as indicated at the top of the gel. (C) (Left) Labeled Est3p obtained by in vitro transcription and translation was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and a PhosphorImager. (Center) Extracts derived from the est3Δ/est3Δ EST2-FLAG3/TAP strain were mixed with 15 μl of mock-translated lysates or Est3p-containing lysates. Telomerases in the mixtures were isolated on IgG-Sepharose and subjected to primer extension using the indicated primers. (Right) Extracts derived from the est3Δ/est3Δ EST2-FLAG3/TAP strain were mixed with increasing amounts of Est3p-containing lysates. Telomerases in the mixtures were isolated on IgG-Sepharose and subjected to primer extension using the P6 primer. The signals were quantified and plotted.
To further test the requirement for functional reconstitution, we examined the ability of in vitro-synthesized Est3p to complement the est3Δ/est3Δ extract. Radioactively labeled Candida Est3p can be identified following a coupled transcription-translation reaction (Fig. 4C, left panel). Addition of an Est3p-containing lysate (but not a control lysate) to the est3Δ/est3Δ extract followed by IgG-Sepharose pulldown resulted in a telomerase preparation that was active on P6 (Fig. 4C, center panel). In addition, the signals were proportional to the amount of Est3p lysate added (Fig. 4C, right panel). Thus, it appears that the only factor missing in the est3Δ/est3Δ extract is Est3p. The ability to incorporate in vitro-synthesized Est3p into the telomerase complex should greatly facilitate future structure-function analysis of Candida Est3p. As far as we are aware, these results represent the development of the first functional reconstitution assays for yeast telomerase regulatory components in vitro.
The association of Est3p with Candida telomerase depends on Est1p.
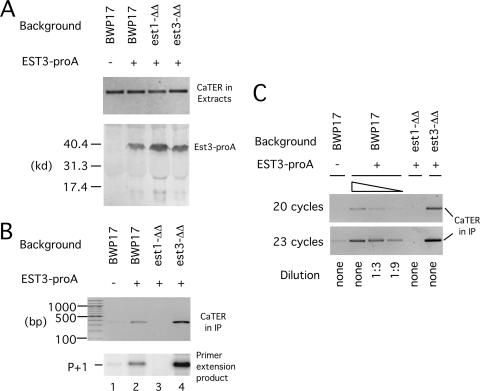
The foregoing results suggest that in C. albicans, either Est1p or Est3p depends on the other regulatory subunit for incorporation into the telomerase complex. Indeed, a recent report demonstrates that in S. cerevisiae, the association of Est3p with telomerase is Est1p dependent, but not vice versa (14). To determine if this is also true for C. albicans, we generated a set of strains containing protein A-tagged Est3p and monitored its association with Candida telomerase RNA (TER) in the presence or absence of Est1p. Southern blot analysis revealed that the tagged EST3 gene (EST3-proA) was capable of maintaining telomeres in the est3Δ/est3Δ background (Fig. 5), indicating that tagging did not abolish function. In addition, we found that a tagged protein of the expected size for Est3-proA was detected in extracts from transformed strains regardless of the presence or absence of Est1p (Fig. 6A). Thus, the stability of Est3p is not compromised in the absence of Est1p, in accordance with the reconstitution analysis. However, the level of Est3p-associated TER as judged by IgG-Sepharose pulldown assays was greatly reduced in the est1Δ/est1Δ strain background in three independent experiments (Fig. 6B and data not shown). Titration studies indicated that the amount of Est3p-associated TER was reduced at least 10-fold in the absence of Est1p (Fig. 6C). In contrast, the levels of TER in the starting extracts were unaffected by deletions of either EST1 or EST3 (Fig. 6A). The association between Est3p and TER likely reflected Est3p binding to active telomerase, because the levels of TER in the precipitates closely paralleled those of telomerase activity (Fig. 6B, compare the two panels). We conclude that, as in S. cerevisiae, the assembly of Est3p into telomerase in C. albicans is Est1p dependent. Notably, the levels of Est3-proA-associated TER and telomerase were reproducibly higher in the est3Δ/est3Δ strain background than in the BWP17 background (Fig. 6B, lanes 2 and 4). This difference was most likely due to the presence of two wild-type copies of EST3 in BWP17, which resulted in the expression of untagged Est3p that could compete with protein A-tagged Est3p for association with telomerase.
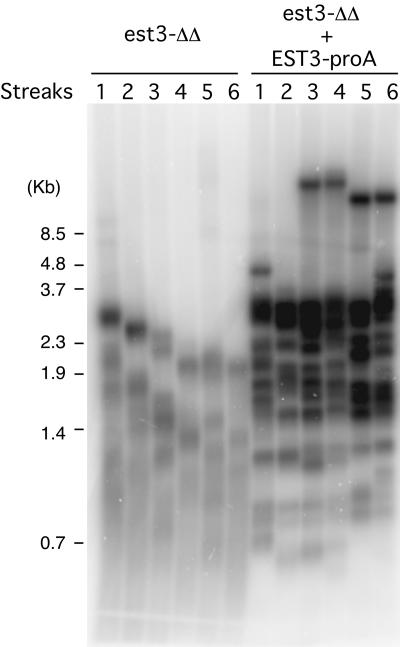
FIG. 5.
The protein A-tagged EST3 gene is functional. Chromosomal DNAs were isolated from successive streaks of the est3Δ/est3Δ (est3-ΔΔ) and est3Δ/est3Δ EST3-proA strains and subjected to telomere length analysis.
FIG. 6.
The association of Candida Est3p with active telomerase is Est1p dependent. (A) (Top) Extracts were prepared from a set of strains with or without protein A-tagged EST3 (EST3-proA). Total RNAs were isolated from the extracts and subjected to RT-PCR using primers designed to amplify a 350-bp fragment of TER. (Bottom) The same extracts were subjected to IgG-Sepharose pulldown followed by Western blot analysis using antibodies directed against protein A. (B) (Top) Extracts from the indicated strains were subjected to IgG-Sepharose pulldown. Total RNAs on the beads were isolated by proteinase K digestion and ethanol precipitation, followed by RT-PCR to detect TER. (Bottom) Est3-proA-associated telomerases from the indicated strains were isolated by IgG-Sepharose pulldown, followed by direct primer extension analysis using P20 as the primer. (C) (Top) Extracts from the indicated strains were subjected to IgG-Sepharose pulldown. Total RNAs on the beads were isolated by proteinase K digestion and ethanol precipitation, followed by RT-PCR to detect TER. To facilitate quantitative comparison, the BWP17 EST3-proA sample was subjected to threefold serial dilutions prior to RT-PCR. Thermocycling was performed for 20 or 23 cycles to ensure the linearity of the signals. est1-ΔΔ, est1Δ/est1Δ; est3-ΔΔ, est3Δ/est3Δ.
The association of Est1p with Candida telomerase depends on Est3p.
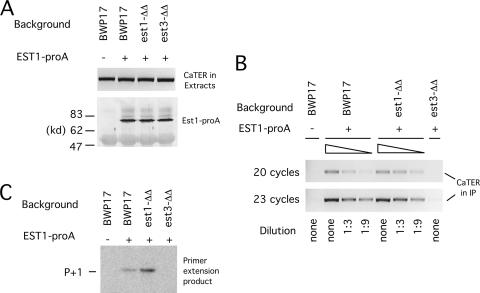
In S. cerevisiae, the association of Est1p with core telomerase is thought to depend primarily on its binding to telomerase RNA, and hence this association is not Est3p dependent (11, 14, 19). To determine if this is also the case for C. albicans, we investigated the requirement for Est1p association with telomerase by using the same strategy as that described above for Est3p. A set of strains containing protein A-tagged Est1p were generated and the association of the tagged protein with TER in cell extracts examined. Western blot analysis indicates that the Est1-proA fusion protein was well expressed in all the strains (Fig. 7A). Moreover, telomere length analysis indicates that the tagged gene was capable of maintaining telomeres in the est1Δ/est1Δ background, indicating that tagging did not abolish function (data not shown). Surprisingly (in light of the S. cerevisiae results), we found in three independent experiments that the level of Est1p-associated TER was reduced >10-fold in the est3Δ/est3Δ strain background (Fig. 7B and data not shown). In contrast, the levels of TER in the starting extracts were unaffected by deletions of either EST1 or EST3 (Fig. 7A). The complex detected between Est1p and TER likely represents active telomerase, because the levels of TER in the precipitates were proportional to those of telomerase activity (Fig. 7B & C). We conclude that, in contrast to S. cerevisiae, the assembly of Est1p into telomerase in C. albicans is Est3p dependent. Thus, the two critical regulatory subunits of Candida telomerase are mutually dependent in their association with the telomerase complex.
FIG. 7.
The association of Candida Est1p with active telomerase is Est3p dependent. (A) (Top) Extracts were prepared from a set of strains with or without protein A-tagged EST1 (EST1-proA). Total RNAs were isolated from the extracts and subjected to RT-PCR using primers designed to amplify a 350-bp fragment of TER. (Bottom) The same extracts were subjected to IgG-Sepharose pulldown followed by Western blot analysis using antibodies directed against protein A. (B) Extracts from the indicated strains were subjected to IgG-Sepharose pulldown. Total RNAs on the beads were isolated by proteinase K digestion and ethanol precipitation, followed by RT-PCR to detect TER. To facilitate quantitative comparison, the BWP17 EST1-proA and est1Δ/est1Δ (est1-ΔΔ) EST1-proA samples were subjected to threefold serial dilutions prior to RT-PCR. Thermocycling was performed for 20 or 23 cycles to ensure the linearity of the signals. (C) Est1-proA-associated telomerases from the indicated strains were isolated by IgG-Sepharose pulldown, followed by direct primer extension analysis using P20 as the primer.
DISCUSSION
Est1p and Est3p are both required for full telomerase function in vitro.
To our knowledge, this is the first instance where deletion of EST3 has been shown to affect the biochemical property of telomerase in vitro. Like Est1p, Est3p is required specifically for reverse transcription through two regions of the Candida telomerase RNA template. The mechanisms for this requirement are not understood, but it is tempting to speculate that the requirement reflects an important in vivo function of these regulatory factors. For example, the telomerase RNP must undergo conformational changes as it catalyzes successive nucleotide additions, because the template is dragged through the protein active site while other regions of RNA are bound by other domains of the protein. At certain template positions, the ability of Est2p to engage in the requisite conformational change may be low and may require the function of regulatory factors. More studies are clearly necessary to understand the mechanistic basis of the stimulatory effects of Est1p and Est3p that we observed in vitro and to determine their functional relevance in vivo.
The nature of the “activation” function for Est1p.
S. cerevisiae Est1p has been proposed to serve a “postrecruitment” or “activation” function based on two lines of evidence. First, even though Est2p was telomere bound in G1 as judged by chromatin immunoprecipitation, telomere extension occurred only in S/G2, when Est1p was recruited to chromosome ends (10, 23). However, because of the limited resolution of chromatin immunoprecipitation, it was unclear if recruitment of telomerase to telomere ends by Est1p in S/G2 was ruled out by this study. Second, certain mutant alleles of EST1 could not be suppressed by the artificial recruitment of telomerase (through a CDC13-EST1 fusion), suggesting that they were defective in a “nonrecruitment” function (8). Based on our earlier finding that Candida telomerase derived from est1Δ/est1Δ strains was impaired in primer utilization, we had proposed that Est1p may directly interact with the 5′ region of substrate DNA and somehow activate the enzyme allosterically (20). This notion was consistent with the DNA-binding activity described for S. cerevisiae Est1p (26). However, the results described in this report as well as those of Osterhage et al. support an alternative hypothesis (14). Because Est1p is required for assembly of Est3p into the telomerase complex, the mutant alleles of EST1 that cannot be suppressed by artificial recruitment may in fact be defective in promoting Est3p assembly. In other words, Est3p is directly responsible for “activating” telomerase, whereas Est1p is indirectly responsible by recruiting Est3p. Further studies will be necessary to address the validity of this hypothesis for S. cerevisiae and C. albicans.
The “mutual assembly” function of Est1p and Est3p.
Our observation that Candida Est3p is required for stable association of Est1p with telomerase is surprising in light of three earlier studies with S. cerevisiae (11, 14, 29), all of which reported that Est3p is dispensable for Est1p assembly. One reasonable possibility is that different tagging and isolation procedures might have affected the results. However, close scrutiny of two of the earlier reports suggests that the difference between the two yeasts may be one of degree rather than kind. Specifically, while it is not abolished, the association of Est1p with core telomerase appears to be significantly reduced by deletion of EST3 in S. cerevisiae (14, 29). Thus, we propose that the “mutual assembly” function of Est1p and Est3p may be conserved in budding yeasts. Interestingly, Est1p in S. cerevisiae is known to bind telomerase RNA (8, 19). Thus, a corollary of our proposal is that this protein-RNA interaction, while specific, must be of relatively low affinity such that it alone is insufficient to recruit all available Est1p into the telomerase complex. A plausible model for the assembly of the full telomerase complex thus entails a network of interactions between multiple telomerase components (Fig. 8). Minimally, we suggest that Est1p interacts with both telomerase RNA and Est3p, while Est3p makes contact with both Est1p and Est2p. Each contact may be of low affinity, but the total free energy of binding may be sufficient to support stable complex formation. This assembly system may have evolved to facilitate the regulation of the enzyme. Indeed, several recent studies have demonstrated that the expression level and assembly of Est1p into telomerase are both cell cycle regulated, peaking in S phase (14, 23). Curiously, the level of Est1p in G1 is reduced only two- to threefold relative to the level in S phase, yet no Est1p-telomerase complex formation is detectable in G1. This drastic concentration dependence can be explained by the multiple low-affinity but cooperative interactions postulated by our model. A future challenge will be to reconstitute the assembly process in vitro and to rigorously determine the energetic contribution of each set of interactions between telomerase components. In this regard, the reconstitution system that we described represents a promising start.
FIG. 8.
Hypothetical model for the contacts between Candida telomerase components, based on the current analysis as well as prior studies with S. cerevisiae. Est1p has been demonstrated to bind telomerase RNA, while Est3p is known to require Est2p for RNA association. In light of the mutual dependence of Est1p and Est3p in telomerase association, we propose that Est1p contacts both Est3p and TER, while Est3p contacts both Est1p and Est2p.
Acknowledgments
We thank Mike McEachern and the Blackburn lab for sharing the template sequence of Candida TER. We also thank Jinan Shaarani for help with the analysis of the Est3p-TER association.
This work was supported by a grant from NIH (GM069507). The Department of Microbiology and Immunology at Weill Cornell Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
Footnotes
Published ahead of print on 1 June 2007.
REFERENCES
- 1.Autexier, C., and N. F. Lue. 2006. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75:493-517. [DOI] [PubMed] [Google Scholar]
- 2.Bianchi, A., S. Negrini, and D. Shore. 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell 16:139-146. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn, E. H. 2005. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 579:859-862. [DOI] [PubMed] [Google Scholar]
- 4.Bosoy, D., Y. Peng, I. Mian, and N. Lue. 2003. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 278:3882-3890. [DOI] [PubMed] [Google Scholar]
- 5.Cech, T. 2004. Beginning to understand the end of the chromosome. Cell 116:273-279. [DOI] [PubMed] [Google Scholar]
- 6.Ciudad, T., E. Andaluz, O. Steinberg-Neifach, N. Lue, N. Gow, R. Calderone, and G. Larriba. 2004. Homologous recombination in Candida albicans: role of CaRad52p in DNA repair, integration of linear DNA fragments and telomere length. Mol. Microbiol. 53:1177-1194. [DOI] [PubMed] [Google Scholar]
- 7.Collins, K. 2006. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 7:484-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, S., and V. Lundblad. 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162:1101-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 10.Fisher, T., A. Taggart, and V. Zakian. 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 11:1198-1205. [DOI] [PubMed] [Google Scholar]
- 11.Hughes, T. R., S. K. Evans, R. G. Weilbaecher, and V. Lundblad. 2000. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 10:809-812. [DOI] [PubMed] [Google Scholar]
- 12.Lundblad, V. 2003. Telomere replication: an Est fest. Curr. Biol. 13:R439-R441. [DOI] [PubMed] [Google Scholar]
- 13.Mozdy, A. D., and T. R. Cech. 2006. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA 12:1721-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osterhage, J. L., J. M. Talley, and K. L. Friedman. 2006. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 13:720-728. [DOI] [PubMed] [Google Scholar]
- 15.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 16.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 17.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 18.Selmecki, A., S. Bergmann, and J. Berman. 2005. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol. Microbiol. 55:1553-1565. [DOI] [PubMed] [Google Scholar]
- 19.Seto, A., A. Livengood, Y. Tzfati, E. Blackburn, and T. Cech. 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 16:2800-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh, S., and N. Lue. 2003. Ever shorter telomere 1 (EST1)-dependent reverse transcription by Candida telomerase in vitro: evidence in support of an activating function. Proc. Natl. Acad. Sci. USA 100:5718-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh, S., O. Steinberg-Neifach, I. Mian, and N. Lue. 2002. Analysis of telomerase in Candida albicans: potential role in telomere end protection. Eukaryot. Cell 1:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinberg-Neifach, O., and N. F. Lue. 2006. Modulation of telomere terminal structure by telomerase components in Candida albicans. Nucleic Acids Res. 34:2710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taggart, A., S. Teng, and V. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023-1026. [DOI] [PubMed] [Google Scholar]
- 24.Taggart, A. K., and V. A. Zakian. 2003. Telomerase: what are the Est proteins doing? Curr. Opin. Cell Biol. 15:275-280. [DOI] [PubMed] [Google Scholar]
- 25.Theimer, C. A., and J. Feigon. 2006. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 16:307-318. [DOI] [PubMed] [Google Scholar]
- 26.Virta-Pearlman, V., D. K. Morris, and V. Lundblad. 1996. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 10:3094-3104. [DOI] [PubMed] [Google Scholar]
- 27.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia, J., Y. Peng, I. S. Mian, and N. F. Lue. 2000. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 20:5196-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou, J., K. Hidaka, and B. Futcher. 2000. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol. 20:1947-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]