Abstract
Cdc37 is a molecular chaperone that has a general function in the biogenesis of protein kinases. We identified mutations within the putative “protein kinase binding domain” of Cdc37 that alleviate the conditional growth defect of a strain containing a temperature-sensitive allele, tpk2(Ts), of the cyclic AMP-dependent protein kinase (PKA). These dominant mutations alleviate the temperature-sensitive growth defect by elevating PKA activity, as judged by their effects on PKA-regulated processes, localization and phosphorylation of the PKA effector Msn2, as well as in vitro PKA activity. Although the tpk2(Ts) growth defect is also alleviated by Cdc37 overproduction, the CDC37 dominant mutants contain wild-type Cdc37 protein levels. In addition, Saccharomyces cerevisiae Ste11 protein kinase has an elevated physical interaction with the altered Cdc37 protein. These results implicate specific amino-terminal residues in the interaction between Cdc37 and client protein kinases and provide further genetic and biochemical support for a model in which Cdc37 functions as a molecular chaperone for protein kinases.
Protein kinases are charged with critical regulatory roles in such diverse cellular processes as cell cycle control, development, stress response, and metabolism. One example is the cyclic AMP (cAMP)-dependent protein kinase (PKA), which is found in all eukaryotes and serves as the biochemical template for all other serine/threonine protein kinases (47). In the yeast Saccharomyces cerevisiae, PKA has been implicated in a myriad of processes including growth, carbon store accumulation, and adaptation to stress (6, 48). Like its mammalian counterpart, yeast PKA is a heterotetramer comprised of two negative regulatory subunits and two catalytic subunits. Activation occurs as regulatory subunits, encoded by BCY1 (Fig. 1), bind the second messenger cAMP and release catalytic subunits redundantly encoded by genes TPK1, TPK2, and TPK3 (8, 49). Strains lacking BCY1 exhibit high cAMP-independent PKA activity, fail to accumulate carbon stores (e.g., glycogen), and are sensitive to stress (7, 8, 50). Strains containing disruptions of any two catalytic genes, tpk1 TPK2 tpk3, are largely indistinguishable from their wild-type parent, whereas mutants in which the single intact catalytic subunit gene has been replaced with a temperature-sensitive allele [tpk1 tpk2(Ts) tpk3] arrest in G1, hyperaccumulate glycogen, and display constitutive resistance to stress (44, 53).
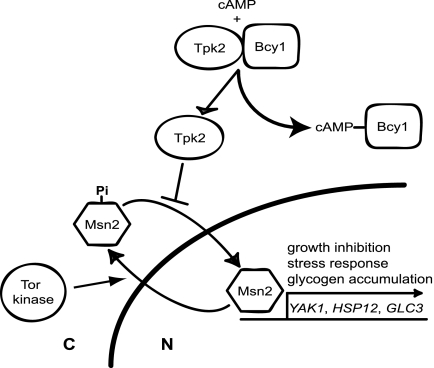
FIG. 1.
PKA and Tor kinase control of the general stress response. Cytoplasmic localization of the general stress response transcription factor Msn2 is independently stimulated by the PKA active subunit Tpk2 and the Tor kinase. Binding of cAMP to the PKA negative regulatory subunit Bcy1 stimulates the release and activation of Tpk2. Phosphorylation of the Msn2 carboxyl terminus by Tpk2 inhibits nuclear import, whereas Tor kinase acts on the Msn2 amino terminus in an unknown way to stimulate efficient nuclear export. Inactivation of either kinase results in the accumulation of Msn2 in the nucleus and the coordinate expression of genes necessary for growth inhibition (YAK1), response to stress (HSP12), and glycogen accumulation (GLC3).
Genetic analyses have identified the general stress response transcription factor Msn2 as well as the protein kinase Yak1 as being potential downstream effectors of PKA function (Fig. 1) (16, 44). Msn2 resides in the cytoplasm of dividing cells and accumulates in the nucleus in response to glucose starvation or exposure to stress (20, 21). The glucose signal is thought to be mediated by the PKA pathway because both glucose starvation and PKA depletion, but not stress, result in the dephosphorylation and nuclear accumulation of a carboxyl-terminal fragment of Msn2 (21). By contrast, nuclear accumulation in response to stress is thought to be mediated by the Tor kinase/PP2A phosphatase signaling pathway, which regulates an amino-terminal export signal (3, 13, 21, 39). As Msn2 accumulates in the nucleus, it stimulates high-level expression of over 150 genes involved in growth arrest (YAK1), glycogen accumulation (GLC3), and stress survival (HSP12, CTT1, DDR2, etc.) (5, 9, 18, 31). Thus, Msn2 regulates several PKA-dependent processes, whereas Yak1 functions exclusively to inhibit growth.
Given the general importance of protein kinases to cell growth, it is not surprising that cells expend significant energy and genetic capacity to maintain protein kinase function. This is achieved, in part, through the action of the multisubunit Hsp90-chaperone complex. Hsp90 is an abundant and highly conserved protein that acts in concert with several cochaperone subunits to maintain a select group of signaling proteins in their functional form by preventing aggregation and facilitating folding (33-35, 37). The importance of Hsp90 is underscored by the observation that yeast viability relies on the presence of at least one of the two highly related Hsp90-encoding genes, HSC82 and HSP82 (4). Although the mechanism of Hsp90 chaperone function is incompletely understood, recent studies have identified a set of cochaperones that are thought to be dedicated to substrate recognition and binding. For example, the cochaperone Sti1 (and its mammalian counterpart Hop) interacts with the carboxyl-terminal tails of Hsp70 and Hsp90, allowing the efficient transfer of select client proteins from the former chaperone to the more exclusive Hsp90 machinery (11, 40). Sti1 binding also inhibits ATP hydrolysis by Hsp90 (29, 36, 37), thereby preventing Hsp90 from completing its ATP-dependent conformational cycle. Interestingly, although Sti1/Hop1 has been shown to be important for Hsp90 chaperone function by both in vitro and in vivo studies, Sti1 is required for normal growth only under limited conditions such as elevated temperature and amino acid depletion (10, 32).
In contrast with Sti1, the Hsp90 cochaperone Cdc37 is essential for growth under all conditions and, with several exceptions, appears to be dedicated to protein kinases. The CDC37 gene was first identified in a cell division cycle screen as a temperature-sensitive mutant, cdc37-1(Ts), that arrested in G1 (38). The G1 arrest is thought to reflect a disruption in the interaction between Cdc37 and the G1-specific kinases Cdc28 (15, 19), Mps1 (42), Kin28 (52), and Cak1 (15); however, analysis of a different conditional allele, cdc37-14(Ts), suggests that G1 arrest may not be a universal property of Cdc37 dysfunction (12). Consistent with the latter notion, Cdc37 has been implicated in the function of an increasing array of yeast protein kinases (1, 2, 12, 28, 45). Although studies of binding between yeast proteins have been hampered by low-affinity interactions, similar studies of mammalian proteins have shown that Hsp90 interacts with the central portion of Cdc37, whereas protein kinases interact with a putative “protein kinase binding domain” within the conserved amino-terminal domain of Cdc37 (22, 43). Those observations have been incorporated into a model in which Cdc37 acts to tether client proteins to the Hsp90-chaperone complex; however, alternative models, such as the transfer of client proteins from Hsp90 to Cdc37, are compatible with the in vitro assignment of chaperone function to Cdc37 itself (25, 26, 51).
Genetic screens to identify downstream effectors of the PKA pathway have focused on genes whose loss of function or overexpression alleviates the conditional growth of a tpk2(Ts) strain (16, 44, 53). To identify novel effectors of this pathway by eliminating recessive suppressors, we isolated temperature-resistant revertants of the conditional growth defect of a homozygous tpk2(Ts)/tpk2(Ts) diploid strain. One class of dominant suppressors falls within the putative protein kinase binding domain of the Hsp90 cochaperone Cdc37. Growth suppression corresponds to the restoration of PKA catalytic function as judged by physiological effects and monitoring of Msn2 localization and function. This conclusion is buttressed by the observation that the dominant CDC37 mutations enhance both wild-type and mutant Tpk2 activity and is consistent with the synthetic lethality of a tpk2(Ts) cdc37(Ts) double mutant.
MATERIALS AND METHODS
Media and growth conditions.
Cells were grown in rich (yeast extract-peptone-dextrose [YEPD]) medium or synthetic complete medium lacking the appropriate amino acids (24). Temperature shifts were achieved by suspending harvested cells in prewarmed media.
Yeast strains.
The yeast strains used in the study are listed in Table 1.
TABLE 1.
Strains of S. cerevisiae used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SGY398 | MATα ura3-52 his3 leu2-3,112 trp1 ade8 tpk1::ADE8 tpk2-63(Ts) tpk3::TRP1 bcy1Δ::LEU2 | 53 |
| SGY446 | MATα ura3-52 his3 leu2-3,112 trp1 ade8 tpk1::ADE8 tpk2-63(Ts) tpk3::TRP1 | 44 |
| SGY448 | MATaura3-52 his3 leu2-3,112 trp1 ade8 tpk1::URA3 tpk2-63(Ts) tpk3::TRP1 | 44 |
| SGY562 | MATα ura3-52 his3 leu2-3,112 trp1 ade8 tpk1::ADE8 tpk2-62(Ts) tpk3::TRP1 | This study |
| SGY559 | SGY446 TPK2 | This study |
| NOY16 | MATα/MATaura3-52/ura3-52 his3/his3 leu2-3,112/leu2-3,112 trp1/trp1 ade8/ade8 tpk1::ADE8/tpk1::URA3 tpk2-63(Ts)/tpk2-63(Ts) tpk3::TRP1/tpk3::TRP1 | This study |
| SGY530 | MATα ura3-52 his3 leu2-3,112 trp1 ade8 tpk1::ADE8 tpk2-63(Ts) tpk3::TRP1 CDC37-101 | This study |
| SGY533 | MATα ura3-52 his3 leu2-3,112 trp1 ade8 tpk1::ADE8 tpk2-63(Ts) tpk3::TRP1 CDC37-106 | This study |
| SGY535 | MATaura3-52 his3 leu2-3,112 trp1 ade8 tpk1::ADE8 tpk2-63(Ts) tpk3::TRP1 CDC37-109 | This study |
| MRY140 | SGY562(pRS316) | This study |
| MRY141 | SGY562(pRS316-CDC37-101) | This study |
| FY70 | MATα ura3-52 his3 leu2-3,112 trp1 ade1 TPK2 | |
| MRY142 | FY70 ura3::pRS306-tpk2-63-myc12 | This study |
| MRY143 | FY70 ura3::pRS306-TPK2-myc12 | This study |
| MRY31 | SGY559(pRS316)[pADH-PKI-MSN2-(576-704)-GFP3] | This study |
| MRY43 | SGY562(pRS316)[pADH-PKI-MSN2-(576-704)-GFP3] | This study |
| MRY45 | SGY562[pRS316-CDC37-101] [pADH-PKI-MSN2-(576-704)-GFP3] | This study |
| MRY4 | MATα cdc37Δ::KanMx lys2 (pRS202-CDC37+) | This study |
| MRY5 | MATα cdc37Δ::KanMx lys2 (pRS316-CDC37-109) | This study |
| MRY8 | MATα cdc37Δ::KanMx met15 (pRS316-CDC37+) | This study |
| MRY11 | MATα cdc37Δ::KanMx lys2 (pRS316-CDC37-101) | This study |
| MRY10 | BRY4742(pRS316-cdc37Δ) | This study |
| MRY47 | SGY562 sti1-1::HIS3 (pRS316-CDC37-101) | This study |
| MRY73 | MATα cdc37Δ::KanMx his3Δ1 leu2Δ1 met15Δ0 ura3Δ1 (pRS315-CDC37+) | This study |
| MRY75 | MATα cdc37Δ::KanMx his3Δ1 leu2Δ1 met15Δ0 ura3Δ1 (pRS315-CDC37-106) | This study |
| MRY79 | MRY73(His6-V5-Ste11ΔN) | This study |
| MRY81 | MRY75(His6-V5-Ste11ΔN) | This study |
| ACY77-3 | MATα ade2 leu2 his3 trp1 ura3 sti1-1::HIS3 | This study |
| Y703 | MATα cdc25-1 ura3-52 leu2-3,112 trp1 ade1 | Jim Broach |
| WX241-12b | MATα mps1-1 ura3-52 his3Δ200 | 42 |
Dominant suppressors of the tpk2-63(Ts) growth defect.
Independent colonies of homozygous tpk2-63(Ts)/tpk2-63(Ts) diploid strain NOY16 were patched onto rich medium (YEPD) agar at 23°C for 36 h and then replicated to 34°C for 2 days. A single, temperature-resistant colony from each patch was purified at 23°C and retested for growth at the nonpermissive temperature. Ten temperature-resistant revertants were subjected to tetrad analysis and backcrosses to determine if suppression was due to a single mutation and if any of the suppressors were linked.
Plasmids used and constructed in this study.
The C-terminal Msn2-green fluorescent protein (GFP) plasmid [pADH-PKI-MSN2(576-704)-GFP3] was described previously (21). The low-copy-number CDC37-101 plasmid pCB4a2 was isolated from a low-copy-number library constructed with genomic DNA from haploid revertant strain SGY530. Briefly, Sau3A fragments of approximately 10 kb were purified by agarose gel electrophoresis separation after partial digestion and cloned into the BamHI site of low-copy-number URA3 vector pRS316. DNA from over 10,000 bacterial transformants was used to transform yeast strain SGY446 to temperature resistance on minimal yeast medium lacking uracil. Plasmid pCB42a contains the full-length CDC37 gene on a 7.4-kbp chromosomal fragment. Subcloning and sequence analysis showed that the CDC37-101 allele was the result of a single amino acid change (L62F). Plasmid pGS224 was constructed by inserting a 6.5-kb KpnI-SpeI fragment from pCB4a2 into the KpnI-XbaI sites of pRS316. Plasmid pGS224 was digested with XbaI, diluted, and then ligated to generate cdc37Δ rescue plasmid pGS226. Wild-type and mutant alleles of CDC37 were rescued by transforming SGY446 (CDC37+), SGY533 (CDC37-106), and SGY535 (CDC37-109) to Ura+ with XbaI-cut pGS226. Corresponding plasmids were shown to contain wild-type (pYZ1 CDC37+) and mutant (pYZ2 CDC37-106; pYZ5 CDC37-109) genes by sequence (Fig. 2C), complementation (see Fig. 5A), and suppression (Fig. 2A) studies. The high-copy-number CDC37+ plasmid pYZ4 was constructed by inserting a 4.5-kb KpnI-SmaI CDC37+ fragment from pYZ1 into the KpnI-Ecl136II sites of URA3 vector pRS202 (19a). Integrating Tpk2-Myc12 (pMR1) and Tpk2-63-Myc12 (pMR3) plasmids were constructed by inserting NotI- and Sal1-digested PCR fragments from SGY559 (TPK2) and SGY446 (tpk2-63) into the corresponding sites of integrating Myc12 vector pRS306-GAL:SWE1-myc12 (30). Those manipulations replaced the GAL:SWE1 fragment with the promoter and coding regions of TPK2 and tpk2-63 and fused the 3′ ends of these genes to the 12 repeats of the Myc epitope. Plasmids were integrated into the chromosome at the ura3-52 locus by digestion with HindIII and selection of Ura+ transformants. The galactose-inducible, His6-tagged Ste11 fusion plasmid His6-V5-Ste11ΔN was described previously (27).
FIG. 2.
The tpk2(Ts) growth defect is suppressed by alterations of the amino-terminal sequence of Cdc37. (A) Suppression of the tpk2(Ts) growth defect by the CDC37-101 mutation. Strains were streaked onto minimal medium and incubated for several days. Wild-type (WT) [SGY559(pRS316)], tpk2-63(Ts) CDC37+ [SGY446(pYZ1)], tpk2-63(Ts) CDC37-101 [SGY446(pGS224)], tpk2-62(Ts) CDC37+ [SGY562 (pYZ1)], and tpk2-62(Ts) CDC37-101 [SGY562(pGS224)] strains were used. (B) Suppression of the tpk2(Ts) growth defect in the absence of Bcy1. Strain SGY398 (tpk2-63 bcy1Δ) was transformed with a low-copy-number vector or the same vector containing wild-type (CDC37+) or mutant (CDC37-101) alleles of CDC37 and incubated on minimal medium for several days at 23°C and 34°C. (C) The dominant CDC37 mutations fall within the amino terminus of Cdc37. The individual mutations are indicated by lines within the putative protein kinase interaction domain (filled box) of Cdc37.
FIG. 5.
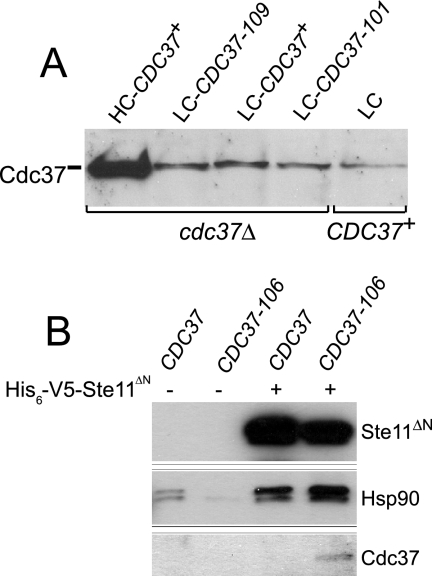
Cdc37 levels and protein interactions in the dominant CDC37 mutants. (A) Protein extracts of the indicated strains were separated by polyacrylamide gel electrophoresis, transferred onto nitrocellulose, and probed with anti-Cdc37 antibody. cdc37Δ (high-copy-number [HC] CDC37+) (MRY4), cdc37Δ (low-copy-number [LC] CDC37-109) (MRY5), cdc37Δ (low-copy-number CDC37+) (MRY8), cdc37Δ (low-copy-number CDC37-101) (MRY11), and CDC37+ (low copy number) (MRY10) strains were used. (B) Interaction of Ste11 with wild-type and mutant Cdc37. Whole-cell extracts were incubated with Ni-nitrilotriacetic acid to purify His6-tagged Ste11 proteins and separated by polyacrylamide gel electrophoresis before sequentially Western blotting with antisera to Ste11, Hsp90, and Cdc37. Extracts were made from CDC37+ (MRY73), CDC37-106 (MRY75), CDC37+ His6-V5-Ste11ΔN (MRY79), and CDC37-106 His6-V5-Ste11ΔN (MRY81) strains.
Plasmid mutagenesis.
The low-copy-number CDC37+ plasmid pYZ1 was transformed into mutD strain LE30 (F− mutD5 rpsL azi galU95) (14), and five mutagenized DNA pools were prepared from independent colonies grown overnight in rich bacterial medium containing ampicillin. Each pool was used to transform yeast strain SGY562 to Ura+, and a single temperature-resistant revertant was selected from each pool for further analysis. Plasmids recovered from two of the temperature-resistant yeast transformants were retransformed into SGY446 and SGY562 to confirm that the temperature resistance was plasmid dependent. Mutations identified by sequencing of the CDC37 coding region in both directions are shown in Fig. 2C.
Fluorescence microscopy.
Cells expressing the carboxyl-terminal Msn2-GFP fusion were grown to mid-log phase before being shifted to 34°C for 60 min. DNA from live cells was visualized by epifluorescence microscopy after incubating cells for 15 min in medium containing Hoechst 33258 stain from Sigma-Aldrich.
Immunoprecipitation and Western blot analysis.
Protein immunoprecipitation of the Myc12-tagged Tpk2 and Tpk2-63 proteins was carried out as described previously (17). Protein quantitation of Tpk2-Myc12, Tpk2-63-Myc12, and Cdc37 was carried out by Western analysis using a 1/2,000 dilution of 9E10B mouse monoclonal anti-Myc antibody (Covance) or a 1/1,000 dilution of mouse monoclonal anti-Cdc37 antibody, respectively. An equal amount of protein extract was loaded onto each lane by normalizing measured protein extract concentrations. In all cases, protein extract concentrations were measured by a modified Bradford assay using the Bio-Rad protein assay reagents (Bio-Rad). Secondary antibodies were goat anti-mouse immunoglobulin G-horseradish peroxidase (Promega) and goat anti-rabbit immunoglobulin G-horseradish peroxidase (Calbiochem) and were used at a 1/1,000 dilution. Cell extracts or immunoprecipitates were separated by polyacrylamide gel electrophoresis, transferred onto nitrocellulose, and probed with appropriate primary and secondary antibodies.
Phosphorylation, coprecipitation, and protein kinase assays.
PKA-dependent phosphorylation of Msn2 was monitored by probing Western blots of cell extracts with an anti-phospho-CREB antibody (Cell Signaling Technology), which was described previously (21). Preparation of cell extracts, polyacrylamide gel electrophoresis, and Western blot analysis were carried out as described previously (17).
Measurements of PKA activity from cell extracts were carried out using a fluorescence-labeled PKA substrate peptide kit (PepTag Assay for Non-Radioactive Detection) that was used according to the supplier's directions (Promega). Cell extracts were prepared to 4 mg/ml and then added to the reaction mix in the indicated amount. Total protein was kept constant by adding bovine serum albumin (Sigma). Reactions were carried out at 23°C for 30 min, stopped by heating to 95°C for 10 min, and then run on an 0.8% agarose gel for 15 min. To determine the activities of purified Tpk2 and Tpk2-63, we immunoprecipitated Tpk2-Myc12 and Tpk2-63-Myc12 from 1.5 mg of cell extract from wild-type and CDC37-106 cells and then incubated 10% of the precipitate with the PKA substrate peptide kit described above. The amount of Tpk2 protein was determined by probing the remainder of the immunoprecipitate with anti-Myc antibody.
Effects of the dominant CDC37 mutations on the binding of Ste11 kinase were monitored as previously described (27). Essentially, strains containing His6-V5-Ste11ΔN were grown in 200 ml of 0.67% yeast nitrogen base plus 2% raffinose to an A600 of 0.2. Galactose was then added to 2% to induce the expression of Ste11ΔN for 12 h. Cells were resuspended in extraction buffer (20 mM HEPES [pH 7.5], 100 mM KCl, and 0.1 mM EDTA plus protease inhibitor cocktail tablets [Boehringer, Indianapolis, IN]), and extracts were prepared by glass bead lysis. His6-V5-Ste11ΔN was isolated after incubating Ni-nitrilotriacetic acid resin with 0.5 ml of whole-cell extracts at 3 mg/ml for 1 h at 4°C. Beads were washed three times with extraction buffer plus 10 mM imidazole, and proteins were eluted by the addition of 0.4 ml extraction buffer plus 150 mM imidazole. Eluted proteins were precipitated with 10% trichloroacetic acid and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer. Samples were resolved by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and proteins were detected by Western blot using specific antisera.
RESULTS
Dominant suppressors of the tpk2(Ts) growth defect.
Previous suppressor analyses of the tpk2(Ts) growth defect identified loss-of-function mutations in the YAK1 and MSN2 genes (16, 44). To identify rare dominant suppressors, we isolated temperature-resistant revertants of a homozygous tpk2-63(Ts)/tpk2-63(Ts) diploid strain. The presence of a second, wild-type allele of the suppressor gene would also alleviate potential growth defects that resulted from an altered function of an essential, PKA-dependent gene. Separate colonies of strain NOY16 were patched onto YEPD agar, incubated for 36 h at 23°C, and then replicated in YEPD agar for 2 days at 34°C. Tetrad analyses of 10 independent revertants gave rise to four-spored ascii that formed colonies at 23°C. In each tetrad, two of the four colonies grew at 34°C, indicating that suppression was due to a single mutation. Tetrads from 7 of the 10 revertants gave rise to two colonies that grew slowly, ranging from very slowly to moderately slowly, at 23°C. Interestingly, these slow-growing colonies corresponded (100% linkage in >20 complete tetrads for each revertant) to the colonies that grew (slowly) at 34°C.
The dominance of each of the suppressors was confirmed by mating temperature-resistant haploid segregants from each revertant with a compatible haploid tpk2-63(Ts) parent (SGY446 or SGY448) and showing that the resulting diploid grew at 34°C. Moreover, diploids generated by mating the slow-growing haploid strains with the tpk2-63(Ts) parent of the opposite mating type grew as well at 23°C as the homozygous tpk2-63(Ts)/tpk2-63(Ts) parent, indicating that, in contrast with suppression, the growth defect conferred by the seven “slow-growth” suppressors was recessive. Subsequent crosses showed that at least four of the dominant suppressors that conferred a slow-growth defect were tightly linked (100% linkage in >10 tetrads) with one another and unlinked with the other three suppressors. The slow-growth suppressors have been tentatively named SOK5 (suppressor of kinase 5). Finally, all three “normal-growth” suppressors were tightly linked (100% linkage in >10 tetrads) with one another and are described below.
tpk2(Ts) suppression by alteration of the kinase-specific chaperone CDC37.
The gene corresponding to one of the “normal-growth” suppressors was identified by transforming tpk2-63(Ts) haploid strain SGY446 to temperature resistance (growth at 34°C) with a low-copy-number plasmid library containing DNA isolated from the corresponding revertant (Fig. 2A) (see Materials and Methods). Suppression was not due to an alteration of the PKA negative regulatory subunit Bcy1 because suppression also occurred in a bcy1Δ derivative (Fig. 2B). Subcloning and sequence analysis showed that the smallest suppressing fragment contained a single open reading frame that, with the exception of a single base transversion (G to T) at the third position of codon 62, was identical with CDC37. A gapped version of the suppressor plasmid was then used to isolate the other two mutations as well as the wild-type CDC37+ gene from the corresponding strains by homologous repair (see Materials and Methods). Sequence analysis showed that each of the three suppressor mutations resulted from a single, and different, base change within a small region toward the 5′ end of the CDC37 coding region (Fig. 2C). The suppressors have been designated CDC37-101, CDC37-106, and CDC37-109. Two additional mutations isolated by random mutagenesis of a CDC37+ plasmid (see Materials and Methods) also fell within the same region of CDC37 and were designated CDC37-111 (Y59C) and CDC37-112 (R65Q). Interestingly, the CDC37-109 suppressor converts a residue (W33R) that is found in most eukaryotic Cdc37 proteins (43).
The CDC37 gene product is thought to function as a protein kinase-specific molecular chaperone that functions in association with Hsp90. Accordingly, the CDC37 dominant mutations may suppress the tpk2-63(Ts) growth defect by restoring function to the temperature-sensitive PKA catalytic subunit. This interpretation is consistent with previously reported observations, which have noted that CDC37+ overexpression can enhance the functions of several conditional yeast protein kinases, including the debilitated spindle pole body duplication protein kinase Mps1 that is found in the mps1-1(Ts) mutant (42). As expected, the conditional growth defects of tpk2-63(Ts) and tpk2-62(Ts) mutants were alleviated, albeit to different degrees, by a high-copy-number plasmid bearing wild-type CDC37+ as well as by low-copy-number plasmids bearing CDC37-101, CDC37-106, and CDC37-109 (Fig. 2A and Table 2). However, a truncated version of CDC37-106 that contained only the protein kinase binding domain and that was deleted for the Hsp90-binding domain was unable to suppress the growth defect of the tpk2(Ts) mutants (data not shown). This finding suggests that the dominant mutants function in association with Hsp90. The dominant CDC37 alleles also alleviated the conditional growth defect of the mps1-1(Ts) mutant (Table 2), suggesting that suppression was neither allele nor gene specific. By contrast, neither high-copy-number CDC37+ nor any of the mutant CDC37 alleles were capable of suppressing the conditional growth defect of the cdc25-1(Ts) mutant (Table 2), which contains a lesion in an upstream component of the PKA pathway.
TABLE 2.
Suppression by CDC37
| Strain | Genotypeb | Growtha
|
||
|---|---|---|---|---|
| 23°C | 30°C | 34°C | ||
| SGY446(pYZ1) | tpk2-63 cdc37+ (LC CDC37+) | + | − | − |
| SGY446(pGS224) | tpk2-63 cdc37+ (LC CDC37-101) | + | + | + |
| SGY446(pYZ2) | tpk2-63 cdc37+ (LC CDC37-106) | + | + | + |
| SGY446(pYZ5) | tpk2-63 cdc37+ (LC CDC37-109) | + | + | + |
| SGY446(pRS202) | tpk2-63 cdc37+ (HC) | + | − | − |
| SGY446(pYZ4) | tpk2-63 cdc37+ (HC CDC37+) | + | + | +/− |
| SGY562(pYZ1) | tpk2-62 cdc37+ (LC CDC37+) | + | − | − |
| SGY562(pGS224) | tpk2-62 cdc37+ (LC CDC37-101) | + | + | + |
| SGY562(pYZ2) | tpk2-62 cdc37+ (LC CDC37-106) | + | + | + |
| SGY562(pYZ5) | tpk2-62 cdc37+ (LC CDC37-109) | + | + | + |
| Y703(pYZ1) | cdc25-1 cdc37+ (LC CDC37+) | + | + | − |
| Y703(pGS224) | cdc25-1 cdc37+ (LC CDC37-101) | + | + | − |
| Y703(pYZ2) | cdc25-1 cdc37+ (LC CDC37-106) | + | + | − |
| Y703(pRS202) | cdc25-1 cdc37+ (HC) | + | + | − |
| Y703(pYZ4) | cdc25-1 cdc37+ (HC CDC37+) | + | + | − |
| WX241-12b(pYZ1) | mps1-1 cdc37+ (LC CDC37+) | + | − | − |
| WX241-12b(pGS224) | mps1-1 cdc37+ (LC CDC37-101) | + | + | − |
| WX241-12b(pYZ2) | mps1-1 cdc37+ (LC CDC37-106) | + | + | − |
| WX241-12b(pRS202) | mps1-1 cdc37+ (HC) | + | − | − |
| WX241-12b(pYZ4) | mps1-1 cdc37+ (HC CDC37+) | + | + | − |
+, growth; −, no growth; +/−, slow growth.
LC, low copy number; HC, high copy number.
To further examine the physiological interaction between Cdc37 and Tpk2, we monitored the growth of strains containing the tpk2-63(Ts) allele and the temperature-sensitive cdc37-1(Ts) mutation, which expresses a truncated version of the Cdc37 protein. Interestingly, none of the 24 predicted tpk2-63(Ts) cdc37-1(Ts) double mutant spores formed colonies at 23°C when a heterozygous TPK2/tpk2-63(Ts) cdc37-1(Ts)/CDC37+ (pRS316) diploid strain was sporulated. By contrast, 15 of 18 predicted double mutants from a heterozygous TPK2/tpk2-63(Ts) cdc37-1(Ts)/CDC37+ (pRS316-TPK2) diploid strain grew at 23°C on both YEPD medium and medium lacking uracil. None of these 15 double mutant colonies grew on minimal medium containing 5-floroorotic acid, which is toxic to cells that are capable of utilizing uracil. Thus, the tpk2-63(Ts) growth defect is sensitive to Cdc37 function. We presume that this synthetic interaction reflects the corollary of suppression; namely, a diminution in functional PKA catalytic activity as a result of the defect in Cdc37.
The dominant CDC37 suppressors alter Msn2 localization and phosphorylation.
The simplest explanation for tpk2(Ts) suppression by the dominant CDC37 mutations is reactivation of the mutant PKA catalytic subunit by the altered chaperone (Fig. 1). Alternative possibilities include interdiction downstream of Msn2 function (e.g., relieving Yak1-specific growth inhibition), direct inactivation of the Msn2 transcriptional activator, and alteration of Msn2 localization by a PKA-independent mechanism (such as through the hyperactivation of the stress/Tor kinase signaling pathway). To differentiate among these four possibilities, we examined the effect of the CDC37 dominant mutations on glycogen accumulation (a growth-independent process) as well as Msn2 localization and phosphorylation (a PKA-dependent process). Whereas the tpk2-62(Ts) CDC37+ strain accumulates much more glycogen than its TPK2 CDC37+ parent, the tpk2-62(Ts) strain containing CDC37-101 (Fig. 3A) or the other suppressors (data not shown) accumulates near-wild-type levels of glycogen. Thus, the suppression of the tpk2(Ts) growth defect is accompanied by the suppression of the glycogen hyperaccumulation defect, implying that the dominant Cdc37 mutations do not act downstream of Msn2. Consistent with this conclusion, Msn2 localization was altered by the CDC37 dominant suppressors, as shown by comparing the cytoplasmic localization of the carboxyl-terminal, PKA-responsive Msn2-GFP fusion (21) in the tpk2-62(Ts) strain containing CDC37-101 (Fig. 3B) or CDC37-106 and CDC37-109 (data not shown) with its nuclear localization in the isogenic tpk2-62(Ts) CDC37+ strain (Fig. 3B). Because the localization of the carboxyl-terminal Msn2-GFP fusion responds to glucose and PKA depletion, but not stress or changes in the Tor kinase pathway (21, 39), these results also rule out the possibility that suppression results from an effect of Cdc37 on the stress-specific Tor kinase pathway (Fig. 1). Finally, we probed extracts of several Msn2-GFP strains with anti-phospho-CREB antibody, which specifically recognizes PKA-dependent phosphorylation of Msn2 (21, 39), to examine the effect of the dominant CDC37 mutations on PKA activity. As shown in Fig. 3C, Msn2 phosphorylation is reduced in the tpk2-62(Ts) CDC37+ strain relative to the wild-type TPK2 CDC37+ strain, and this phosphorylation is partially restored by the presence of the CDC37-101 mutation. Msn2 protein levels remained the same, as shown by reprobing stripped blots with anti-GFP antibody (Fig. 3C). Together, these results suggest that tpk2(Ts) suppression results from an altered form of Cdc37 impinging upon PKA.
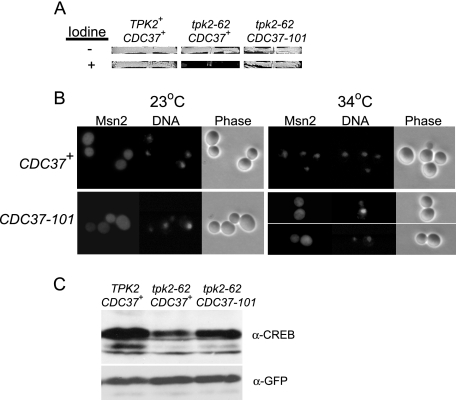
FIG. 3.
Mutant Cdc37 restores Msn2 function, localization, and phosphorylation to a tpk2(Ts) strain. (A) A tpk2(Ts) CDC37-101 strain accumulates wild-type levels of glycogen. Strains were patched onto rich medium agar and incubated at 23°C for several days before (−) and after (+) exposure to iodine vapors. TPK2 CDC37+ [SGY559(pRS316)], tpk2-62 CDC37+ [SGY562(pYZ1)], and tpk2-62 CDC37-101 [SGY562(pGS224)] strains were used. (B) The CDC37-101 mutation blocks Msn2-GFP from accumulating in the nucleus of a tpk2-62(Ts) strain. Cells were grown at 23°C to mid-log phase before shifting to 34°C for 2 h. Fifteen minutes before harvesting, Hoechst 33342 stain was added (50-μg/ml final concentration) to visualize DNA. Strains were derivatives of tpk2-62(Ts) strain SGY562 containing the MSN2-GFP3 fusion plasmid pADH-PKI-MSN2-(576-704)-GFP3. CDC37+ (MRY43) and CDC37-101 (MRY45) strains were used. (C) The CDC37-101 mutation partially restores Msn2 phosphorylation in a tpk2-62(Ts) strain. Strains were grown at 23°C to mid-log phase and then shifted to 34°C for 2 h. Extracts were separated by polyacrylamide gel electrophoresis, transferred onto nitrocellulose, and then exposed sequentially to anti-phospho-CREB antibody (α-CREB) to monitor PKA-dependent phosphorylation and to anti-GFP antibody (α-GFP) to monitor Msn2 levels. TPK2 CDC37+ (MRY31), tpk2-62 CDC37+ (MRY43), and tpk2-62 CDC37-101 (MRY45) strains were used.
The CDC37 mutations restore PKA-specific activity but not stability.
The stability of most yeast protein kinases is affected by Cdc37 function (28). To compare PKA stability in the presence of wild-type and dominant Cdc37 proteins, we examined levels of Myc-tagged Tpk2 and Tpk2-63 proteins in extracts of CDC37+ and CDC37-106 strains after treatment with cycloheximide. In the wild-type CDC37+ strain, both Tpk2-Myc and Tpk2-63-Myc fusions were stable for at least 4 h at 34°C (Fig. 4A). Moreover, levels of both tagged proteins were identical in the CDC37+ and CDC37-106 strains (Fig. 4B), indicating that dominant suppression does not result from Tpk2-63 accumulation.
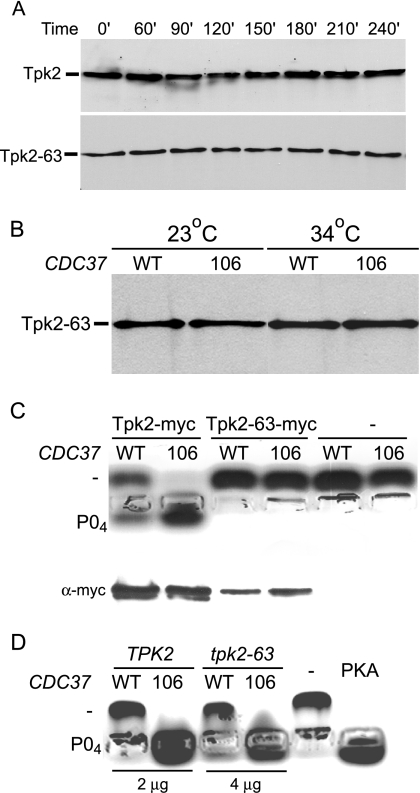
FIG. 4.
The CDC37 dominant mutations alter Tpk2 activity. (A) Stability of wild-type and temperature-sensitive Tpk2 proteins. Strains containing Myc-tagged wild-type Tpk2 and mutant Tpk2-63 fusion proteins were grown at 23°C and shifted to 34°C medium containing 100 mg/ml cycloheximide. Protein extracts were separated by polyacrylamide gel electrophoresis, transferred onto nitrocellulose, and probed with anti-Myc antibody. Tpk2 (MRY142) and Tpk2-63 (MRY143) strains were used. (B) Dominant CDC37 mutations do not alter Tpk2 levels. Levels of the Myc-tagged temperature-sensitive Tpk2-63 fusion protein were determined after shifting derivatives of strain MRY142 from 23°C to 34°C for 3 h. Derivatives contained plasmids with either wild-type (WT) (CDC37+) (pYZ1) or mutant 106 (CDC37-106) (pGS224) alleles of CDC37. Extracts were separated by polyacrylamide gel electrophoresis, transferred onto nitrocellulose, and probed with anti-Myc antibody to measure Tpk2 levels. (C) Dominant CDC37 mutations alter the activity of purified Tpk2. Tpk2 fusion proteins were immunopurified and used in an in vitro peptide kinase assay. After 30 min, peptide from each assay was separated by agarose gel electrophoresis to determine if the peptide was unphosphorylated (−) or phosphorylated (PO4). Aliquots of the immunoprecipitate were also separated by polyacrylamide gel electrophoresis, transferred onto nitrocellulose, and probed with anti-Myc antibody (α-myc) to determine if similar levels of protein kinase were present in each reaction. Strains were derivatives of Tpk2-Myc (MRY143) or Tpk2-63-Myc (MRY42) strains containing plasmids with WT (CDC37+ pYZ1) or 106 (CDC37 pGS224) alleles of CDC37. (D) Dominant CDC37 mutations alter the activity of wild-type and mutant PKA. Cell extracts from the indicated strains were used in the in vitro peptide kinase assay. TPK2 CDC37+ [SGY559(pRS316)], TPK2 CDC37-106 [SGY559(pYZ2)], tpk-63(Ts) CDC37+ [SGY446(pRS316)], and tpk-63(Ts) CDC37-106 [SGY446(pYZ2)] strains were used.
To measure PKA activity directly, we purified Myc-tagged wild-type (Tpk2) and mutant (Tpk2-63) proteins from strains grown at 23°C. Purified PKA was then used in an in vitro peptide phosphorylation assay. As shown in Fig. 4C, wild-type Tpk2-Myc exhibited greater activity when it was purified from the CDC37-106 strain than when it was purified from the CDC37+ wild-type strain. Unfortunately, the Tpk2-63-Myc fusion protein was devoid of measurable activity regardless of whether it was purified from the wild-type CDC37+ or CDC37-106 strain (Fig. 4C) or if it was immunoprecipitated from 10 times as much extract (data not shown). This defect is consistent with our previous observation that the tpk2-63(Ts) allele confers a partial PKA defect even at the permissive temperature (23, 44, 53). Using a fluorescent peptide assay, we also monitored PKA activities of whole-cell extracts made from strains containing either a wild-type or mutant allele of CDC37. Once again, PKA activity of the TPK2 CDC37-106 strain was greater than that of the TPK2 strain containing the wild-type CDC37+ allele (Fig. 4D). Moreover, extracts of the tpk2-63(Ts) cdc37-106 strain exhibited greater PKA activity than extracts of the isogenic tpk2-63(Ts) CDC37 strain (Fig. 4D). Thus, at least one dominant CDC37 mutation enhances the specific activity of both wild-type and mutant forms of the PKA catalytic subunit Tpk2.
Dominant mutations alter kinase affinity, but not levels, of Cdc37.
One trivial explanation for the suppression of the tpk2-63(Ts) growth defect is that each of the CDC37 mutations increases the amount of Cdc37 available for chaperone function. Cdc37 levels of strains containing wild-type and mutant CDC37 alleles, as well as a strain containing CDC37+ on a high-copy-number plasmid, were monitored by probing extracts with anti-Cdc37 antibody. Interestingly, none of the three dominant mutants examined (CDC37-101, CDC37-106, and CDC37-109) contained elevated levels of Cdc37 protein compared with those of the wild-type strain (Fig. 5A and data not shown). This was not due to an inherent insensitivity of the assay because the strain containing the high-copy-number CDC37+ plasmid exhibited significantly higher levels of Cdc37 protein than strains bearing either the wild-type or mutant CDC37 allele on the low-copy-number plasmid. Moreover, suppression did not correlate with Cdc37 levels because the tpk2-63(Ts) growth defect was more efficiently alleviated by the dominant mutants than the CDC37+ high-copy-number plasmid (Table 2).
All of the dominant CDC37 mutations fall within the conserved protein kinase interaction domain within the amino terminus of Cdc37 (Fig. 2C). Thus, the mutations might alter the physical interaction between Cdc37 and its client protein kinase subunit. We were unable to observe a physical interaction between Tpk2-Myc and either Cdc37 or Cdc37-106 (data not shown), consistent with previous conclusions that yeast Cdc37 binds protein kinases much more weakly than does its mammalian counterpart. Instead, we exploited a previously documented interaction between yeast Cdc37 and the catalytic domain of the pheromone-sensing pathway kinase Ste11 (1, 27). Although we failed to observe wild-type Cdc37 binding to the Ste11 catalytic domain (Ste11ΔN) in our strain background, we did observe the interaction of Ste11ΔN with Cdc37-106 in immunoprecipitation experiments (Fig. 5B), suggesting that the dominant mutant form of Cdc37 has increased affinity for its kinase clients.
Synthetic interaction between tpk2-63(Ts) and mutations in Hsp90 complex genes.
Although Cdc37 is generally thought to serve as a cochaperone of the Hsp90 complex, several observations are consistent with a model in which Cdc37 is capable of functioning alone (25-27). To examine if Hsp90 or its cochaperones contribute to PKA function, we combined the conditional tpk2-62(Ts) mutation with an sti1Δ deletion. The STI1 gene encodes an Hsp90 regulatory factor that is dispensable for viability at 30°C but that becomes essential for normal growth in cells shifted to elevated temperatures (10, 32). Interestingly, STI1 could not be deleted from the tpk2-62(Ts) strain unless the strain also contained CDC37-101 (Fig. 6A) or one of the other dominant alleles (data not shown). Thus, the tpk2-62(Ts) allele is synthetically lethal with mutations in two (cdc37-1 and sti1Δ) of the Hsp90 complex component genes. Although, Sti1 contributes to PKA function, it is not essential, because the tpk2-62(Ts) sti1Δ synthetic growth defect can be suppressed by the CDC37 dominant mutations. Nevertheless, the sti1Δ mutation blocks the suppression of the tpk2-62(Ts) conditional growth defect by CDC37-101 (Fig. 6B), as well as CDC37-106 and CDC37-109 (data not shown), indicating that Cdc37 and the Hsp90-chaperone complex may have interdependent roles in maintaining PKA function.
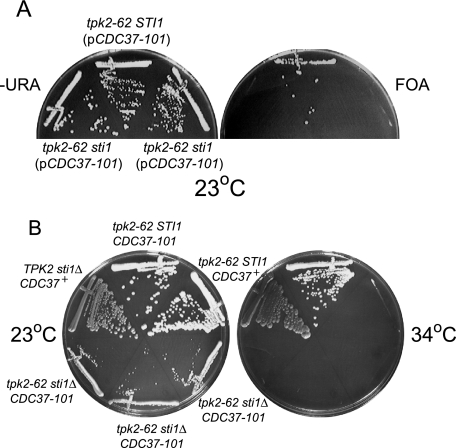
FIG. 6.
Functional interaction between PKA and the Hsp90-chaperone complex. (A) Synthetic lethal interaction between Hsp90 subunit gene STI1 and TPK2. The indicated strains were streaked onto minimal medium agar lacking uracil (−URA) or minimal medium agar containing uracil and 5-fluoroorotic acid (FOA) to select against plasmids expressing URA3 and incubated at 23°C. tpk2-62 sti1Δ (pCDC37-101) (MRY47) and tpk2-62 STI1 (pCDC37-101) [SGY562(pGS224)] strains were used. (B) Sti1 is essential for tpk2(Ts) suppression by CDC37-101. Strains were streaked onto minimal medium agar lacking uracil and incubated at 23°C and 34°C for 2 days. TPK2 sti1Δ CDC37+ (ACY77-3), tpk2-62 sti1Δ CDC37-101 [SGY562(pGS224)], tpk2-62 STI1 CDC37+ [SGY562(pRS316)], and tpk2-62 sti1Δ CDC37-101 (MRY47) strains were used.
DISCUSSION
We report here the isolation of mutations in the Hsp90 cochaperone Cdc37 that alleviate the growth defect of a conditional PKA mutant. These mutations identify residues of the N-terminal domain that play critical roles in the physical and functional interaction between Cdc37 and its protein kinase clients. Our results also suggest that the suppression of the conditional growth defect results from the restoration of PKA activity.
General consensus holds that Cdc37 functions as the kinase-specific targeting subunit of the Hsp90-chaperone complex. Biochemical support for this role comes from binding studies, which have assigned the protein kinase interaction domain of mammalian Cdc37 to the N-terminal 126 residues and the Hsp90-binding domain to a region between residues 127 and 283 (22, 43). Alanine-scanning mutagenesis has shown that several N-terminal residues are necessary for protein kinase binding (2, 43); however, it has been difficult to determine if those residues are directly involved in protein-protein interactions or if their effect on protein binding is a result of global changes within the protein. For example, although residues 2, 3, 4, and 7 are critical for the interaction between mammalian Cdc37 and heme-regulated HRI kinase, the W7A alteration also affected the interaction between Cdc37 and Hsp90, implying that relatively minor changes can have global effects (43). By contrast, residues identified in our suppressor screen fall within a small region (residues 33, 58, 59, 62, and 65) of the N-terminal domain and enhance, rather than abrogate, Cdc37 client binding and function. Of the five residues altered, W33 is the most evolutionarily conserved. It is found in Cdc37 proteins from such diverse organisms as humans, mice, fruit flies, and the yeasts Schizosaccharomyces pombe and Candida albicans, with only a limited number of fungi such as Neurospora crassa and Magnaporthe grisea containing an A at the same position. Neither of the other residues is as conserved, although L62 is found within all fungi with sequenced genomes, including S. pombe, C. albicans, N. crassa, and M. grisea. Because the screen was not saturating, it will be interesting to see if other conserved residues can be affected and if each residue has only a limited spectrum of acceptable changes.
The yeast CDC37 gene has been isolated in several screens for high-copy-number suppressors of growth defects conferred by conditional protein kinases. This is consistent with the conclusion that Cdc37, not Hsp90, is the limiting component of the Hsp90-chaperone complex (2, 25, 41, 46). Although Cdc37 overproduction can suppress the tpk2(Ts) growth defect, at least three of the five dominant mutations suppressed without elevating Cdc37 levels. Thus, the limiting nature of Cdc37 can be overcome by either increased levels or enhanced function. Because at least one of the mutant Cdc37 alleles enhanced protein kinase binding, we speculate that the interaction between Cdc37 and its client protein is a limiting factor in Cdc37 function.
This result is also intriguing because all five mutations were dominant and in different, although clustered, residues. The fact that all five mutations are different would imply that there are many ways in which the amino-terminal region might be altered to increase the binding affinity between Cdc37 and the kinase client. Although the interaction between Cdc37 and its large number of kinase clients must be sufficiently flexible to accommodate sequence and structural variation found in both Ser/Thr and Tyr kinases, it is still intriguing that all five mutations were in different amino acids. One way to reconcile this apparent paradox would be to imagine that the interaction between Cdc37 and the kinase is actually mediated by a domain of the amino-terminal region that is linked to, but separate from, the domain identified by the dominant mutations that fall between residues 33 and 65. In this scenario, the domain identified by the suppressor mutations would function as an intramolecular kinase-binding inhibitory domain, much like the autoinhibitory domain found in many protein kinases. Alteration of this domain would decrease its affinity for the kinase interaction domain within Cdc37, thereby increasing the relative affinity between Cdc37 and its client kinase. One prediction of the model is that Cdc37-kinase binding could be enhanced by relatively frequent alterations in the autoinhibitory domain (as exemplified by the mutations shown in Fig. 1) or relatively rare alterations of the (unknown) kinase interaction domain. Of course, both mutations would be dominant. Unfortunately, while the amino terminus is the most structurally conserved region of Cdc37, the absence of a three-dimensional structure for this portion of Cdc37 makes it hard to predict how these changes would affect this interaction. Nevertheless, these mutations should prove useful in future studies of Cdc37 binding and function.
Although Cdc37 is often described as a client-specific targeting subunit of the Hsp90-chaperone complex, several observations are consistent with Cdc37 possessing Hsp90-independent chaperone function. For example, purified yeast Cdc37 is capable of maintaining unfolded proteins in a reactivation-competent state (25), and yeast viability is supported by elevated levels of a Cdc37 truncation fragment that lacks the carboxyl-terminal Hsp90 interaction domain (26, 51). These results imply that Hsp90 interaction and function are not absolutely essential for Cdc37 function. Interestingly, while our genetic results suggest that Sti1 (and hence Hsp90) contributes to PKA function, they also show that Cdc37 function is not completely dependent upon Sti1 for function. These results are compatible with a model in which Cdc37 and Sti1/Hsp90 play interdependent roles in PKA maintenance.
The dominant CDC37 mutations ameliorate the tpk2(Ts) growth defect by enhancing the function of the mutant PKA catalytic subunit as judged by their effects on Msn2 localization and function as well as an increase in both in vivo and in vitro PKA activities. These results add PKA to a growing list of protein kinases whose functions can be influenced by Cdc37. Interestingly, this influence is not limited to PKA subunits that have been inactivated by extreme conditions or mutation, because the activity of the wild-type PKA subunit is also enhanced. Thus, Cdc37 may optimize proper folding during PKA synthesis as well as ensure that PKA catalytic activity remains elevated during periods of stress. The notion that Cdc37 plays a physiological role in PKA function is further supported by the synthetic growth defect exhibited by strains containing the cdc37-1(Ts) and tpk2-63(Ts) mutations.
In addition to the rare, dominant suppressors in CDC37, our selection allowed us to identify a second, intriguing class of suppressors that combined dominant suppression with a recessive growth defect. We have not characterized this second class of suppressors; however, most, if not all, fall within a single linkage group. The simplest explanation for the combined phenotype is that the same modification that alleviates the loss of PKA activity compromises an essential function. In the diploid strain, this growth defect is masked by the complementing wild-type allele. An alternative explanation is that both dominant suppression and the recessive growth defect result from a loss of function. According to this alternative model, diploid strains lacking one copy of the suppressor gene would have insufficient gene product to inhibit PKA-dependent growth but enough product to support viability, whereas haploid strains containing only the inactive allele would exhibit slow or no growth. This issue will be resolved by molecular characterization of the suppressors; however, in the meantime, we favor the former explanation because both dominant suppressor classes were isolated much less frequently (at least 100-fold) than loss-of-function mutations in YAK1 and MSN2 (16, 44).
Acknowledgments
We thank Chris Barry and Yan Zhang for excellent technical assistance.
This work was supported by NIH grants GM44666 (S.G.) and GM 70596 (A.C.). P.L. was supported by predoctoral training grant NIH 5T32DK07645.
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Abbas-Terki, T., O. Donze, and D. Picard. 2000. The molecular chaperone Cdc37 is required for Ste11 function and pheromone-induced cell cycle arrest. FEBS Lett. 467:111-116. [DOI] [PubMed] [Google Scholar]
- 2.Bandhakavi, S., R. O. McCann, D. E. Hanna, and C. V. Glover. 2003. A positive feedback loop between protein kinase CKII and Cdc37 promotes the activity of multiple protein kinases. J. Biol. Chem. 278:2829-2836. [DOI] [PubMed] [Google Scholar]
- 3.Beck, T., and M. N. Hall. 1999. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 4.Borkovich, K. A., F. W. Farrelly, D. B. Finkelstein, J. Taulien, and S. Lindquist. 1989. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boy-Marcotte, E., M. Perrot, F. Bussereau, H. Boucherie, and M. Jacquet. 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broach, J. R., and R. J. Deschenes. 1990. The function of ras genes in Saccharomyces cerevisiae. Adv. Cancer Res. 54:79-139. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, S., L. Levin, M. Zoller, and M. Wigler. 1988. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell 53:555-566. [DOI] [PubMed] [Google Scholar]
- 8.Cannon, J. F., and K. Tatchell. 1987. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 7:2653-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, H. C., D. F. Nathan, and S. Lindquist. 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17:318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, S., V. Prapapanich, R. A. Rimerman, B. Honore, and D. F. Smith. 1996. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins hsp90 and hsp70. Mol. Endocrinol. 10:682-693. [DOI] [PubMed] [Google Scholar]
- 12.Dey, B., J. J. Lightbody, and F. Boschelli. 1996. CDC37 is required for p60v-src activity in yeast. Mol. Biol. Cell 7:1405-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Düvel, K., A. Santhanam, S. Garrett, L. Schneper, and J. R. Broach. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11:1467-1478. [DOI] [PubMed] [Google Scholar]
- 14.Enquist, L., and R. A. Weisberg. 1977. A genetic analysis of the att-int-xis region of coliphage lambda. J. Mol. Biol. 111:97-120. [DOI] [PubMed] [Google Scholar]
- 15.Farrell, A., and D. O. Morgan. 2000. Cdc37 promotes the stability of protein kinases Cdc28 and Cak1. Mol. Cell. Biol. 20:749-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett, S., and J. Broach. 1989. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 3:1336-1348. [DOI] [PubMed] [Google Scholar]
- 17.Garrett, S., M. M. Menold, and J. R. Broach. 1991. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol. Cell. Biol. 11:4045-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerber, M. R., A. Farrell, R. J. Deshaies, I. Herskowitz, and D. O. Morgan. 1995. Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proc. Natl. Acad. Sci. USA 92:4651-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Gimeno, C. J., and G. R. Fink. 1994. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 14:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorner, W., E. Durchschlag, J. Wolf, E. L. Brown, G. Ammerer, H. Ruis, and C. Schuller. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21:135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grammatikakis, N., J.-H. Lin, A. Grammatikakis, P. N. Tsichlis, and B. H. Cochran. 1999. p50cdc37 acting in concert with Hsp90 is required for Raf-1 function. Mol. Cell. Biol. 19:1661-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartley, A. D., M. P. Ward, and S. Garrett. 1994. The Yak1 protein kinase of Saccharomyces cerevisiae moderates thermotolerance and inhibits growth by an Sch9 protein kinase-independent mechanism. Genetics 136:465-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Kimura, Y., S. L. Rutherford, Y. Miyata, I. Yahara, B. C. Freeman, L. Yue, R. I. Morimoto, and S. Lindquist. 1997. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 11:1775-1785. [DOI] [PubMed] [Google Scholar]
- 26.Lee, P., J. Rao, A. Fliss, E. Yang, S. Garrett, and A. J. Caplan. 2002. The Cdc37 protein kinase-binding domain is sufficient for protein kinase activity and cell viability. J. Cell Biol. 159:1051-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, P., A. Shabbir, C. Cardozo, and A. J. Caplan. 2004. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol. Biol. Cell 15:1785-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandal, A. K., P. Lee, J. A. Chen, N. Nillegoda, A. Heller, S. DiStasio, H. Oen, J. Victor, D. M. Nair, J. L. Brodsky, and A. J. Caplan. 2007. Cdc37 has distinct roles in protein kinase quality control that protect nascent chains from degradation and promote posttranslational maturation. J. Cell Biol. 176:319-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaughlin, S. H., H. W. Smith, and S. E. Jackson. 2002. Stimulation of the weak ATPase activity of human hsp90 by a client protein. J. Mol. Biol. 315:787-798. [DOI] [PubMed] [Google Scholar]
- 30.McMillan, J. N., R. A. Sia, and D. J. Lew. 1998. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol. 142:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moskvina, E., C. Schuller, C. T. Maurer, W. H. Mager, and H. Ruis. 1998. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 14:1041-1050. [DOI] [PubMed] [Google Scholar]
- 32.Nicolet, C. M., and E. A. Craig. 1989. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 9:3638-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearl, L. H., and C. Prodromou. 2001. Structure, function, and mechanism of the Hsp90 molecular chaperone. Adv. Protein Chem. 59:157-186. [DOI] [PubMed] [Google Scholar]
- 34.Picard, D. 2002. Heat shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. 59:1640-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pratt, W. B., and D. O. Toft. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228:111-133. [DOI] [PubMed] [Google Scholar]
- 36.Prodromou, C., G. Siligardi, R. O'Brien, D. N. Woolfson, L. Regan, B. Panaretou, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prodromou, C., and L. H. Pearl. 2003. Structure and functional relationships of Hsp90. Curr. Cancer Drug Targets 3:301-323. [DOI] [PubMed] [Google Scholar]
- 38.Reed, S. I. 1980. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics 95:561-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santhanam, A., A. Hartley, K. Düvel, J. R. Broach, and S. Garrett. 2004. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot. Cell 3:1261-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F. U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199-210. [DOI] [PubMed] [Google Scholar]
- 41.Scholz, G., S. D. Hartson, K. Cartledge, N. Hall, J. Shao, A. R. Dunn, and R. L. Matts. 2000. p50Cdc37 can buffer the temperature-sensitive properties of a mutant of Hck. Mol. Cell. Biol. 20:6984-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schutz, A. R., T. H. Giddings, Jr., E. Steiner, and M. Winey. 1997. The yeast CDC37 gene interacts with MPS1 and is required for proper execution of spindle pole body duplication. J. Cell Biol. 136:969-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao, J., T. Prince, S. D. Hartson, and R. L. Matts. 2003. Phosphorylation of serine 13 is required for the proper function of the Hsp90 co-chaperone, Cdc37. J. Biol. Chem. 278:38117-38120. [DOI] [PubMed] [Google Scholar]
- 44.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stepanova, L., X. Leng, S. B. Parker, and J. W. Harper. 1996. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 10:1491-1502. [DOI] [PubMed] [Google Scholar]
- 46.Stepanova, L., M. Finegold, F. DeMayo, E. V. Schmidt, and J. W. Harper. 2000. The oncoprotein kinase chaperone CDC37 functions as an oncogene in mice and collaborates with both c-myc and cyclin D1 in transformation of multiple tissues. Mol. Cell. Biol. 20:4462-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, S. S., J. Yang, J. Wu, N. M. Haste, E. Radzio-Andzelm, and G. Anand. 2004. PKA: a portrait of protein kinase dynamics. Biochim. Biophys. Acta 11:259-269. [DOI] [PubMed] [Google Scholar]
- 48.Thevelein, J. M., and J. H. De Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 49.Toda, T., S. Cameron, P. Sass, M. Zoller, and M. Wigler. 1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50:277-287. [DOI] [PubMed] [Google Scholar]
- 50.Toda, T., S. Cameron, P. Sass, M. Zoller, J. D. Scott, B. McMullen, M. Hurwitz, E. G. Krebs, and M. Wigler. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turnbull, E. L., I. V. Martin, and P. A. Fantes. 2005. Cdc37 maintains cellular viability in Schizosaccharomyces pombe independently of interactions with heat-shock protein 90. FEBS J. 272:4129-4140. [DOI] [PubMed] [Google Scholar]
- 52.Valay, J. G., M. Simon, M. F. Dubois, O. Bensaude, C. Facca, and G. Faye. 1995. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol. 249:535-545. [DOI] [PubMed] [Google Scholar]
- 53.Ward, M. P., and S. Garrett. 1994. Suppression of a yeast cyclic AMP-dependent protein kinase defect by overexpression of SOK1, a yeast gene exhibiting sequence similarity to a developmentally regulated mouse gene. Mol. Cell. Biol. 14:5619-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]