Abstract
Moraxella catarrhalis ETSU-9 was subjected to random transposon insertion mutagenesis to identify genes encoding products involved in the ability of the organism to form biofilms in vitro. Screening of approximately 3,000 transposon insertion mutants in the crystal violet-based biofilm assay system yielded six mutants that exhibited greatly reduced abilities to form biofilms. Three of these mutants had transposon insertions in the uspA2H gene, which encodes a surface protein previously shown to be involved in the ability of M. catarrhalis to both attach to human cell lines in vitro and resist killing by normal human serum. Random insertion mutagenesis of the uspA2H gene, involving the introduction of a 15-nucleotide fragment encoding 5 amino acids, was used to attempt to identify the domain(s) necessary for biofilm formation. Most of these insertions adversely affected biofilm formation, whereas the abilities of these same mutants to attach to Chang conjunctival epithelial cells in vitro were usually not reduced. Gain-of-function experiments showed that introduction of the M. catarrhalis ETSU-9 uspA2H gene into Escherichia coli conferred biofilm formation ability on this recombinant strain. Two of the other three M. catarrhalis ETSU-9 transposon insertion mutants that had greatly reduced abilities to form biofilms were shown to have insertions in genes encoding products predicted to be directly or indirectly involved in cell wall metabolism.
Moraxella catarrhalis is a gram-negative, unencapsulated coccobacillus that can colonize the human nasopharynx. This colonization event occurs very frequently in infants and very young children (12), in whom the organism is a significant cause of acute otitis media (10, 43). Colonization of the nasopharynx in adults is less common, but in persons with chronic obstructive pulmonary disease, M. catarrhalis originating from the nasopharynx is a significant cause of infectious exacerbations of the disease (32). Less frequently, the bacterium can also cause other diseases, most of which involve the respiratory tract (31).
There are limited data available about the interaction of M. catarrhalis with the mucosal surface in the host environment. Several M. catarrhalis cell surface proteins that function as adhesins for human cells, at least in vitro, have been described (20, 22, 45, 48). In the human nasopharynx, it is likely that M. catarrhalis exists in a biofilm together with commensal bacteria on the mucosa, and there is a recent report describing M. catarrhalis biofilms detected on the mucosa of the middle ear in children with otitis media (15). To date, there have been only a few studies of biofilm formation by M. catarrhalis in vitro (8, 41, 42). In one of these, the UspA1 adhesin protein was shown to be important for biofilm formation in vitro by the O46E strain of M. catarrhalis (42). UspA1 forms a filamentous projection on the surface of M. catarrhalis (41) and binds several types of human cells, as well as fibronectin (47), the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) (17, 18), and both C3 (36) and C4B-binding protein (35). A uspA1 gene is apparently present in most, if not all, strains of M. catarrhalis (29), uspA1 mRNA has been detected in saliva from children colonized with the organism (28), and expression of UspA1 has been reported to be affected by temperature (16). In addition to UspA1, many strains of M. catarrhalis produce the UspA2 protein, which has been shown to confer serum resistance on some strains of the pathogen (1, 4, 5). The UspA1 and UspA2 proteins have in common an epitope that binds the monoclonal antibody (MAb) 17C7 (2). UspA2 forms relatively short projections on the surface of the organism (41), is produced in much larger quantities than UspA1, and has been reported to bind vitronectin (5, 27), fibronectin (47), C3 (36), and C4B-binding protein (35).
Approximately 20% of M. catarrhalis strains produce a UspA2H protein in place of a UspA2 protein (22). The UspA2H protein possesses properties of both UspA1 and UspA2 in that UspA2H proteins can confer both attachment ability and serum resistance on M. catarrhalis (22). Like UspA2, the UspA2H protein is much more abundant than UspA1 in strains that produce the former macromolecule. In the present study, transposon-mediated mutagenesis revealed that the UspA2H protein and at least three other gene products of M. catarrhalis strain ETSU-9 are involved in the ability of the organism to produce a biofilm in the crystal violet-based biofilm system in vitro.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The M. catarrhalis strains used in this study are described in Table 1, with the exception of the numerous mutants with small insertion mutations in the uspA2H gene. M. catarrhalis ETSU-9 produces a UspA2H protein in place of a UspA2 protein (22). The M. catarrhalis ETSU-9 strain used in this study lacked the ability to express a Hag protein (41) because of apparent slipped-strand mispairing in the poly(G) tract located at the very start of the hag open reading frame (ORF) (41). This strain also possessed nine G residues in the poly(G) tract of its uspA1 gene and consequently expressed a relatively low level of UspA1 (23). M. catarrhalis strains were grown as described previously using brain heart infusion (BHI) (Difco, Detroit, MI) or Todd-Hewitt (TH) medium (Difco) with antibiotic supplementation as necessary (41, 42). M. catarrhalis biofilm formation assays were performed without antibiotics. Escherichia coli strains were routinely grown on LB agar (Difco) supplemented with kanamycin (50 μg/ml) or chloramphenicol (20 μg/ml) as appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain | Description | Source or reference |
|---|---|---|
| M. catarrhalis | ||
| O35E | Wild-type isolate | 2 |
| ETSU-9 | Wild-type isolate | Steven Berk |
| ETSU-9.Strr | Spontaneous streptomycin-resistant rpsL mutant of ETSU-9 | 42 |
| ETSU-9.1 | uspA1 null mutant of ETSU-9; kanamycin resistant | This study |
| ETSU-9.1Strr | Streptomycin-resistant ETSU-9.1 | This study |
| ETSU-9.2 | uspA2H null mutant of ETSU-9 constructed using pELU244SPEC as the source of the mutant allele; spectinomycin resistant | This study |
| ETSU-9.12 | uspA1 uspA2H mutant of ETSU-9; kanamycin and spectinomycin resistant | This study |
| ETSU-9.12Strr | Streptomycin-resistant ETSU-9.12 | This study |
| ETSU-9.2H | uspA2H null mutant of ETSU-9 constructed using pMPGTG3spec as the source of the mutant allele; spectinomycin resistant | This study |
| ETSU-9.mut92 | ETSU-9 mutant with 418-aa deletion in the UspA2H protein | This study |
| ETSU-9.mut92.2 | uspA2H null mutant of ETSU-9mut.92 | This study |
| ETSU-9.698 | ETSU-9 with a transposon insertion in the uspA2H gene; Kanr | This study |
| ETSU-9.724 | ETSU-9 with a transposon insertion in the uspA2H gene; Kanr | This study |
| ETSU-9.2311 | ETSU-9 with a transposon insertion in the uspA2H gene; Kanr | This study |
| ETSU-9.222 | ETSU-9 with a transposon insertion in the putative lytic murein transglycosylase D gene; Kanr | This study |
| ETSU-9.222cat | Lytic murein transglycosylase D gene mutant of ETSU-9; Cmr | This study |
| ETSU-9.714 | ETSU-9 with a transposon insertion in the putative aminoglycoside phosphotransferase gene; Kanr | This study |
| ETSU-9.714cat | Aminoglycoside phosphotransferase mutant of ETSU-9; Cmr | This study |
| ETSU-9.988 | ETSU-9 with a transposon insertion in the putative ampD gene; Kanr | This study |
| ETSU-9.988cat | ampD mutant of ETSU-9; Cmr | This study |
| E. coli | ||
| EPI3000 | Host for CopyControl pCC1 vector | Epicentre |
| DH5α | Host for cloning experiments | Invitrogen |
| E. coli INVαF′ | Host for cloning experiments | Invitrogen |
| Plasmids | ||
| pCR-Blunt | Cloning vector; Kanr Zeocinr | Invitrogen |
| pMPGTG3 | pCR-Blunt containing the majority of the ETSU-9 uspA2H ORF | This study |
| pMPGTG3spec | pMPGTG3 with a spectinomycin resistance cartridge inserted in the uspA2H ORF | This study |
| pELU244SPEC | pBluescript containing a truncated uspA2 gene from M. catarrhalis P44 into which a spectinomycin resistance cartridge was inserted | 22 |
| pCR2.1 | Cloning vector; Kanr Ampr | Invitrogen |
| pMP222 | pCR2.1 containing the putative M. catarrhalis lytic murein transglycosylase D gene | This study |
| pMP222cat | pMP222 with a promoterless cat cartridge inserted into the NdeI site in the putative lytic murein transglycosylase D gene; Kanr Ampr Cmr | This study |
| pMP714 | pCR2.1 containing the putative M. catarrhalis aminoglycoside phosphotransferase gene | This study |
| pMP714cat | pMP714 with a promoterless cat cartridge inserted into the AccI site in the putative aminoglycoside phosphotransferase gene; Kanr Ampr Cmr | This study |
| pMP988 | pCR2.1 containing the putative M. catarrhalis ampD gene | This study |
| pMP988cat | pMP988 with a promoterless cat cartridge inserted into the AccI site in the ampD gene; Kanr Ampr Cmr | This study |
| pUSPA1KAN | pBluescript with a uspA1 gene fragment containing a kan cartridge | 2 |
| pCC1 | CopyControl vector; Cmr | Epicentre |
| pCC1kan | pCC1 containing a cloned kan cartridge | This study |
| pMPE9U2H | pCC1 containing the wild-type ETSU-9 uspA2H gene | This study |
Preparation of antigens and MAbs and Western blot analysis.
Whole-cell lysates of agar plate-grown bacteria were prepared as described previously (40). Outer membrane vesicles were isolated according to the broth culture supernatant method of Murphy and Loeb (33). Lipooligosaccharide (LOS) was detected in proteinase K-treated whole-cell lysates by silver staining as described previously (4, 49). MAb 17C7, which is reactive with both the UspA1 and the UspA2/UspA2H proteins of M. catarrhalis, has been described previously (2, 22). The goat anti-mouse secondary antibody and chemiluminescence detection method have been described previously (41).
Measurement of β-lactamase activity using nitrocefin.
β-Lactamase activity in bacterial culture supernatant fluid was measured by using the chromogenic cephalosporin nitrocefin as the substrate in a method adapted from that of Angus and colleagues (3). M. catarrhalis cultures were grown in 10 ml BHI to a final Klett reading of 300, as determined by the use of a Klett colorimeter (VWR Scientific). The cells in each culture were then pelleted by centrifugation (12,000 × g for 15 min at 4°C), and the resultant supernatant fluid was removed and used in the assay. Serial 10-fold dilutions of the supernatant fluid (375 μl), 10 mM HEPES buffer, pH 7.0 (75 μl), and nitrocefin (50 μl of a 1-mg/ml stock) were added to plastic tubes. The negative control had sterile BHI in place of culture supernatant fluid. The mixture in each tube was aliquoted in duplicate into a 96-well microtiter plate (200 μl per well), which was incubated at 37°C for 30 min, after which the optical density of the solution at 485 nm was determined.
Transposon mutagenesis.
M. catarrhalis strain O35E was mutagenized using the EZ::TN<KAN-2> transposome kit (Epicentre, Madison, WI). Briefly, TH broth-grown cells of strain O35E were electroporated with the transposome, and transposon-containing mutants were selected on TH agar containing kanamycin. Approximately 5,000 transposon insertion mutant colonies were pooled and grown in 500 ml TH broth for 4 h. Chromosomal DNA containing transposon insertions was isolated from the cells in this culture. M. catarrhalis strain ETSU-9, which cannot be mutagenized directly with this transposome system, was transformed with 10 μl (∼10 μg) of the chromosomal DNA using the plate transformation protocol as described previously (42), and kanamycin-resistant ETSU-9 transformants containing transposon insertions were selected on TH agar containing kanamycin.
Crystal violet-based assay for biofilm-deficient mutants.
Approximately 3,000 ETSU-9 transposon mutants were patched onto TH agar plates containing kanamycin and grown overnight. A swatch of bacterial growth from each patch was used to inoculate a well of a 24-well tissue culture plate containing 2 ml TH broth. The tissue culture plates were incubated at 37°C for 19 h. These plates were washed and stained with crystal violet to detect biofilm formation as described previously (42) using a technique adapted from that described by O'Toole and Kolter (37). Briefly, the broth culture medium containing bacterial growth was removed from each well, and then 2 ml of medium 199 tissue culture medium containing Earle's balanced salt solution, l-glutamine, and HEPES (Fisher Scientific) plus 100 μl 0.7% (wt/vol) crystal violet (Sigma) was added. The fluid content of each well was decanted after 15 min, three washes were performed with deionized water, and then a 2-ml volume of 95% ethanol was added to each well. After 15 min of gentle agitation at room temperature, a 1.5-ml volume of this solution was removed and its absorbance at 570 nm was measured.
Mutants that were apparently deficient in pellicle or biofilm formation but which appeared to grow well otherwise were retained for further analysis. To confirm that these ETSU-9 mutants were biofilm deficient as a result of the transposon insertion, chromosomal DNA was isolated from each transposon-bearing, biofilm-negative ETSU-9 mutant and used to transform ETSU-9 again (i.e., a genetic backcross); the resultant kanamycin-resistant transformants (e.g., ETSU-9.698) were used in all subsequent studies.
Chang cell attachment assay.
M. catarrhalis strains were evaluated for their abilities to attach to Chang conjunctival epithelial cells (ATCC CCL 20.2) in vitro as described previously (50).
PCR and nucleotide sequence analysis.
PCR amplification was achieved by use of ExTaq DNA polymerase (PanVera, Madison, WI). M. catarrhalis chromosomal DNA templates were prepared using the Easy-DNA kit (Invitrogen, Carlsbad, CA). Plasmid DNA templates were prepared using the QIAprep Spin miniprep kit (QIAGEN, Valencia, CA). M. catarrhalis chromosomal DNA for direct sequencing was isolated using the UltraClean Microbial DNA isolation kit (MoBio Laboratories, Inc., Solana Beach, CA). Nucleotide sequences obtained from automated sequencing systems were analyzed using the MacVector analysis package (version 6.5; Oxford Molecular Group, Campbell, CA). The design of oligonucleotide primers for use in the PCR-based amplification of selected regions of ETSU-9 chromosomal DNA was accomplished using data obtained from the nucleotide sequence of the genome of M. catarrhalis ATCC 43617 as deposited at NCBI under patent WO0078968.
Construction of M. catarrhalis mutants.
Inactivation of the uspA2H gene was accomplished by deleting part of the gene in M. catarrhalis ETSU-9. A 2.6-kb fragment encompassing all of the ETSU-9 uspA2H ORF except the extreme 5′ end was amplified by PCR using the primers 5′-AAATGCCGCAGGTCATTCGG-3′ and 5′-CTTCTAGAGCTTTTATCCATCACTCAC-3′ (the XbaI site is underlined) and was cloned by using the Zero Blunt TOPO PCR cloning kit (Invitrogen) to construct plasmid pMPGTG3. The plasmid was then digested with both EcoRI and NheI to remove an ∼1-kb fragment from the uspA2H ORF, and the spectinomycin resistance cartridge from pSPECR (52) was ligated into the deletion site to construct pMPGTG3spec. ETSU-9 was transformed with the plasmid to obtain the spectinomycin-resistant uspA2H mutant ETSU-9.2H. A second uspA2H mutant was constructed by transforming ETSU-9 with pELU244SPEC (22) and selection with spectinomycin; the resultant uspA2H mutant was designated ETSU-9.2. The uspA1 gene was inactivated in M. catarrhalis strains by using the recombinant plasmid pUSPA1KAN (1) to transform the target strain by either electroporation or plate transformation. The uspA1 uspA2H mutant ETSU-9.12 was obtained by transforming the uspA1 mutant ETSU-9.1 with pELU244SPEC (22).
A fragment lacking the ATG translational start codon of the putative lytic murein transglycosylase D gene from M. catarrhalis ETSU-9 was amplified by PCR using the primers 5′-AAACACACCAAATCTTCG-3′ and 5′-TTATCGATTTGGTTCGGC-3′ with ETSU-9 chromosomal DNA as the template. This fragment was cloned into pCR2.1, and the resultant plasmid, designated pMP222, was digested with NdeI, which cuts within the ORF. This linearized plasmid was ligated to the cat cartridge from pSL1 (25) and used to transform E. coli DH5α. The plasmid from a chloramphenicol-resistant transformant was designated pMP222cat and was used to transform ETSU-9 to obtain the mutant ETSU-9.222cat.
A 2.5-kb fragment encompassing the gene encoding the predicted aminoglycoside phosphotransferase was PCR amplified using the primers 5′-TCCAGTTTTGGGGTGGTAGG-3′ and 5′-TGATCTTGGACATAAGGCTTCG-3′ with ETSU-9 chromosomal DNA as the template and cloned into pCR2.1 to obtain pMP714. The plasmid was digested with AccI, which cuts within the predicted ORF. The cat cartridge from pSL33 (24) was ligated with the linearized pMP714 and used to transform E. coli strain DH5α. Plasmid pMP714cat was purified from a chloramphenicol-resistant transformant and used to transform M. catarrhalis ETSU-9 to construct the mutant ETSU-9.714cat.
A 2.7-kb fragment encompassing the putative ampD gene was amplified by PCR from ETSU-9 chromosomal DNA using the primers 5′-CAATCGCCACACCAATGAGTC-3′ and 5′-AAATCGCTGCAATGCGTGAGGG-3′ and cloned into pCR2.1 to obtain pMP988. After digestion with AccI, which cuts within the ampD ORF, the cat cartridge from pCWnpcat1 (51) was ligated with the linearized pMP988 plasmid and used to transform E. coli strain DH5α. The plasmid from a chloramphenicol-resistant transformant was designated pMP988cat and used to transform M. catarrhalis strain ETSU-9 to obtain the mutant ETSU-9.988cat.
Insertional mutagenesis of the uspA2H gene.
Random 15-nucleotide (nt) insertions in the ETSU-9 uspA2H gene were obtained by use of the GPS-LS Linker Scanning System (New England Biolabs, Beverly, MA) to mutagenize pMPGTG3. After excision of the transposon to leave a 15-nt insertion in pMPGTG3, the plasmids were transformed into the spectinomycin-resistant M. catarrhalis uspA2H mutant ETSU-9.2 by using the plate transformation method. This uspA2H mutant was constructed by transforming ETSU-9 with pELU244SPEC (22). Because the pELU244SPEC plasmid was derived by using the uspA2 gene of M. catarrhalis strain P44 (22), some nucleotides in the mutated ETSU-9.2 uspA2H gene were derived from the P44 uspA2 gene. Consequently, some of the uspA2H insertion mutants also contained these P44-derived uspA2 nucleotides. These mutants (10, 28, 32, 49, 93, and 109) were therefore reconstructed using the ETSU-9.2H strain (described above). However, most insertion mutants derived from ETSU-9.2 were confirmed by nucleotide sequence analysis to contain only ETSU-9 uspA2H nucleotides. M. catarrhalis transformants possessing 15-nt insertions in uspA2H were enriched by using congression as described previously (42) and were screened for loss of spectinomycin resistance. The presence of the 15-nt insertion in the uspA2H gene in the chromosome of each mutant was confirmed by nucleotide sequence analysis.
Cloning and expression of uspA2H genes in E. coli.
The uspA2H gene from M. catarrhalis strain ETSU-9 was amplified by PCR using the primers 5′-ATCTCGGGAGCTAAGCTTGAGGGTTTGG-3′ and 5′-ATCTCGGGCTTTTATCCATCACTCAC-3′, ligated into the plasmid vector pCC1, and used to electroporate E. coli EPI300 as described in the CopyControl PCR Cloning Kit (Epicenter) instructions. Transformant colonies were inoculated into LB broth containing both chloramphenicol and CopyControl induction solution according to the manufacturer's instructions. Western blot analysis of whole-cell lysates was performed as described for M. catarrhalis, except that proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore Corp., Bedford, MA) instead of nitrocellulose. The recombinant plasmid expressing the ETSU-9 UspA2H protein was designated pMPE9U2H.
Crystal violet-based assay for E. coli biofilm formation.
Recombinant E. coli EPI300 strains were grown overnight in 5 ml LB broth supplemented with chloramphenicol. A 0.5-ml portion of this culture was used to inoculate 4.5 ml LB (with chloramphenicol as necessary) containing 5 μl CopyControl induction solution. Cultures were grown with vigorous agitation for 5 h, after which all cultures were adjusted to the same density by the addition of LB broth. A 20-μl portion of each induced culture was used to inoculate 2 ml LB broth containing 2 μl CopyControl induction solution in a 24-well tissue culture plate. The remainder of the crystal violet-based assay was performed as described above for M. catarrhalis.
Nucleotide sequence accession number.
The nucleotide sequence of the M. catarrhalis ETSU-9 uspA2H gene has been deposited at GenBank (accession number DQ811779).
RESULTS
Screening for biofilm-negative mutants of ETSU-9.
Approximately 3,000 kanamycin-resistant M. catarrhalis ETSU-9 transposon insertion mutants were screened for loss of the ability to form biofilms in vitro using the crystal violet-based assay. These transposon mutants were obtained by transforming M. catarrhalis strain ETSU-9 with chromosomal DNA from M. catarrhalis strain O35E mutants that contained random transposon insertions as described in Materials and Methods. These screening experiments and subsequent backcross efforts led to the identification of six kanamycin-resistant ETSU-9 mutants that were consistently biofilm negative or markedly attenuated for biofilm formation.
Characterization of biofilm-negative transposon insertion mutants.
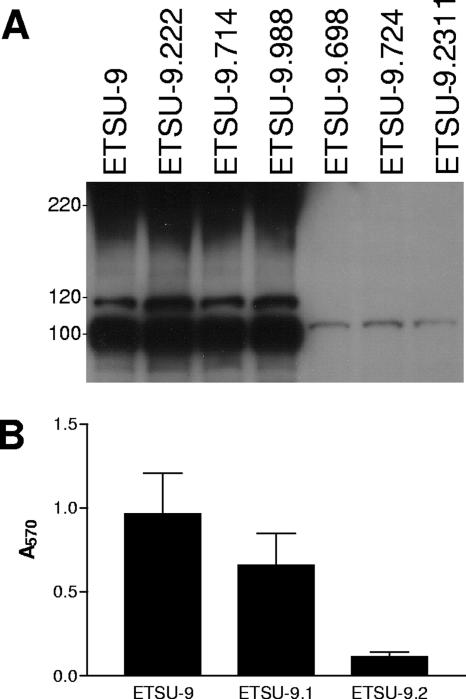
The UspA1 protein has been shown to play a role in biofilm formation by M. catarrhalis strain O46E (42), so whole-cell lysates of these six mutants were first subjected to Western blot analysis to determine whether the mutants still expressed the UspA1 protein or the closely related UspA2H protein (22). All six of the strains still expressed UspA1 (data not shown). However, three of the six transposon insertion mutants (ETSU-9.698, ETSU-9.724, and ETSU-9.2311) lacked expression of UspA2H (Fig. 1A). The involvement of UspA2H in biofilm formation by ETSU-9 was confirmed by measuring biofilm formation by the strain and both its uspA1 mutant, ETSU-9.1, and its uspA2H mutant, ETSU-9.2 (Fig. 1B). The uspA2H mutation in ETSU-9.2 had a much more profound effect on biofilm formation than did the uspA1 mutation in ETSU-9.1 (Fig. 1B). The other three biofilm-negative transposon insertion mutants (ETSU-9.222, ETSU-9.714, and ETSU-9.988) still expressed a UspA2H protein and are discussed below. Because mutations in LOS have been shown to have an effect on biofilm formation by other bacterial species (46), proteinase K-treated whole-cell lysates from all six mutants were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 15% (wt/vol) polyacrylamide gel and stained with silver to detect LOS. There were no differences observed between LOS from the transposon insertion mutants and LOS from the ETSU-9 parent strain (data not shown).
FIG. 1.
Characterization of backcross-derived, biofilm-negative M. catarrhalis ETSU-9 mutants. (A) Detection of UspA2H expression by biofilm-negative ETSU-9 transposon mutants. Whole-cell lysates of the ETSU-9 parent strain and the six biofilm-negative transposon mutants were probed in Western blot analysis for UspA2H expression using MAb 17C7 as the primary antibody probe. The UspA2H protein appears as a smear around the 220-kDa position marker and as several smaller MAb-reactive species with apparent molecular masses of 118 kDa and 100 kDa in the Western blot. The UspA1 protein is visible in the last three lanes as a faint band just above the 100-kDa position marker. Molecular mass position markers (in kDa) are on the left. (B) Biofilm formation by wild-type and mutant strains of ETSU-9. Biofilm formation by the ETSU-9 parent strain, the uspA1 mutant ETSU-9.1, and the uspA2H mutant ETSU-9.2 was measured using the crystal violet-based assay. Each strain was grown in three different wells per experiment, and the data represent the means and standard errors from three independent experiments.
Insertional mutagenesis of the ETSU-9 uspA2H gene.
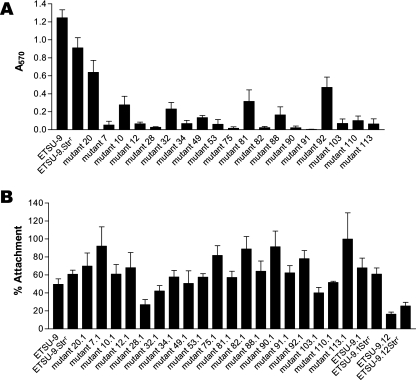
The data described above indicated that the UspA2H protein had a significant role in allowing biofilm development by ETSU-9 in the crystal violet-based assay. To attempt to identify the domain(s) of the ETSU-9 UspA2H protein that might be necessary for biofilm formation, a 15-nt insertion sequence was randomly introduced into the cloned ETSU-9 uspA2H ORF by means of a transposon-based insertion/excision system. These insertion mutations in uspA2H were then introduced into strain ETSU-9 by plate transformation. Because there is no selectable marker in the 15-nt insertion after the transposon has been excised, the probability of obtaining the desired transformants was increased by employing the technique of congression (34), using a mutated M. catarrhalis rpsL gene conferring streptomycin resistance (42). Thus, all of these new insertion mutants were also streptomycin resistant, and a streptomycin-resistant mutant of ETSU-9, designated ETSU-9.Strr (42), was used as a positive control in subsequent experiments measuring biofilm formation. (This streptomycin-resistant mutant was the source of the mutated rpsL gene used for the congression experiments.) All mutated uspA2H genes were subjected to nucleotide sequence analysis to confirm the presence of the 15-nt insertion. The position of each of the resultant 5-amino-acid (aa) insertions within the ETSU-9 UspA2H protein is depicted in Fig. 2.
FIG. 2.
Schematic of the amino acid sequence of the M. catarrhalis ETSU-9 UspA2H protein with the positions of the 5-aa insertions introduced by transposon-mediated mutagenesis indicated by inverted triangles. The numbers flanking each triangle refer to the UspA2H amino acids that flank the insertion site, using the complete UspA2H protein, including the leader peptide, for the numbering scheme. Three insertions (i.e., 81, 92, and 103) resulted in subsequent in-frame deletions of uspA2H DNA; the extent of each deletion is indicated. Mutant ETSU-9.mut20 contained a 15-nt insertion in the DNA immediately 3′ of the translational termination codon of the uspA2H gene.
Each of these insertion mutants was tested for its ability to form biofilms using the crystal violet-based assay. Surprisingly, the majority of these mutants were biofilm negative (Fig. 3A). To confirm that all of the insertion mutants were producing a UspA2H protein, Western blot analysis was performed. Most of the mutants were positive for UspA2H protein production detectable by Western blotting with MAb 17C7, with the exceptions of ETSU-9.mut3, ETSU-9.mut86, and ETSU-9.mut92 (data not shown). ETSU-9.mut3 was found to have a 15-nt insertion at the very end of the uspA2H ORF (Fig. 2), which likely affected the membrane-anchoring region of the protein. ETSU-9.mut86 had a 15-nt insertion near the 3′ end of the uspA2H gene, which resulted in a premature translational stop codon (Fig. 2). Three of the mutants (ETSU-9.mut81, ETSU-9.mut103, and ETSU-9.mut92) had internal deletions, with ETSU-9.mut92 having the largest in-frame deletion in the middle of its uspA2H gene (Fig. 2). These deletions likely occurred during allelic exchange between the mutated copy of the uspA2H gene and the wild-type allele in the ETSU-9 chromosome, as the deleted portions of uspA2H contained several highly conserved repeating sequences. In ETSU-9.mut92, the internal deletion resulted in the loss of 418 of the 875 aa in the UspA2H protein, including the epitope recognized by MAb 17C7 (2).
FIG. 3.
Biofilm formation and Chang cell attachment ability of mutants with 5-aa insertions in the UspA2H protein. (A) Biofilm formation by these uspA2H insertion mutants as measured in the crystal violet-based assay. (B) Attachment to Chang cells by the uspA2H mutants that also possessed a uspA1 mutation. All insertion mutants in panel B are streptomycin resistant and do not express UspA1. ETSU-9.mut20.1 is both streptomycin resistant and a uspA1 mutant that possesses a 15-nt insertion immediately downstream of the uspA2H ORF (see Fig. 2); it is included as a control for the 15-nt insertion. These data represent the means and standard errors from three independent experiments.
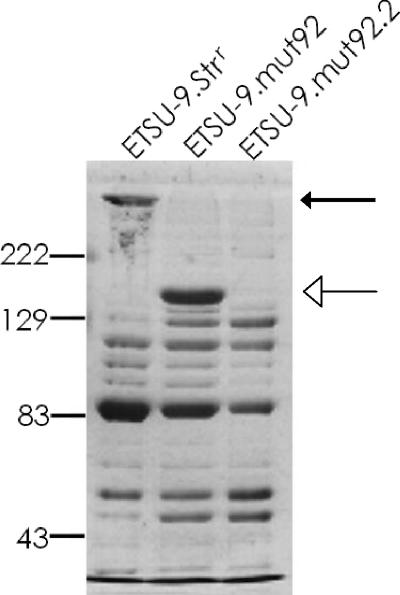
To determine whether the ETSU-9.mut92 mutant with the large internal deletion was actually expressing any UspA2H protein, the uspA2H gene in ETSU-9.mut92 was inactivated by insertional mutagenesis, yielding ETSU-9.mut92.2. Outer membrane vesicles were isolated from ETSU-9.Strr ETSU-9.mut92, and ETSU-9.mut92.2, and the outer membrane proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue. ETSU-9.mut92 produced a much smaller form of UspA2H than did the ETSU-9 parent strain (Fig. 4), consistent with the internal deletion in the uspA2H gene of the former strain. This smaller band was absent when the uspA2H null mutant ETSU-9.mut92.2 was analyzed (Fig. 4).
FIG. 4.
ETSU-9.mut92 expresses a UspA2H protein with an internal deletion. Proteins in outer membrane vesicles isolated from ETSU-9.Strr, ETSU-9.mut92, and the uspA2H null mutant ETSU-9.mut92.2 were resolved by SDS-PAGE and stained with Coomassie blue. The wild-type UspA2H protein expressed by ETSU-9.Strr is indicated by the black arrow, and the much smaller form of UspA2H expressed by ETSU-9.mut92 is indicated by the white arrow. Molecular mass position markers (in kDa) are on the left.
Abilities of the uspA2H insertion mutants to attach to Chang cells.
An attachment assay involving Chang conjunctival epithelial cells was used to test for functional, surface-expressed UspA2H protein in the biofilm-negative mutants (Fig. 3A). To accomplish this, the uspA1 gene first had to be inactivated by insertion of a kan cartridge, because both the UspA1 and UspA2H proteins are adhesins for Chang cells (22). All of these newly constructed uspA1 mutants derived from the uspA2H insertion mutants readily attached to Chang cells (Fig. 3B), with the exception of mutant ETSU-9.mut28.1, which exhibited an attachment level that was similar to that obtained with the uspA1 uspA2H mutant ETSU-9.12Strr, which was used as the negative control strain. Western blot analysis of outer membrane vesicles from the latter mutant indicated that it expressed a UspA2H protein at a level equivalent to that of its parent strain (data not shown).
Biofilm formation by recombinant E. coli expressing UspA2H.
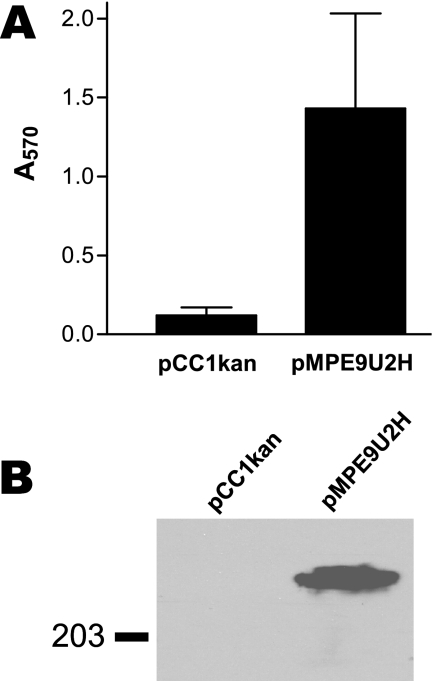
We next performed gain-of-function experiments to determine whether expression of the ETSU-9 UspA2H protein was sufficient to confer biofilm formation ability on a biofilm-negative E. coli strain. The PCR-amplified ETSU-9 uspA2H gene was cloned into pCC1 to obtain pMPE9U2H. When analyzed by Western blotting, E. coli EPI300(pMPE9U2H) (Fig. 5B) was shown to express the UspA2H protein, and the same recombinant strain readily formed a biofilm in the crystal violet-based assay (Fig. 5A). The recombinant E. coli strain EPI300(pCC1kan), containing the same plasmid vector with a kan gene cloned in place of the uspA2H gene, was, as expected, unreactive with MAb 17C7 in Western blot analysis (Fig. 5B) and had little or no ability to form a biofilm in this assay system (Fig. 5A).
FIG. 5.
Biofilm formation by a recombinant E. coli strain expressing the M. catarrhalis UspA2H protein. (A) E. coli EPI300 cells containing a cloned wild-type ETSU-9 uspA2H gene (in pMPE9U2H) or a kan gene (in pCC1kan) were used in the crystal violet-based biofilm assay. These data represent the means and standard errors from three independent experiments. (B) Whole-cell lysates of the same recombinant strains were probed in Western blot analysis with MAb 17C7 to detect UspA2H protein expression. A molecular mass position marker (in kDa) is on the left.
Characterization of other biofilm-negative transposon insertion mutants.
As stated above, transposon insertions in three other genes in addition to uspA2H were found to cause a drastic reduction in biofilm formation by strain ETSU-9. The identification of the ORFs disrupted by the transposon was accomplished by direct sequencing of chromosomal DNA purified from these three mutants, using oligonucleotide primers that bound to the extreme ends of the transposon. ETSU-9.714 had an insertion in a gene predicted to encode a protein with significant identity to the predicted aminoglycoside phosphotransferase of Psychrobacter cryohalolentis (GenBank accession no. YP_580958). ETSU-9.222 was found to have a transposon insertion in a gene encoding a protein that was very similar to a predicted lytic murein transglycosylase D from Psychrobacter arcticus (GenBank accession no. AAZ19123). The insertion in ETSU-9.988 disrupted a gene predicted to encode a protein that most closely resembled a probable N-acetylmuramoyl-l-alanine amidase (AmpD) from P. arcticus (GenBank accession no. AAZ19605).
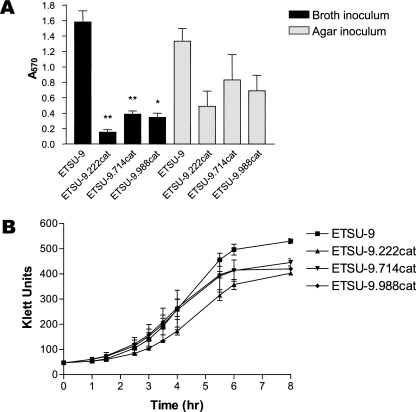
New M. catarrhalis mutants were constructed by insertion of a promoterless cat cartridge into each of these three ORFs in ETSU-9, and all three of the newly constructed mutants were found to be attenuated for biofilm formation (Fig. 6A) when a broth culture was used as the inoculum for the 24-well tissue culture plate. These differences were statistically significant (Fig. 6A). However, when a swatch of bacterial growth from an agar plate was used as the inoculum, the defect in biofilm formation was much less pronounced for all three mutants, and the differences between the mutants and the parent strain, while readily visible, were not statistically significant (Fig. 6A). Analysis of the abilities of these three mutants to grow in broth culture revealed that all three of the mutants had a modest growth defect relative to the ETSU-9 parent strain (Fig. 6B).
FIG. 6.
Biofilm formation by additional ETSU-9 mutant strains. (A) The ETSU-9 parent strain and the ETSU-9.222cat, ETSU-9.714cat, and ETSU-9.988cat mutants were grown in broth (black columns) or on agar plates (gray columns) and then used to inoculate a 24-well plate. Biofilm development was measured as described in Materials and Methods. These data represent the means and standard errors from three independent experiments. A two-tailed t test was used to compare biofilm formation by each of the mutants to biofilm formation by the parent strain with a given inoculum (i.e., broth grown or agar plate grown); *, P < 0.05; **, P < 0.01. (B) Growth of the same four strains in TH broth.
DISCUSSION
UspA1, UspA2, and UspA2H have been classified as members of the YadA family of nonfimbrial, nonpilus adhesins, which reach the bacterial surface by means of an autotransporter mechanism (9, 19, 21). The UspA2H protein has been shown to function in attachment to Chang conjunctival epithelial cells in vitro (22) and can also confer serum resistance on some strains of M. catarrhalis (22). In the present study, it was shown that the UspA2H protein plays a role in biofilm formation by M. catarrhalis strain ETSU-9 in vitro, as evaluated with the crystal violet-based assay. Studies of the Yersinia YadA protein, the prototype of this autotransporter family, have indicated that this nonfimbrial adhesin appears to form a very dense layer on the surfaces of Yersinia species and possesses an architecture that involves a “head-stalk-anchor” structure, with the “head” component binding various eukaryotic proteins (reviewed in reference 21). Both the UspA1 and UspA2 proteins of M. catarrhalis O35E were also shown to form structures with a similar “lollipop” appearance (19), with the UspA2 protein being much more abundant (41). UspA2H proteins are hybrid macromolecules, involving a leader peptide and an N-terminal region that resemble those of UspA1 proteins, together with a C-terminal half that most closely resembles that of a UspA2 protein (22).
Mutant analysis revealed that lack of expression of the UspA1 protein by M. catarrhalis ETSU-9 had a relatively modest effect on the ability of the strain to form a biofilm in the crystal violet-based assay system (Fig. 1B). In contrast, a uspA2H mutant of ETSU-9 had a much reduced ability to form a biofilm in this system relative to both the uspA1 mutant and the ETSU-9 parent strain (Fig. 1B). This result is perhaps not surprising in view of the fact that the relative level of UspA2H expression by ETSU-9 is apparently much greater than that of UspA1 (Fig. 1A). What was unexpected was that the introduction of a 15-nt insertion in frame into numerous different sites within the uspA2H gene of ETSU-9 would have such a negative effect on the ability of the UspA2H protein to mediate biofilm formation by this strain (Fig. 3A). This insertion mutagenesis technique was intended to identify domains within the UspA2H protein that were directly (or indirectly) involved in the ability of the strain to form biofilms. Instead, it appeared that insertion of this 5-aa sequence at any one of many different sites in the UspA2H protein resulted in loss of function, at least with regard to biofilm formation in the crystal violet-based assay. Interestingly, nearly all of these mutant UspA2H proteins were still able to mediate attachment of the organism to Chang conjunctival epithelial cells (Fig. 3B), indicating that these 5-aa insertions were likely not causing a gross conformational change in the UspA2H protein or at least did not occur in the region(s) essential for bacterial attachment. These results also suggest that UspA2H-dependent epithelial cell attachment and biofilm formation mediated by the M. catarrhalis ETSU-9 UspA2H protein are separate and distinguishable activities.
Even though a number of different 5-aa insertions adversely affected biofilm formation ability, a mutant (ETSU-9.mut92) in which 418 aa was deleted from the UspA2H protein still formed a biofilm, albeit at a slightly reduced level relative to its parent strain (Fig. 3A). The same deletion mutant also attached at wild-type levels to Chang cells in vitro (Fig. 3B). These data indicate that the portion of the UspA2H protein deleted in this particular mutant is not essential for biofilm development or attachment to Chang cells. This 418-aa section is located within what would be predicted to be the “stalk” region of the “lollipop” structure proposed for UspA1 and UspA2 proteins by Hoiczyk and colleagues (19). It is possible that this internal deletion resulted in the formation of a shorter “stalk” that still allowed proper orientation of the “head” region predicted to be involved in attachment of UspA1 to cell membranes (19). This internal deletion does not include the predicted autotransporter domain (21), which is consistent with the functional ability of this UspA2H mutant protein, which is apparently present on the surface of the outer membrane. This internal deletion does include the region which binds MAb 17C7 (2) and therefore may also include a region with homology to the closely related UspA1 protein, which was recently shown to bind CEACAM (17) and fibronectin (47).
The “lollipop” model proposed by Hoiczyk et al. (19) might explain why biofilm formation by ETSU-9 is so sensitive to perturbations in UspA2H. In this model, the N-terminal “head” region would be the domain directly involved in biofilm formation, so insertions in this region would prevent biofilms. The “stalk,” or coiled-coil, region of UspA2H is composed of six repeating amino acid sequences and would be expected to present the “head” domain in the correct orientation. Deletion of some or all of the repeating units of the “stalk” (mutants 81 and 92) resulted in only minor reductions in biofilm formation. However, interruption of the repeating units, either by deletion (mutant 103) or by insertion, could result in a kink in the “stalk” so that the “head” domain is no longer in the correct orientation to mediate biofilm formation.
Mutations in three other genes in addition to uspA2H were shown to adversely affect the biofilm formation ability of M. catarrhalis ETSU-9 in the crystal violet-based assay. These genes encoded a predicted aminoglycoside phosphotransferase, a predicted lytic murein transglycosylase D, and a predicted N-acetylmuramyl-l-alanine amidase (AmpD). The last two gene products likely have roles in bacterial cell wall metabolism in M. catarrhalis, and mutations in ampD may have an indirect effect on biofilm formation. More specifically, a transposon insertion in the ampD gene of Pseudomonas aeruginosa resulted in high-level expression of a chromosomally encoded β-lactamase (6), and expression of some β-lactamases has been reported to inhibit biofilm formation by both E. coli and P. aeruginosa, perhaps by affecting peptidoglycan remodeling (13). M. catarrhalis also has a chromosomally encoded β-lactamase (7), and we found that the level of β-lactamase activity in culture supernatant fluid from the ampD mutant ETSU-9.988 was greater than that obtained with the parent strain (data not shown). As for the lytic murein transglycosylase, it is known that E. coli recycles almost 50% of its murein in each generation (14, 38). Although the role of murein recycling has not yet been fully elucidated, Park (39) has suggested that this function may be one method for sensing the condition of the cell wall. It is possible that a disruption of the normal murein-cycling pathway could have an adverse affect on biofilm formation.
One unexpected finding from this study was that the ability of the ETSU-9.222cat, ETSU-9.714cat, and ETSU-9.988cat mutants (described immediately above) to form biofilms in the crystal violet-based assay was affected by the type of inoculum (i.e., broth-grown cells versus agar-grown cells). Wells inoculated with mutants that had been grown on agar plates had more biofilm development than did wells that were inoculated with broth-grown cells (Fig. 6A). It has been suggested that inoculating these wells in the tissue culture plate with a cluster of cells scraped from the surface of an agar plate may be akin to seeding the wells with a preformed biofilm (J. W. Costerton, personal communication); this could account for these inoculum-dependent results. It can also be inferred from these data that future screens for biofilm-negative M. catarrhalis mutants should be carried out using broth-grown inocula to minimize the probability of missing mutants attenuated for biofilm development in this in vitro system.
It must also be noted that the crystal violet-based biofilm assay used in the present study, while very useful for efficiently screening large numbers of mutants, does have some limitations. First, many workers consider this assay appropriate for measuring early events in biofilm development on abiotic surfaces (37, 44). Second, data obtained on the biofilm formation abilities of wild-type strains and mutants in the crystal violet-based assay may be different from or similar to results obtained with the same strains in continuous-flow biofilm systems (11, 26, 46). Third, there are obvious issues involving depletion of nutrients and potential lack of aeration in the crystal violet-based assay system (30). In the present study, the biofilm-deficient M. catarrhalis mutants were not tested in alternative biofilm systems.
Based on the data presented in this report, the UspA2H protein of M. catarrhalis ETSU-9 is necessary for biofilm formation by this strain in the crystal violet-based assay system. Moreover, the gain-of-function experiments involving recombinant E. coli expressing UspA2H reinforce the functional role of this M. catarrhalis gene product in biofilm development in this particular assay system. However, the existence of the other three biofilm-deficient mutants described above (i.e., ETSU-9.222, ETSU-9.714, and ETSU-9.988) indicate that UspA2H expression by itself is not sufficient for biofilm formation by ETSU-9 in this model system and that other gene products, including some that affect different aspects of cell wall synthesis, are also important for normal biofilm development in the system.
Acknowledgments
This study was supported by U.S. Public Health Service grant no. AI36344 to E.J.H. M.M.P. was supported by U.S. Public Health Service training grant no. 5-T32-AI007520.
We thank John Nelson and Steven Berk for supplying the isolates of M. catarrhalis used in this study and Jennifer Sedillo for expert technical assistance.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 11 June 2007.
REFERENCES
- 1.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. R. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis O35E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65:4367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angus, B. L., A. M. Carey, D. A. Caron, A. M. Kropinski, and R. E. Hancock. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob. Agents Chemother. 21:299-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attia, A. S., E. R. Lafontaine, J. L. Latimer, C. Aebi, G. A. Syrogiannopoulos, and E. J. Hansen. 2005. The UspA2 protein of Moraxella catarrhalis is directly involved in the expression of serum resistance. Infect. Immun. 73:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attia, A. S., S. Ram, P. A. Rice, and E. J. Hansen. 2006. Binding of vitronectin by the Moraxella catarrhalis UspA2 protein interferes with late stages of the complement cascade. Infect. Immun. 74:1597-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagge, N., O. Ciofu, M. Hentzer, J. I. Campbell, M. Givskov, and N. Hoiby. 2002. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bootsma, H. J., H. van Dijk, P. Vauterin, J. Verhoef, and F. R. Mooi. 2000. Genesis of BRO beta-lactamase-producing Moraxella catarrhalis: evidence for transformation-mediated horizontal transfer. Mol. Microbiol. 36:93-104. [DOI] [PubMed] [Google Scholar]
- 8.Budhani, R. K., and J. K. Struthers. 1998. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of beta-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob. Agents Chemother. 42:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotter, S. E., N. K. Surana, and J. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13:199-205. [DOI] [PubMed] [Google Scholar]
- 10.Cripps, A. W., D. C. Otczyk, and J. M. Kyd. 2005. Bacterial otitis media: a vaccine preventable disease? Vaccine 23:2304-2310. [DOI] [PubMed] [Google Scholar]
- 11.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faden, H., L. Duffy, R. Wasielewski, J. Wolf, D. Krystofik, Y. Tung, et al. 1997. Relationship between nasopharyngeal colonization and the development of otitis media in children. J. Infect. Dis. 175:1440-1445. [DOI] [PubMed] [Google Scholar]
- 13.Gallant, C. V., C. Daniels, J. M. Leung, A. S. Ghosh, K. D. Young, L. P. Kotra, and L. L. Burrows. 2005. Common beta-lactamases inhibit bacterial biofilm formation. Mol. Microbiol. 58:1012-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heiniger, N., R. Troller, P. S. Meier, and C. Aebi. 2005. Cold shock response of the UspA1 outer membrane adhesin of Moraxella catarrhalis. Infect. Immun. 73:8247-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill, D. J., A. M. Edwards, H. A. Rowe, and M. Virji. 2005. Carcinoembryonic antigen-related cell adhesion molecule (CEACAM)-binding recombinant polypeptide confers protection against infection by respiratory and urogenital pathogens. Mol. Microbiol. 55:1515-1527. [DOI] [PubMed] [Google Scholar]
- 18.Hill, D. J., and M. Virji. 2003. A novel cell-binding mechanism of Moraxella catarrhalis ubiquitous surface protein UspA: specific targeting of the N-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol. Microbiol. 48:117-129. [DOI] [PubMed] [Google Scholar]
- 19.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm, M. M., S. L. Vanlerberg, I. M. Foley, D. D. Sledjeski, and E. R. Lafontaine. 2004. The Moraxella catarrhalis porin-like outer membrane protein CD is an adhesin for human lung cells. Infect. Immun. 72:1906-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koretke, K. K., P. Szczesny, M. Gruber, and A. N. Lupas. 2006. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J. Struct. Biol. 155:154-161. [DOI] [PubMed] [Google Scholar]
- 22.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukomski, S., N. P. Hoe, I. Abdi, J. Rurangirwa, P. Kordari, M. Liu, S. J. Dou, G. G. Adams, and J. M. Musser. 2000. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect. Immun. 68:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukomski, S., R. A. Hull, and S. I. Hull. 1996. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J. Bacteriol. 178:240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mampel, J., T. Spirig, S. S. Weber, J. A. Haagensen, S. Molin, and H. Hilbi. 2006. Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 72:2885-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMichael, J. C., M. J. Fiske, R. A. Fredenburg, D. N. Chakravarti, K. R. VanDerMeid, V. Barniak, J. Caplan, E. Bortell, S. Baker, R. Arumugham, and D. Chen. 1998. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect. Immun. 66:4374-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier, P. S., S. Freiburghaus, A. Martin, N. Heiniger, R. Troller, and C. Aebi. 2003. Mucosal immune response to specific outer membrane proteins of Moraxella catarrhalis in young children. Pediatr. Infect. Dis. J. 22:256-262. [DOI] [PubMed] [Google Scholar]
- 29.Meier, P. S., R. Troller, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2002. The outer membrane proteins UspA1 and UspA2 of Moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 20:1754-1760. [DOI] [PubMed] [Google Scholar]
- 30.Merritt, J. H., Daniel E. Kadouri, and George A. O'Toole. 2005. Growing and analyzing static biofilms, p. 1B.1.1-1B.1.17. In Current protocols in microbiology. John Wiley & Sons, Inc., New York, NY. [DOI] [PMC free article] [PubMed]
- 31.Murphy, T. F. 2005. Moraxella (Branhamella) catarrhalis and other gram-negative cocci, p. 2529. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious disease. Elsevier Inc., Philadelphia, PA.
- 32.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172:195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy, T. F., and M. R. Loeb. 1989. Isolation of the outer membrane of Branhamella catarrhalis. Microb. Pathog. 6:159-174. [DOI] [PubMed] [Google Scholar]
- 34.Nester, E. W., M. Schafer, and J. Lederberg. 1963. Gene linkage in DNA transfer: a cluster of genes concerned with aromatic biosynthesis in Bacillus subtilis. Genetics 48:529-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordstrom, T., A. M. Blom, A. Forsgren, and K. Riesbeck. 2004. The emerging pathogen Moraxella catarrhalis interacts with complement inhibitor C4b binding protein through ubiquitous surface proteins A1 and A2. J. Immunol. 173:4598-4606. [DOI] [PubMed] [Google Scholar]
- 36.Nordstrom, T., A. M. Blom, T. T. Tan, A. Forsgren, and K. Riesbeck. 2005. Ionic binding of C3 to the human pathogen Moraxella catarrhalis is a unique mechanism for combating innate immunity. J. Immunol. 175:3628-3636. [DOI] [PubMed] [Google Scholar]
- 37.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 38.Park, J. T. 1996. The murein sacculus, p. 48-57. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, et al. (ed.), Escherichia coli and Salmonella. Cellular and molecular biology. ASM Press, Washington, DC.
- 39.Park, J. T. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 17:421-426. [DOI] [PubMed] [Google Scholar]
- 40.Patrick, C. C., S. E. Pelzel, P. A. Gulig, C. J. McCracken, J. D. Radolf, and E. J. Hansen. 1989. Antigenic evidence for the synthesis of two different lipooligosaccharides by some strains of Haemophilus influenzae type b. Infect. Immun. 57:1971-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St. Geme III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 70:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson, M. M., C. A. Laurence, S. E. Guinn, and E. J. Hansen. 2006. Biofilm formation by Moraxella catarrhalis in vitro: roles of the UspA1 adhesin and the Hag hemagglutinin. Infect. Immun. 74:1588-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelton, S. I. 2005. Otitis media: re-evaluation of diagnosis and treatment in the era of antimicrobial resistance, pneumococcal conjugate vaccine, and evolving morbidity. Pediatr. Clin. N. Am. 52:711-728, v-vi. [DOI] [PubMed] [Google Scholar]
- 44.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 45.Reddy, M. S., T. F. Murphy, H. S. Faden, and J. M. Bernstein. 1997. Middle ear mucin glycoprotein; purification and interaction with nontypeable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol. Head Neck Surg. 116:175-180. [DOI] [PubMed] [Google Scholar]
- 46.Swords, W. E., M. L. Moore, L. Godzicki, G. Bukofzer, M. J. Mitten, and J. Von Cannon. 2004. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect. Immun. 72:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, T. T., T. Nordstrom, A. Forsgren, and K. Riesbeck. 2005. The respiratory pathogen Moraxella catarrhalis adheres to epithelial cells by interacting with fibronectin through ubiquitous surface proteins A1 and A2. J. Infect. Dis. 192:1029-1038. [DOI] [PubMed] [Google Scholar]
- 48.Timpe, J. M., M. M. Holm, S. L. Vanlerberg, V. Basrur, and E. R. Lafontaine. 2003. Identification of a Moraxella catarrhalis outer membrane protein exhibiting both adhesin and lipolytic activities. Infect. Immun. 71:4341-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai, C.-M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharide in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 50.Wang, W., A. S. Attia, L. Liu, T. Rosche, N. J. Wagner, and E. J. Hansen. 2006. Development of a shuttle vector for Moraxella catarrhalis. Plasmid 55:50-57. [DOI] [PubMed] [Google Scholar]
- 51.Ward, C. K., J. R. Mock, and E. J. Hansen. 2004. The LspB protein is involved in the secretion of the LspA1 and LspA2 proteins by Haemophilus ducreyi. Infect. Immun. 72:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitby, P. W., D. J. Morton, and T. L. Stull. 1998. Construction of antibiotic resistance cassettes with multiple paired restriction sites for insertional mutagenesis of Haemophilus influenzae. FEMS Microbiol. Lett. 158:57-60. [DOI] [PubMed] [Google Scholar]