Abstract
The Lyme disease spirochete, Borrelia burgdorferi, is largely resistant to being killed by its hosts’ alternative complement activation pathway. One possible resistance mechanism of these bacteria is to coat their surfaces with host complement regulators, such as factor H. Five different B. burgdorferi outer surface proteins having affinities for factor H have been identified: complement regulator-acquiring surface protein 1 (BbCRASP-1), encoded by cspA; BbCRASP-2, encoded by cspZ; and three closely related proteins, BbCRASP-3, -4, and -5, encoded by erpP, erpC, and erpA, respectively. We now present analyses of the recently identified BbCRASP-2 and cspZ expression patterns throughout the B. burgdorferi infectious cycle, plus novel analyses of BbCRASP-1 and erp-encoded BbCRASPs. Our results, combined with data from earlier studies, indicate that BbCRASP-2 is produced primarily during established mammalian infection, while BbCRASP-1 is produced during tick-to-mammal and mammal-to-tick transmission stages but not during established mammalian infection, and Erp-BbCRASPs are produced from the time of transmission from infected ticks into mammals until they are later acquired by other feeding ticks. Transcription of cspZ and synthesis of BbCRASP-2 were severely repressed during cultivation in laboratory medium relative to mRNA levels observed during mammalian infection, and cspZ expression was influenced by culture temperature and pH, observations which will assist identification of the mechanisms employed by B. burgdorferi to control expression of this borrelial infection-associated protein.
Lyme disease spirochetes are maintained in nature by a cycle of alternately infecting vertebrate hosts and Ixodes species ticks. As an infected tick feeds on its host, Borrelia burgdorferi is transmitted directly into the blood pool at the tick bite site. Bacteria then spread via the bloodstream and by invasion of host tissues to establish a chronic, disseminated infection (21, 64, 81). Spirochetes may later be acquired by additional ticks as they take a blood meal from the infected host. Like many other blood-borne pathogens, B. burgdorferi is naturally resistant to the innate immune system of its hosts: as few as 20 organisms can efficiently infect immunocompetent animals (11). The alternative pathway of complement activation is an important arm of vertebrate innate immunity, rapidly clearing susceptible microorganisms from the host in the absence of antibody or other aspects of acquired immunity (37). In culture, most infectious isolates of B. burgdorferi are resistant to the alternative pathway of complement activation (12, 13, 38, 74), which has been associated with binding the host serum complement regulator factor H, enhanced breakdown of C3b and C3 convertase, and prevention of membrane attack complex formation (6, 44). Serum-resistant strains of B. burgdorferi produce several distinct outer-surface proteins, termed “BbCRASPs” (B. burgdorferi complement regulator-acquiring surface proteins), that are able to bind host factor H (43, 44). The ability of B. burgdorferi to bind host factor H to its surface is apparently not the only mechanism by which the Lyme disease spirochete evades host complement in vivo, since mice deficient in factor H are infected to the same degree as wild-type animals (80). Nevertheless, at least two BbCRASPs contribute to complement resistance in vitro (14, 29) and have therefore been hypothesized to play important roles in the multitiered defense system that protects the pathogen from clearance by its host.
The B. burgdorferi type strain, B31, produces five distinct BbCRASPs. BbCRASP-1 is encoded by cspA, located on an ∼54-kb linear DNA element named lp54 (40). All examined Lyme disease spirochetes carry multiple paralogous genes related to cspA, although only the cspA gene product is capable of binding factor H (19, 27, 42, 79). The gene encoding BbCRASP-2, cspZ, was recently identified as being located on a separate linear DNA element, lp28-3, and unlike all the other BbCRASP-encoding genes, B. burgdorferi does not carry any additional genes paralogous to cspZ (19, 27, 29). BbCRASP-3, -4, and -5 are each members of the Erp paralog family, named ErpP, ErpC, and ErpA, respectively (4, 5, 19, 33, 39, 41, 51, 66, 69). All Lyme disease spirochetes naturally maintain 6 to 11 distinct episomal prophages, known as cp32s, each of which carries a mono- or bicistronic erp locus (65, 71). Strain B31 carries three identical copies of erpA, on prophages cp32-1, cp32-5, and cp32-8, and one copy each of erpC and erpP, on cp32-2 and cp32-9, respectively (19, 20, 69). Erp paralogs of other B. burgdorferi strains known to bind factor H have been given various names, including OspE, p21, and Erp41 (2, 5, 33, 46, 67). Some publications have referred to the strain B31 BbCRASPs by the open reading frame (ORF) numbers assigned to genes following sequencing and annotation of the genome of a strain B31 subculture, which are presented here to aid cross-referencing: cspA is ORF BBA68, cspZ is ORF BBH06, the cp32-1 erpA gene is ORF BBP38, the cp32-8 erpA gene is ORF BBL39, and erpP is ORF BBN38 (19, 27). The sequenced B31 subculture had lost cp32-2 and cp32-5, so erpC and the cp32-5 erpA gene do not have ORF numbers (19).
Factor H consists of 20 repeated motifs, termed short consensus repeats (SCRs) (83). BbCRASP-1 and -2 both bind primarily to SCR 7, while the Erp-BbCRASPs bind only to the carboxy-terminal SCR 20 (29, 33, 40, 43-45). These different affinities may have important consequences: factor H in solution has a compact structure, with only the carboxy-terminal ligand-binding sites exposed, but binding of factor H via the carboxy terminus unfurls the protein to permit interactions between internal SCRs and their ligands (7, 58). Thus, Erp-BbCRASPs may provide initial binding of factor H, while BbCRASP-1 and/or -2 then facilitates additional binding of the host protein. Cultured B. burgdorferi that lacks cspA is sensitive to killing by the alternative pathway of complement activation, even when such bacteria carry cspZ and one or more BbCRASP-encoding erp genes (14, 59). Moreover, complementation of a cspA deletion mutant with a copy of the wild-type gene restored in vitro complement resistance (14). In studies of cultivated B. burgdorferi, Erp proteins by themselves do not provide complement resistance: a mutant of strain B31, B31-e2, lacks all BbCRASP-encoding genes except cspA plus one copy of erpA but is as resistant to complement as its wild-type parent, whereas a sibling cspA cspZ mutant, B313, carries erpC and one copy of erpA yet is extremely sensitive to killing by complement (references 9, 29, and 85 and our unpublished results). Transformation of mutant B313 with a wild-type copy of cspZ provided partial resistance to complement in vitro (29). However, studies have yet to be performed on erp-deficient bacteria to examine the ability of BbCRASP-1 or -2 to function in the complete absence of Erp-BbCRASPs, so the possibility of cooperation between those borrelial surface proteins cannot be ruled out. As an additional caveat, the relative importance of each gene during infection processes is unknown, since neither cspA, cspZ, nor all the erp genes have been specifically deleted from an otherwise infectious bacterium.
Why does B. burgdorferi encode multiple distinct but apparently redundant proteins that can bind host factor H? It is well known that B. burgdorferi produces different proteins during the various stages of its mammal-tick infectious cycle, which suggested to us that BbCRASPs may function at different times. The expression patterns of BbCRASP-1 and Erp proteins during the infectious cycle are reasonably well characterized (53-56, 65, 70, 75). With the recent identification of cspZ as encoding BbCRASP-2 and the subsequent development of BbCRASP-2-specific antisera (29), we were able to examine the transcription of this gene and the synthesis of its protein throughout the B. burgdorferi mammal-tick infectious cycle. In vitro studies of cspZ and other BbCRASP-encoding genes were also performed to help elucidate mechanisms by which BbCRASP levels are controlled. Results from these studies indicate unique regulatory mechanisms for each class of BbCRASP that result in distinct in vivo expression profiles.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. burgdorferi strain B31-MI-16 is a clonal derivative of B31-MI, the nonclonal culture of type strain B31, whose complete genome sequence has been determined (19, 27, 55). B31-MI-16 contains all the plasmids of culture B31-MI and is fully infectious for both mice and ticks (55). Strain B31-A3 is a distinct clonal derivative of strain B31 (25). Strains B31-A3ntrA and B31-A3rpoS are ΔrpoN (ntrA) and ΔrpoS derivatives, respectively, of strain B31-A3 (25, 26). All B. burgdorferi strains were grown in modified Barbour-Stoenner-Kelly (BSK-II) medium (84). Media used to study the effect of environmental pH were supplemented with 25 mM HEPES and buffered to pH values of either 6.5, 7.0, or 8.0 (18), and the pH values of the media were again measured following cell harvesting. Bacterial cultures were grown at either 34°C or 23°C or shifted from 23 to 34°C, as required (68).
To test the effects of the culture pH, temperature, or sigma factor mutations on gene expression, three independent cultures for each condition or bacterial strain were grown to densities of approximately 107 bacteria per ml, harvested by centrifugation, and washed twice with phosphate-buffered saline (PBS). cDNAs and whole-cell lysates were produced from each culture as described below.
Infection of mice and ticks.
Eight female BALB/c mice were infected by subcutaneous injection of 104 B31-MI-16 bacteria from a mid-exponential-phase 34°C culture. Seven days later, the mouse infection status was assessed by inoculation of a 1-mm2 ear biopsy specimen from each animal into BSK-II medium containing antibiotics (phosphomycin and rifampin) and antifungal agents (amphotericin B) (Sigma, St. Louis, MO). The biopsy cultures were examined 10 days later by dark-field microscopy. These mice then served to infect Ixodes scapularis larvae as described below.
Egg masses laid by pathogen-free I. scapularis ticks were obtained from the Department of Entomology, Oklahoma State University (Stillwater), and held in a humidified chamber until they hatched. For B. burgdorferi acquisition studies, approximately 200 naive larvae were placed on each of the above-described B. burgdorferi-infected mice. For studies of B. burgdorferi acquisition by ticks, some feeding larvae were removed 72 h after placement on the infected mice. Some of these partially fed larvae were immediately dissected and examined microscopically by indirect immunofluorescence analysis (IFA), and pools of 50 to 70 larvae from three independent feedings were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction and analysis by quantitative reverse transcription-PCR (Q-RT-PCR). After 96 h, the remaining ticks had engorged fully and naturally dropped off the mice. At that time, some ticks were dissected immediately for IFA, while the remaining ticks were returned to the humidified chamber. A cohort of larvae were dissected for IFA 12 days postattachment, and the remaining ticks were allowed to molt to the nymphal stage. Two weeks after ecdysis, some unfed nymphs were dissected and analyzed by IFA, three independent pools of 20 to 30 were frozen for Q-RT-PCR analysis, and the remainder were fed upon uninfected female BALB/c mice. Some nymphs were allowed to complete engorgement and drop off naturally, while others were forcibly removed after only 72 h of feeding. These 72-hour-fed nymphs were dissected and examined by IFA, or pools of 20 to 30 ticks from three independent experiments were frozen for analysis by Q-RT-PCR. During nymph removal, a piece of mouse skin often remained attached to the hypostome of the feeding tick, in which case the bite site skin samples were dissected away from the ticks for a separate IFA. Eight of the mice infected through feeding by the infected nymphs were killed 2 weeks after completion of feeding, and their ear pinnae, hearts, and tibiotarsal joints were collected and frozen for RNA extraction and Q-RT-PCR.
All infection studies were performed under protocols approved by the University of Kentucky Institutional Animal Care and Use Committee and the University of Kentucky Institutional Biosafety Committee.
Analysis of B. burgdorferi mRNA levels.
Total RNA was extracted from cultured bacteria or tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA). Frozen mouse tissue samples were first ground with a mortar and pestle, followed by homogenization with a Tissue Tearor (Biospec Products, Bartlesville, OK) in TRIzol reagent at 4°C. RNA was resuspended in RNAsecure reagent (Ambion, Austin, TX) and treated with DNase I (Ambion) to remove contaminating DNA. The DNase was inactivated using DNase Inactivation Reagent (Ambion). A 1-μg aliquot of each DNA-free RNA preparation was reverse transcribed using First Strand cDNA synthesis kits (Roche Applied Science, Indianapolis, IN) with random hexamers and avian myeloblastosis virus reverse transcriptase enzyme (RTase). As controls, mixtures containing all components except RTase were prepared and treated similarly. Primers and templates were annealed for 10 min at room temperature, followed by cDNA synthesis at 42°C for 1 h. RTase was inactivated by heating it for 5 min at 99°C, followed by 10 min at 4°C. All cDNAs and control reaction mixtures were diluted 10-fold with water before being used as templates for Q-RT-PCR.
Quantitative PCR was performed using a LightCycler thermal cycler (Roche Applied Science), as previously described (52, 53). Briefly, cDNA or diluted genomic DNA (see below) was added to an 8-μl master mixture containing 1× PCR buffer (Idaho Technology, Salt Lake City, UT), deoxynucleoside triphosphates (Idaho Technology), Platinum Taq polymerase (Invitrogen; final dilution, 1:10 in enzyme diluent [Idaho Technology]), SYBR green (Invitrogen; final dilution, 1:10,000 in Tris-EDTA), oligonucleotide primers (0.4-mM final concentration), and nuclease-free water (Promega, Madison, WI). All cDNA samples were analyzed in triplicate. Each LightCycler run included negative controls of RNA processed without RTase (see above) to test for DNA contamination of each RNA preparation and samples that lacked template to test for DNA contamination of reagents.
Oligonucleotide primers used for amplification of cspZ, cspA, erpA, erpP, flaB, ospC, and ospA cDNAs were CSPZF-3 plus CSPZR-4, CSPAF-3 plus CSPAR-4, ERPAF-1 plus ERPAR-2, ERPPF-3 plus ERPPR-2, FLA3 plus FLA4, OSPCF-7 plus OSPCR-8, and OSPAF-1 plus OSPAR-2, respectively (Table 1). All amplicons were approximately 150 bp in size. Reaction conditions consisted of a 2-minute initial 94°C denaturation, followed by 45 cycles of 94°C for 5 s; 55°C (for cspZ, flaB, ospC, and ospA amplicons), 50°C (for cspA and erpA amplicons), or 48°C (for erpP amplicons) for 5 s; and 72° for 30 s. Tenfold serial dilutions of B31-MI-16 genomic DNA (100 ng to 100 fg) were included in every assay for each primer set. This enabled the generation of standard curves, from which the amount of transcript present in each cDNA sample could be calculated using Light Cycler software v.3.5.3 (Roche Applied Science). The same software package was also used for melting-curve analysis. To verify amplicon sizes and purities, all products were separated by agarose gel electrophoresis, and DNA was visualized with ethidium bromide (28). Representative amplicons generated with each primer set from each cDNA template were completely sequenced to confirm their identities (Davis Sequencing, Davis, CA). Average expression values obtained from triplicate runs of each cDNA sample for all the genes of interest were calculated relative to the average triplicate value for the B. burgdorferi housekeeping gene flaB from the same cDNA preparation (52). Statistical analyses of data were performed by one-way analysis of variance.
TABLE 1.
Oligonucleotides used in this work
| Targeted gene | Primer name | Sequence (5′ to 3′) |
|---|---|---|
| cspZ | CSPZF-3 | AGACGCTATTTATAACGAATGTACAGGAGC |
| CSPZR-4 | CAGCAACATGTCTGGCATTAGACAC | |
| cspA | CSPAF-3 | CTAAAAGCAATTGGTAAGGAACTG |
| CSPAR-4 | TCAATAAGATCGTAAGGACCAACT | |
| erpA | ERPAF-1 | AAAGCAATGGAGAGGTAAAGGTC |
| ERPAR-2 | GCTTTTATAAAGTTATTAATTTCTTCCTCTTC | |
| erpP | ERPPF-3 | CGGCTACATTCTTTTCATTAAAAGAATC |
| ERPPR-2 | ACGCAATATGTTCAGCACCATTAATAC | |
| flaB | FLA3 | GGGTCTCAAGCGTCTTGG |
| FLA4 | GAACCGGTGCAGCCTGAG | |
| ospC | OSPCF-7 | CAGGGAAAGATGCGAATACATCTGC |
| OSPCR-8 | TAAGCTAAAGCTAACAATGATCC | |
| ospA | OSPAF-1 | CAAAATGTTAGCAGCCTTGACGAG |
| OSPAR-2 | CTTCAAGTGTGGTTTGACCTAGATCG |
IFA.
Mouse and tick tissues were dissected in 10 μl of PBS on glass slides and allowed to air dry overnight. The tissues were then fixed and permeabilized by immersion in acetone for 15 min. The slides were air dried and then blocked overnight at 4°C in PBS containing 0.2% bovine serum albumin and 10% goat serum. After being washed in PBS-0.2% bovine serum albumin, the slides were incubated for 1 h at room temperature in polyclonal mouse antiserum specific for BbCRASP-2 (29) diluted 1:50. The slides were then washed and incubated for 1 h at room temperature in a 1:50,000 dilution of rabbit polyclonal antiserum raised against B. burgdorferi total membrane proteins (55). The slides were washed again and then incubated in 1:1,000 dilutions of both Alexa Fluor 488-labeled goat anti-mouse immunoglobulin G and Alexa Fluor 594-labeled goat anti-rabbit immunoglobulin G (Invitrogen) for 45 min at room temperature. The slides were then washed, dried, and mounted with ProLong Anti-Fade Mounting Medium (Invitrogen). The slides were viewed with an Olympus BX51 epifluorescence microscope using a 100× objective lens, and images were captured with a Retiga 2000R Fast 1394 system and QCapture Pro software 5.0.1.26 (both from QImaging, Surrey, BC, Canada). Bacteria within 25 random fields were counted to determine the proportions of bacteria containing detectable levels of BbCRASP-2 (i.e., positive for anti-BbCRASP-2 labeling) relative to the total number of bacteria present in a given field (as assessed by anti-B. burgdorferi labeling). A total of 88 bacteria were observed in approximately 20 skin samples examined. Slides of dissected tissues were incubated with either polyclonal anti-BbCRASP-2 or polyclonal anti-B. burgdorferi antibody alone, or only the secondary antibodies, to serve as negative fluorescence controls.
Immunoblot analyses.
Cultured bacteria were collected by centrifugation, washed twice with PBS, suspended in sodium dodecyl sulfate loading buffer, and then lysed in a boiling-water bath for 5 min. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes. The membranes were incubated with either mouse polyclonal antiserum directed against BbCRASP-2 (29), murine monoclonal antibody RH1 directed against BbCRASP-1 (40), rabbit polyclonal antiserum specific for ErpA or ErpP (23), murine monoclonal antibody B5 directed against OspC (49), or murine monoclonal antibody H9724, which recognizes the constitutively expressed FlaB (flagellin) protein (10). Bound polyclonal antibodies were detected with horseradish peroxidase-linked goat anti-mouse or goat anti-rabbit antibodies (GE Healthcare Bio-Sciences, Piscataway, NJ) and visualized by chemiluminescence (Pierce, Rockford, IL).
RESULTS
Analyses of cspZ and cspA mRNA levels during mouse and tick infections.
During mammalian infection, B. burgdorferi bacteria are widely dispersed at low densities throughout their hosts’ bodies, making it very difficult to examine levels of protein expression by visual techniques, such as IFA. However, B. burgdorferi gene expression levels during mammalian infection can often be assessed by the highly sensitive method of Q-RT-PCR (52). To do so, eight mice were infected with B. burgdorferi through feeding by infectious I. scapularis nymphs, and the infection status of each mouse was confirmed by cultivation of ear biopsy specimens. Two weeks after completion of tick feeding, the mice were killed, and total RNA was purified from the ears, heart, and tibiotarsal joints. cspZ mRNA was detected in tissues from seven of the eight infected mice (Fig. 1A and data not shown). Wide ranges of cspZ expression levels were detected in the infected mice, with cspZ transcript in all tissues of mouse no. 5 falling below the threshold of detection. All mouse tissues contained detectable levels of flaB mRNA.
FIG. 1.
(A and B) Temporal analyses of cspZ (A) and cspA (B) gene expression levels during mammalian and tick infections. (C and D) Control analyses of the previously characterized ospC and ospA genes. Illustrated are Q-RT-PCR results from three independently collected and processed pools of 20 to 30 unfed infected nymphs, three independent pools of 20 to 30 infected nymphs that had fed on naïve mice for 72 h, ear pinna tissues from eight different mice that had been infected 2 weeks earlier by tick bite, and three independent pools of 50 to 70 larvae that had fed for 72 h on infected mice. The black bars represent RNA samples extracted from ticks, and the gray bars represent RNA obtained from mouse tissues. Gene expression levels were calculated as nanograms of the target gene transcript per nanogram of the constitutively expressed B. burgdorferi flaB gene. The threshold of detection (no amplicon produced after 45 PCR cycles) is indicated on the y axes as “t.d”.
None of the 2-week-infected mouse tissues contained detectable levels of cspA transcript (Fig. 1B), consistent with results from previous studies of mice infected for 4 or more weeks (47, 50, 78). B. burgdorferi in mouse tissues exhibited a range of ospC transcript levels, with mouse no. 5 being the lowest (Fig. 1C and data not shown). Expression of ospC was consistent with previously published data for that gene (34, 48, 57, 70, 73). Q-RT-PCR of the other experimental control gene, ospA, did not detect transcript in any tissue, also consistent with published data (34, 61, 70).
Expression levels of cspZ and cspA during stages of tick colonization were next assessed by Q-RT-PCR of RNAs extracted from pools of unfed infected nymphs, infected nymphs that had fed on mice for 72 h, and previously uninfected larvae that had fed on infected mice for 72 h. The levels of cspZ transcripts present in feeding nymphs and larvae were between 2- and 20-fold lower than the levels detected in infected mouse tissues (Fig. 1A). cspZ mRNA was not detected in unfed tick nymphs. In contrast, cspA mRNA was detected during all stages of tick colonization (Fig. 1B). Transcript levels of the control ospC and ospA genes corresponded to previously published data (61, 62) (Fig. 1C and D).
IFA of BbCRASP-2 production during tick and mammalian infection.
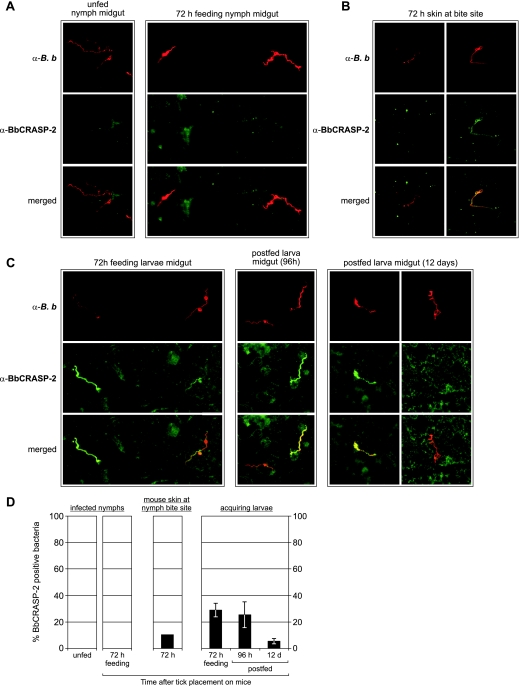
B. burgdorferi is abundant within the midguts of infected tick larvae and nymphs. In addition, the spirochetes can often be microscopically visualized within mammalian skin biopsy specimens taken from the site of tick feeding. We therefore used IFA to assess expression of the BbCRASP-2 protein by B. burgdorferi during stages of tick colonization/transmission and during transmission into mouse skin by infected, feeding ticks. No B. burgdorferi bacteria produced a detectable level of BbCRASP-2 protein during colonization of unfed tick nymphs (Fig. 2A and D), consistent with the inability to detect cspZ mRNA at that stage. Likewise, none of the bacteria within midguts of feeding nymphs produced detectable levels of BbCRASP-2, indicating either that the low levels of cspZ mRNA found in such ticks was insufficient for synthesis of detectable amounts of BbCRASP-2 or that the protein is regulated posttranscriptionally. Approximately 10% of B. burgdorferi bacteria observed in mouse skin at the site of nymph feeding produced a detectable level of BbCRASP-2, further demonstrating induction of BbCRASP-2 expression during mammalian infection (Fig. 2B and D). As naïve larvae acquired B. burgdorferi by feeding on infected mice, 20 to 30% of those bacteria produced detectable levels of BbCRASP-2 (Fig. 2C and D). Twelve days after completion of larval feeding, the number of spirochetes producing detectable levels of BbCRASP-2 had dropped to only 5 to 10%.
FIG. 2.
IFA of BbCRASP-2 in ticks and mouse skin at tick bite sites. Bacteria were simultaneously labeled with mouse polyclonal antiserum specific for BbCRASP-2 (green channel) and rabbit polyclonal antibodies raised against B. burgdorferi membrane extract (red channel). The bottom rows represent merged images. (A) Representative images of B. burgdorferi within the midguts of unfed and partially fed tick nymphs. (B) Representative images of B. burgdorferi in the skin of mice being fed upon by infected tick nymphs. (C) Representative images of B. burgdorferi within the midguts of partially fed, completely fed, and postfed tick larvae that acquired spirochetes during feeding on infected mice. (D) Quantification of IFA results. The bars indicate mean percentages of bacteria producing detectable levels of BbCRASP-2 in nymphs, mouse biopsy specimens at nymph bite sites, and larvae. The error bars represent 1 standard deviation of the means. For ticks, not less than three to five ticks per time point were examined, with 25 random fields viewed per tick. For skin samples, the results are from 88 bacteria detected in a total of 20 biopsies. α-B. b., anti-B. burgdorferi.
Sigma factor utilization for transcription of BbCRASP-encoding genes.
The data described above indicate that cspZ transcription increases during transmission from feeding ticks to vertebrate hosts. Previous studies by our laboratory and others demonstrated that expression of BbCRASP-1 and Erp proteins also increases during tick-to-mammal transmission (22, 32, 55, 70, 75). Several other B. burgdorferi genes that are induced during transmission from feeding ticks, including ospC, are controlled at the level of transcription by the alternative RNA polymerase sigma factor RpoS (16, 26, 36). The apparent role of RpoS in regulating the expression of some proteins involved in mammalian infection led us to examine whether that sigma subunit is involved in transcription of the BbCRASP-encoding cspZ, cspA, erpA, and erpP genes. B. burgdorferi encodes three sigma factors: the housekeeping RpoD (σ70) and the alternative sigma factors RpoS (σS) and RpoN (NtrA; σ54) (27). Transcription of rpoS is dependent upon RpoN-containing RNA polymerase holoenzyme (63). Q-RT-PCR analyses of wild-type B. burgdorferi and isogenic ΔrpoS and ΔrpoN mutants indicated that neither of the mutants differed from the wild type in their abilities to express cspZ, cspA, erpA, and erpP (Fig. 3A). Immunoblot analyses demonstrated that BbCRASP protein expression was also unchanged by the rpoS and rpoN mutations (Fig. 3B). Analyses of the control gene ospC and its protein showed significant inhibition by both sigma mutations, as had been reported previously (36). These data indicate that all of the BbCRASP-encoding genes are transcribed using RpoD and that neither of the alternative sigma factors directly influences BbCRASP production.
FIG. 3.
None of the BbCRASP-encoding genes requires an alternative sigma factor for transcription. Wild-type (wt) and isogenic strains lacking either rpoN or rpoS were grown at 23°C and subsequently diluted in fresh medium, grown at 34°C, and harvested at late exponential growth phase. (A) Q-RT-PCR analyses of cspZ, cspA, erpA, and erpP. The ospC gene was also analyzed as a control, since its expression is known to be dependent upon both RpoN and RpoS. Each experiment was performed three times, and the error bars represent 1 standard deviation from the means. The expression level for each gene by wild-type bacteria was set to 1, and the values of each mutant were calculated relative to the wild-type standard. **, statistically significant differences (P < 0.01). (B) Equal masses of total protein lysates from wild-type, rpoN, and rpoS cultures were analyzed by immunoblotting using specific antibodies directed against each of the BbCRASPs and the control protein, OspC. Blots were also analyzed using antibodies directed against the constitutively expressed FlaB protein to confirm equal loading of protein lysates in all lanes. Immunoblot experiments were performed at least two times, and representative images are presented.
Effects of environmental conditions on cspZ expression.
B. burgdorferi, like many other pathogens, regulates gene expression in response to various environmental stimuli, which serve as cues for the bacterium to determine its location in the vertebrate-tick infectious cycle (70). Insight regarding the signaling pathways controlling gene expression levels may be obtained by studying alterations in culture conditions, such as temperature and pH. Shifting B. burgdorferi from 23 to 34°C in culture medium held at pH 7.0 led to a statistically significant twofold drop in cspZ transcript levels (Fig. 4A). That temperature change at pH 8.0 led to only a slight, statistically insignificant decrease in cspZ. At a constant 34°C, bacteria grown at pH 8.0 contained levels of cspZ mRNA that were a significant twofold greater than those of bacteria grown at pH 7.0. Increasing the pH of cultures grown at 23°C did not significantly change cspZ transcript levels. The effects of culture pH and temperature on BbCRASP-2 protein levels were generally less obvious (Fig. 4B). When using IFA, no appreciable differences could be detected with B. burgdorferi cultured at either 23 or 34°C or any tested pH (Fig. 4C).
FIG. 4.
Effects of culture conditions on cspZ transcript and BbCRASP-2 protein levels. (A) Q-RT-PCR analysis of cspZ mRNA levels in bacteria cultured at either 23 or 34°C in medium buffered to remain at either pH 7.0 or 8.0. The results are illustrated relative to levels determined for the constitutively expressed gene flaB. The graphs represent the means of three independent experiments ± standard errors. Statistically significant differences (P < 0.001) are indicated by asterisks. (B) Immunoblot analyses of bacterial lysates. The blots were stripped and reprobed with a murine monoclonal antibody specific for the constitutively expressed FlaB protein to confirm that equal quantities of total protein were loaded in all lanes. (C) IFA of B. burgdorferi cultured in normal BSK-II medium (pH 7.4) at either 23 or 34°C. The bacteria were incubated simultaneously with mouse polyclonal antiserum specific for BbCRASP-2 (green channel) and rabbit polyclonal antibodies raised against B. burgdorferi membrane extract (red channel). The bottom row shows merged images. α-B. b., anti-B. burgdorferi.
In addition to the above-noted effects of culture temperature and pH, it is obvious that cultivation itself significantly represses cspZ transcript levels. Maximum cspZ levels were achieved at 23°C and pH 8.0, with an average of 0.025 ng cspZ mRNA per 1.0 ng flaB mRNA (Fig. 4A). In contrast, cspZ levels in infected mouse tissues were as high as 0.44 ng cspZ mRNA per 1.0 ng flaB mRNA (Fig. 1A). The repressive effect of culture medium is also evident when comparing IFA images of B. burgdorferi during mammal or tick infection with those grown in vitro. During infection, BbCRASP-2 directed antibodies uniformly labeled the bacteria (Fig. 2). When the same antibody preparations and methods were used, cultured bacteria were very sparsely labeled (Fig. 4C).
DISCUSSION
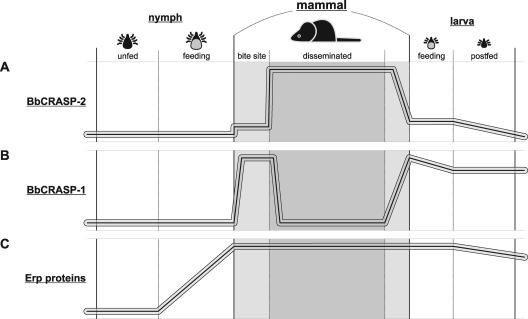
The B. burgdorferi type strain B31 produces five distinct BbCRASPs, with BbCRASP-1, BbCRASP-2, and the Erp proteins (which include BbCRASP-3, -4, and -5) being expressed at different times throughout the spirochete's infectious cycle (Fig. 5). Bacteria that produce at least one Erp-BbCRASP plus either BbCRASP-1 or BbCRASP-2 are resistant to complement-mediated killing during laboratory cultivation (14, 29). Erp-BbCRASPs by themselves are unable to protect against complement in vitro, while the abilities of BbCRASP-1 and -2 to defend against complement in the absence of Erp-BbCRASPs has yet to be addressed (references 14, 29, and 59 and our unpublished results). B. burgdorferi produces high levels of BbCRASP-1 during transmission from feeding ticks to naïve hosts and from infected hosts to feeding, naïve ticks, stages of the infectious cycle when B. burgdorferi is directly exposed to vertebrate blood (75). However, both the present study and others indicated that cspA transcription becomes undetectable within 2 weeks of establishing mammalian infection (47, 50, 78). Infected humans and laboratory mice produce limited antibody responses to BbCRASP-1, consistent with brief exposure of that protein to host immune systems (50, 60). Our studies indicate that transcription of cspZ increases dramatically during mammalian infection, the same time at which cspA is repressed. Humans and laboratory animals infected with B. burgdorferi produce robust antibody responses to BbCRASP-2, also indicating substantial production of that borrelial protein during vertebrate infection (reference 29 and our unpublished results). Thus, BbCRASP-1 may help overcome host defenses during transmission stages, while BbCRASP-2 could serve that purpose during established mammalian infection. Additional functional studies of these proteins are ongoing in our laboratories to determine why B. burgdorferi differentially expresses the two proteins instead of continuously producing only a single protein.
FIG. 5.
Comparison of B. burgdorferi BbCRASP expression profiles during the mammal-tick infectious cycle, as determined by the present and other studies (22, 24, 31, 32, 47, 50, 53-56, 60, 65, 70, 75, 78). Relative expression levels were determined by IFA for each protein during tick infection stages and in mouse skin during tick-to-mammal transmission and by Q-RT-PCR for each gene during disseminated mammalian infection. BbCRASP-2 appears to perform its function(s) primarily during established mammalian infection, BbCRASP-1 likely functions only during stages when bacteria are moving from mammal to tick or vice versa, and Erp proteins perform their functions during all stages between transmission from infected ticks and acquisition by subsequently feeding ticks. (A) BbCRASP-2 is absent from B. burgdorferi within the midguts of unfed tick nymphs. Feeding by infected nymphs appears to stimulate cspZ expression somewhat, but only a fraction of bacteria express detectable levels of BbCRASP-2 protein at the time of transmission into the skin of their vertebrate host. Within 2 weeks of establishing mammalian infection, B. burgdorferi transcribes cspZ at high levels, suggesting that BbCRASP-2 is likewise synthesized during established mammalian infection. Repression of cspZ appears to precede acquisition of B. burgdorferi from mammals by feeding tick larvae, since less than half of the bacteria within feeding tick larvae produce detectable levels of BbCRASP-2 and cspZ mRNA levels in those ticks were lower than those found in infected mouse tissues. (B) BbCRASP-1 is poorly expressed during tick colonization but is produced at detectable levels by essentially all bacteria transmitted from feeding infected tick nymphs to mammalian hosts. Q-RT-PCR analyses indicate that cspA mRNA is undetectable in mouse tissues by 2 weeks after transmission, suggesting that the BbCRASP-1 protein is also down regulated during established mammalian infection. BbCRASP-1 is again induced during transmission from infected mammals to feeding ticks, with approximately 100% of those bacteria producing BbCRASP-1. (C) Erp proteins, such as BbCRASP-3, -4, and -5, are also unexpressed during tick colonization. Feeding by infected nymphs stimulates Erp production by B. burgdorferi while still in the tick midgut, and essentially all of the spirochetes produce Erp proteins by the time they are transmitted into their mammalian host's skin. erp transcription remains elevated throughout mammalian infection, and when naïve tick larvae feed on infected mammals, all acquired B. burgdorferi bacteria produce every examined Erp protein.
Growth of B. burgdorferi in culture medium led to cspZ transcription levels that were significantly lower than those seen during mammalian infection. Repression of cspZ during cultivation may explain why cspZ+ cspA bacteria are sensitive to killing by complement in vitro (14). As has been previously noted, artificial culture medium provides a mixture of signals to B. burgdorferi, causing the bacterium to simultaneously produce mammal and tick infection-specific proteins (70). The results of our studies indicate that a substance(s) present in or absent from conventional culture medium prevents maximal expression of cspZ. We also note that cultured B. burgdorferi does not express Erp proteins at the high levels achieved during mammalian infection (1, 3, 32, 72, 77). We have initiated searches for the roots of in vitro cspZ repression on the premise that signaling pathways that affect transcription during culture will also affect expression during infection processes. Cultured B. burgdorferi regulates the transcription of cspZ and a variety of other genes in response to changes in culture temperature and pH, although neither the mechanisms nor the significance of those changes is understood yet. Many B. burgdorferi genes (e.g., ospC and erp genes) that are induced during transmission from tick to mammal are also induced by culture temperatures close to that of the mammalian body, although some mammal-specific genes are instead induced by culture at cooler room temperature (15, 62, 68, 70). The tick-to-vertebrate transmission process includes exposure of the bacteria to acidification in the feeding tick's midgut and alkalinization in the tick's salivary glands, and some mammal-specific genes are induced by acidic culture pH while others, such as cspZ, are induced by alkaline culture conditions (15, 17, 18, 70, 76, 82). Neither cspZ, cspA, nor the examined erp genes were affected by rpoS or rpoN mutations, indicating that, unlike some other B. burgdorferi proteins involved in mammalian infection, none of the BbCRASP-encoding genes is directly affected by either of the borrelial alternative sigma factors. Those results suggest that interactions between borrelial transcription factors and DNA of the cspZ locus are responsible for regulation of the gene's expression. The observed pattern of cspZ transcription in vivo is distinct from those of all previously investigated B. burgdorferi genes, suggesting that the regulatory factors controlling cspZ expression are different from those controlling the production of other mammalian infection-associated genes. Substantial headway has been made toward identifying regulatory factors controlling erp expression (8, 9), and similar techniques may also identify the means by which B. burgdorferi controls cspZ.
These studies also provided new insight into mechanisms controlling BbCRASP-1 production. A previous study used IFA to examine the production of that protein by B. burgdorferi during various stages of tick colonization (75). When naïve tick larvae acquired B. burgdorferi through feeding on infected mice, essentially all of the bacteria produced detectable levels of BbCRASP-1, consistent with detection of cspA mRNA in such bacteria during the present study. However, levels of BbCRASP-1 protein declined after the ticks completed blood feeding and molted to the nymphal stage, so that no bacteria within unfed nymphs produced a detectable level of BbCRASP-1 (75). In contrast, cspA mRNA levels remained constant or increased following the larva-to-nymph molt (Fig. 1B). Thus, while the protein was evidently turned over and not replaced, either the mRNA was not degraded or the gene continued to be transcribed during that time. Transcript levels in midguts remained high during nymph feeding, a stage at which BbCRASP-1 protein was rarely detected (75). These data suggest that B. burgdorferi may control BbCRASP-1 content at a posttranscriptional level during tick colonization. As a caveat, it is possible that differences in sensitivities of the techniques used in these studies may be partly responsible for the observed differences in results. Noting that essentially 100% of B. burgdorferi bacteria transmitted to mice from feeding nymphs express BbCRASP-1 (75) and the significant role BbCRASP-1 can perform in protecting bacteria from complement-mediated killing (14), such posttranscriptional regulation of BbCRASP-1 expression could allow the bacteria to rapidly produce large quantities of the protein as soon as the tick vector begins feeding.
In conclusion, this study, together with previously published data, revealed distinct expression patterns for the three types of B. burgdorferi CRASPs. BbCRASP-1 is produced during both tick-to-mammal and mammal-to-tick transmission but apparently not during established mammalian infection. BbCRASP-2 is produced poorly during transmission stages but at high levels during established infection. Erp proteins, which include BbCRASP-3, -4, and -5, are produced during transmission steps and throughout mammalian infection. All of these proteins share the ability to bind host factor H, although, since factor H-deficient mice can be infected at levels essentially identical to those of wild-type mice, the possible coating of the B. burgdorferi surface with factor H via BbCRASPs is redundant with at least one additional mechanism of complement resistance (80). Related to that observation, many of the BbCRASPs have been found to bind additional host proteins (references 30, 35, and 50 and our unpublished results). What purpose do those additional interactions serve? Are the various B. burgdorferi BbCRASPs functionally redundant but temporally or spatially distinct? Or does each class of BbCRASP perform an overlapping set of functions, with one set necessary only during persistent mammalian infection, another set used during transmission stages only, and the third set required during both transmission and persistent infection? Additional studies designed to answer these questions regarding this genetically distinct but functionally related group of B. burgdorferi outer-surface lipoproteins will undoubtedly give significant insight into the infectious properties of Lyme disease spirochetes.
Acknowledgments
This work was funded by U.S. National Institutes of Health grant R01-AI44254 to B. Stevenson, Deutsche Forschungsgemeinschaft grant Kr3383/1-1 to P. Kraiczy, and Deutsche Forschungsgemeinschaft grant Wa533/7-1 to R. Wallich.
We thank Patricia Rosa and Mark Fisher for providing bacterial strains and Logan Burns, Jennifer Miller, Sean Riley, Ashutosh Verma, and Kate von Lackum for their assistance and helpful comments on this work.
Editor: D. L. Burns
Footnotes
Published ahead of print on 11 June 2007.
REFERENCES
- 1.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 101:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. D. Radolf. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 4.Alitalo, A., T. Meri, T. Chen, H. Lankinen, Z.-Z. Cheng, T. S. Jokiranta, I. J. T. Seppälä, P. Lahdenne, P. S. Hefty, D. R. Akins, and S. Meri. 2004. Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J. Immunol. 172:6195-6201. [DOI] [PubMed] [Google Scholar]
- 5.Alitalo, A., T. Meri, H. Lankinen, I. Seppälä, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. [DOI] [PubMed] [Google Scholar]
- 6.Alitalo, A., T. Meri, L. Rämö, T. S. Jokiranta, T. Heikkilä, I. J. T. Seppälä, J. Oksi, M. Viljanen, and S. Meri. 2001. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 69:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aslam, M., and S. J. Perkins. 2001. Folded-back solution structure of monomeric factor H of human complement by sychrotron X-ray and neutron scattering, analytical ultracentrifugation and constrained molecular modeling. J. Mol. Biol. 309:1117-1138. [DOI] [PubMed] [Google Scholar]
- 8.Babb, K., T. Bykowski, S. P. Riley, M. C. Miller, E. DeMoll, and B. Stevenson. 2006. Borrelia burgdorferi EbfC, a novel, chromosomally encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete's resident cp32 prophages. J. Bacteriol. 188:4331-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babb, K., J. D. McAlister, J. C. Miller, and B. Stevenson. 2004. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 186:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbour, A. G., S. F. Hayes, R. A. Heiland, M. E. Schrumpf, and S. L. Tessier. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect. Immun. 52:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barthold, S. W. 1991. Infectivity of Borrelia burgdorferi relative to route of inoculation and genotype in laboratory mice. J. Infect. Dis. 163:419-420. [DOI] [PubMed] [Google Scholar]
- 12.Brade, V., I. Kleber, and G. Acker. 1992. Differences of two Borrelia burgdorferi strains in complement activation and serum resistance. Immunobiology 185:453-465. [DOI] [PubMed] [Google Scholar]
- 13.Breitner-Ruddock, S., R. Würzner, J. Schulze, and V. Brade. 1997. Heterogeneity in the complement-dependent bacteriolysis within the species of Borrelia burgdorferi. Med. Microbiol. Immunol. 185:253-260. [DOI] [PubMed] [Google Scholar]
- 14.Brooks, C. S., S. R. Vuppala, A. M. Jett, A. Alitalo, S. Meri, and D. R. Akins. 2005. Complement regulator-acquiring surface protein 1 imparts resistance to human serum in Borrelia burgdorferi. J. Immunol. 175:3299-3308. [DOI] [PubMed] [Google Scholar]
- 15.Bykowski, T., K. Babb, K. von Lackum, S. P. Riley, S. J. Norris, and B. Stevenson. 2006. Transcriptional regulation of the Borrelia burgdorferi antigenically variable VlsE surface protein. J. Bacteriol. 188:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caimano, M. J., C. H. Eggers, K. R. O. Hazlett, and J. D. Radolf. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 72:6433-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll, J. A., R. M. Cordova, and C. F. Garon. 2000. Identification of eleven pH-regulated genes in Borrelia burgdorferi localized to linear plasmids. Infect. Immun. 68:6677-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carroll, J. A., C. F. Garon, and T. G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 20.Casjens, S., R. van Vugt, K. Tilly, P. A. Rosa, and B. Stevenson. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassatt, D. R., N. K. Patel, N. D. Ulbrandt, and M. S. Hanson. 1998. DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect. Immun. 66:5379-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das, S., S. W. Barthold, S. Stocker Giles, R. R. Montgomery, S. R. Telford, and E. Fikrig. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Investig. 99:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Hage, N., K. Babb, J. A. Carroll, N. Lindstrom, E. R. Fischer, J. C. Miller, R. D. Gilmore, Jr., M. L. Mbow, and B. Stevenson. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147:821-830. [DOI] [PubMed] [Google Scholar]
- 24.El-Hage, N., and B. Stevenson. 2002. Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J. Bacteriol. 184:4536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher, M. A., D. Grimm, A. K. Henion, A. F. Elias, P. E. Stewart, P. A. Rosa, and F. C. Gherardini. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl. Acad. Sci. USA 102:5162-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 28.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 29.Hartmann, K., C. Corvey, C. Skerka, M. Kirschfink, M. Karas, V. Brade, J. C. Miller, B. Stevenson, R. Wallich, P. F. Zipfel, and P. Kraiczy. 2006. Functional characterization of BbCRASP-2, a distinct outer membrane protein of Borrelia burgdorferi that binds host complement regulators factor H and FHL-1. Mol. Microbiol. 61:1220-1236. [DOI] [PubMed] [Google Scholar]
- 30.Haupt, K., P. Kraiczy, R. Wallich, V. Brade, C. Skerka, and P. F. Zipfel. 2007. Binding of human factor H-related protein 1 to serum-resistant Borrelia burgdorferi is mediated by borrelial complement regulator-acquiring surface proteins. J. Infect. Dis. 196:124-133. [DOI] [PubMed] [Google Scholar]
- 31.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, and D. R. Akins. 2002. Changes in the temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 70:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hellwage, J., T. Meri, T. Heikkilä, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppälä, and S. Meri. 2001. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 34.Hodzic, E., S. Feng, K. J. Freet, and S. W. Barthold. 2003. Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect. Immun. 71:5042-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hovis, K. M., E. Tran, C. M. Sundy, E. Buckles, J. V. McDowell, and R. T. Marconi. 2006. Selective binding of Borrelia burgdorferi OspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis. Infect. Immun. 74:1967-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hübner, A., X. Yang, D. M. Nolen, T. G. Popova, P. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 98:12724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janeway, C. A., P. Travers, M. Walport, and J. D. Capra. 1999. Immunobiology, 4th ed. Elsevier Science Ltd., New York, NY.
- 38.Kochi, S. K., R. C. Johnson, and A. P. Dalmasso. 1991. Complement-mediated killing of the Lyme disease spirochete Borrelia burgdorferi: role of antibody in formation of an effective membrane attack complex. J. Immunol. 146:3964-3970. [PubMed] [Google Scholar]
- 39.Kraiczy, P., K. Hartmann, J. Hellwage, C. Skerka, V. Brade, P. F. Zipfel, R. Wallich, and B. Stevenson. 2004. Immunological characterization of the complement regulator factor H-binding CRASP and Erp proteins of Borrelia burgdorferi. Int. J. Med. Microbiol. 37:152-157. [DOI] [PubMed] [Google Scholar]
- 40.Kraiczy, P., J. Hellwage, C. Skerka, H. Becker, M. Kirschfink, M. M. Simon, V. Brade, P. F. Zipfel, and R. Wallich. 2004. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 279:2421-2429. [DOI] [PubMed] [Google Scholar]
- 41.Kraiczy, P., J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, and R. Wallich. 2003. Immune evasion of Borrelia burgdorferi: mapping of a complement inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 33:697-707. [DOI] [PubMed] [Google Scholar]
- 42.Kraiczy, P., E. Rossmann, V. Brade, M. M. Simon, C. Skerka, P. F. Zipfel, and R. Wallich. 2006. Binding of human complement regulators FHL-1 and factor H to CRASP-1 orthologs of Borrelia burgdorferi. Wien. Klin. Wochenschr. 118:669-676. [DOI] [PubMed] [Google Scholar]
- 43.Kraiczy, P., C. Skerka, V. Brade, and P. F. Zipfel. 2001. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69:7800-7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kraiczy, P., C. Skerka, M. Kirschfink, V. Brade, and P. F. Zipfel. 2001. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and factor H. Eur. J. Immunol. 31:1674-1684. [DOI] [PubMed] [Google Scholar]
- 45.Kraiczy, P., C. Skerka, P. F. Zipfel, and V. Brade. 2002. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: a new protein family involved in complement resistance. Wien. Klin. Wochenschr. 114:568-573. [PubMed] [Google Scholar]
- 46.Lam, T. T., T.-P. K. Nguyen, R. R. Montgomery, F. S. Kantor, E. Fikrig, and R. A. Flavell. 1994. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect. Immun. 62:290-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lederer, S., C. Brenner, T. Stehle, L. Gern, R. Wallich, and M. M. Simon. 2005. Quantitative analysis of Borrelia burgdorferi gene expression in naturally (tick) infected mouse strains. Med. Microbiol. Immunol. 194:81-90. [DOI] [PubMed] [Google Scholar]
- 48.Liang, F. T., M. B. Jacobs, L. C. Bowers, and M. T. Philipp. 2002. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mbow, M. L., R. D. Gilmore, Jr., and R. G. Titus. 1999. An OspC-specific monoclonal antibody passively protects mice from tick-transmitted infection by Borrelia burgdorferi B31. Infect. Immun. 67:5470-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDowell, J. V., K. M. Hovis, H. Zhang, E. Tran, J. Lankford, and R. T. Marconi. 2006. Evidence that BBA68 protein (BbCRASP-1) of the Lyme disease spirochetes does not contribute to factor H-mediated immune evasion in humans and other animals. Infect. Immun. 74:3030-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metts, M. S., J. V. McDowell, M. Theisen, P. R. Hansen, and R. T. Marconi. 2003. Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection. Infect. Immun. 71:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller, J. C. 2005. Example of real-time quantitative reverse transcription-PCR (Q-RT-PCR) analysis of bacterial gene expression during mammalian infection: Borrelia burgdorferi in mouse tissues, p. 1D.3. In R. T. Coico, T. F. Kowalik, J. Quarles, B. Stevenson, and R. Taylor (ed.), Current protocols in microbiology. J. Wiley and Sons, Hoboken, NJ. [DOI] [PubMed]
- 53.Miller, J. C., K. Narayan, B. Stevenson, and A. R. Pachner. 2005. Expression of Borrelia burgdorferi erp genes during infection of non-human primates. Microb. Pathog. 39:27-33. [DOI] [PubMed] [Google Scholar]
- 54.Miller, J. C., and B. Stevenson. 2006. Borrelia burgdorferi erp genes are expressed at different levels within tissues of chronically infected mammalian hosts. Int. J. Med. Microbiol. 296:185-194. [DOI] [PubMed] [Google Scholar]
- 55.Miller, J. C., K. von Lackum, K. Babb, J. D. McAlister, and B. Stevenson. 2003. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect. Immun. 71:6943-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller, J. C., K. von Lackum, M. E. Woodman, and B. Stevenson. 2006. Detection of Borrelia burgdorferi gene expression during mammalian infection using transcriptional fusions that produce green fluorescent protein. Microb. Pathog. 41:43-47. [DOI] [PubMed] [Google Scholar]
- 57.Montgomery, R. R., S. E. Malawista, K. J. M. Feen, and L. K. Bockenstedt. 1996. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J. Exp. Med. 183:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oppermann, M., T. Manuelian, M. Józsi, E. Brandt, T. S. Jokiranta, S. Heinen, S. Meri, C. Skerka, O. Götze, and P. F. Zipfel. 2006. The C-terminus of compement regulator Factor H mediates target recognition: evidence for a compact conformation of the native protein. Clin. Exp. Immunol. 144:342-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patarakul, K., M. F. Cole, and C. A. N. Hughes. 1999. Complement resistance in Borrelia burgdorferi strain 297: outer membrane proteins prevent MAC formation at lysis susceptible sites. Microb. Pathog. 27:25-41. [DOI] [PubMed] [Google Scholar]
- 60.Rossmann, E., V. Kitiratschky, H. Hofmann, P. Kraiczy, M. M. Simon, and R. Wallich. 2006. Borrelia burgdorferi complement regulator-acquiring surface protein 1 of the Lyme disease spirochetes is expressed in humans and induces antibody responses restricted to nondenatured structural determinants. Infect. Immun. 74:7024-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, A. H., J. S. Blevins, G. N. Bachlani, X. F. Yang, and M. V. Norgard. 2007. Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN). J. Bacteriol. 189:2139-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stanek, G., and F. Strle. 2003. Lyme borreliosis. Lancet 362:1639-1647. [DOI] [PubMed] [Google Scholar]
- 65.Stevenson, B., T. Bykowski, A. E. Cooley, K. Babb, J. C. Miller, M. E. Woodman, K. von Lackum, and S. P. Riley. 2006. The Lyme disease spirochete Erp lipoprotein family: structure, function and regulation of expression, p. 354-372. In F. C. Cabello, H. P. Godfrey, and D. Hulinska (ed.), Molecular biology of spirochetes. IOS Press, Amsterdam, The Netherlands.
- 66.Stevenson, B., N. El-Hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stevenson, B., and J. C. Miller. 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 57:309-324. [DOI] [PubMed] [Google Scholar]
- 68.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stevenson, B., K. von Lackum, S. P. Riley, A. E. Cooley, M. E. Woodman, and T. Bykowski. 2006. Evolving models of Lyme disease spirochete gene regulation. Wien. Klin. Wochenschr. 118:643-652. [DOI] [PubMed] [Google Scholar]
- 71.Stevenson, B., W. R. Zückert, and D. R. Akins. 2001. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species, p. 87-100. In M. H. Saier and J. García-Lara (ed.), The spirochetes: molecular and cellular biology. Horizon Press, Oxford, United Kingdom. [PubMed]
- 72.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 92:4269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tilly, K., J. G. Krum, A. Bestor, M. W. Jewett, D. Grimm, D. Bueschel, R. Byram, D. Dorward, M. J. Vanraden, P. Stewart, and P. Rosa. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74:3554-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Dam, A. P., A. Oei, R. Jaspars, C. Fijen, B. Wilske, L. Spanjaard, and J. Dankert. 1997. Complement-mediated serum sensitivity among spirochetes that cause Lyme disease. Infect. Immun. 65:1228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von Lackum, K., J. C. Miller, T. Bykowski, S. P. Riley, M. E. Woodman, V. Brade, P. Kraiczy, B. Stevenson, and R. Wallich. 2005. Borrelia burgdorferi regulates expression of complement regulator-acquiring surface protein 1 during the mammal-tick infection cycle. Infect. Immun. 73:7398-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von Lackum, K., K. M. Ollison, T. Bykowski, A. J. Nowalk, J. L. Hughes, J. A. Carroll, W. R. Zückert, and B. Stevenson. 2007. Regulated synthesis of the Borrelia burgdorferi inner-membrane lipoprotein IpLA7 (P22, P22-A) during the Lyme disease spirochaete's mammal-tick infectious cycle. Microbiology 153:1361-1371. [DOI] [PubMed] [Google Scholar]
- 77.Wallich, R., C. Brenner, M. D. Kramer, and M. M. Simon. 1995. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect. Immun. 63:3327-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wallich, R., O. Jahraus, T. Stehle, T. T. T. Tran, C. Brenner, H. Hofmann, L. Gern, and M. M. Simon. 2003. Artificial-infection protocols allow immunodetection of novel Borrelia burgdorferi antigens suitable as vaccine candidates against Lyme disease. Eur. J. Immunol. 33:708-719. [DOI] [PubMed] [Google Scholar]
- 79.Wallich, R., J. Pattathu, V. Kitiratschky, C. Brenner, P. F. Zipfel, V. Brade, M. M. Simon, and P. Kraiczy. 2005. Identification and functional characterization of complement regulator-acquiring surface protein 1 of the Lyme disease spirochetes Borrelia afzelii and Borrelia garinii. Infect. Immun. 73:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woodman, M. E., A. E. Cooley, J. C. Miller, J. J. Lazarus, K. Tucker, T. Bykowski, M. Botto, J. Hellwage, R. M. Wooten, and B. Stevenson. 2007. Borrelia burgdorferi binding of host complement regulator factor H is not required for efficient mammalian infection. Infect. Immun. 75:3131-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wormser, G. P. 2006. Hematogenous dissemination in early Lyme disease. Wien. Klin. Wochenschr. 118:634-637. [DOI] [PubMed] [Google Scholar]
- 82.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470-1479. [DOI] [PubMed] [Google Scholar]
- 83.Zipfel, P. F., C. Skerka, J. Hellwage, S. T. Jokiranta, S. Meri, V. Brade, P. Kraiczy, M. Noris, and G. Remuzzi. 2002. Factor H family proteins: on complement, microbes and human diseases. Biochem. Soc. Trans. 30:971-978. [DOI] [PubMed] [Google Scholar]
- 84.Zückert, W. R. 2007. Laboratory maintenance of Borrelia burgdorferi, p. 12C.1.1-12C.1.10. In R. T. Coico, T. F. Kowalik, J. Quarles, B. Stevenson, and R. Taylor (ed.), Current protocols in microbiology. J. Wiley and Sons, Hoboken, NJ. [DOI] [PubMed]
- 85.Zückert, W. R., J. Meyer, and A. G. Barbour. 1999. Comparative analysis and immunological characterization of the Borrelia Bdr protein family. Infect. Immun. 67:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]