Abstract
Aggregatibacter (Actinobacillus) actinomycetemcomitans is a gram-negative oral pathogen that is the etiologic agent of localized aggressive periodontitis and systemic infections. A. actinomycetemcomitans produces leukotoxin (LtxA), which is a member of the RTX (repeats in toxin) family of secreted bacterial toxins and is known to target human leukocytes and erythrocytes. To better understand how LtxA functions as a virulence factor, we sought to detect and study potential A. actinomycetemcomitans proteins that interact with LtxA. We found that Cu,Zn superoxide dismutase (SOD) interacts specifically with LtxA. Cu,Zn SOD was purified from A. actinomycetemcomitans to homogeneity and remained enzymatically active. Purified Cu,Zn SOD allowed us to isolate highly specific anti-Cu,Zn SOD antibody and this antibody was used to further confirm protein interaction. Cu,Zn SOD-deficient mutants displayed decreased survival in the presence of reactive oxygen and nitrogen species and could be complemented with wild-type Cu,Zn SOD in trans. We suggest that A. actinomycetemcomitans Cu,Zn SOD may protect both bacteria and LtxA from reactive species produced by host inflammatory cells during disease. This is the first example of a protein-protein interaction involving a bacterial Cu,Zn SOD.
Aggregatibacter (Actinobacillus) actinomycetemcomitans is a gram-negative oral pathogen that is the etiologic agent of localized aggressive periodontitis that occurs in adolescents (23, 37, 77). Localized aggressive periodontitis is a destructive disease of the periodontal ligament and surrounding bone and often results in the loss of teeth when untreated (77). A. actinomycetemcomitans can also cause several systemic diseases, including infective endocarditis, and is a member of the Haemophilus, Actinobacillus, Cardiobacterium, Eikenella, and Kingella group of bacteria (6, 14).
Among other virulence factors, A. actinomycetemcomitans produces an RTX (repeat in toxin) leukotoxin (LtxA). LtxA is known to kill specifically leukocytes of humans and Old World Primates (67-69). The toxins of the RTX family are exemplified by Escherichia coli α-hemolysin (12, 55, 74), Bordetella pertussis adenylate cyclase (29, 34), and Mannheimia haemolytica leukotoxin (7, 30). They are large secreted proteins that contain glycine-rich repeats. The repeats are responsible for binding calcium, which is required for toxin activity (12, 54, 59). These toxins are modified with fatty acid moieties attached to internal lysine residues, which is a unique characteristic of RTX toxins (66). RTX toxins destroy target cells by inserting into membranes to form pores, causing membrane disruption and cell death (74).
We have recently found that, in addition to leukotoxic activity, LtxA from A. actinomycetemcomitans can also destroy erythrocytes (1). Purified leukotoxin was able to lyse sheep and human erythrocytes in vitro, and the production of this toxin resulted in beta-hemolytic colonies on solid medium. We also found that the secretion of LtxA was completely inhibited by free iron in a manner independent of gene regulation (2). Therefore, A. actinomycetemcomitans leukotoxin may play an additional role in disease by releasing iron from erythrocytes and making it available for the invading pathogen, as has been shown for other hemolysins (11, 41, 53). LtxA is secreted into culture supernatants of A. actinomycetemcomitans under normal growth conditions (16, 44, 45), similar to RTX toxins of other bacteria, but can also be found in the outer membrane and lipid vesicles (15, 16, 43). The secretion of LtxA requires ltxB and ltxD (32; M. P. Isaza, M. S. Duncan, and S. C. Kachlany, unpublished data), as well as TdeA, a TolC-like protein (13).
Most of the studies on LtxA have focused on the interactions between the toxin and host cells. It was shown that LtxA binds to LFA-1 (lymphocyte function-associated antigen 1) on HL-60 cells and then kills the cells by inducing apoptosis or necrosis (50). However, very little is known about the interactions between LtxA and other proteins in A. actinomycetemcomitans. In our search for interacting proteins, we found that LtxA interacts with Cu,Zn SOD from A. actinomycetemcomitans. We show here that Cu,Zn SOD from A. actinomycetemcomitans protects the bacterium and the toxin from oxidative damage, and we suggest that Cu,Zn SOD may also play a role in heme transport. To our knowledge, this is the first report of an interaction between a bacterial Cu,Zn SOD and another protein.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A. actinomycetemcomitans strain JP2 is a nonadherent, smooth, laboratory isolate (71), while strain DF2200 is an adherent, rough, clinical isolate (47). A. actinomycetemcomitans growth medium (AAGM) has been described previously (22). Hemolytic activity was detected on Columbia agar with 5% sheep blood (PML Microbiologicals, Inc., Wilsonville, OR). After bacteria were streaked on solid media, the plates were incubated at 37°C in the presence of 10% CO2 for 2 to 3 days. Colonies were inoculated into AAGM broth and incubated for 24 h unless indicated otherwise. E. coli TOP10 was grown in Luria-Bertani (LB) broth and agar (63) at 37°C.
Protein interaction on AminoLink column.
Purified leukotoxin (0.4 mg) was immobilized on an AminoLink column using the AminoLink Plus immobilization kit (Pierce, Rockford, IL). Two ml of strain JP2 extract (2.6 mg/ml) was then passed through the column. After the column was washed with 20 ml of wash buffer (4.29 mM Na2HPO4, 14.7 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, 0.1% Tween 20, 0.002% sodium azide, pH 7.3), proteins were eluted with 1 ml 100 mM citric acid, pH 2.2, and neutralized with 150 μM 200 mM Tris base, pH 10.4. The samples from each fraction were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by staining with GelCode blue stain reagent (Pierce, Rockford, IL).
Purification of LtxA from A. actinomycetemcomitans.
LtxA was isolated from JP2 as previously described (44), with some modifications (16). The purity of LtxA was determined on a 4 to 20% SDS-PAGE gel, and the concentration was measured with a bicinchoninic acid assay according to the manufacturer's protocol (Pierce, Rockford, IL).
Purification of Cu,Zn SOD from A. actinomycetemcomitans.
Pelleted cells obtained from 400 ml of strain JP2 culture were washed once with phosphate-buffered saline (PBS), pH 7.4. The cells were resuspended in 6 ml of PBS, pH 7.4, and sonicated three times for 30 sec. Cell debris was precipitated by centrifuging the mixture at 10,000 × g for 10 min. The supernatant was heated at 60°C for 1 h, and coagulated proteins were precipitated by centrifugation at 10,000 × g for 10 min. The resulting supernatant was passed through 1 ml of Talon resin (BD Biosciences, Palo Alto, CA) prewashed with 20 ml 50 mM phosphate buffer, pH 7.4, with 300 mM NaCl. The resin was then washed with another 20 ml of the same buffer. Cu,Zn SOD was eluted with 1 ml of elution buffer (150 mM imidazole, 300 mM NaCl, pH 7.0). The eluted samples were applied to a PD-10 column (Amersham Biosciences, Uppsala, Sweden) prewashed with PBS, pH 7.4. The desalted protein was then eluted with the same buffer, aliquoted, and frozen at −80°C. The purity of the Cu,Zn SOD was determined on a 4 to 20% SDS-PAGE gel, and the concentration was measured by a bicinchoninic acid assay.
SOD activity assay.
Cu,Zn SOD activity was visualized in nondenaturing PAGE as a clear zone that did not stain upon photochemical reduction of nitroblue tetrazolium (NBT) to formazan blue (5). A Cu,Zn SOD activity kit (Sigma, St. Louis, MO) was used for the quantitative measurement of enzyme activity. Cu,Zn SOD from bovine erythrocytes (Alexis USA, San Diego, CA) was used as a control.
Isolation of polyclonal antibody.
To isolate specific anti-Cu,Zn SOD antibody, we used antiserum obtained from injecting A. actinomycetemcomitans extract into a rabbit (gift of Daniel Fine). Two mg of purified Cu,Zn SOD from A. actinomycetemcomitans were immobilized on an AminoLink column (Pierce, Rockford, IL). Two ml of the rabbit antiserum was passed through the column with immobilized Cu,Zn SOD. Nonspecifically interacting proteins were washed, and Cu,Zn SOD-specific antibody was eluted as described for LtxA.
Overlay and dot blot assay.
Purified proteins (0.5 μg) were resolved by SDS-PAGE and transferred to a nitrocellulose membrane (overlay assay) or loaded directly onto the membrane (dot blot assay). The membrane was incubated with purified LtxA or Cu,Zn SOD (50 μg/ml) for 2 h. Another membrane that served as a negative control was treated the same way but without LtxA or Cu,Zn SOD. The membranes were subjected to Western blot analysis with anti-LtxA or anti-Cu,Zn SOD antibody as previously described (16).
MALDI-TOF MS.
Individual protein bands were excised and digested with trypsin, and peptides were extracted for matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis using a Voyager-DE PRO MALDI-TOF mass spectrometer (PerSeptive Biosystems, Ramsey, MN). A search for the peptide mass fingerprint was carried out with the Profound database. All procedures were carried out by the New Jersey Medical School Proteomics Core Facility.
Isolation of sodC mutants.
The sodC gene mutants were isolated using an allelic exchange method that has been previously described (13, 62). Gene disruption mutants were generated by using pMB78 vector (M. K. Bhattacharjee, unpublished data) that contains A. actinomycetemcomitans uptake sequences (70). The sodC gene, including the 500-bp upstream and downstream flanking regions, was first PCR amplified from strain DF2200 genomic DNA by using an Expand high fidelity PCR system (Roche, Mannheim, Germany). PCR amplification was performed for 30 cycles using an annealing temperature of 58°C for 1 min and primer extension at 72°C for 2 min. The primers used were SOD-LFW 5′-GAAGCTTGTGAGCATCGCCTCGTTAATC-3′ and SOD-LRV 5′-GAAGCTTCACACGAAGTAAGTCATTCAACG-3′. The resulting ∼1.5-kb PCR fragment was first cloned into pCR2.1 vector using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) and then subcloned into pMB78. The EZ-Tn5 <KAN-2> (Epicenter Biotechnologies, Madison, WI) transposition reaction was carried out in vitro according to the manufacturer's instructions. Transposon insertions within sodC were mapped using the primers SODFW 5′-CAAGCTTATGAAAATCAAAACTATTTTAGCC-3′ and SODRV 5′-GCAAGCTTTTTAATTACACCACACGCC-3′. DF2200 was transformed with the Pvu II-digested recombinant pMB78 vector using the procedure described previously (73). Allelic exchange mutants were selected on AAGM with kanamycin (40 μg/ml). Gene disruption was confirmed by PCR using the SODFW and SODRV primers. The protein expression levels were determined by Western blot analysis using anti-Cu,Zn SOD antibody.
Complementation of the sodC mutation.
For complementation studies, sodC was amplified from strain DF2200 using an Expand high fidelity PCR system (Roche, Mannheim, Germany) and the SODFW and SODRV primers (see above). The ∼500-bp product was cloned into pCR2.1 using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA). The sodC gene was then subcloned into pJAK16 as described previously (46). The new plasmid containing sodC or the empty vector (pJAK16) was mobilized into A. actinomycetemcomitans as previously described (46). The expression of sodC was carried out in liquid AAGM containing 0.1 mM isopropyl-β-d-thiogalactopyranoside.
In vitro reactive oxygen species (ROS) and reactive nitrogen species (RNS) susceptibility assay.
The superoxide killing assay was performed by using a xanthine/xanthine oxidase system to generate superoxide (75). Bacterial cells (1 × 106 cells/ml) were exposed to superoxide generated by combining 250 μM xanthine (Sigma, St. Louis, MO) and 0.1 U of xanthine oxidase (Sigma, St. Louis, MO). The percent survival was determined at different times by plating serial dilutions of bacteria on AAGM plates after 0 and 15 min of exposure. The CFU were counted to determine the number of viable A. actinomycetemcomitans cells. Purified LtxA (2 mg) was exposed to superoxide generated together with nitric oxide (NO). NO was produced by using 1 mM Spermine NONOate (Alexis biochemicals, San Diego, CA). After 40 min of exposure, LtxA was isolated on a PD-10 column (Amersham Biosciences, Uppsala, Sweden) and eluted with PBS, pH 7.4. This desalted sample of LtxA was used for the HL-60 killing assay.
HL-60 killing assay.
The human promyelocytic leukemia HL-60 cell line was purchased from ATCC and grown in RPMI medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 2 mM l-glutamine. All cells were maintained at a density of 0.5 to 1 × 106 cells/ml in a culture incubator at 37°C with 5% CO2. Cell counts were performed by using a VI-CELL cell viability analyzer (Beckman Coulter, Hialeah, FL), which employs trypan blue staining.
Fractionation of bacteria.
Bacteria were fractionated into cytosol, inner membrane, and outer membrane by using sarkosyl as previously described (2, 8, 33). This method of fractionation has previously been carried out with A. actinomycetemcomitans for the separation of cytosolic and membrane proteins (8, 16, 33). With this method, we have not observed cross-contamination of fractions, as indicated by the successful identification of membrane protein markers (16).
RESULTS
A. actinomycetemcomitans proteins that interact with LtxA.
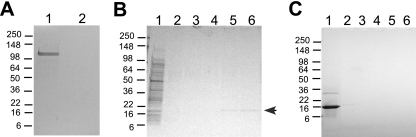
To identify proteins in A. actinomycetemcomitans that interact with LtxA, we first immobilized purified LtxA to resin beads through amino linkages (AminoLink resin; Pierce, Rockford, IL). To prevent the cell extract protein from binding, the remaining unreacted groups were blocked with 1 M Tris-HCl. Nearly all of the LtxA loaded onto the AminoLink column (0.4 mg) covalently bound to the beads, as the toxin was not detected in the flowthrough wash (Fig. 1A). We next passed cell extract (∼5 mg of protein) from strain JP2 through the column to allow the cellular proteins to bind immobilized LtxA (Fig. 1B). The column was washed, and proteins that remained bound to LtxA were eluted with 100 mM citric acid and neutralized (Fig. 1B). A single ∼18-kDa protein band was consistently detected in the eluted fractions when the proteins were resolved using SDS-PAGE and stained with Coomassie blue. To identify the nature of the protein, the band was excised from the gel and its tryptic-digest peptides were analyzed by MALDI-TOF MS. A search of the peptide masses in the Profound database revealed that the 18-kDa protein was Cu,Zn SOD (EC 1.15.1.1).
FIG. 1.
Protein interaction on AminoLink column. (A) LtxA was immobilized on an AminoLink column. Lanes: 1, input LtxA (4 μg); 2, column flowthough. (B) Interaction between immobilized LtxA and proteins from A. actinomycetemcomitans strain JP2 extract. Lanes: 1, input JP2 cell extract (10 μg); 2 and 3, column wash; 4 to 6, elutions. The arrow indicates the 18-kDa protein band. (C) Bovine Cu,Zn SOD interaction control. Lanes: 1, input bovine Cu,Zn SOD (2 μg); 2 and 3, column wash; 4 to 6, elutions. All gels were stained with Coomassie blue. Molecular mass standards are in kDa.
To confirm that the interaction between LtxA and Cu,Zn SOD from A. actinomycetemcomitans was specific, we tested whether bovine Cu,Zn SOD could bind immobilized LtxA. We passed 0.6 mg bovine Cu,Zn SOD through the column and found that all of the protein eluted in the wash fractions and not in the citric acid elution fractions (Fig. 1C). The absence of interaction between bovine Cu,Zn SOD and A. actinomycetemcomitans LtxA suggests that Cu,Zn SOD from A. actinomycetemcomitans interacts specifically with LtxA.
Purification and activity of native A. actinomycetemcomitans Cu,Zn SOD.
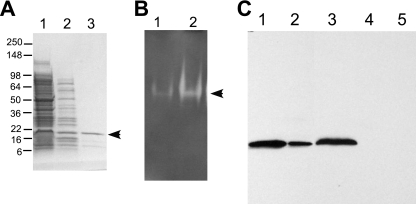
To further study the interaction, we purified Cu,Zn SOD from A. actinomycetemcomitans. We took advantage of two unique properties of prokaryotic Cu,Zn SOD proteins to purify the protein. First, Cu,Zn SOD proteins have a natural resistance to high temperature (52). Thus, during the first step of our purification, we heated the bacterial extract at 60°C for 1 h. Most proteins coagulated after heating and were precipitated by centrifugation (Fig. 2A). Another feature of prokaryotic Cu,Zn SOD proteins is that they contain a histidine-rich N-terminal region reminiscent of engineered histidine-tagged proteins. These natural histidine residues are sufficient to purify the protein by metal affinity chromatography (4). Therefore, we used cobalt affinity resin (Talon column; BD Biosciences, Palo Alto, CA) for our second purification step, and A. actinomycetemcomitans Cu,Zn SOD was eluted from the resin using imidazole (Fig. 2A). We then detected Cu,Zn SOD activity in two ways: on native polyacrylamide gel as a clear zone that did not stain upon photochemical reduction of NBT to formazan blue (Fig. 2B) (5) and quantitatively by formazan dye formation in the reaction of soluble tetrazolium salt (WST-1) with a superoxide anion (Sigma, St. Louis, MO). As shown in our purification scheme in Table 1, A. actinomycetemcomitans Cu,Zn SOD was purified approximately 60-fold and the purified protein remained active.
FIG. 2.
Cu,Zn SOD purification and activity. (A) Purification of Cu,Zn SOD from A. actinomycetemcomitans cell extract. Lanes: 1, strain JP2 cell extract after sonication (4 μg); 2, JP2 soluble extract (4 μg) after incubation at 60°C for 1 h; 3, Talon column eluate (2.5 μg). The gel was stained with Coomassie blue. (B) NBT SOD activity assay. Protein samples were resolved by native PAGE and stained with NBT. Lanes: 1, strain JP2 cell extract (2 μg); 2, purified Cu,Zn SOD (2.5 μg). (C) Isolation of and Western blot analysis with anti-Cu,Zn SOD antibody. Lanes: 1, purified Cu,Zn SOD (5 μg); 2, purified Cu,Zn SOD (0.5 μg); 3, JP2 cell extract (4 μg); 4, E. coli cell extract (5 μg); 5, bovine Cu,Zn SOD (1 μg). Molecular mass standards are in kDa. The arrows indicate the position of Cu,Zn, SOD.
TABLE 1.
Cu,Zn SOD purification from A. actinomycetemcomitans
| Purification step | Total protein (mg) | Sp act (% inhibition rate per μg of protein)a | Fold purification |
|---|---|---|---|
| Sonicated cell extract | 15.6 | 0.3 | 1.0 |
| Heated soluble extract | 3.7 | 12.3 | 4.2 |
| Talon column eluate (desalted) | 0.3 | 84.1 | 62.4 |
Inhibition rate of formazan dye reduction with a superoxide anion.
Isolation of anti-Cu,Zn SOD antibody.
To further study the interaction between LtxA and Cu,Zn SOD, we isolated anti-Cu,Zn SOD antibody. An alternate approach to injecting a protein into rabbits to stimulate an immune response is to pull out specific antibody from antiserum generated against a bacterial extract. To do this, we immobilized purified A. actinomycetemcomitans Cu,Zn SOD on AminoLink resin as described above for LtxA (data not shown). We then passed anti-A. actinomycetemcomitans polyclonal antiserum through the Cu,Zn SOD-conjugated resin. The resin was washed with high-salt buffer, and specifically bound antibodies were eluted with low-pH buffer followed by neutralization. Figure 2C shows that we successfully obtained highly specific antibody. This antibody preparation recognized a single Cu,Zn SOD band in A. actinomycetemcomitans cell extract in a Western blot analysis (Fig. 2C). Anti-Cu,Zn SOD antibody did not detect E. coli proteins and did not cross-react with Cu,Zn SOD from bovine erythrocytes (Fig. 2C).
Confirmation of LtxA-SOD interaction.
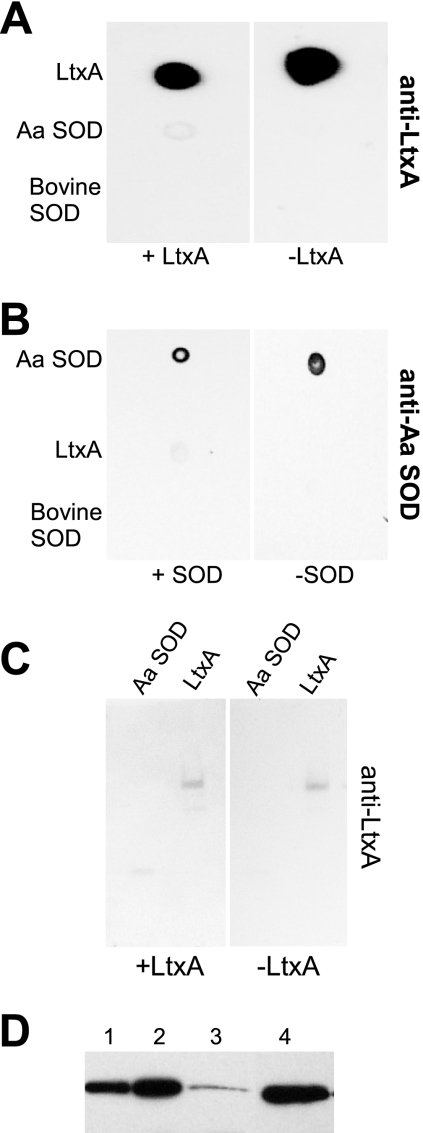
With purified Cu,Zn SOD and anti-Cu,Zn SOD antibody available, we wished to confirm the interaction between A. actinomycetemcomitans LtxA and Cu,Zn SOD. We first performed a dot blot assay by spotting A. actinomycetemcomitans LtxA, Cu,Zn SOD, and bovine Cu,Zn SOD onto a nitrocellulose membrane. The membrane was then incubated in purified LtxA, washed to remove unbound LtxA, and probed with anti-LtxA antibody to detect LtxA that remained bound to the membrane. We also included a control membrane that was not incubated with LtxA prior to probing with anti-LtxA antibody. We found that when the membrane was incubated with LtxA and then probed with anti-LtxA antibody, we were able to detect LtxA in the spots where purified LtxA and purified A. actinomycetemcomitans Cu,Zn SOD were spotted (Fig. 3A, left). In contrast, no signal was detected in the spot where bovine Cu,Zn SOD was spotted. On our control membrane that was not incubated in LtxA, only the LtxA spot was detected after probing with anti-LtxA antibody (Fig. 3A, right). These results indicate that soluble LtxA interacts with immobilized A. actinomycetemcomitans Cu,Zn SOD, but not with immobilized bovine Cu,Zn SOD.
FIG. 3.
Protein interaction assays. (A and B) Dot blot assays. Purified proteins (0.5 μg) were spotted onto a nitrocellulose membrane. The membrane was blocked and then incubated with purified LtxA (50 μg/ml) (A) or Cu,Zn SOD (B). Membranes that were not treated with LtxA (A) or Cu,Zn SOD (B) served as negative controls. The membranes were subjected to Western blot analysis with anti-LtxA (A) or anti-Cu,Zn SOD (B) antibody. (C) Overlay assay. Purified proteins (0.5 μg) were resolved by SDS-PAGE and then transferred to a nitrocellulose membrane. The membrane was incubated with purified LtxA (50 μg/ml). A membrane that was not incubated with LtxA served as a negative control. The membranes were subjected to Western blot analysis with anti-LtxA antibody. (D) Localization of Cu,Zn SOD. Cells were fractionated with the detergent sarkosyl. Lanes: 1, periplasm; 2, cytosol; 3, membrane; 4, purified Cu,Zn SOD (2.5 μg). Aa SOD, A. actinomycetemcomitans Cu,Zn SOD; Bovine SOD, bovine Cu,Zn SOD.
We repeated the dot blot experiment, except that we incubated the membrane in purified A. actinomycetemcomitans Cu,Zn SOD and probed with anti-Cu,Zn SOD antibody (Fig. 3B). We found that anti-Cu,Zn SOD antibody reacted with A. actinomycetemcomitans Cu,Zn SOD in both membranes, but only reacted with LtxA when the membrane was first incubated in A. actinomycetemcomitans Cu,Zn SOD. Bovine Cu,Zn SOD did not bind A. actinomycetemcomitans anti-Cu,Zn SOD antibody in either membrane. These results indicate that the interaction between LtxA and Cu,Zn SOD from A. actinomycetemcomitans can be detected in both directions.
Because our purified preparation of Cu,Zn SOD could have contaminating proteins that are interacting with LtxA, we wished to further confirm the interaction by first separating proteins by SDS-PAGE. Thus, purified A. actinomycetemcomitans Cu,Zn SOD and LtxA were resolved by SDS-PAGE and then transferred to a nitrocellulose membrane. The membrane was incubated in LtxA, washed, and then probed with anti-LtxA antibody. A membrane that was not incubated in LtxA was included as a control. Only after incubating the membrane with LtxA could we detect a band in the lane where A. actinomycetemcomitans Cu,Zn SOD was loaded that corresponded to the molecular weight of A. actinomycetemcomitans Cu,Zn SOD (Fig. 3C). In contrast, A. actinomycetemcomitans LtxA could be detected in both gels, regardless of whether the gel was incubated in LtxA prior to probing with anti-LtxA antibody (Fig. 3C). These results indicate that the only protein in our purified Cu,Zn SOD preparation that is interacting with LtxA is Cu,Zn SOD.
Localization of A. actinomycetemcomitans Cu,Zn SOD.
To identify the cellular localization of Cu,Zn SOD in A. actinomycetemcomitans, we fractionated cells into periplasm, cytosol, and membrane. This fractionation technique is based on the differential solubilization of membranes in the detergent sarkosyl that has previously been used for the localization of other proteins in A. actinomycetemcomitans (2, 8, 33). Equivalent amounts of protein from each fraction were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to Western blot analysis using anti-Cu,Zn SOD antibody. A significant fraction of Cu,Zn SOD was found in the periplasmic and cytosolic fractions (Fig. 3D). In addition, a minor fraction of Cu,Zn SOD was detected in the membrane fraction of the cells.
Construction of an sodC mutant.
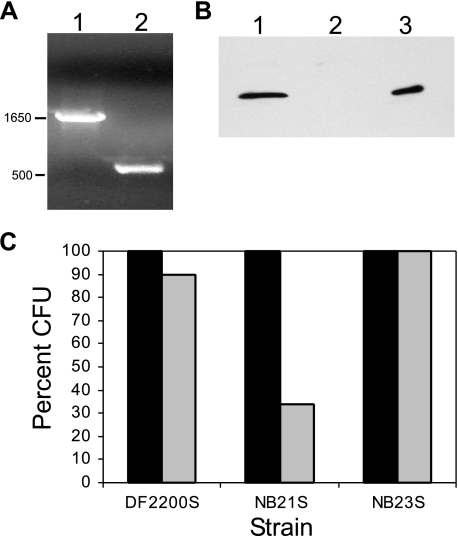
To determine the function of Cu,Zn SOD in A. actinomycetemcomitans, we constructed an A. actinomycetemcomitans mutant defective in Cu,Zn SOD production. The mutation was constructed in the adherent clinical isolate DF2200 (47). In this mutant (strain NB9), sodC was disrupted by the insertion of a kanamycin resistance gene via homologous recombination (Fig. 4A). Rough clinical isolates of A. actinomycetemcomitans aggregate and form clumps when grown in liquid culture (21). Therefore, to facilitate studies that required consistent cell densities, we obtained nonadherent (S) variants of the original mutants.
FIG. 4.
Site-directed mutagenesis of sodC. (A) PCR product of sodC gene. Lanes: 1, sodC::EZ-Tn5; 2, wild-type sodC. The sizes on the left are in bp. (B) Western blot analysis of Cu,Zn SOD in A. actinomycetemcomitans. Lanes: 1, strain DF2200; 2, sodC mutant strain NB9; 3, Cu,Zn SOD (2.5 μg). (C) Survival of A. actinomycetemcomitans after exposure to ROS. The values represent the means of the results of duplicate experiments. The data are expressed as percentages of the time zero values. Black bars, 0 min; gray bars, 15 min.
Strain NB9S did not produce Cu,Zn SOD (Fig. 4B) and expressed less SOD activity than the wild-type strain (Table 2). The residual activity seen in the mutant is likely due to Mn SOD that is present in A. actinomycetemcomitans (http://www.genome.ou.edu/act.html). The sodC gene mutant was genetically complemented with wild-type sodC cloned into the IncQ pJAK16 vector (strain NB23S). When induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside, this strain expressed SOD activity at the level detected in parental strain DF2200S (Table 2). In contrast, the mutant harboring the vector control (strain NB21S) expressed much-reduced levels of SOD activity compared to the levels in the wild-type strain (Table 2). These results show that the mutation in sodC is responsible for the lack of SOD activity.
TABLE 2.
Cu,Zn SOD activity in A. actinomycetemcomitans
| Strain | Description | SOD activity (% inhibition rate per μg of protein)a |
|---|---|---|
| DF2200S | wild type | 1.5 |
| NB9S | sodC::EZ-Tn5 | 0.7 |
| NB21S | sodC::EZ-Tn5 (pJAK16-sodC) | 1.3 |
| NB23S | sodC::EZ-Tn5 (pJAK16) | 0.6 |
Inhibition rate of formazan dye reduction with a superoxide anion.
We wished to determine if a sodC mutation affects LtxA expression or activity. The sodC gene mutants were still able to produce LtxA and were beta-hemolytic on Columbia agar plates with 5% sheep blood (data not shown). Thus, a mutation in sodC does not affect the expression or activity of genes in the ltx operon.
Sensitivity of the sodC mutant to superoxide.
The mutation in sodC was not essential for the viability of A. actinomycetemcomitans in AAGM, and no difference between the growth rates of the mutant and wild-type strains (data not shown) was observed. To determine the sensitivity of the mutant to exogenous superoxide, we challenged bacteria with superoxide generated with the xanthine/xanthine oxidase system. The most efficient killing was observed during the first 15 min of exposure to superoxide. After this time of exposure to superoxide, there was an approximately 10% decrease in the viability of strain DF2200S, while the viable cell count of the sodC mutant with the vector control (strain NB21S) decreased by more than 70% (Fig. 4C). To regenerate the Cu,Zn SOD protective system in the sodC mutant, we used the mutant that was genetically complemented with wild-type sodC in trans (NB23S). In the sodC-complemented strain, resistance to superoxide was restored to wild-type levels. These results indicate that Cu,Zn SOD plays an important role in the protection of A. actinomycetemcomitans from exogenous superoxide.
Sensitivity of LtxA to ROS and RNS.
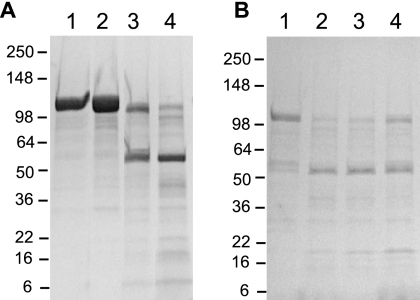
To determine if LtxA is sensitive to an oxidative burst, we exposed the purified protein to ROS and RNS. RNS were generated by adding NO to a xanthine/xantine oxidase system. We found that a 20-min exposure to ROS and RNS caused significant degradation of LtxA which continued for up to 40 min (Fig. 5A). When the ROS- and RNS-generating system was first boiled, no effect was seen on LtxA (data not shown). After 40 min, we separated LtxA from residual ROS and RNS by using a PD-10 desalting column. This degraded LtxA sample was unable to kill HL-60 cells, in contrast to the LtxA control sample that was not exposed to ROS and RNS (data not shown). To determine if Cu,Zn SOD can provide protection for LtxA, we added purified Cu,Zn SOD from A. actinomycetemcomitans to LtxA in a dose-dependent manner prior to exposure to ROS and RNS. After examination on SDS-PAGE, we observed an increase in LtxA stability with increasing concentrations of Cu,Zn SOD (Fig. 5B). Thus, Cu,Zn SOD may prevent LtxA degradation when LtxA is exposed to reactive species produced by host inflammatory cells.
FIG. 5.
Exposure of A. actinomycetemcomitans leukotoxin to ROS and RNS. (A) LtxA (4 μg) treatment at 37°C. Lanes: 1, LtxA alone after 20 min; 2, LtxA alone after 40 min; 3, LtxA with ROS and RNS after 20 min; 4, LtxA with ROS and RNS after 40 min. (B) LtxA (2 μg) treatment for 40 min at 37°C. Lanes: 1, Purified LtxA; 2, LtxA with ROS, RNS, and Cu,Zn SOD (0.3 μg); 3, LtxA with ROS, RNS, and Cu,Zn SOD (0.6 μg); 4, LtxA with ROS, RNS, and Cu,Zn SOD (1.2 μg). Molecular mass standards are in kDa.
DISCUSSION
The production of ROS and RNS by inflammatory cells is a major component of host antimicrobial defenses (35). Bacteria produce numerous factors to help defend themselves from the host immune response. One important bacterial factor involved in the detoxification of superoxide radicals is SOD. We have shown here that A. actinomycetemcomitans produces Cu,Zn SOD and this enzyme interacts with A. actinomycetemcomitans leukotoxin.
Superoxide dismutases are a class of enzymes that neutralize superoxide generated as a by-product of aerobic metabolism. Highly reactive superoxide can damage proteins (25-27), DNA (38), and lipids (19, 57). Superoxide dismutases use metal cofactors (Mn, Fe, Cu, and Zn) to dismutase superoxide to hydrogen peroxide and molecular oxygen (58). Cu,Zn SOD proteins are widely distributed among bacteria and are located in the periplasm or in the outer membrane (3, 18, 49). This enzyme may be important for the survival of bacterial pathogens in the host environment by protecting against ROS and RNS produced by host inflammatory cells (3). This hypothesis is supported by several studies demonstrating that mutants deficient in Cu,Zn SOD production (sodC mutants) are attenuated in animal models for disease. However, the role that Cu,Zn SOD plays in bacterial virulence is ambiguous—it clearly contributes to disease in some organisms (20, 75), but not in others (9, 64). This discrepancy may be due to differences in experimental models.
In addition to protecting bacteria from exogenous superoxide, Cu,Zn SOD may also play an important self-protective role against superoxide formed endogenously. It has recently been found that substantial superoxide is released into the periplasm of E. coli, apparently due to the spontaneous oxidation of menaquinone (48). This endogenous superoxide generated during respiration may in fact be the primary substrate for Cu,Zn SOD in gram-negative bacteria.
Herein, we report the physical interaction between A. actinomycetemcomitans LtxA, an RTX toxin, and Cu,Zn SOD. Toxins of the RTX family contain glycine-rich repeats that are responsible for binding calcium (12, 56, 59) and are required for toxin activity (17, 51). Cu,Zn SOD proteins from pathogenic bacteria are characterized by histidine-rich N-terminal extensions that may be involved in metal uptake under conditions of metal starvation in vivo (4). Thus, one possibility is that Cu,Zn SOD may bind the calcium-rich regions of LtxA through imidazole side chains of histidine. While the molecular mechanism of LtxA-Cu,Zn SOD interaction remains to be investigated, it will be of significant interest to determine if Cu,Zn SOD from other bacteria interact with similar toxins.
Cu,Zn SOD from A. actinomycetemcomitans has previously not been studied. We found that the sodC mutant was more sensitive than the wild-type strain to superoxide generated in vitro. This result indicates that Cu,Zn SOD may play an important role in protection against an oxidative burst generated by the host defense system during infection. Macrophages and neutrophils can produce superoxide simultaneously with nitric oxide, yielding significantly more reactive species, such as peroxynitrite (40). Consistent with previous findings (10, 61, 78), we found that ROS and RNS generated together were more toxic to A. actinomycetemcomitans than superoxide alone (data not shown).
Superoxide has been shown to affect primarily proteins containing iron-sulfur clusters such as dehydratases (25-27). The action of superoxide can result in peptide bond cleavage, modification of amino acid side chains, and conversion of the protein to derivatives that are highly sensitive to proteolytic degradation (65, 72). Peroxynitrite and other RNS can nonspecifically oxidize proteins at a variety of sites (39). Interestingly, it is known that subnanomolar concentrations of LtxA can stimulate an oxidative burst in host inflammatory cells (76), and our results show that purified LtxA rapidly degrades and is rendered inactive in the presence of ROS and RNS. We demonstrated here that Cu,Zn SOD protected LtxA from ROS- and RNS-induced damage. Therefore, interaction with Cu,Zn SOD may protect both bacteria and secreted LtxA during infection.
To better understand the role of the LtxA-Cu,Zn SOD interaction, it is important to identify the cellular localization of the proteins. We have shown that all A. actinomycetemcomitans strains examined were able to secrete active LtxA into the culture supernatant (2, 23). LtxA is also found in the outer membrane (15) with a portion of it exposed to the extracellular environment (2). Cu,Zn SOD from gram-negative bacteria is often located in the periplasm (3); however, it was shown that Cu,Zn SOD from Mycobacterium tuberculosis is a membrane-associated protein (18, 61). In addition, Fletcher et al. (24) reported that A. actinomycetemcomitans Cu,Zn SOD is a secreted surface-associated protein. To further confirm the location of Cu,Zn SOD in A. actinomycetemcomitans, we fractionated cells by using a technique based on the differential solubility of membranes in the detergent sarkosyl. Our data show that A. actinomycetemcomitans Cu,Zn SOD is located in the periplasm and cytosol. In addition, we also detected a small fraction of Cu,Zn COD in the membrane fraction, indicating that Cu,Zn SOD is associated with the cell membrane, consistent with the results of Fletcher et al. (24). Therefore, it is possible that Cu,Zn SOD and LtxA interact on the surface of bacterial cells. Furthermore, interaction between secreted LtxA and Cu,Zn SOD may occur when Cu,Zn SOD is released from lysed bacterial cells, as would take place when an immune cell, such as a macrophage, attacks invading pathogens.
Iron limitation in vivo is a major obstacle that infecting bacteria must overcome to proliferate and cause disease. It was shown that heme and hemoglobin, but not transferrin and lactoferrin, can be used by A. actinomycetemcomitans as iron and heme sources (31, 36). We have recently found that LtxA is able to lyse erythrocytes (1), and hemolysis may be an important strategy for A. actinomycetemcomitans. Therefore, hemoglobin and/or heme, produced as a result of the LtxA-mediated hemolysis, may be significant sources of iron during infection (36). A recent study revealed a new function for Cu,Zn SOD from Haemophilus ducreyi. It was shown that this enzyme is a heme-binding protein and may serve to accumulate heme from the environment and supply the cell with heme and iron. It was also suggested that under certain conditions, Cu,Zn SOD may protect bacteria from toxic oxyradicals formed from the reaction between heme iron and oxygen (60). Thus, we suggest that Cu,Zn SOD from A. actinomycetemcomitans may potentially play a role in iron and heme acquisition, especially during systemic disease.
Based on the data presented here and the results of other studies, we propose the following model for the biological role of A. actinomycetemcomitans Cu,Zn SOD during infection. LtxA and other A. actinomycetemcomitans virulence factors stimulate ROS and RNS production in host inflammatory cells (76). Cu,Zn SOD inactivates superoxide generated by host white blood cells to protect bacteria, while Cu,Zn SOD also interacts with LtxA, either in the extracellular environment or at the cell membrane, to provide protection from protein degradation. In addition, Cu,Zn SOD from A. actinomycetemcomitans may be involved in heme and iron transport (60), as LtxA-mediated hemolysis would cause the release of hemoglobin from erythrocytes. The spontaneous and enzymatic oxidation of hemoglobin results in heme accumulation in the environment (28, 42). In turn, heme may bind to Cu,Zn SOD and serve as a source of iron and heme for A. actinomycetemcomitans, as occurs for Haemophilus ducreyi. In further support of this model, we have recently reported that iron represses the secretion of LtxA and consequently results in decreased lysis of erythrocytes (2). This model describing the interplay between a bacterial SOD and a toxin may represent a new paradigm in bacterial pathogenesis.
Acknowledgments
We thank Daniel Fine and David Furgang for providing A. actinomycetemcomitans polyclonal antiserum. We thank Amy Le for preparing the HL-60 cells. We are grateful to Juan Crosby for thoughtful comments and suggestions throughout the project.
This work was generously supported by grants from the National Institute of Dental and Craniofacial Research (R01 DE16133 to S.C.K., 1F32DE017828-01 to N.V.B., and R01 DE014713 to D.H.F.).
Editor: V. J. DiRita
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Balashova, N. V., J. A. Crosby, L. Al Ghofaily, and S. C. Kachlany. 2006. Leukotoxin confers beta-hemolytic activity to Actinobacillus actinomycetemcomitans. Infect. Immun. 74:2015-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balashova, N. V., R. Diaz, S. V. Balashov, J. A. Crosby, and S. C. Kachlany. 2006. Regulation of Aggregatibacter (Actinobacillus) actinomycetemcomitans leukotoxin secretion by iron. J. Bacteriol. 188:8658-8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battistoni, A. 2003. Role of prokaryotic Cu,Zn superoxide dismutase in pathogenesis. Biochem. Soc. Trans. 31:1326-1329. [DOI] [PubMed] [Google Scholar]
- 4.Battistoni, A., F. Pacello, A. P. Mazzetti, C. Capo, J. S. Kroll, P. R. Langford, A. Sansone, G. Donnarumma, P. Valenti, and G. Rotilio. 2001. A histidine-rich metal binding domain at the N terminus of Cu,Zn-superoxide dismutases from pathogenic bacteria: a novel strategy for metal chaperoning. J. Biol. Chem. 276:30315-30325. [DOI] [PubMed] [Google Scholar]
- 5.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 6.Berbari, E. F., F. R. Cockerill III, and J. M. Steckelberg. 1997. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin. Proc. 72:532-542. [DOI] [PubMed] [Google Scholar]
- 7.Berggren, K. A., C. S. Baluyut, R. R. Simonson, W. J. Bemrick, and S. K. Maheswaran. 1981. Cytotoxic effects of Pasteurella haemolytica on bovine neutrophils. Am. J. Vet. Res. 42:1383-1388. [PubMed] [Google Scholar]
- 8.Bhattacharjee, M. K., S. C. Kachlany, D. H. Fine, and D. H. Figurski. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bong, C. T., K. R. Fortney, B. P. Katz, A. F. Hood, L. R. San Mateo, T. H. Kawula, and S. M. Spinola. 2002. A superoxide dismutase C mutant of Haemophilus ducreyi is virulent in human volunteers. Infect. Immun. 70:1367-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunelli, L., J. P. Crow, and J. S. Beckman. 1995. The comparative toxicity of nitric oxide and peroxynitrite to Escherichia coli. Arch. Biochem. Biophys. 316:327-334. [DOI] [PubMed] [Google Scholar]
- 11.Chan, E. C., and R. McLaughlin. 2000. Taxonomy and virulence of oral spirochetes. Oral Microbiol. Immunol. 15:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Coote, J. G. 1992. Structural and functional relationships among the RTX toxin determinants of gram-negative bacteria. FEMS Microbiol. Rev. 8:137-161. [DOI] [PubMed] [Google Scholar]
- 13.Crosby, J. A., and S. C. Kachlany. 2007. TdeA, a TolC-like protein required for toxin and drug export in Aggregatibacter (Actinobacillus) actinomycetemcomitans. Gene 388:83-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, M., A. D. Badley, F. R. Cockerill, J. M. Steckelberg, and W. R. Wilson. 1997. Infective endocarditis caused by HACEK microorganisms. Annu. Rev. Med. 48:25-33. [DOI] [PubMed] [Google Scholar]
- 15.Demuth, D. R., D. James, Y. Kowashi, and S. Kato. 2003. Interaction of Actinobacillus actinomycetemcomitans outer membrane vesicles with HL60 cells does not require leukotoxin. Cell. Microbiol. 5:111-121. [DOI] [PubMed] [Google Scholar]
- 16.Diaz, R., L. A. Ghofaily, J. Patel, N. V. Balashova, A. C. Freitas, I. Labib, and S. C. Kachlany. 2006. Characterization of leukotoxin from a clinical strain of Actinobacillus actinomycetemcomitans. Microb. Pathog. 40:48-55. [DOI] [PubMed] [Google Scholar]
- 17.Dobereiner, A., A. Schmid, A. Ludwig, W. Goebel, and R. Benz. 1996. The effects of calcium and other polyvalent cations on channel formation by Escherichia coli alpha-hemolysin in red blood cells and lipid bilayer membranes. Eur. J. Biochem. 240:454-460. [DOI] [PubMed] [Google Scholar]
- 18.D'Orazio, M., S. Folcarelli, F. Mariani, V. Colizzi, G. Rotilio, and A. Battistoni. 2001. Lipid modification of the Cu,Zn superoxide dismutase from Mycobacterium tuberculosis. Biochem. J. 359:17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esterbauer, H., R. J. Schaur, and H. Zollner. 1991. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 11:81-128. [DOI] [PubMed] [Google Scholar]
- 20.Farrant, J. L., A. Sansone, J. R. Canvin, M. J. Pallen, P. R. Langford, T. S. Wallis, G. Dougan, and J. S. Kroll. 1997. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol. Microbiol. 25:785-796. [DOI] [PubMed] [Google Scholar]
- 21.Fine, D. H., D. Furgang, J. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 44:1063-1076. [DOI] [PubMed] [Google Scholar]
- 22.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 23.Fine, D. H., J. B. Kaplan, S. C. Kachlany, and H. C. Schreiner. 2006. How we got attached to Actinobacillus actinomycetemcomitans: a model for infectious diseases. Periodontol. 2000 42:114-157. [DOI] [PubMed] [Google Scholar]
- 24.Fletcher, J. M., S. P. Nair, J. M. Ward, B. Henderson, and M. Wilson. 2001. Analysis of the effect of changing environmental conditions on the expression patterns of exported surface-associated proteins of the oral pathogen Actinobacillus actinomycetemcomitans. Microb. Pathog. 30:359-368. [DOI] [PubMed] [Google Scholar]
- 25.Flint, D. H., M. H. Emptage, M. G. Finnegan, W. Fu, and M. K. Johnson. 1993. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J. Biol. Chem. 268:14732-14742. [PubMed] [Google Scholar]
- 26.Gardner, P. R., and I. Fridovich. 1991. Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J. Biol. Chem. 266:1478-1483. [PubMed] [Google Scholar]
- 27.Gardner, P. R., and I. Fridovich. 1991. Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 266:19328-19333. [PubMed] [Google Scholar]
- 28.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 29.Glaser, P., H. Sakamoto, J. Bellalou, A. Ullmann, and A. Danchin. 1988. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 7:3997-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez, C. T., and S. K. Maheswaran. 1993. The role of induced virulence factors produced by Pasteurella haemolytica in the pathogenesis of bovine pneumonic pasteurellosis: review and hypotheses. Br. Vet. J. 149:183-193. [DOI] [PubMed] [Google Scholar]
- 31.Grenier, D., A. Leduc, and D. Mayrand. 1997. Interaction between Actinobacillus actinomycetemcomitans lipopolysaccharides and human hemoglobin. FEMS Microbiol. Lett. 151:77-81. [DOI] [PubMed] [Google Scholar]
- 32.Guthmiller, J. M., D. Kolodrubetz, and E. Kraig. 1995. Mutational analysis of the putative leukotoxin transport genes in Actinobacillus actinomycetemcomitans. Microb. Pathog. 18:307-321. [DOI] [PubMed] [Google Scholar]
- 33.Haase, E. M., J. L. Zmuda, and F. A. Scannapieco. 1999. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 67:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackett, M., C. B. Walker, L. Guo, M. C. Gray, S. Van Cuyk, A. Ullmann, J. Shabanowitz, D. F. Hunt, E. L. Hewlett, and P. Sebo. 1995. Hemolytic, but not cell-invasive activity, of adenylate cyclase toxin is selectively affected by differential fatty-acylation in Escherichia coli. J. Biol. Chem. 270:20250-20253. [DOI] [PubMed] [Google Scholar]
- 35.Halliwell, B., and J. M. Gutteridge. 1984. Free radicals, lipid peroxidation, and cell damage. Lancet 2:1095. [DOI] [PubMed] [Google Scholar]
- 36.Hayashida, H., K. Poulsen, and M. Kilian. 2002. Differences in iron acquisition from human haemoglobin among strains of Actinobacillus actinomycetemcomitans. Microbiology 148:3993-4001. [DOI] [PubMed] [Google Scholar]
- 37.Henderson, B., S. P. Nair, J. M. Ward, and M. Wilson. 2003. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annu. Rev. Microbiol. 57:29-55. [DOI] [PubMed] [Google Scholar]
- 38.Inoue, S., and S. Kawanishi. 1995. Oxidative DNA damage induced by simultaneous generation of nitric oxide and superoxide. FEBS Lett. 371:86-88. [DOI] [PubMed] [Google Scholar]
- 39.Ischiropoulos, H., and A. B. al-Mehdi. 1995. Peroxynitrite-mediated oxidative protein modifications. FEBS Lett. 364:279-282. [DOI] [PubMed] [Google Scholar]
- 40.Ischiropoulos, H., L. Zhu, and J. S. Beckman. 1992. Peroxynitrite formation from macrophage-derived nitric oxide. Arch. Biochem. Biophys. 298:446-451. [DOI] [PubMed] [Google Scholar]
- 41.Janda, J. M., and S. L. Abbott. 1993. Expression of an iron-regulated hemolysin by Edwardsiella tarda. FEMS Microbiol. Lett. 111:275-280. [DOI] [PubMed] [Google Scholar]
- 42.Jeney, V., J. Balla, A. Yachie, Z. Varga, G. M. Vercellotti, J. W. Eaton, and G. Balla. 2002. Pro-oxidant and cytotoxic effects of circulating heme. Blood 100:879-887. [DOI] [PubMed] [Google Scholar]
- 43.Johansson, A., R. Claesson, L. Hanstrom, and S. Kalfas. 2003. Serum-mediated release of leukotoxin from the cell surface of the periodontal pathogen Actinobacillus actinomycetemcomitans. Eur. J. Oral Sci. 111:209-215. [DOI] [PubMed] [Google Scholar]
- 44.Kachlany, S. C., D. H. Fine, and D. H. Figurski. 2002. Purification of secreted leukotoxin (LtxA) from Actinobacillus actinomycetemcomitans. Protein Expr. Purif. 25:465-471. [DOI] [PubMed] [Google Scholar]
- 45.Kachlany, S. C., D. H. Fine, and D. H. Figurski. 2000. Secretion of RTX leukotoxin by Actinobacillus actinomycetemcomitans. Infect. Immun. 68:6094-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaplan, J. B., H. C. Schreiner, D. Furgang, and D. H. Fine. 2002. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. J. Clin. Microbiol. 40:1181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korshunov, S., and J. A. Imlay. 2006. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J. Bacteriol. 188:6326-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kroll, J. S., P. R. Langford, and B. M. Loynds. 1991. Copper-zinc superoxide dismutase of Haemophilus influenzae and H. parainfluenzae. J. Bacteriol. 173:7449-7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lally, E. T., R. B. Hill, I. R. Kieba, and J. Korostoff. 1999. The interaction between RTX toxins and target cells. Trends Microbiol. 7:356-361. [DOI] [PubMed] [Google Scholar]
- 51.Lally, E. T., I. R. Kieba, N. S. Taichman, J. Rosenbloom, C. W. Gibson, D. R. Demuth, G. Harrison, and E. E. Golub. 1991. Actinobacillus actinomycetemcomitans leukotoxin is a calcium-binding protein. J. Periodontal Res. 26:268-271. [DOI] [PubMed] [Google Scholar]
- 52.Langford, P. R., B. M. Loynds, and J. S. Kroll. 1996. Cloning and molecular characterization of Cu,Zn superoxide dismutase from Actinobacillus pleuropneumoniae. Infect. Immun. 64:5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis, J. P., J. A. Dawson, J. C. Hannis, D. Muddiman, and F. L. Macrina. 1999. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 181:4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ludwig, A., F. Garcia, S. Bauer, T. Jarchau, R. Benz, J. Hoppe, and W. Goebel. 1996. Analysis of the in vivo activation of hemolysin (HlyA) from Escherichia coli. J. Bacteriol. 178:5422-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ludwig, A., and W. Goebel. 2000. Dangerous signals from E. coli toxin. Nat. Med. 6:741-742. [DOI] [PubMed] [Google Scholar]
- 56.Ludwig, A., T. Jarchau, R. Benz, and W. Goebel. 1988. The repeat domain of Escherichia coli haemolysin (HlyA) is responsible for its Ca2+-dependent binding to erythrocytes. Mol. Gen. Genet. 214:553-561. [DOI] [PubMed] [Google Scholar]
- 57.Marnett, L. J. 1999. Lipid peroxidation-DNA damage by malondialdehyde. Mutat. Res. 424:83-95. [DOI] [PubMed] [Google Scholar]
- 58.McCord, J. M., and I. Fridovich. 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244:6049-6055. [PubMed] [Google Scholar]
- 59.Ostolaza, H., A. Soloaga, and F. M. Goni. 1995. The binding of divalent cations to Escherichia coli alpha-haemolysin. Eur. J. Biochem. 228:39-44. [PubMed] [Google Scholar]
- 60.Pacello, F., P. R. Langford, J. S. Kroll, C. Indiani, G. Smulevich, A. Desideri, G. Rotilio, and A. Battistoni. 2001. A novel heme protein, the Cu,Zn-superoxide dismutase from Haemophilus ducreyi. J. Biol. Chem. 276:30326-30334. [DOI] [PubMed] [Google Scholar]
- 61.Piddington, D. L., F. C. Fang, T. Laessig, A. M. Cooper, I. M. Orme, and N. A. Buchmeier. 2001. Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect. Immun. 69:4980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rhodes, E. R., A. P. Tomaras, G. McGillivary, P. L. Connerly, and L. A. Actis. 2005. Genetic and functional analyses of the Actinobacillus actinomycetemcomitans AfeABCD siderophore-independent iron acquisition system. Infect. Immun. 73:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 64.Sheehan, B. J., P. R. Langford, A. N. Rycroft, and J. S. Kroll. 2000. [Cu,Zn]-superoxide dismutase mutants of the swine pathogen Actinobacillus pleuropneumoniae are unattenuated in infections of the natural host. Infect. Immun. 68:4778-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stadtman, E. R. 2004. Role of oxidant species in aging. Curr. Med. Chem. 11:1105-1112. [DOI] [PubMed] [Google Scholar]
- 66.Stanley, P., V. Koronakis, and C. Hughes. 1998. Acylation of Escherichia coli hemolysin: a unique protein lipidation mechanism underlying toxin function. Microbiol. Mol. Biol. Rev. 62:309-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taichman, N. S., R. T. Dean, and C. J. Sanderson. 1980. Biochemical and morphological characterization of the killing of human monocytes by a leukotoxin derived from Actinobacillus actinomycetemcomitans. Infect. Immun. 28:258-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taichman, N. S., D. L. Simpson, S. Sakurada, M. Cranfield, J. DiRienzo, and J. Slots. 1987. Comparative studies on the biology of Actinobacillus actinomycetemcomitans leukotoxin in primates. Oral Microbiol. Immunol. 2:97-104. [DOI] [PubMed] [Google Scholar]
- 69.Taichman, N. S., and J. M. Wilton. 1981. Leukotoxicity of an extract from Actinobacillus actinomycetemcomitans for human gingival polymorphonuclear leukocytes. Inflammation 5:1-12. [DOI] [PubMed] [Google Scholar]
- 70.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 181:7298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai, C. C., B. J. Shenker, J. M. DiRienzo, D. Malamud, and N. S. Taichman. 1984. Extraction and isolation of a leukotoxin from Actinobacillus actinomycetemcomitans with polymyxin B. Infect. Immun. 43:700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valko, M., C. J. Rhodes, J. Moncol, M. Izakovic, and M. Mazur. 2006. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 160:1-40. [DOI] [PubMed] [Google Scholar]
- 73.Wang, Y., S. D. Goodman, R. J. Redfield, and C. Chen. 2002. Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J. Bacteriol. 184:3442-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Welch, R. A. 2001. RTX toxin structure and function: a story of numerous anomalies and few analogies in toxin biology. Curr. Top. Microbiol. Immunol. 257:85-111. [DOI] [PubMed] [Google Scholar]
- 75.Wilks, K. E., K. L. Dunn, J. L. Farrant, K. M. Reddin, A. R. Gorringe, P. R. Langford, and J. S. Kroll. 1998. Periplasmic superoxide dismutase in meningococcal pathogenicity. Infect. Immun. 66:213-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamaguchi, N., I. R. Kieba, J. Korostoff, P. S. Howard, B. J. Shenker, and E. T. Lally. 2001. Maintenance of oxidative phosphorylation protects cells from Actinobacillus actinomycetemcomitans leukotoxin-induced apoptosis. Cell. Microbiol. 3:811-823. [DOI] [PubMed] [Google Scholar]
- 77.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]
- 78.Zhu, L., C. Gunn, and J. S. Beckman. 1992. Bactericidal activity of peroxynitrite. Arch. Biochem. Biophys. 298:452-457. [DOI] [PubMed] [Google Scholar]